Aedes aegypti Piwi4 Structural Features Are Necessary for RNA Binding and Nuclear Localization
Abstract
:1. Introduction
2. Results
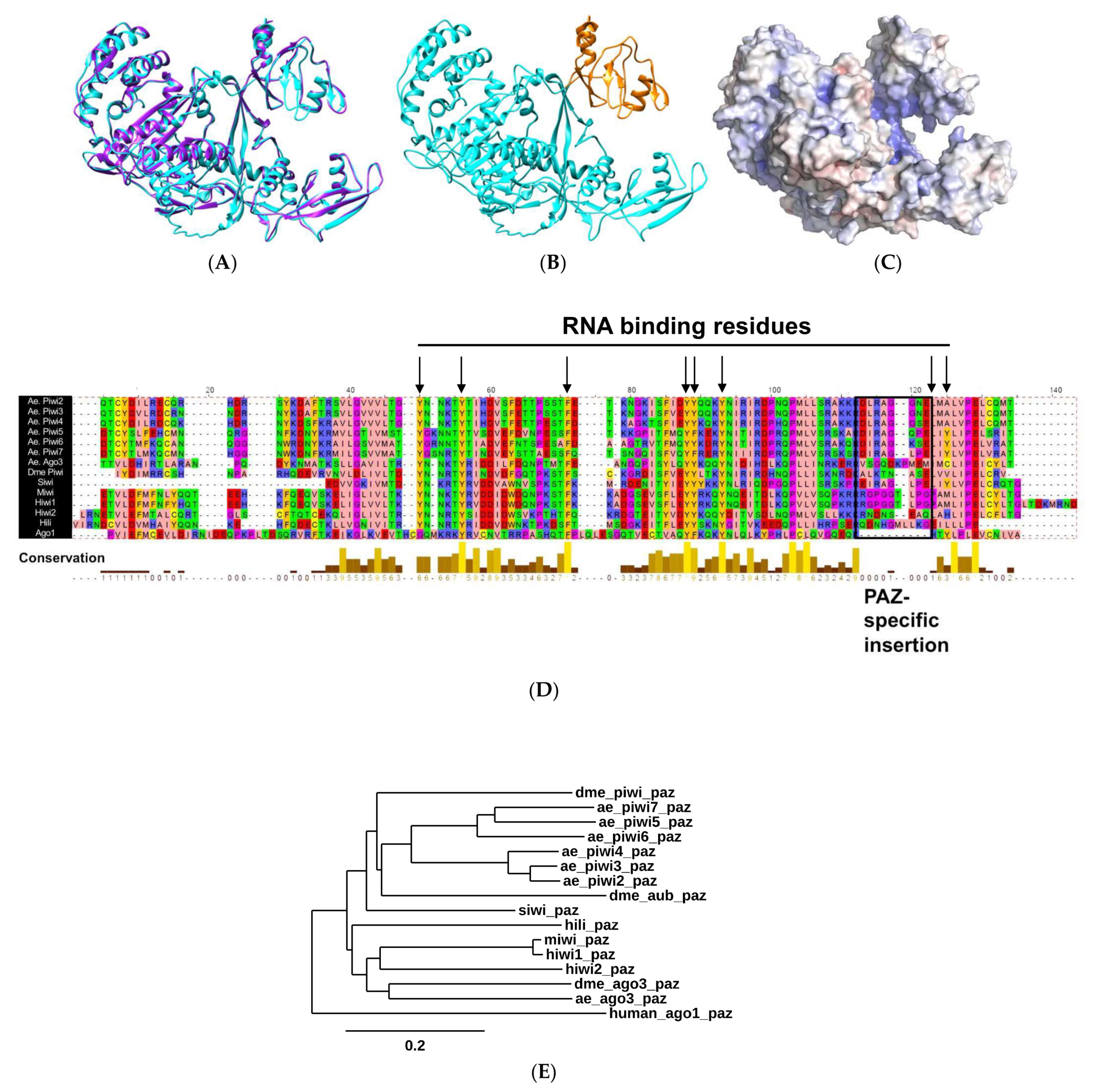
2.1. Biophysical Properties of AePiwi4 by Structural Modeling and Alignment
2.2. A. aegypti Piwi4 PAZ Binds 3′ 2′ O-Methylated and Non-Methylated piRNAs in a Sequence-Independent Manner
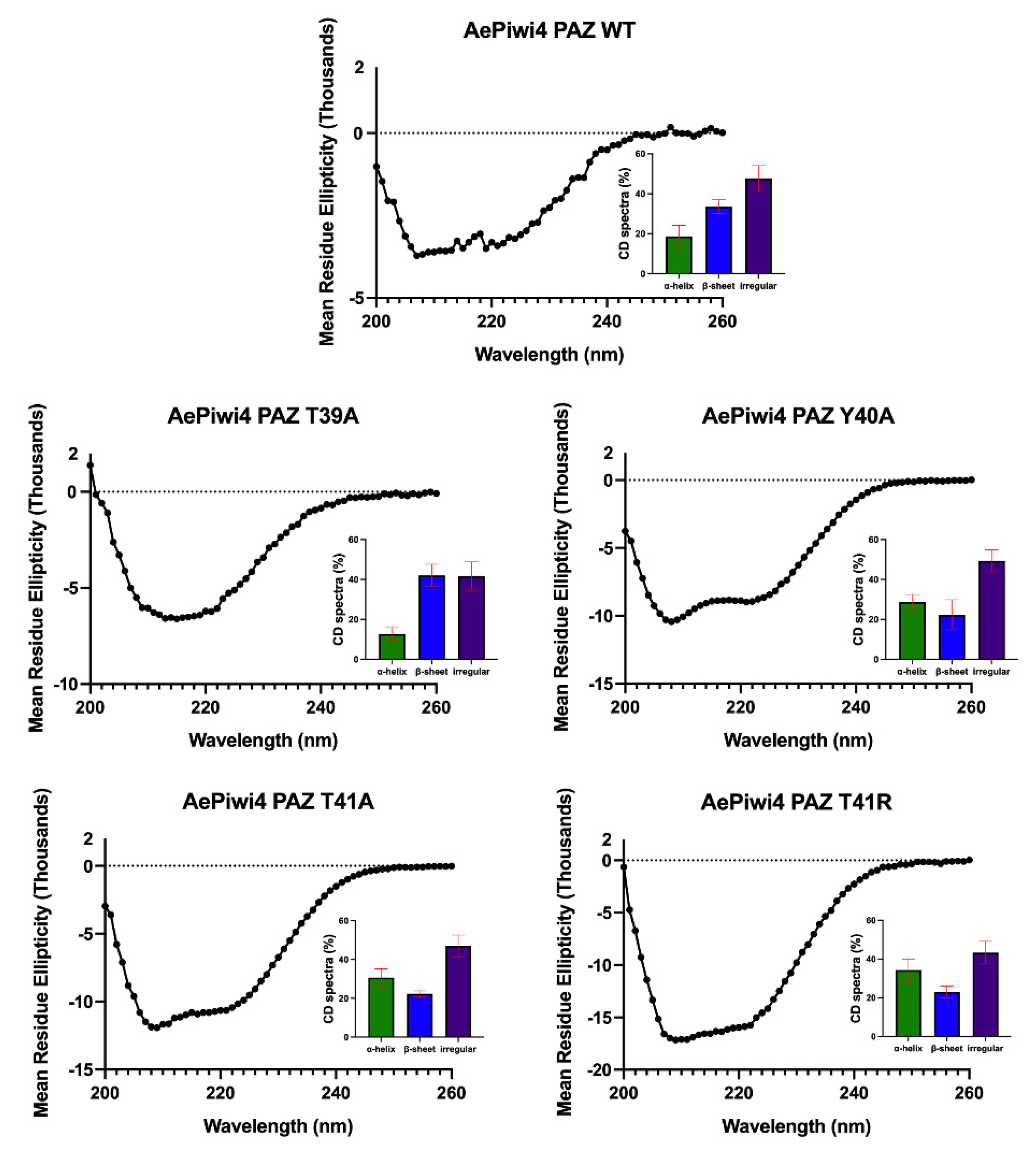
2.3. A. aegypti Piwi4 PAZ Mutants Reveal the Amino Acids Necessary for piRNA Binding
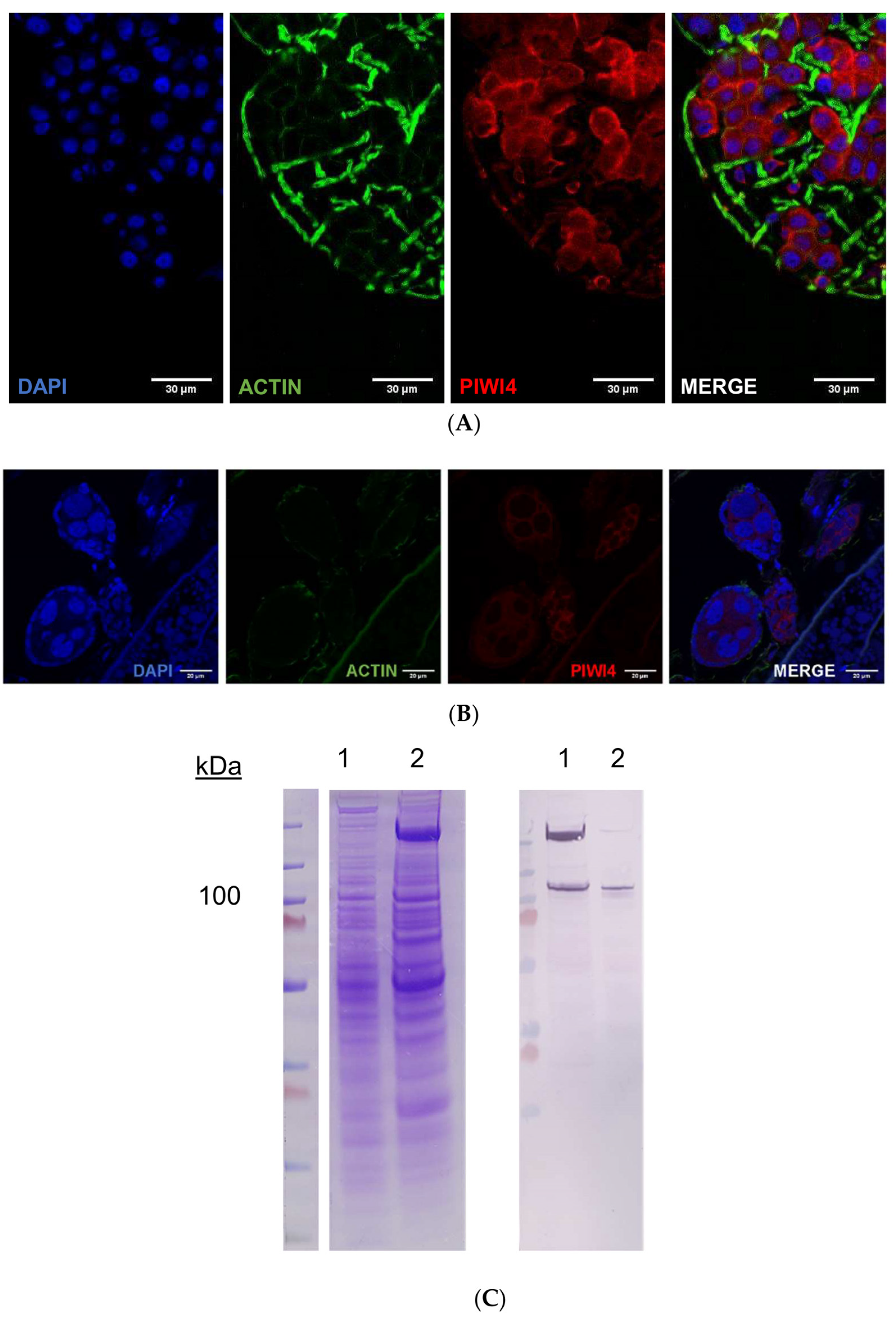
2.4. AePiwi4 Co-Localizes in the Cytoplasm and Nucleus in A. aegypti Tissues
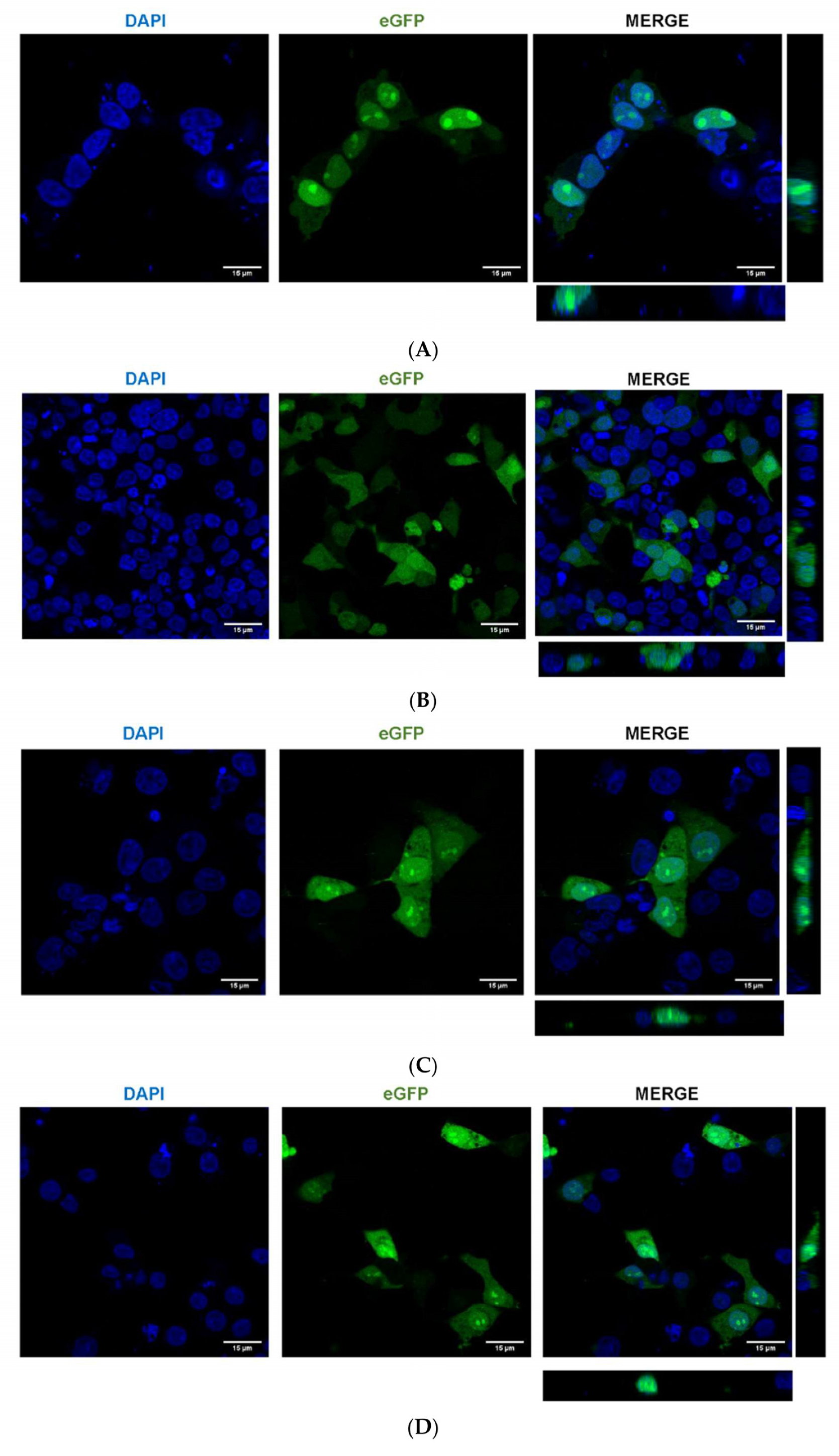
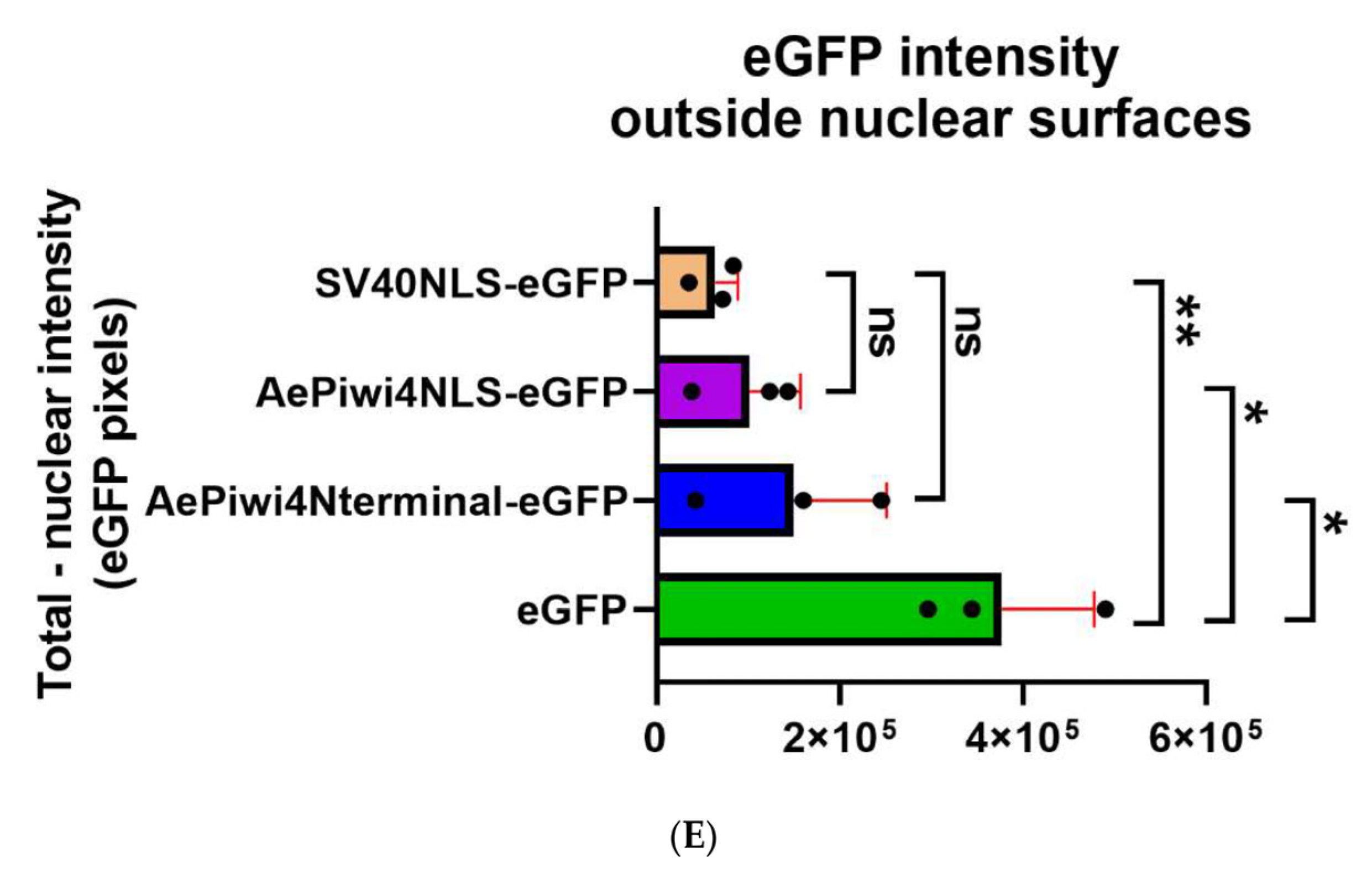
2.5. A. aegypti Piwi4 Expresses a Nuclear Localization Signal in the N-Terminal Region of the Protein
3. Discussion
4. Materials and Methods
4.1. A. aegypti Piwi4 Structure Model Prediction
4.2. Cloning
4.3. Recombinant Protein Expression
4.4. Recombinant Protein Purification
4.5. SDS-PAGE
4.6. Western Blot
4.7. Mosquito Rearing
4.8. Sequence Alignment
4.9. Site Directed Mutagenesis
4.10. Surface Plasmon Resonance (SPR)
4.11. Circular Dichroism
4.12. RNA Synthesis
4.13. Mouse Polyclonal Antibody Production
4.14. Mass Spectrometry
4.15. IFA
4.16. HEK293 Cell Culture and Transfection
4.17. Statistics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Aravin, A.A.; Sachidanandam, R.; Girard, A.; Fejes-Toth, K.; Hannon, G.J. Developmentally Regulated piRNA Clusters Implicate MILI in Transposon Control. Science 2007, 316, 744–747. [Google Scholar] [CrossRef] [Green Version]
- O’Donnell, K.A.; Boeke, J. Mighty Piwis Defend the Germline against Genome Intruders. Cell 2007, 129, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Senti, K.A.; Brennecke, J. The piRNA pathway: A fly’s perspective on the guardian of the genome. Trends Genet. 2010, 26, 499–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tóth, K.F.; Pezic, D.; Stuwe, E.; Webster, A. The piRNA Pathway Guards the Germline Genome Against Transposable Elements. Adv. Exp. Med. Biol. 2016, 886, 51–77. [Google Scholar] [CrossRef] [Green Version]
