Dysregulated MicroRNAs as Biomarkers or Therapeutic Targets in Cisplatin-Induced Nephrotoxicity: A Systematic Review
Abstract
:1. Introduction
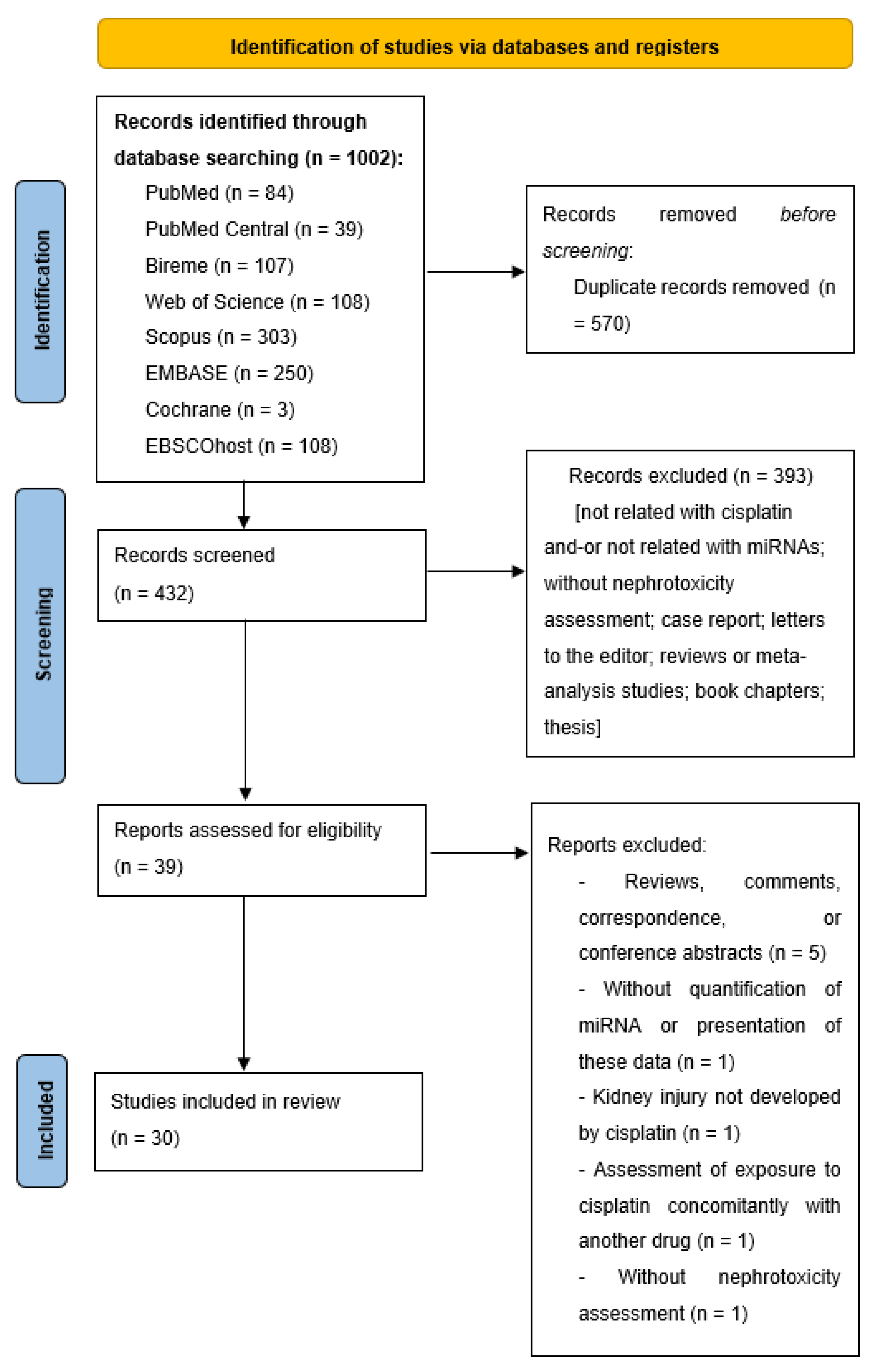
2. Methods
2.1. Search Strategy
2.2. Study Selection
2.3. Data Extraction and Analysis
2.4. Bioinformatics Analysis
3. Results and Discussion
3.1. Methods of Included Studies
3.2. miRNAs as Biomarkers or Therapeutic Agents or Targets of AKI
3.3. Future Perspectives
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jelinek, M.J.; Lee, S.M.; Wyche Okpareke, A.; Wing, C.; Koyner, J.L.; Murray, P.T.; Stadler, W.M.; O’ Donnell, P.H. Predicting Acute Renal Injury in Cancer Patients Receiving Cisplatin Using Urinary Neutrophil Gelatinase-Associated Lipocalin and Cystatin C. Clin. Transl. Sci. 2018, 11, 420–427. [Google Scholar] [CrossRef] [Green Version]
- Pabla, N.; Dong, Z. Cisplatin nephrotoxicity: Mechanisms and renoprotective strategies. Kidney Int. 2008, 73, 994–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waikar, S.S.; Betensky, R.A.; Emerson, S.C.; Bonventre, J.V. Imperfect gold standards for kidney injury biomarker evaluation. J. Am. Soc. Nephrol. 2012, 23, 13–21. [Google Scholar] [CrossRef]
- Van Meer, L.; Moerland, M.; Cohen, A.F.; Burggraaf, J. Urinary kidney biomarkers for early detection of nephrotoxicity in clinical drug development. Br. J. Clin. Pharmacol. 2014, 77, 947–957. [Google Scholar] [CrossRef] [Green Version]
- Waikar, S.S.; Sabbisetti, V.S.; Bonventre, J.V. Normalization of urinary biomarkers to creatinine during changes in glomerular filtration rate. Kidney Int. 2010, 78, 486–494. [Google Scholar] [CrossRef] [Green Version]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Zhang, W.; Duan, N.; Zhang, Q.; Song, T.; Li, Z.; Zhang, C.; Chen, X.; Wang, K. DNA methylation mediated down-regulation of miR-370 regulates cell growth through activation of the Wnt/β-catenin signaling pathway in human osteosarcoma cells. Int. J. Biol. Sci. 2017, 13, 561–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambros, V. MicroRNAs and Developmental Timing. Curr. Opin. Genet. Dev. 2011, 21, 511–517. [Google Scholar] [CrossRef] [Green Version]
- Small, E.M.; Olson, E.N. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011, 469, 336–342. [Google Scholar] [CrossRef] [Green Version]
- Chong, M.M.W.; Zhang, G.; Cheloufi, S.; Neubert, T.A.; Hannon, G.J.; Littman, D.R. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010, 24, 1951–1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kellum, J.A.; Lameire, N.; Aspelin, P.; Barsoum, R.S.; Burdmann, E.A.; Goldstein, S.L.; Herzog, C.A.; Joannidis, M.; Kribben, A.; Levey, A.S.; et al. KDIGO clinical practice guideline for acute kidney injury. Kidney Int. Suppl. 2012, 2, 1–138. [Google Scholar] [CrossRef] [Green Version]
- Fan, P.C.; Chen, C.C.; Chen, Y.C.; Chang, Y.S.; Chu, P.H. MicroRNAs in acute kidney injury. Hum. Genomics 2016, 10, 29. [Google Scholar] [CrossRef] [Green Version]
- Bellinger, M.A.; Bean, J.S.; Rader, M.A.; Heinz-Taheny, K.M.; Nunes, J.S.; Haas, J.V.; Michael, L.F.; Rekhter, M.D. Concordant changes of plasma and kidney microRNA in the early stages of acute kidney injury: Time course in a mouse model of bilateral renal ischemia-reperfusion. PLoS ONE 2014, 9, e93297. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.F.; Yi, Z.F.; Li, H.W.; Wang, F.; Bian, Q.; Lai, X.L.; Yu, G. Screening plasma miRNAs as biomarkers for renal ischemia-reperfusion injury in rats. Med. Sci. Monit. 2014, 20, 283–289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguado-Fraile, E.; Ramos, E.; Conde, E.; Rodríguez, M.; Martín-Gómez, L.; Lietor, A.; Candela, Á.; Ponte, B.; Liaño, F.; García-Bermejo, M.L. A pilot study identifying a set of microRNAs as precise diagnostic biomarkers of acute kidney injury. PLoS ONE 2015, 10, e0127175. [Google Scholar] [CrossRef]
- Xu, D.; Li, W.; Zhang, T.; Wang, G. miR-10a overexpression aggravates renal ischemia-reperfusion injury associated with decreased PIK3CA expression. BMC Nephrol. 2020, 21, 248. [Google Scholar] [CrossRef]
- Li, Y.F.; Jing, Y.; Hao, J.; Frankfort, N.C.; Zhou, X.; Shen, B.; Liu, X.; Wang, L.; Li, R. MicroRNA-21 in the pathogenesis of acute kidney injury. Protein Cell 2013, 4, 813–819. [Google Scholar] [CrossRef] [Green Version]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Koturbash, I.; Tolleson, W.H.; Guo, L.; Yu, D.; Chen, S.; Hong, H.; Mattes, W.; Ning, B. MicroRNAs as pharmacogenomic biomarkers for drug efficacy and drug safety assessment. Biomark. Med. 2015, 9, 1153–1176. [Google Scholar] [CrossRef] [Green Version]
- Roderburg, C.; Luedde, T. Circulating microRNAs as markers of liver inflammation, fibrosis and cancer. J. Hepatol. 2014, 61, 1434–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rupaimoole, R.; Slack, F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017, 16, 203–221. [Google Scholar] [CrossRef] [PubMed]
- Tricco, A.C.; Lillie, E.; Zarin, W.; Al, E. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.Y.; Liu, M.Y.; Hong, Q.; Zhang, D.; Geng, W.J.; Xie, Y.S.; Chen, X.M. Role of microRNA-181a in the apoptosis of tubular epithelial cell induced by cisplatin. Chin. Med. J. 2012, 125, 523–526. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.; Yuan, X.; Wang, L.; Xiao, Y. MicroRNA-205 inhibits renal cells apoptosis via targeting CMTM4. Iran. J. Basic Med. Sci. 2015, 18, 1020–1026. [Google Scholar] [CrossRef]
- Harrill, A.H.; Lin, H.; Tobacyk, J.; Seely, J.C. Mouse population-based evaluation of urinary protein and miRNA biomarker performance associated with cisplatin renal injury. Exp. Biol. Med. 2017, 243, 237–247. [Google Scholar] [CrossRef]
- Wolenski, F.S.; Shah, P.; Sano, T.; Shinozawa, T.; Bernard, H.; Gallacher, M.J.; Wyllie, S.D.; Varrone, G.; Cicia, L.A.; Carsillo, M.E.; et al. Identification of microRNA biomarker candidates in urine and plasma from rats with kidney or liver damage. J. Appl. Toxicol. 2017, 37, 278–286. [Google Scholar] [CrossRef]
- Glineur, S.F.; Hanon, E.; Dremier, S.; Snelling, S.; Berteau, C.; De Ron, P.; Nogueira da Costa, A. Assessment of a Urinary Kidney MicroRNA Panel as Potential Nephron Segment-Specific Biomarkers of Subacute Renal Toxicity in Preclinical Rat Models. Toxicol. Sci. 2018, 166, 409–419. [Google Scholar] [CrossRef]
- Kagawa, T.; Zárybnický, T.; Omi, T.; Shirai, Y.; Toyokuni, S.; Oda, S.; Yokoi, T. A scrutiny of circulating microRNA biomarkers for drug-induced tubular and glomerular injury in rats. Toxicology 2019, 415, 26–36. [Google Scholar] [CrossRef]
- El Magdoub, H.M.; Schaalan, M.F.; Rahmo, R.M.; Farag, D.B.; Khedr, L.H. Implications of miRNAs on TGF-β/TAK1/mTOR pathway in mediating the renoprotective effects of pentoxifylline against cisplatin-induced nephrotoxicity in rats. Toxicol. Appl. Pharmacol. 2020, 404, 115184. [Google Scholar] [CrossRef]
- Huang, S.J.; Huang, J.; Yan, Y.B.; Qiu, J.; Tan, R.Q.; Liu, Y.; Tian, Q.; Guan, L.; Niu, S.S.; Zhang, Y.; et al. The renoprotective effect of curcumin against cisplatin-induced acute kidney injury in mice: Involvement of miR-181a/PTEN axis. Ren. Fail. 2020, 42, 350–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Li, C.; Li, Q.; Li, J.; Lu, X. Puerarin alleviates cisplatin-induced acute renal damage and upregulates microRNA-31-related signaling. Exp. Ther. Med. 2020, 20, 3122–3129. [Google Scholar] [CrossRef]
- Okamoto, M.; Ichii, O.; Mie, K.; Nishida, H.; Akiyoshi, H. Altered clinicopathology and renal pathology in dogs treated with a clinical dose of cisplatin. Jpn. J. Vet. Res. 2021, 69, 109–124. [Google Scholar] [CrossRef]
- Bhatt, K.; Zhou, L.; Mi, Q.S.; Huang, S.; She, J.X.; Dong, Z. MicroRNA-34a is induced via p53 during cisplatin nephrotoxicity and contributes to cell survival. Mol. Med. 2010, 16, 409–416. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.G.; Kim, J.G.; Kim, H.J.; Kwon, H.K.; Cho, I.J.; Choi, D.W.; Lee, W.H.; Kim, W.D.; Hwang, S.J.; Choi, S.; et al. Discovery of an integrative network of microRNAs and transcriptomics changes for acute kidney injury. Kidney Int. 2014, 86, 943–953. [Google Scholar] [CrossRef] [Green Version]
- Qin, W.; Xie, W.; Yang, X.; Xia, N.; Yang, K. Inhibiting microrna-449 attenuates cisplatin-induced injury in nrk-52e cells possibly via regulating the sirt1/p53/bax pathway. Med. Sci. Monit. 2016, 22, 818–823. [Google Scholar] [CrossRef]
- Du, B.; Dai, X.M.; Li, S.; Qi, G.L.; Cao, G.X.; Zhong, Y.; Yang, X.S. MiR-30c regulates cisplatin-induced apoptosis of renal tubularc epithelial cells by targeting Bnip3L and Hspa5. Cell Death Dis. 2017, 8, e2987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, J.; Lou, Q.; Wei, Q.; Mei, S.; Li, L.; Wu, G.; Mi, Q.S.; Mei, C.; Dong, Z. MicroRNA-375 is induced in cisplatin nephrotoxicity to repress hepatocyte nuclear factor 1-β. J. Biol. Chem. 2017, 292, 4571–4582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, W.; Fu, Z.; Zou, Y.; Wen, D.; Ma, H.; Zhou, F.; Chen, Y.; Zhang, M.; Zhang, W. MicroRNA-140-5p attenuated oxidative stress in Cisplatin induced acute kidney injury by activating Nrf2/ARE pathway through a Keap1-independent mechanism. Exp. Cell Res. 2017, 360, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ni, J.; Chen, S.; Bai, M.; Lin, J.; Ding, G.; Zhang, Y.; Sun, P.; Jia, Z.; Huang, S.; et al. MicroRNA-709 Mediates Acute Tubular Injury through Effects on Mitochondrial Function. J. Am. Soc. Nephrol. 2018, 29, 449–461. [Google Scholar] [CrossRef] [Green Version]
- Yang, A.; Liu, F.; Guan, B.; Luo, Z.; Lin, J.; Fang, W.; Liu, L.; Zuo, W. p53 induces miR-199a-3p to suppress mechanistic target of rapamycin activation in cisplatin-induced acute kidney injury. J. Cell. Biochem. 2019, 120, 17625–17634. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, Z.L.; Wang, F.M.; Tang, R.N.; Tu, Y.; Liu, H. MicroRNA26a inhibits cisplatin-induced renal tubular epithelial cells apoptosis through suppressing the expression of transient receptor potential channel 6 mediated dynamin-related protein 1. Cell Biochem. Funct. 2019, 38, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fan, X.; Wang, Q.; Gong, Y.; Guo, L. Long noncoding RNA PRNCR1 reduces renal epithelial cell apoptosis in cisplatin-induced AKI by regulating miR-182-5p/EZH1. Kidney Blood Press. Res. 2021, 46, 162–172. [Google Scholar] [CrossRef]
- Xiong, X.; Tang, B.; Ji, T.; Li, X.; Bai, S. Ameliorative effects of miR-186 on cisplatin-Triggered acute kidney injury via targeting ZEB1. Am. J. Transl. Res. 2021, 13, 4296–4308. [Google Scholar]
- Quintanilha, J.C.F.; Cursino, M.A.; Borges, J.B.; Torso, N.G.; Bastos, L.B.; Oliveira, J.M.; Cobaxo, T.S.; Pincinato, E.C.; Hirata, M.H.; Geraldo, M.V.; et al. MiR-3168, miR-6125, and miR-4718 as potential predictors of cisplatin-induced nephrotoxicity in patients with head and neck cancer. BMC Cancer 2021, 21, 575. [Google Scholar] [CrossRef]
- Pavkovic, M.; Robinson-Cohen, C.; Chua, A.S.; Nicoara, O.; Cárdenas-González, M.; Bijol, V.; Ramachandran, K.; Hampson, L.; Pirmohamed, M.; Antoine, D.J.; et al. Detection of drug-induced acute kidney injury in humans using urinary KIM-1, miR-21, -200c, and -423. Toxicol. Sci. 2016, 152, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Suter-Dick, L.; Mauch, L.; Ramp, D.; Caj, M.; Vormann, M.K.; Hutter, S.; Lanz, H.L.; Vriend, J.; Masereeuw, R.; Wilmer, M.J. Combining Extracellular miRNA Determination with Microfluidic 3D Cell Cultures for the Assessment of Nephrotoxicity: A Proof of Concept Study. AAPS J. 2018, 20, 86. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, X.Q.; Ma, Q.; Yang, Q.; Gao, L.; Li, D.H.; Wang, J.N.; Wei, B.; Wen, J.; Li, J.; et al. hsa-miR-500a-3P alleviates kidney injury by targeting MLKL-mediated necroptosis in renal epithelial cells. FASEB J. 2019, 33, 3523–3535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Li, D.D.; Li, J.Y.; Yin, Y.C.; Li, P.C.; Qiu, L.; Chen, L.M. Identification of microRNA-mRNA networks involved in cisplatin-induced renal tubular epithelial cells injury. Eur. J. Pharmacol. 2019, 851, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, H.; Kong, W.; Zhang, B. Functional role of microRNA-500a-3P-loaded liposomes in the treatment of cisplatininduced AKI. IET Nanobiotechnol. 2020, 14, 465–469. [Google Scholar] [CrossRef]
- Kanki, M.; Moriguchi, A.; Sasaki, D.; Mitori, H.; Yamada, A.; Unami, A.; Miyamae, Y. Identification of urinary miRNA biomarkers for detecting cisplatin-induced proximal tubular injury in rats. Toxicology 2014, 324, 158–168. [Google Scholar] [CrossRef]
- Pavkovic, M.; Riefke, B.; Ellinger-Ziegelbauer, H. Urinary microRNA profiling for identification of biomarkers after cisplatin-induced kidney injury. Toxicology 2014, 324, 147–157. [Google Scholar] [CrossRef]