- Höck, J.; Meister, G. The Argonaute protein family. Genome Biol. 2008, 9, 210. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Munafo, M.; Ciabrelli, F.; Eastwood, E.L.; Fabry, M.H.; Kneuss, E.; Hannon, G.J. piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu. Rev. Genet. 2018, 52, 131–157. [Google Scholar] [CrossRef] [PubMed]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef] [Green Version]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [Green Version]
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA Pathways Act in Germline and Somatic Tissues of the Drosophila Ovary. Cell 2009, 137, 522–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théron, E.; Maupetit-Mehouas, S.; Pouchin, P.; Baudet, L.; Brasset, E.; Vaury, C. The interplay between the Argonaute proteins Piwi and Aub within Drosophila germarium is critical for oogenesis, piRNA biogenesis and TE silencing. Nucleic Acids Res. 2018, 46, 10052–10065. [Google Scholar] [CrossRef]
- Le Thomas, A.; Rogers, A.K.; Webster, A.; Marinov, G.K.; Liao, S.E.; Perkins, E.M.; Hur, J.K.; Aravin, A.A.; Tóth, K.F. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013, 27, 390–399. [Google Scholar] [CrossRef] [Green Version]
- Senti, K.-A.; Jurczak, D.; Sachidanandam, R.; Brennecke, J. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 2015, 29, 1747–1762. [Google Scholar] [CrossRef] [Green Version]
- Fabry, M.H.; Ciabrelli, F.; Munafo, M.; Eastwood, E.L.; Kneuss, E.; Falciatori, I.; Falconio, F.A.; Hannon, G.J.; Czech, B. piRNA-guided co-transcriptional silencing coopts nuclear export factors. eLife 2019, 8, e47999. [Google Scholar] [CrossRef]
- Cox, D.N.; Chao, A.; Baker, J.; Chang, L.; Qiao, D.; Lin, H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998, 12, 3715–3727. [Google Scholar] [CrossRef] [Green Version]
- Aravin, A.A.; Naumova, N.M.; Vagin, V.V.; Rozovsky, Y.M.; Gvozdev, V.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001, 11, 1017–1027. [Google Scholar] [CrossRef] [Green Version]
- Klattenhoff, C.; Bratu, D.P.; McGinnis-Schultz, N.; Koppetsch, B.S.; Cook, H.A.; Theurkauf, W.E. Drosophila rasiRNA Pathway Mutations Disrupt Embryonic Axis Specification through Activation of an ATR/Chk2 DNA Damage Response. Dev. Cell 2007, 12, 45–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Vagin, V.V.; Lee, S.; Xu, J.; Ma, S.; Xi, H.; Seitz, H.; Horwich, M.D.; Syrzycka, M.; Honda, B.M.; et al. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 2009, 137, 509–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muerdter, F.; Guzzardo, P.M.; Gillis, J.; Luo, Y.; Yu, Y.; Chen, C.; Fekete, R.; Hannon, G.J. A Genome-wide RNAi Screen Draws a Genetic Framework for Transposon Control and Primary piRNA Biogenesis in Drosophila. Mol. Cell 2013, 50, 736–748. [Google Scholar] [CrossRef] [Green Version]
- Handler, D.; Meixner, K.; Pizka, M.; Lauss, K.; Schmied, C.; Gruber, F.S.; Brennecke, J. The Genetic Makeup of the Drosophila piRNA Pathway. Mol. Cell 2013, 50, 762–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theron, E.; Dennis, C.; Brasset, E.; Vaury, C. Distinct features of the piRNA pathway in somatic and germ cells: From piRNA cluster transcription to piRNA processing and amplification. Mob. DNA 2014, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.R.; Juliano, C.E. Untangling the web: The diverse functions of the PIWI/piRNA pathway. Mol. Reprod. Dev. 2013, 80, 632–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weick, E.-M.; Miska, E.A. piRNAs: From biogenesis to function. Development 2014, 141, 3458–3471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.-N.; Li, Y.; Xia, S.-Q.; Zhang, Y.-Y.; Zheng, J.-H.; Li, W. PIWI Proteins and PIWI-Interacting RNA: Emerging Roles in Cancer. Cell. Physiol. Biochem. 2017, 44, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Lewis, S.; Quarles, K.; Yang, Y.; Tanguy, M.; Frezal, L.; Smith, S.; Sharma, P.; Cordaux, R.; Gilbert, C.; Giraud, I.; et al. Pan-arthropod analysis reveals somatic piRNAs as an ancestral defence against transposable elements. Nat. Ecol. Evol. 2018, 2, 174–181. [Google Scholar] [CrossRef] [Green Version]