- Cho, Y.E.; Kim, S.H.; Lee, B.H.; Baek, M.C. Circulating plasma and exosomal microRNAs as indicators of drug-induced organ injury in rodent models. Biomol. Ther. 2017, 25, 367–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehta, R.L.; Kellum, J.A.; Shah, S.V.; Molitoris, B.A.; Ronco, C.; Warnock, D.G.; Levin, A.; Bagga, A.; Bakkaloglu, A.; Bonventre, J.V.; et al. Acute kidney injury network: Report of an initiative to improve outcomes in acute kidney injury. Crit. Care 2007, 11, R31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, G.M. Biomarkers in Nephrology. Am. J. Kidney Dis. 2013, 62, 165–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortez, M.A.; Calin, G.A. MicroRNA identification in plasma and serum: A new tool to diagnose and monitor diseases. Expert Opin. Biol. Ther. 2009, 9, 703–711. [Google Scholar] [CrossRef]
- Melkonyan, H.S.; Feaver, W.J.; Meyer, E.; Scheinker, V.; Shekhtman, E.M.; Xin, Z.; Umansky, S.R. Transrenal nucleic acids: From proof of principle to clinical tests. Ann. N. Y. Acad. Sci. 2008, 1137, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Rana, T.M. Therapeutic targeting of microRNAs: Current status and future challenges. Nat. Rev. Drug Discov. 2014, 13, 622–638. [Google Scholar] [CrossRef]
- Hermeking, H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010, 17, 193–199. [Google Scholar] [CrossRef]
- Chang, T.C.; Wentzel, E.A.; Kent, O.A.; Ramachandran, K.; Mullendore, M.; Lee, K.H.; Feldmann, G.; Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J.; et al. Transactivation of miR-34a by p53 Broadly Influences Gene Expression and Promotes Apoptosis. Mol. Cell 2007, 26, 745–752. [Google Scholar] [CrossRef] [Green Version]
- Manohar, S.; Leung, N. Cisplatin nephrotoxicity: A review of the literature. J. Nephrol. 2018, 31, 15–25. [Google Scholar] [CrossRef]
- Ong, A.L.C.; Ramasamy, T.S. Role of Sirtuin1-p53 regulatory axis in aging, cancer and cellular reprogramming. Ageing Res. Rev. 2018, 43, 64–80. [Google Scholar] [CrossRef]
- Zhu, S.; Pabla, N.; Tang, C.; He, L.; Dong, Z. DNA damage response in cisplatin-induced nephrotoxicity. Arch. Toxicol. 2015, 89, 2197–2205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, A.C.K.; Yu, X.; Lan, H.Y. MicroRNA and nephropathy: Emerging concepts. Int. J. Nephrol. Renovasc. Dis. 2013, 6, 169–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endale, M.; Kim, T.H.; Kwak, Y.S.; Kim, N.M.; Kim, S.H.; Cho, J.Y.; Yun, B.S.; Rhee, M.H. Torilin Inhibits Inflammation by Limiting TAK1-Mediated MAP Kinase and NF- B Activation. Mediators Inflamm. 2017, 2017, 7250968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- Saal, S.; Harvey, S.J. MicroRNAs and the kidney: Coming of age. Curr. Opin. Nephrol. Hypertens. 2009, 18, 317–323. [Google Scholar] [CrossRef]
- Ma, L.; Qu, L. The function of MicroRNAs in renal development and pathophysiology. J. Genet. Genomics 2013, 40, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Cao, X.; Zou, L.; Chen, Y.; Guo, J.; Chen, Z.; Hu, S.; Zheng, Z. MicroRNA-21 and Risk of Severe Acute Kidney Injury and Poor Outcomes after Adult Cardiac Surgery. PLoS ONE 2013, 8, e63390. [Google Scholar] [CrossRef] [PubMed]
- Zununi Vahed, S.; Omidi, Y.; Ardalan, M.; Samadi, N. Dysregulation of urinary miR-21 and miR-200b associated with interstitial fibrosis and tubular atrophy (IFTA) in renal transplant recipients. Clin. Biochem. 2017, 50, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Chung, A.C.K.; Chen, H.Y.; Dong, Y.; Meng, X.M.; Li, R.; Yang, W.; Hou, F.F.; Lan, H.Y. MiR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia 2013, 56, 663–674. [Google Scholar] [CrossRef]
- Thum, T.; Catalucci, D.; Bauersachs, J. MicroRNAs: Novel regulators in cardiac development and disease. Cardiovasc. Res. 2008, 79, 562–570. [Google Scholar] [CrossRef] [Green Version]
- Godwin, J.G.; Ge, X.; Stephan, K.; Jurisch, A.; Tullius, S.G.; Iacomini, J. Identification of a microRNA signature of renal ischemia reperfusion injury. Proc. Natl. Acad. Sci. USA 2010, 107, 14339–14344. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Friggeri, A.; Yang, Y.; Milosevic, J.; Ding, Q.; Thannickal, V.J.; Kaminski, N.; Abraham, E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010, 207, 1589–1597. [Google Scholar] [CrossRef]
- Comoglio, P.M.; Trusolino, L.; Boccaccio, C. Known and novel roles of the MET oncogene in cancer: A coherent approach to targeted therapy. Nat. Rev. Cancer 2018, 18, 341–358. [Google Scholar] [CrossRef] [PubMed]
- Gharaibeh, L.; Elmadany, N.; Alwosaibai, K.; Alshaer, W. Notch1 in cancer therapy: Possible clinical implications and challenges. Mol. Pharmacol. 2020, 98, 559–576. [Google Scholar] [CrossRef]
- Peres, L.A. lbert. B.; da Cunha, A.D. anta. Acute nephrotoxicity of cisplatin: Molecular mechanisms. J. Bras. Nefrol. 2013, 35, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.C.M.; Thomas, D.M.; Choong, P.F.M.; Dass, C.R. RECK—A newly discovered inhibitor of metastasis with prognostic significance in multiple forms of cancer. Cancer Metastasis Rev. 2007, 26, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Backes, C.; Meese, E.; Keller, A. Specific miRNA Disease Biomarkers in Blood, Serum and Plasma: Challenges and Prospects. Mol. Diagn. Ther. 2016, 20, 509–518. [Google Scholar] [CrossRef] [PubMed]

| Cells | |||||
| Author(s)/Year | Country | Aim | Population | Cisplatin Treatment/Exposure | Nephrotoxicity Markers |
| Zhu et al./2012 [24] | China | To investigate the role of miR-181a in the apoptosis of tubular epithelial cell induced by CIS. | HK-2 cells. | 50 µmol/L for 24 h. | Apoptosis. |
| Zhang et al./2015 [25] | China | To elucidate the role of miR-205 in CIS-induced renal cell apoptosis and explored the molecular mechanisms. | HK-2 cells. | 100 μg/mL for 6 h. | Apoptosis. |
| Jiang et al./2019 [48] | China | To evaluate miRNAs that potentially target MLKL and evaluate their function in human tubular epithelial cells in response to toxic and ischemic insults. | HK-2 cells. | 20 μM | Programmed cell death and the relative mRNA levels of KIM-1. |
| Wu et al./2019 [49] | China | To examine the changes of the miRNA and mRNA expression profiles in CIS treated HK-2 cells. | HK-2 cells. | 10 µM for 24 h. | Cell viability and apoptosis. |
| Zhang et al./2020 [50] | China | To improve the therapeutic efficacy in CIS-induced AKI. | HK-2 cells. | 15 µM for up to 48 h. | Apoptosis, inflammatory markers (TNF-a and IL-8) and necroptosis. |
| Suter-Dick et al./2018 [47] | Switzerland | To investigate changes in miRNA released to the cell culture medium as potential markers for nephrotoxicity. | HPTECs cells line overexpressing the OAT1. | 0–30 μM for 24 h or 48 h. | Cell viability and NAG. |