- Miesen, P.; Girardi, E.; van Rij, R.P. Distinct sets of PIWI proteins produce arbovirus and transposon-derived piRNAs in Aedes aegypti mosquito cells. Nucleic Acids Res. 2015, 43, 6545–6556. [Google Scholar] [CrossRef] [Green Version]
- Joosten, J.; Taşköprü, E.; Jansen, P.W.; Pennings, B.; Vermeulen, M.; Van Rij, R.P. PIWI proteomics identifies Atari and Pasilla as piRNA biogenesis factors in Aedes mosquitoes. Cell Rep. 2021, 35, 109073. [Google Scholar] [CrossRef]
- Schnettler, E.; Donald, C.; Human, S.; Watson, M.; Siu, R.W.C.; McFarlane, M.; Fazakerley, J.; Kohl, A.; Fragkoudis, R. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J. Gen. Virol. 2013, 94, 1680–1689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miesen, P.; Joosten, J.; Van Rij, R.P. PIWIs Go Viral: Arbovirus-Derived piRNAs in Vector Mosquitoes. PLoS Pathog. 2016, 12, e1006017. [Google Scholar] [CrossRef] [PubMed]
- Varjak, M.; Dietrich, I.; Sreenu, V.B.; Till, B.E.; Merits, A.; Kohl, A.; Schnettler, E. Spindle-E Acts Antivirally Against Alphaviruses in Mosquito Cells. Viruses 2018, 10, 88. [Google Scholar] [CrossRef] [Green Version]
- Varjak, M.; Leggewie, M.; Schnettler, E. The antiviral piRNA response in mosquitoes? J. Gen. Virol. 2018, 99, 1551–1562. [Google Scholar] [CrossRef]
- Lee, W.-S.; Webster, J.A.; Madzokere, E.T.; Stephenson, E.B.; Herrero, L.J. Mosquito antiviral defense mechanisms: A delicate balance between innate immunity and persistent viral infection. Parasites Vectors 2019, 12, 165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tassetto, M.; Kunitomi, M.; Whitfield, Z.J.; Dolan, P.T.; Sánchez-Vargas, I.; Knight, M.A.G.; Ribiero, I.; Chen, T.E.; Olson, K.; Andino, R. Control of RNA viruses in mosquito cells through the acquisition of vDNA and endogenous viral elements. eLife 2019, 8, 8. [Google Scholar] [CrossRef]
- Blair, C.D. Deducing the Role of Virus Genome-Derived PIWI-Associated RNAs in the Mosquito–Arbovirus Arms Race. Front. Genet. 2019, 10, 1114. [Google Scholar] [CrossRef] [Green Version]