| Qin et al./2016 [36] | China | To explore the expression and function of miR-449 in CIS-induced AKI. | NRK-52E cells. | 20 μg/mL for 24 h. | Cell viability and apoptosis. |
| Animals | |||||
| Author(s)/Year | Country | Aim | Population | Cisplatin Treatment/Exposure | Nephrotoxicity Markers |
| Cho et al./2017 [53] | Republic of Korea | To evaluate if miRNAs circulating exosomes may serve as biomarkers of drug-induced kidney injury. | Male Balb/C mice. | Single injection, 10 mg/kg, IP. | Histological analyses. |
| Huang et al./2020 [31] | China | To investigate the role of miRNAs against CIS-induced AKI in mice. | Male C57BL/6 mice | 20 mg/kg/day, for three, IP. | BUN and histological analyses |
| Harrill et al./2017 [26] | USA | To investigate performance of urinary kidney biomarkers against classical preclinical kidney injury biomarkers. | Female DO mice | Single injection, 5 mg/kg, IP. | BUN, serum CRE, histological analyses, and urine b2-microglobulin, interferon gamma-induced protein 10, KIM-1, renin, and osteopontin. |
| Glineur et al./2018 [28] | Belgica | To evaluate the performance of a panel of 68 urinary miRNAs as potential nephron segment-specific biomarkers before and after treatment with nephrotoxic drugs. | Male Hannover Wistar rats. | Single dose, 2.5 mg/kg, IP. | Urinary KIM-1, albumin, and clusterin, and histological analyses. |
| Kanki et al./2014 [51] | Japan | To identify urinary miRNAs markers suitable for use in detecting CIS-induced nephrotoxicity. | Male Sprague-Dawley rats. | Study 1: single injection, 6 mg/kg, IP. Study 2: single injection, 0, 1, 3 and 6 mg/kg, IP. | BUN, serum CRE, urinary KIM-1 and clusterin, and histopathological analysis. |
| Wolenski et al./2017 [27] | USA | To dose rats with toxicants that cause nephrotoxicity, and then identify changes within the miRNA expression profiles in urine, plasma, and tissue. | Male Sprague-Dawley rats | Single dose, 2 or 5 mg/kg, IV. | Serum BUN, urinary CRE and KIM-1, and histological analyses. |
| Kagawa et al./2019 [29] | Japan | To identify plasma miRNAs that may enable early and specific detection of drug-induced tubular and glomerular injury through next-generation sequencing analysis. | Male Sprague-Dawley rats. | Single dose, 6 mg/kg, IV. | Plasma CRE and BUN, urinary albumin, and histological analyses. |
| Wu et al./2020 [32] | China | To investigate the function of puerarin in a CIS-induced AKI rat model via RT-qPCR and western blot analyses. | Female Sprague-Dawley rats. | 20 mg/kg, IP. | BUN, serum CRE, and histological analyses. |
| Pavkovic et al./2014 [52] | Germany | To evaluate whether urinary miRNAs could serve as biomarkers for CIS-induced kidney injury. | Male Wistar rats. | Single dose, 1 or 3 mg/kg, IP. | BUN, serum CRE, urinary KIM-1 and alpha-GST, and histopathological analysis. |
| El Magdoub et al./2020 [30] | Egypt | To investigate the involvement of miRNAs let-7b, 26b, and 34a on CIS-induced nephrotoxicity. | Male Wistar rats. | Single dose on day 4, 5 mg/kg, IP. | Plasma urea, CRE and KIM-1, and histological analyses. |
| Okamoto et al./2021 [33] | Japan | To investigate clinicopathological changes in dogs treated with a clinical dose of CIS. | Female beagle dogs | 70 mg/m2 over 20 min, IV (via cephalic vein). | Urinary protein, CRE, NAG, albumin, and L-FABP; blood CRE, BUN, magnesium, calcium, inorganic phosphorus, sodium, potassium, and chloride; histopathological analysis. |
| Cells and Animals | |||||
| Author(s)/Year | Country | Aim | Population | Cisplatin Treatment/exposure | Nephrotoxicity Markers |
| Bhatt et al./2010 [34] | USA | To examine the regulation of miR-34a and its role in CIS-induced AKI and nephrotoxicity. | BUMPT-306 cells. | 40 μmol/L for 0–12 h. | Apoptosis. |
| Male wild type and p53-deficient C57BL/6 mice. | Single injection of CIS 30 mg/kg, IP. | BUN. | |||
| Du et al./2017 [37] | China | To investigate the mechanism of CIS-induced renal injury. | HK-2 and NRK-52E cells. | 10 μM for 12, 24, 48 or 72 h. | Apoptosis. |
| Wistar rats. | 10 mg/kg, IP. | Apoptosis and histological analyses. | |||
| Liao et al./2017 [39] | China | To investigate the levels of miR-140-5p and its functional role in pathogenesis of CIS-induced AKI. | HK-2 cells. | 20 μM for 6 h. | ROS and LDH levels, MnSOD activity, and cell viability. |
| Male mice. | Single injection, 20 mg/kg, IP. | BUN and serum CRE. | |||
| Yang et al./2019 [41] | China | To probe into the role of p53 in CIS-induced AKI. | HK-2 cells. | 100 μM for 24 h. | Apoptosis. |
| C57/BL male mice. | Single dose, 20mg/kg, IP. | BUN, serum CRE, and histological analyses. | |||
| Yang et al./2019 [42] | China | To study on whether miR-26a plays an anti-apoptotic role through regulating the expression of TRPC6. | HK-2 cells. | 0, 1, 2, 4 or 8 μM. | Apoptosis. |
| Male C57BL/6 mice. | 20 mg/kg, IP. | BUN, serum CRE, and histological analyses | |||
| Li et al./2021 [43] | China | To examine the role of long noncoding RNA PRNCR1 in CIS-induced AKI in vitro and in vivo. | HK-2 cells. | 10−9, 10−8, 10−7 and 10−6 M for 24 h. | Cell viability and apoptosis. |
| Male BALB/c mice. | 20 mg/kg, IP. | Serum CRE and histological analyses. | |||
| Guo et al./2018 [40] | China | To investigate the pathogenic role of miR-709 in mediating mitochondrial impairment and tubular cell death in AKI. | mPTC cells. | 0–20 mM CIS for 0–24 h. | Apoptosis and determination of mitochondrial function. |
| Male C57BL/6 mice. | 20 mg/kg, IP. | BUN, serum CRE, histological analyses, and determination of mitochondrial function. | |||
| Lee et al./2014 [35] | Korea | To evaluate an integrative network of miRNAs and mRNA data to discover a possible master regulator of AKI. | NRK-52E cells. | 30 mmol/L for 48 h. | Cell viability. |
| Male C57BL/6 mice. | Single dose, 15 mg/kg, IP. | BUN, serum CRE, KIM-1, neutrophil elastase, F4/80, CD3e, CD19, TNF alfa, ICAM-1, and histopathological and immunohistochemical analysis. | |||
| Xiong et al./2021 [44] | China | To investigate the roles of miR-186 in cisplatin-induced AKI. | NRK-52E cells. | 6 μM. | Cell viability and apoptosis. |
| Male Wistar rats. | 6 mg/kg, IP. | Plasma BUN and CRE, and histological analyses. | |||
| Hao et al./2017 [38] | USA andChina | To examine the role of miRNAs in CIS-induced nephrotoxicity. | RPTC cells. | 20 μM for 16 h. | Apoptosis. |
| Male C57 mice. | 30 mg/kg, IP. | BUN and serum CRE. | |||
| Humans | |||||
| Author(s)/Year | Country | Aim | Population | Cisplatin Treatment/Exposure | Nephrotoxicity Markers |
| Quintanilha et al./2021 [45] | Brazil | To identify circulating plasma miRNAs as biomarkers of cisplatin-induced nephrotoxicity using the patients’ samples. | Patients with primary squamous cell carcinoma of the head and neck. | One cycle of 80 or 100 mg/m2. | Serum CRE, BUN, CRE clearance, and Common Toxicity Criteria for Adverse Events version 4. |
| Humans and Cells | |||||
| Author(s)/Year | Country | Aim | Population | Cisplatin Treatment/Exposure | Nephrotoxicity Markers |
| Pavkovic et al./2016 [46] | USA | To evaluate KIM-1 in conjunction with miRNAs as biomarkers for drug-induced AKI. | HPTEC cells. | 85 μM, for 24 h. | Cell viability. |
| Patients with malignant mesothelioma. | Intraoperative CIS therapy. | AKI defined by AKI Network criteria (serum CRE dependent status) [54]. | |||
| Cells | ||||||
| Author(s)/Year | Sample | Methods Used to Identify miRNAs | Time When the miRNAs Were Analyzed (after CIS Exposure/Administration) | miRNAs Differentially Expressed in Nephrotoxicity | Pathophysiological Implications of Dysregulated miRNAs (Related to the Authors’ Own Results) | Role of miRNAs as Biomarkers of Nephrotoxicity |
| Zhu et al./2012 [24] | Cells (HK-2) | RT-qPCR | 24 h | Upregulated: miR-181a | CIS upregulated miR-181a expression leading to negative regulation of Bcl-2 (anti-apoptotic gene) and positive regulation of BAX (pro-apoptotic gene). Thus, miR-181a expression is associated with cell apoptosis. | CIS may play a role in tubular epithelial cell apoptosis by suppressing Bcl-2 expression, which is achieved by regulating the target gene of microRNA-181a. These findings pave a novel approach to the enhancement of prevention treatment of CIS-induced nephrotoxicity. |
| Zhang et al./2015 [25] | Cells (HK-2) | RT-qPCR | 6 h | Downregulated: miR-205 miR-21 miR-29 | MiR-205 could be an anti-apoptotic molecule, because it coordinates the expression of this target gene CMTM4 to module the renal cells apoptosis. | MiR-205 was revealed as an important inhibitor in the regulation of apoptosis in renal cells. |
| Jiang et al./2019 [48] | NR | RT-PCR | 12, 24, 36, and 48 h | a. Upregulated: miR-194-5p miR-577 b. Downregulated: miR-500a-3p | Overexpression of miR-500a-3P had a protective role in CIS-induced kidney injury, as it showed: - To limit programmed cell death; - To decreased chemokine MCP-1 and proinflammatory cytokines TNF-a and IL-8; - To decrease phosphorylation and membrane translocation of MLKL, a key index for detecting necroptosis. | Considering the antinecroptotic and anti-inflammatory merits, miR-500a-3P may be a novel therapeutic agent for AKI treatment. |
| Wu et al./2019 [49] | Cells (HK-2) | MiRNA microarray and RT-qPCR | 24 h | 1. Microarray results: 26 miRNAs upregulated and 21 miRNAs downregulated. Top 5: a. Upregulated: miR-4793-3p miR-4299 miR-4440 miR-297 miR-4485 b. Downregulated: miR- 6841-5p miR-3605-5p miR-27b-5p miR-1236-5p miR-23b-5p 2. RT-qPCR results (validation of 5 miRNAs): Upregulated: miR-4299 miR- 297 miR- 3135b miR-9-3p miR-371b-5p | HIPK2, a key regulator of kidney fibrosis, was predicted as the common target gene of miR-9-3p and miR-371b-5p. | An integrative network approach encompassing miRNAs, target genes, and bioinformatics analysis showed that miR-9-3p and miR-371b-5p could be critical miRNAs in CIS-induced renal tubular cell injury. |
| Zhang et al./2020 [50] | Cells (HK-2) | RT-qPCR | 6, 12, 24, 36, and 48 h | Downregulated: miR-500a-3P | The downregulation of miR-500a-3P in CIS-induced kidney injury was related to apoptosis, NF-kB-based inflammation (increased expression of TNF-a, IL-8, and P-P65), and necroptosis by RIPK3-based downstream signaling pathway (increased expression of P-MLKL and RIPK3 proteins). | MiR-500a-3P is effective in controlling the AKI and may be an appropriate miRNA therapeutics. |
| Suter-Dick et al./2018 [47] | Supernatant of cells culture (HPTECs cells line overexpressing the OAT1). | RT-PCR | 0, 24, and 48 h | Upregulated: Cells treated with 5 uM CIS in 48h: miR-34a miR-192 miR-29a miR-21 Cells treated with 15 uM CIS in 24h: miR-34a miR-192 miR-29a | No explanations were provided. | The data suggest that mir-21, mir-29a, mir-34a and mir-192 are early and sensitive biomarkers of damage to renal proximal tubule cells. |
| Qin et al./2016 [36] | Cells (NRK-52E) | RT-qPCR | 72 h | Upregulated: miR-449 | SIRT1/P53/BAX pathway is related with apoptosis. The study results showed that SIRT1 was inhibited and acetylated p53 and BAX were promoted when miR-449 was upregulated by CIS. Thus, miR-499 expression inhibits cell viability and intensifies apoptosis. | The data suggest that there may exist a pro-apoptotic role of miR-449 in CIS-induced AKI via regulating the SIRT1/P53/BAX pathway. Therefore, it is suggested that miR-449 be a potential therapeutic target for treating AKI. |
| Animals | ||||||
| Author(s)/Year | Sample | Methods Used to Identify miRNAs | Time When the miRNAs Were Analyzed (after CIS Exposure/Administration) | miRNAs Differentially Expressed in Nephrotoxicity | Pathophysiological Implications of Dysregulated miRNAs (Related to the Authors’ Own Results) | Role of miRNAs as Biomarkers of Nephrotoxicity |
| Cho et al./2017 [53] | Plasma. | RT-qPCR. | NR | Upregulated: miR-146a | No explanations were provided. | MiR-146a is a potential biomarker for drug-induced kidney injury. |
| Huang et al./2020 [31] | Kidney | RT-qPCR | Day 3. | Upregulated: miR-181a | MiR-181a can be involved in inhibit PTEN protein expression. | PTEN is one of the miR-181a targets. |
| Harrill et al./2017 [26] | Urine. | Microarray with RT-qPCR. | 54 h. | Of the 335 unique miRNAs assayed on the array platform, 10 miRNA species were significantly affected by treatment (all upregulated) and differed in abundance based on presence of tubule necrosis (grade 2): miR-130a miR-151-3p miR-218 miR-320 miR-680 miR-138 miR-152 miR-221 miR-328 miR-685 | No explanations were provided. | These urinary miRNAs are biomarker candidates, as they were elevated alongside BUN and protein biomarkers in animals with grade 2 proximal tubular cell necrosis. |