- Joosten, J.; Miesen, P.; Taşköprü, E.; Pennings, B.; Jansen, P.W.T.C.A.; Huynen, M.; Vermeulen, M.; Van Rij, R.P. The Tudor protein Veneno assembles the ping-pong amplification complex that produces viral piRNAs in Aedes mosquitoes. Nucleic Acids Res. 2019, 47, 2546–2559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, Y.; Baidaliuk, A.; Miesen, P.; Frangeul, L.; Crist, A.B.; Merkling, S.H.; Fontaine, A.; Lequime, S.; Moltini-Conclois, I.; Blanc, H.; et al. Non-retroviral Endogenous Viral Element Limits Cognate Virus Replication in Aedes aegypti Ovaries. Curr. Biol. 2020, 30, 3495–3506. [Google Scholar] [CrossRef] [PubMed]
- Petit, M.; Mongelli, V.; Frangeul, L.; Blanc, H.; Jiggins, F.; Saleh, M.-C. piRNA pathway is not required for antiviral defense in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2016, 113, E4218–E4227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vodovar, N.; Bronkhorst, A.W.; Van Cleef, K.W.R.; Miesen, P.; Blanc, H.; Van Rij, R.P.; Saleh, M.-C. Arbovirus-Derived piRNAs Exhibit a Ping-Pong Signature in Mosquito Cells. PLoS ONE 2012, 7, e30861. [Google Scholar] [CrossRef]
- Morazzani, E.M.; Wiley, M.R.; Murreddu, M.G.; Adelman, Z.N.; Myles, K.M. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012, 8, e1002470. [Google Scholar] [CrossRef]
- Halbach, R.; Miesen, P.; Joosten, J.; Taskopru, E.; Rondeel, I.; Pennings, B.; Vogels, C.B.F.; Merkling, S.H.; Koenraadt, C.J.; Lambrechts, L.; et al. A satellite repeat-derived piRNA controls embryonic development of Aedes. Nature 2020, 580, 274–277. [Google Scholar] [CrossRef]
- Betting, V.; Joosten, J.; Halbach, R.; Thaler, M.; Miesen, P.; Van Rij, R.P. A piRNA-lncRNA regulatory network initiates responder and trailer piRNA formation during mosquito embryonic development. RNA 2021, 27, 1155–1172. [Google Scholar] [CrossRef]
- Arensburger, P.; Hice, R.H.A.; Wright, J.; Craig, N.L.; Atkinson, P.W. The mosquito Aedes aegypti has a large genome size and high transposable element load but contains a low proportion of transposon-specific piRNAs. BMC Genom. 2011, 12, 606. [Google Scholar] [CrossRef] [Green Version]