| Glineur et al./2018 [28] | Urine | PCR | Before and days 1, 3, 6, 13, 20, and 27 after. | FC > 3.0-Upregulated: miR-34c-5p FC < 3.0-Upregulated: miR-183-5p miR-15b-5p miR-193a-3p miR-423-3p miR-182-5p miR-210-3p miR-155-5p | Urinary miR-34c-5p after CIS administration probably reveals CIS-induced DNA damage leading to p53 transcription factor activation. Moreover, the miR-34c-5p urinary release could attest the activation of a cellular apoptotic process. miR-34c-5p outperformed KIM-1 under subacute conditions (100% of accurate classification vs 75% with KIM-1). | MiR-34c-5p could complement urinary protein biomarkers to detect subacute CIS-induced kidney injury. |
| Kanki et al./2014 [51] | Urine and kidney tissue. | Microarray and RT-qPCR. | Study 1: day 5. Study 2: days 1, 3 and 7. | Levels of 25 miRNAs were elevated in urine concurrently with the appearance of the necrosis of proximal tubules (most of them were conversely decreased in the cortex or outer medulla of kidney): miR-335 miR-378a-5p miR-183-5p miR-328a-3p miR-1839-5p let-7a-1-3p miR-93-5p miR-532-3p miR-192-5p miR-20b-5p miR-17-5p miR-140-3p miR-25-3p miR-340-5p miR-191a-5p let-7g-5p miR-193-5p miR-218a-5p miR-7a-1-3p miR-130b-3p miR-30a-5p miR-26b-3p miR-744-5p miR-320-3p | No explanations were provided. | These 25 urinary miRNAs biomarkers could potentially be used to detect proximal tubular injury due to CIS exposure. They were considered to have sensitivities comparable to BUN, serum CRE, and urinary protein biomarkers. |
| Wolenski et al./2017 [27] | Plasma, urine, and kidney tissue. | NGS | 24 and 72 h. | FC > 2,0: 1. Kidney tissue (upregulated): miR-34a 2. Urine (upregulated): miR-378a miR-1839 miR-140 miR-26b let-7g 3.Plasma (upregulated): miR-34c miR-128 miR-34a miR-130b | No explanations were provided. | MiR-378a is a novel urinary biomarker of kidney damage. |
| Kagawa et al./2019 [29] | Plasma | NGS and RT-qPCR | Days 0, 1, 2, 3, and 5. | 1. Early, downregulated: miR-122-5p miR-143-3p miR-26a-5p miR-215 let-7c-5p miR-146b-5p miR-192-5p miR-191a-5p 2. Middle, downregulated: miR-122-5p miR-143-3p let-7i-5p miR-140-3p let-7g-5p let-7f-5p miR-378a-3p miR-148a-3p miR-25-3p miR-23a-3p 3. Late, downregulated: miR-122-5p miR-143-3p let-7i-5p miR-140-3p let-7g-5p let-7f-5p miR-148a-3p miR-23a-3p miR-26a-5p miR-215 let-7c-5p miR-30a-5p miR-486 | No explanations were provided. | MiR-143-3p and miR-122-5p may be potential biomarkers for the early detection of tubular damage. The downregulation of these miRNAs was earlier than the changes in the traditional biomarkers, such as plasma CRE and BUN. |
| Wu et al./2020 [32] | Kidney | RT-qPCR | Days 0, 1, 3, and 5. | Upregulated: miR-31 | The study hypothesized that the negative regulation of Numb affects Notch signaling via miR-31 in CIS-induced AKI, because Notch signaling is associated with the balance among the cell proliferation and apoptosis that influence the process of various organ injuries. | MiR-31 expression is upregulated in CIS-induced AKI. |
| Pavkovic et al./2014 [52] | Kidney tissue and urine. | Microarray with RT-qPCR. | Kidney tissue: days 3, 5, 8, and 26. Urine: days 3, 5, 8, 15, and 26. | 1. Urine: a. CIS 3mg/kg: 136 miRNAS appeared significantly affected by CIS. 11 upregulated selected by >20-fold change: miR-15b miR-16 miR-20a miR-20b miR-21 miR-34a miR-185 miR-192 miR-200b miR-210 miR-339-3p b. CIS 1mg/kg or 3 mg/kg (day 3 and day 5). Upregulated: miR-15 miR-16 miR-20a miR-192 miR-193 miR-210 2. Tissue: a. CIS 1 mg/Kg. Upregulated: miR-21 miR-34a miR-184 miR-327 b. Donwregulated: miR-15b miR-16 miR-20a miR-20b miR-141 miR-146a miR-185 miR-192 miR-193 miR-196c.rno miR-200b miR-210 miR-223 | These miRNAs are associated with pathways, as DNA damage response, apoptosis, cell cycle regulation, and inflammation. The top canonical pathway affected were p53 and PI3K/AKT pathways. Also, mRNAs predicted as targets of the altered miRNAs in the kidney were associated with DNA damage response, apoptosis, and cell cycle regulation. | These miRNAs are potential urinary biomarkers for CIS-induced kidney injury. |
| El Magdoub et al./2020 [30] | Kidney tissue | RT-PCR | Day 3. | Downregulated: miRNA-let-7b miRNA-26b Upregulated: miRNA-34a | With the alteration of these miRNAs expressions, CIS induced TGF-β1. TGFβR-1, TAK1, and mTOR levels were increased, while LC3-II level was decreased. | Potential involvement of those 3 miRNAs in the pathogenesis of CIS-induced nephrotoxicity. |
| Okamoto et al./2021 [33] | Serum and urine | RT-qPCR | 0, 6, 12, 24, 72, and 168 h. | Serum-Upregulated: miR-21 miR-26a miR-10a Urine-Upregulated: miR-21 | Serum miR-21 was correlated with dynamics of blood calcium, inorganic phosphorus, and magnesium. Increased serum miR-21 levels might indicate kidney injury, as well as biological alerts from a damaged vascular system or other organs. | Increased serum miR-21 levels might indicate kidney injury. Altered urinary levels of miR-21 and miR-26a (or miR-10a) might reflect tubulointerstitial and glomerular lesions by CIS, respectively. |
| Cells and Animals | ||||||
| Author(s)/Year | Sample | Methods Used to Identify miRNAs | Time When the miRNAs Were Analyzed (after CIS Exposure/Administration) | miRNAs Differentially Expressed in Nephrotoxicity | Pathophysiological Implications of Dysregulated miRNAs (Related to the Authors’ Own Results) | Role of miRNAs as Biomarkers of Nephrotoxicity |
| Bhatt et al./2010 [34] | Cells (BUMPT-306) | RT-qPCR | 0, 4, 8, and 12 h. | Upregulated: miR-34a | This study showed that miR-34a induction during CIS nephrotoxicity was mediated by p53. However, blockage of miR-34a increased cell injury and death. The authors speculated that miR-34a may regulate or repress proapoptotic genes. | There is evidence for a cytoprotective role of induced miR-34a against CIS-induced apoptosis in renal cells. |
| Kidney tissue | Days 0, 1, 2, and 3. | |||||
| Du et al./2017 [37] | Cells (HK-2 and NRK-52E). | RT-qPCR | 24, 48, 72 h (HK2 cells) or 12, 24, 48 h (NRK-52E). | Downregulated-Both cells: miR-30a miR-30b miR-30c miR-30d miR-30e | The downregulation of miR-30c induced by CIS positively regulated the expression of its Bnip3L and Hspa5 target genes, which resulted in significant increase on apoptosis. | MiR-30c might be involved in regulating CIS-induced cell apoptosis, and it might supply a new strategy to minimize CIS-induced nephrotoxicity. |
| Kidney tissue | Days 1, 3, and 7. | |||||
| Liao et al./2017 [39] | Cells (HK-2) | RT-qPCR | 42 h. | Upregulated: miR-140-5p | CIS exposure upregulated miR-140-5p in response to oxidative stress induced by CIS. It was also showed that MnSOD activity and cell vitality were increased, and LDH leakage was reduced in miR-140-5p overexpression. In fact, miR-140-5p directly targets the 3′-UTR of Nrf2 mRNA and increases the Nrf2 expression. The activation of Nrf2 pathway is a mechanism involved in ROS-protection by increased expression of antioxidant genes thus attenuating oxidative stress. | The overexpression of miR-140-5p after exposure to CIS may protect against CIS induced oxidative stress by activating Nrf2-dependent antioxidant pathway and provides a potentially therapeutic target in AKI. |
| Kidney tissue | Days 1, 3, 5, 7, and 14. | |||||
| Yang et al./2019 [41] | Cells (HK-2) | RT-qPCR | 24 h. | Upregulated: miR-199a-3p | MiR-199a-3p directly bound to mTOR 3′-untranslated region and reduced the expression and phosphorylation of mTOR. Moreover, p53 inhibited mTOR activation through activating miR-199a-3p. Blockade of miR-199a-3p significantly reduced CIS-induced cell apoptosis and inhibited caspase-3 activity. | P53 promoted miR-199a-3p expression both in vivo and in vitro, which subsequently inhibited mTOR signaling. So, it might provide a promising therapeutic target of AKI. |
| Kidney tissue | Day 3. | |||||
| Yang et al./2019 [42] | Cells (HK-2) | RT-qPCR | 24 h. | Downregulated: miR-26a | The upregulation of miR-26a (using miR- 26a mimics) alleviated the CIS-induced injury via the downregulation of TRPC6. Overexpression of miR-26a could attenuate CIS-induced cell injury. Upregulation of miR-26a could restrain Drp1 expression (an important mediator in regulating mitochondrial fission), which was consistent with the changes in TRPC6 expression. This means that the renoprotective effects of miR-26a against CIS-induced cells injury were inhibited through the mitochondrial apoptosis pathway. | MiR-26a can protect CIS-induced HK2 cell apoptosis via negatively regulating TRPC6 expression and may be targeted for the prevention and treatment of drug-related AKI. |
| Kidney tissues | Day 3. | |||||
| Li et al./2021 [43] | Cells (HK-2) | RT-qPCR | 24h. | Cells and kidney—Upregulated: miR-182-5p | Bioinformatic analysis predicted that miR-182-5p is a target gene for PRNCR1, and EZH1 was predicted to be a target gene for miR-182-5p. The study showed that miR-182-5p was negatively regulated by PRNCR1 and leed to upregulation of EZH1 expression. Overexpression of PRNCR1 attenuated CIS-induced apoptosis by downregulating the miR-182-5p/EZH1 axis. | The expression level of miR-182-5p was raised in mouse kidney and HK-2 cells after cisplatin treatment. miR-182-5p was the target gene of PRNCR1. |
| Kidney | Days 1,3, and 7. | |||||
| Guo et al./2018 [40] | Cells (mPTC) | Microarray and qPCR | 0, 2, 6, 12, and 24 h. | Upregulated: miR-709 | The renal tubular mitochondrial dysfunction and cell apoptosis induced by cisplatin insult was almost completely blocked by anti–miR-709 management both in vitro and in vivo, suggesting a pathogenic role of miR-709 through mitochondrial damage in this kidney toxic injury model. MiR-709 targets the critical mitochondrial protective protein TFAM and impairs the biogenesis of the mitochondria in the renal tubular cells after acute insult. | Upregulation of renal tubular miR-709 after AKI mediates mitochondrial dysfunction and cell apoptosis by depressing TFAM expression. Targeting miR-709 may serve as a new approach to preserving mitochondrial function and preventing cell death in AKI. |
| Kidney tissue | Day 3. | |||||
| Lee et al./2014 [35] | Cell (NRK-52E) | Microarray and RT-qPCR | 24, 48, and 72 h. | Downregulated: miR-122 (the most downregulated) miR-10b miR-30e miR-193 miR-26a Upregulated: miR-34a (the most upregulated) let-7g | MiR-122 can be a direct suppressor of Foxo3 mRNA translation, while miR-34a activates Foxo3 by suppressing SIRT1. Increased expression and activation of Foxo3 has a role in triggering the p53 signaling pathway, culminating in cell apoptosis. Therefore, miR-122 and miR-34a dysregulation induces and actives Foxo3 contributing to CIS-induced acute tubular injury by fortifying the p53 signaling pathway. | The modulation of miR-122 and miR-34a could be a mechanism with which to prevent or treat AKI-induced by CIS. |
| Kidney tissue | Days 1, 3, and 5. | |||||
| Xiong et al./2021 [44] | Cells (NRK-52E) | RT-qPCR | 0, 12, 24, 36, 48 h | Cells and animals - Downregulated: miR-186 | Overexpressing miR-186 could reverse the effects of cisplatin on NRK-52E cells proliferation and apoptosis. Moreover, inflammatory cytokines (IL-6, IL-1β, TNF-α, and Cox-2) expression was elevated by CIS; the increase of miR-186 reversed it, implying that increase of miR-186 repressed cell inflammatory response induced by CIS. ZEB1 was identified as miR-186 downstream target, which was found to be increased in AKI rat models. Knockdown of ZEB1 increased NRK-52E cell proliferation and restrained the apoptosis induced by CIS. | Loss of miR-186 expression contributed to CIS-induced AKI, partly through targeting ZEB1. MiR-186 might be provided an effective biomarker of AKI and a potential therapeutic target for its treatment. |
| Serum and kidney | Days 0, 1, 3, and 5. | |||||
| Hao et al./2017 [38] | Cells (RPTC) | Microarray and RT-qPCR | 0, 4, 8, 12, 16 h. | Kidney tissue-Microarray: both 1 and 3 days of CIS, FC ≥ 3 (all upregulated): miR-375 miR-503 miR-547 miR-212 miR-31* miR-743a | CIS treatment induces the activation of P53 and NF-κB, which collaboratively induce the expression of miR-375, which then represses the anti-apoptotic gene HNF-1β contributing to renal tubular cell injury and death (P53/NF-κB/miR-375/HNF-1β pathway in CIS-induced apoptosis). | It is suggested that miR-375 is an injurious miRNA. It may contribute to tubular cell injury and death during CIS nephrotoxicity. Delineation of P53/NF- κB/miR-375/HNF-1β pathway may provide novel therapeutic targets for kidney protection during CIS chemotherapy in cancer patients. |
| Kidney tissues | Days 1 and 3. | |||||
| Humans | ||||||
| Author(s)/Year | Sample | Methods Used to Identify miRNAs | Time When the miRNAs Were Analyzed (after CIS Exposure/Administration) | miRNAs Differentially Expressed in Nephrotoxicity | Pathophysiological Implications of Dysregulated miRNAs (Related to the Authors’ Own Results) | Role of miRNAs as Biomarkers of Nephrotoxicity |