- Girardi, E.; Miesen, P.; Pennings, B.; Frangeul, L.; Saleh, M.-C.; Van Rij, R.P. Histone-derived piRNA biogenesis depends on the ping-pong partners Piwi5 and Ago3 in Aedes aegypti. Nucleic Acids Res. 2017, 45, 4881–4892. [Google Scholar] [CrossRef] [PubMed]
- Varjak, M.; Maringer, K.; Watson, M.; Sreenu, V.B.; Fredericks, A.C.; Pondeville, E.; Donald, C.; Sterk, J.; Kean, J.; Vazeille, M.; et al. Aedes aegypti Piwi4 Is a Noncanonical PIWI Protein Involved in Antiviral Responses. mSphere 2017, 2, e00144-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Simanshu, D.K.; Ma, J.-B.; Patel, D.J. Structural basis for piRNA 2′-O-methylated 3′-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains. Proc. Natl. Acad. Sci. USA 2011, 108, 903–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, S.; Oe, A.; Nishida, K.M.; Yamashita, K.; Kajiya, A.; Hirano, S.; Matsumoto, N.; Dohmae, N.; Ishitani, R.; Saito, K.; et al. Crystal structure of Drosophila Piwi. Nat. Commun. 2020, 11, 858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, B.; Kirkpatrick, J.P.; Eckhardt, S.; Reuter, M.; Rocha, E.A.; Andrade, M.; Sehr, P.; Pillai, R.S.; Carlomagno, T. Recognition of 2′-O-Methylated 3′-End of piRNA by the PAZ Domain of a Piwi Protein. Structure 2011, 19, 172–180. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, N.; Nishimasu, H.; Sakakibara, K.; Nishida, K.M.; Hirano, T.; Ishitani, R.; Siomi, H.; Siomi, M.C.; Nureki, O. Crystal Structure of Silkworm PIWI-Clade Argonaute Siwi Bound to piRNA. Cell 2016, 167, 484–497. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.-B.; Ye, K.; Patel, D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nat. Cell Biol. 2004, 429, 318–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Q.; Srivastav, S.P.; Gamez, S.; Dayama, G.; Feitosa-Suntheimer, F.; Patterson, E.I.; Johnson, R.M.; Matson, E.M.; Gold, A.S.; Brackney, D.E.; et al. A mosquito small RNA genomics resource reveals dynamic evolution and host responses to viruses and transposons. Genome Res. 2021, 31, 512–528. [Google Scholar] [CrossRef]
- Wiedemann, C.; Bellstedt, P.; Görlach, M. CAPITO—A web server-based analysis and plotting tool for circular dichroism data. Bioinformatics 2013, 29, 1750–1757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariotti, N.; Hall, T.; Rae, J.; Ferguson, C.; McMahon, K.-A.; Martel, N.; Webb, R.E.; Webb, R.I.; Teasdale, R.; Parton, R.G. Modular Detection of GFP-Labeled Proteins for Rapid Screening by Electron Microscopy in Cells and Organisms. Dev. Cell 2015, 35, 513–525. [Google Scholar] [CrossRef] [Green Version]
- Gamez, S.; Srivastav, S.; Akbari, O.S.; Lau, N.C. Diverse Defenses: A Perspective Comparing Dipteran Piwi-piRNA Pathways. Cells 2020, 9, 2180. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Han, B.; Tipping, C.; Ge, D.T.; Zhang, Z.; Weng, Z.; Zamore, P.D. Slicing and Binding by Ago3 or Aub Trigger Piwi-Bound piRNA Production by Distinct Mechanisms. Mol. Cell 2015, 59, 819–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Hu, H.; Webster, A.; Zou, F.; Du, J.; Patel, D.J.; Sachidanandam, R.; Toth, K.F.; Aravin, A.A.; Li, S. Binding of guide piRNA triggers methylation of the unstructured N-terminal region of Aub leading to assembly of the piRNA amplification complex. Nat. Commun. 2021, 12, 4061. [Google Scholar] [CrossRef]
- Yashiro, R.; Murota, Y.; Nishida, K.M.; Yamashiro, H.; Fujii, K.; Ogai, A.; Yamanaka, S.; Negishi, L.; Siomi, H.; Siomi, M.C. Piwi Nuclear Localization and Its Regulatory Mechanism in Drosophila Ovarian Somatic Cells. Cell Rep. 2018, 23, 3647–3657. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dou, M.; Song, X.; Dong, Y.; Liu, S.; Liu, H.; Tao, J.; Li, W.; Yin, X.; Xu, W. The emerging role of the piRNA/piwi complex in cancer. Mol. Cancer 2019, 18, 123. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Pan, Y.; Fang, Y.; Zhang, J.; Xie, M.; Yang, F.; Yu, T.; Ma, P.; Li, W.; Shu, Y. The Biogenesis and Functions of piRNAs in Human Diseases. Mol. Ther.-Nucleic Acids 2020, 21, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; López, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin-Martin, I.; Paige, A.; Leon, P.C.V.; Gittis, A.G.; Kern, O.; Bonilla, B.; Chagas, A.C.; Ganesan, S.; Smith, L.B.; Garboczi, D.N.; et al. ADP binding by the Culex quinquefasciatus mosquito D7 salivary protein enhances blood feeding on mammals. Nat. Commun. 2020, 11, 2911. [Google Scholar] [CrossRef] [PubMed]