| Quintanilha et al./2021 [45] | Plasma | NGS and RT-qPCR | Before and 5 days after. | Upregulated (before CIS administration in patients with grade ≥ 2 increased serum CRE): miR-3168 miR-4718 miR-6125 | Bioinformatics analysis showed that upregulated miR-3168 targeting genes of the ErbB signaling pathway, which target PDK, could downregulate the pathway, leading to CIS-induced apoptosis in renal cells. The regulation of genes involved in the mitochondrial apoptosis pathway may also contribute to higher nephrotoxicity, suggested by a decrease in the activity of the anti-apoptotic protein Bcl-2 by miR-3168 and miR-6125. Genes of the CIS detoxification pathway, which includes the conjugation of CIS with glutathione, are also shown to be target of miR-3168 and miR-6125, which could reduce the content of glutathione S-transferase and reduced glutathione. | The evidence suggests the baseline plasmatic expression of miR-3168, miR-6125, and miR-4718 as potential predictors of CIS-induced nephrotoxicity, with miR-4718 being the most promising marker. |
| Humans and Cells | ||||||
| Author(s)/Year | Sample | Methods Used to Identify miRNAs | Time When the miRNAs Were Analyzed (after CIS Exposure/Administration) | miRNAs Differentially Expressed in Nephrotoxicity | Pathophysiological Implications of Dysregulated miRNAs (Related to the Authors’ Own Results) | Role of miRNAs as Biomarkers of Nephrotoxicity |
| Pavkovic et al./2016 [46] | Cells (HPTEC) | qPCR | 24 h. | Upregulated in human urine e downregulated in cells: miR-21 miR-200c miR-423 (Obs.: in humans, the miRNAs were high in patients with AKI diagnosis and also in patients without clinically proven AKI.) | Target prediction analysis of these miRNAs showed that the top pathway and associated pathological condition was found to be MYC-mediated apoptosis signaling and renal necrosis/cell death, respectively. In addition, they have several lapping targets including genes well-known in apoptosis as p21. | MiR-21, miR-200c, and miR-423 can be non-invasive and specific urinary biomarkers for the detection of drug-induced AKI in patients. |
| Urine | Prior therapy and on subsequent time points: 4, 8, 12, 24, 48, 72, 96, 120, and 144 h. | |||||
| miRNAs | Cells | Animals | Human | Total Number of Studies Where the miRNA Was Dysregulated | |||||
|---|---|---|---|---|---|---|---|---|---|
| Urine/Plasma | Kidney Tissue | Urine/Plasma | |||||||
| Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | ||
| miR-34a | Suter-Dick et al.,2018 [47]; Bhatt et al./2010 [34]; Lee at al./2014 [35] | Pavkovic et al., 2014 [52]; Wolenski et al./2017 [27] | Pavkovic et al., 2014 [52]; Wolenski et al./2017 [27]; El Magdoub et al., 2020 [30]; Bhatt et al./2010 [34]; Lee at al./2014 [35] | 6 | |||||
| miR-21 | Suter-Dick et al.,2018 [47] | Zhang et al., 2015 [25]; Pavkovic et al., 2016 [46] | Pavkovic et al., 2014 [52]; Okamoto et al./2021 [33] | Pavkovic et al., 2014 [52] | Pavkovic et al., 2016 [46] | 5 | |||
| let-7g-5p | Kanki et al., 2014 [51]; Wolenski et al., 2017 [27] | Kagawa et al., 2019 [29] | Lee at al./2014 [35] | 4 | |||||
| miR-26a-5p | Yang et al., 2019 [42] | Okamoto et al./2021 [33] | Kagawa et al., 2019 [29] | Lee at al./2014 [35]; Yang et al., 2019 [42] | 4 | ||||
| miR-192-5p | Suter-Dick et al./2018 [47] | Kanki et al., 2014 [51]; Pavkovic et al., 2014 [52] | Kagawa et al., 2019 [29] | Pavkovic et al., 2014 [52] | 4 | ||||
| miR-30a-5p | Du et al., 2017 [37] | Kanki et al., 2014 [51] | Kagawa et al., 2019 [29] | Du et al., 2017 [37] | 3 | ||||
| miR-15b-5p | Pavkovic et al., 2014 [52]; Glineur et al., 2018 [28] | Pavkovic et al., 2014 [52] | 2 | ||||||
| miR-20b-5p | Kanki et al., 2014 [51]; Pavkovic et al., 2014 [52] | Pavkovic et al., 2014 [52] | 2 | ||||||
| miR-25-3p | Kanki et al., 2014 [51] | Kagawa et al., 2019 [29] | 2 | ||||||
| miR-26b-5p | Wolenski et al., 2017 [27] | El Magdoub et al., 2020 [30] | 2 | ||||||
| miR-30e | Du et al., 2017 [37] | Lee at al./2014 [35]; Du et al., 2017 [37] | 2 | ||||||
| miR-34c-5p | Wolenski et al., 2017 [27]; Glineur et al., 2018 [28] | 2 | |||||||
| miR-122-5p | Lee at al./2014 [35] | Kagawa et al., 2019 [29] | Lee at al./2014 [35] | 2 | |||||
| miR-130b-3p | Kanki et al., 2014 [51]; Wolenski et al., 2017 [27] | 2 | |||||||
| miR-140-3p | Kanki et al., 2014 [51] | Kagawa et al., 2019 [29] | 2 | ||||||
| miR-146a | Cho et al.,2017 [53] | Pavkovic et al., 2014 [52] | 2 | ||||||
| miR-181a | Zhu et al., 2012 [24] | Huang et al.,2020 [31] | 2 | ||||||
| miR-182-5p | Li et al., 2021 [43] | Glineur et al., 2018 [28]; Li et al., 2021 [43] | 2 | ||||||
| miR-183-5p | Kanki et al., 2014 [51]; Glineur et al., 2018 [28] | 2 | |||||||
| miR-191a-5p | Kanki et al., 2014 [51] | Kagawa et al., 2019 [29] | 2 | ||||||
| miR-193 | Pavkovic et al., 2014 [52] | Pavkovic et al., 2014 [52]; Lee at al./2014 [35] | 2 | ||||||
| miR-210-3p | Pavkovic et al., 2014 [52]; Glineur et al., 2018 [28] | Pavkovic et al., 2014 [52] | 2 | ||||||
| miR-320-3p | Kanki et al., 2014 [51]; Harrill et al., 2017 [26] | 2 | |||||||
| miR-423-3p | Pavkovic et al., 2016 [46] | Glineur et al., 2018 [28] | Pavkovic et al., 2016 [46] | 2 | |||||
| miR-500a-3p | Jiang et al.,2019 [48]; Zhang et al., 2020 [50] | 2 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Godoy Torso, N.; Pereira, J.K.N.; Visacri, M.B.; Vasconcelos, P.E.N.S.; Loren, P.; Saavedra, K.; Saavedra, N.; Salazar, L.A.; Moriel, P. Dysregulated MicroRNAs as Biomarkers or Therapeutic Targets in Cisplatin-Induced Nephrotoxicity: A Systematic Review. Int. J. Mol. Sci. 2021, 22, 12765. https://doi.org/10.3390/ijms222312765
de Godoy Torso N, Pereira JKN, Visacri MB, Vasconcelos PENS, Loren P, Saavedra K, Saavedra N, Salazar LA, Moriel P. Dysregulated MicroRNAs as Biomarkers or Therapeutic Targets in Cisplatin-Induced Nephrotoxicity: A Systematic Review. International Journal of Molecular Sciences. 2021; 22(23):12765. https://doi.org/10.3390/ijms222312765
Chicago/Turabian Stylede Godoy Torso, Nadine, João Kleber Novais Pereira, Marília Berlofa Visacri, Pedro Eduardo Nascimento Silva Vasconcelos, Pía Loren, Kathleen Saavedra, Nicolás Saavedra, Luis A. Salazar, and Patricia Moriel. 2021. "Dysregulated MicroRNAs as Biomarkers or Therapeutic Targets in Cisplatin-Induced Nephrotoxicity: A Systematic Review" International Journal of Molecular Sciences 22, no. 23: 12765. https://doi.org/10.3390/ijms222312765
APA Stylede Godoy Torso, N., Pereira, J. K. N., Visacri, M. B., Vasconcelos, P. E. N. S., Loren, P., Saavedra, K., Saavedra, N., Salazar, L. A., & Moriel, P. (2021). Dysregulated MicroRNAs as Biomarkers or Therapeutic Targets in Cisplatin-Induced Nephrotoxicity: A Systematic Review. International Journal of Molecular Sciences, 22(23), 12765. https://doi.org/10.3390/ijms222312765