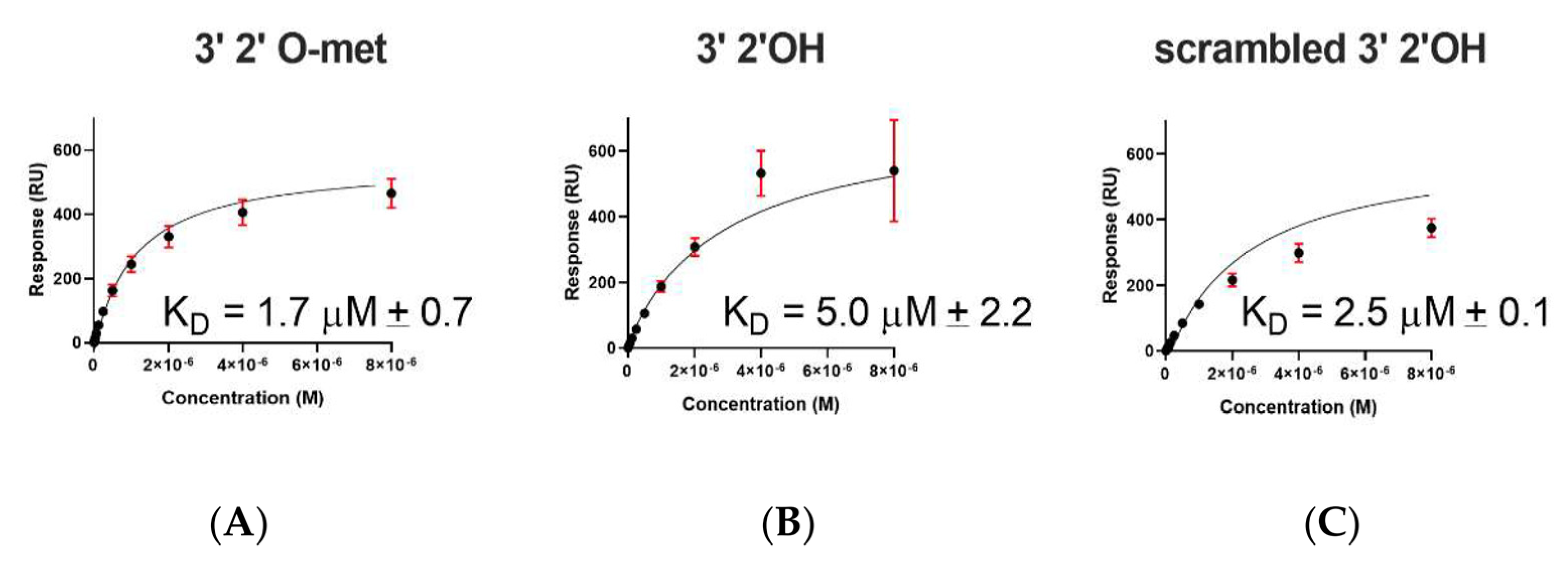
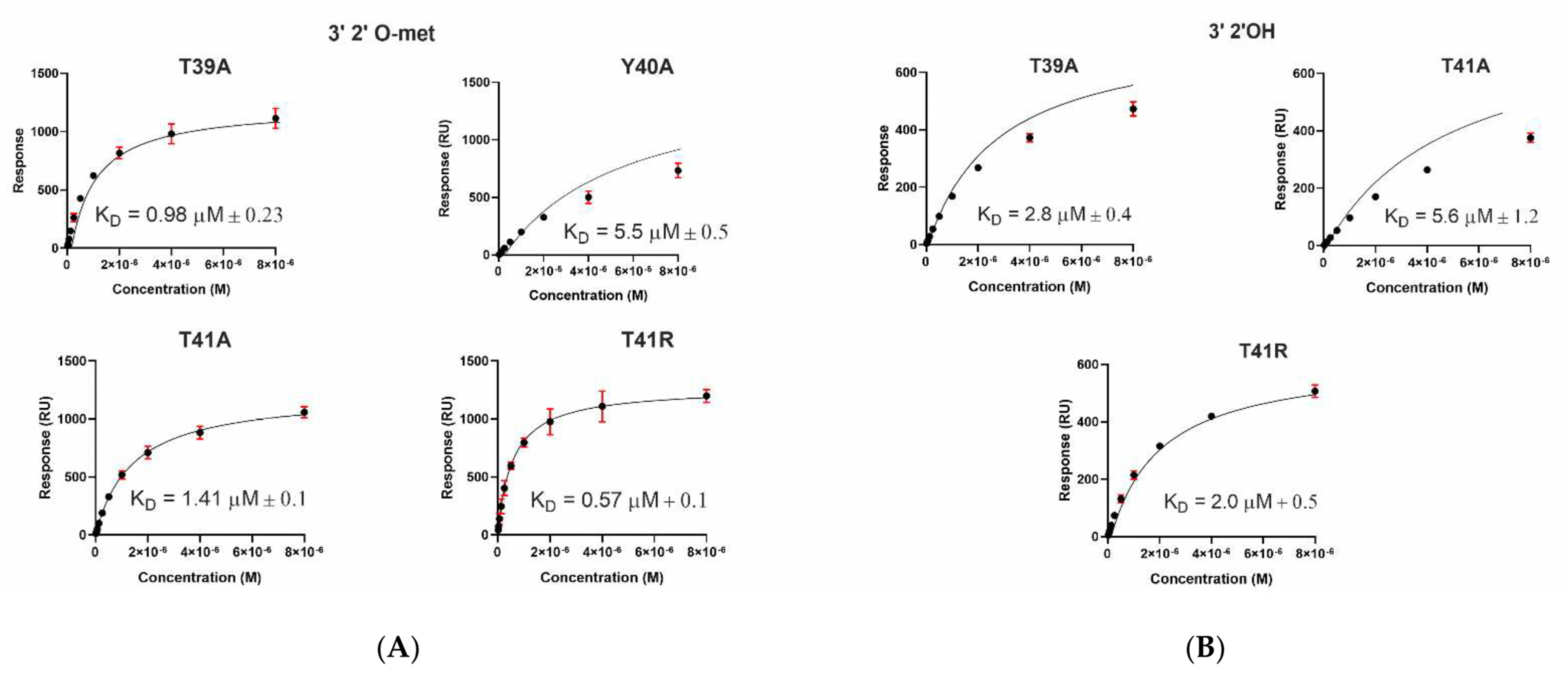
| Immobilized Ligand | Binding AePiwi4 PAZ | KD Values (μM) | Rmax (RU) |
|---|---|---|---|
| 3′ 2′met piRNA | WT | 1.7 ± 0.7 | 1364 ± 3 |
| T39A | 0.98 ± 0.2 | 1343 ± 2 | |
| Y40A | 5.5 ± 0.5 * | 1103 ± 62 | |
| T41A | 1.4 ± 0.1 | 1272 ± 14 | |
| T41R | 0.57 ± 0.1 | 1388 ± 18 | |
| F55A | No binding | ||
| 3′ 2′nmet piRNA | WT | 5 ± 2.2 | 585 ± 3 |
| T39A | 2.8 ± 0.4 * | 596 ± 5 | |
| Y40A | No binding | ||
| T41A | 5.6 ± 1.2 | 580 ± 8 | |
| T41R | 2.0 ± 0.5 * | 590 ± 3 | |
| F55A | No binding |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Williams, A.E.; Shrivastava, G.; Gittis, A.G.; Ganesan, S.; Martin-Martin, I.; Valenzuela Leon, P.C.; Olson, K.E.; Calvo, E. Aedes aegypti Piwi4 Structural Features Are Necessary for RNA Binding and Nuclear Localization. Int. J. Mol. Sci. 2021, 22, 12733. https://doi.org/10.3390/ijms222312733
Williams AE, Shrivastava G, Gittis AG, Ganesan S, Martin-Martin I, Valenzuela Leon PC, Olson KE, Calvo E. Aedes aegypti Piwi4 Structural Features Are Necessary for RNA Binding and Nuclear Localization. International Journal of Molecular Sciences. 2021; 22(23):12733. https://doi.org/10.3390/ijms222312733
Chicago/Turabian StyleWilliams, Adeline E., Gaurav Shrivastava, Apostolos G. Gittis, Sundar Ganesan, Ines Martin-Martin, Paola Carolina Valenzuela Leon, Ken E. Olson, and Eric Calvo. 2021. "Aedes aegypti Piwi4 Structural Features Are Necessary for RNA Binding and Nuclear Localization" International Journal of Molecular Sciences 22, no. 23: 12733. https://doi.org/10.3390/ijms222312733
APA StyleWilliams, A. E., Shrivastava, G., Gittis, A. G., Ganesan, S., Martin-Martin, I., Valenzuela Leon, P. C., Olson, K. E., & Calvo, E. (2021). Aedes aegypti Piwi4 Structural Features Are Necessary for RNA Binding and Nuclear Localization. International Journal of Molecular Sciences, 22(23), 12733. https://doi.org/10.3390/ijms222312733








