Identification of microRNAs in the Lyme Disease Vector Ixodes scapularis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Ticks and Tissue Dissections
2.3. RNA Isolation, cDNA Synthesis, and PCR-Based B. burgdorferi Detection in Tick Tissues
2.4. Small RNA Sequencing (RNA-Seq)
2.5. Data Analysis
2.6. ISE6 Cell Culture, Infection with B. burgdorferi, and Validation of Differentially Expressed miRNAs by qRT-PCR
2.7. Normalization, Differential Expression (DE), and Statistical Analysis of miRNAs between Uninfected and Infected Salivary Glands
2.8. In Silico Mapping of B. burgdorferi-Infected Small RNA Sequences to the Borrelia burgdorferi Genome
2.9. Prediction of Target Genes, Proteome Re-Annotation, Gene Ontology (GO), and KEGG Enrichment Analyses
3. Results and Discussion
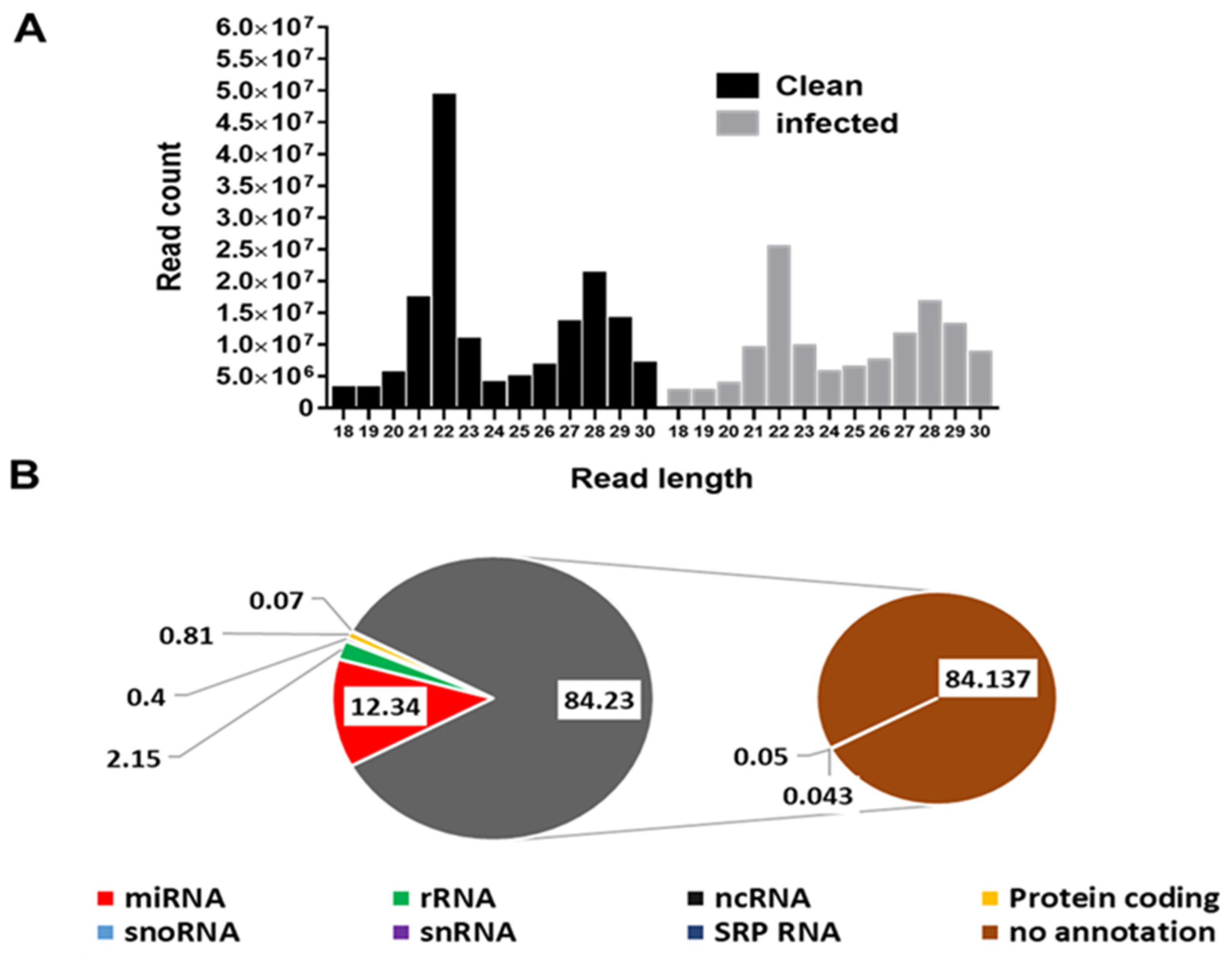
3.1. Profile Characteristics of Small RNA Libraries
3.2. Other Small RNA Categories
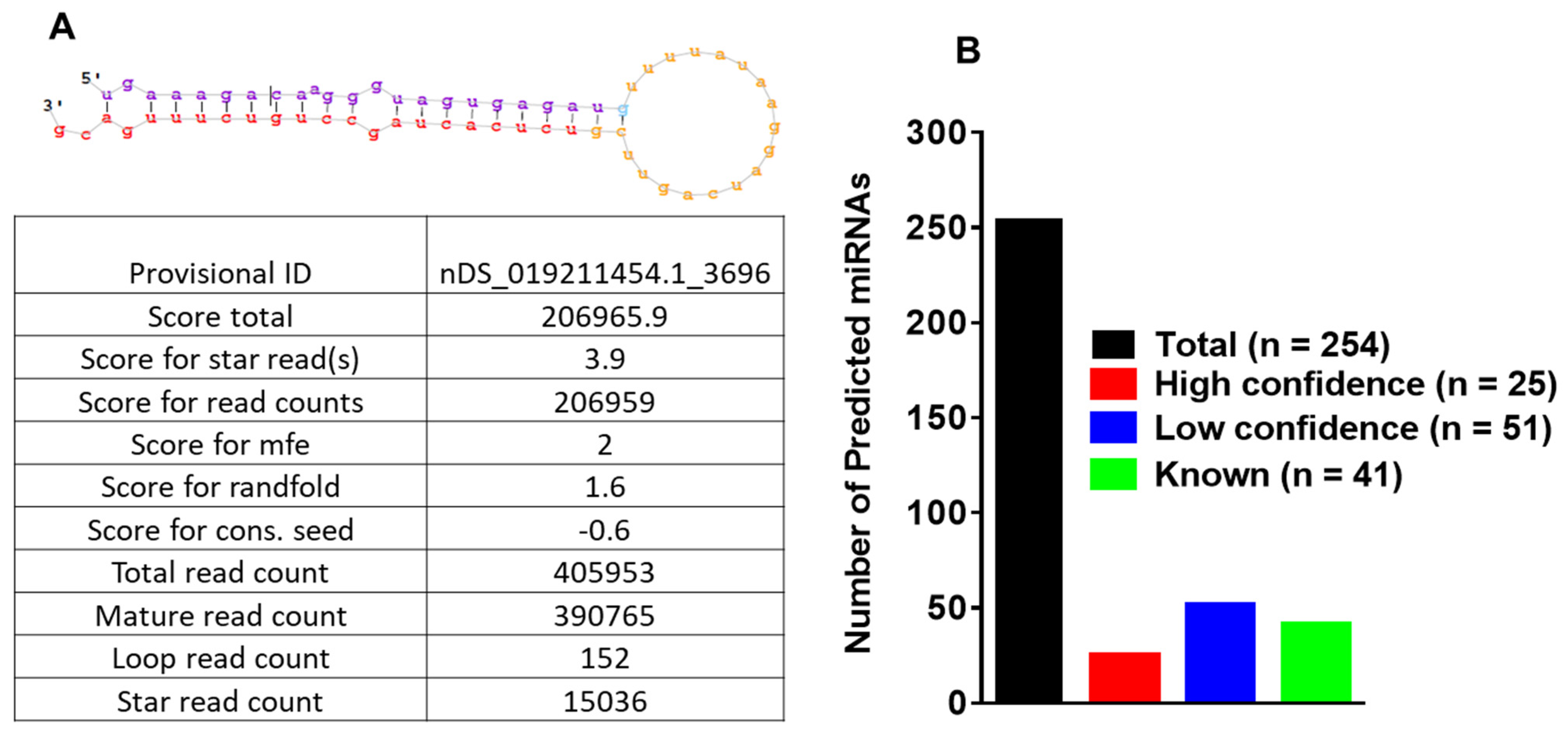
3.3. MicroRNA Profiling of Infected and Uninfected Salivary Glands and Identification of Novel Tick miRNAs
3.4. In Silico DE Analysis of miRNAs in B. burgdorferi-Infected Salivary Glands
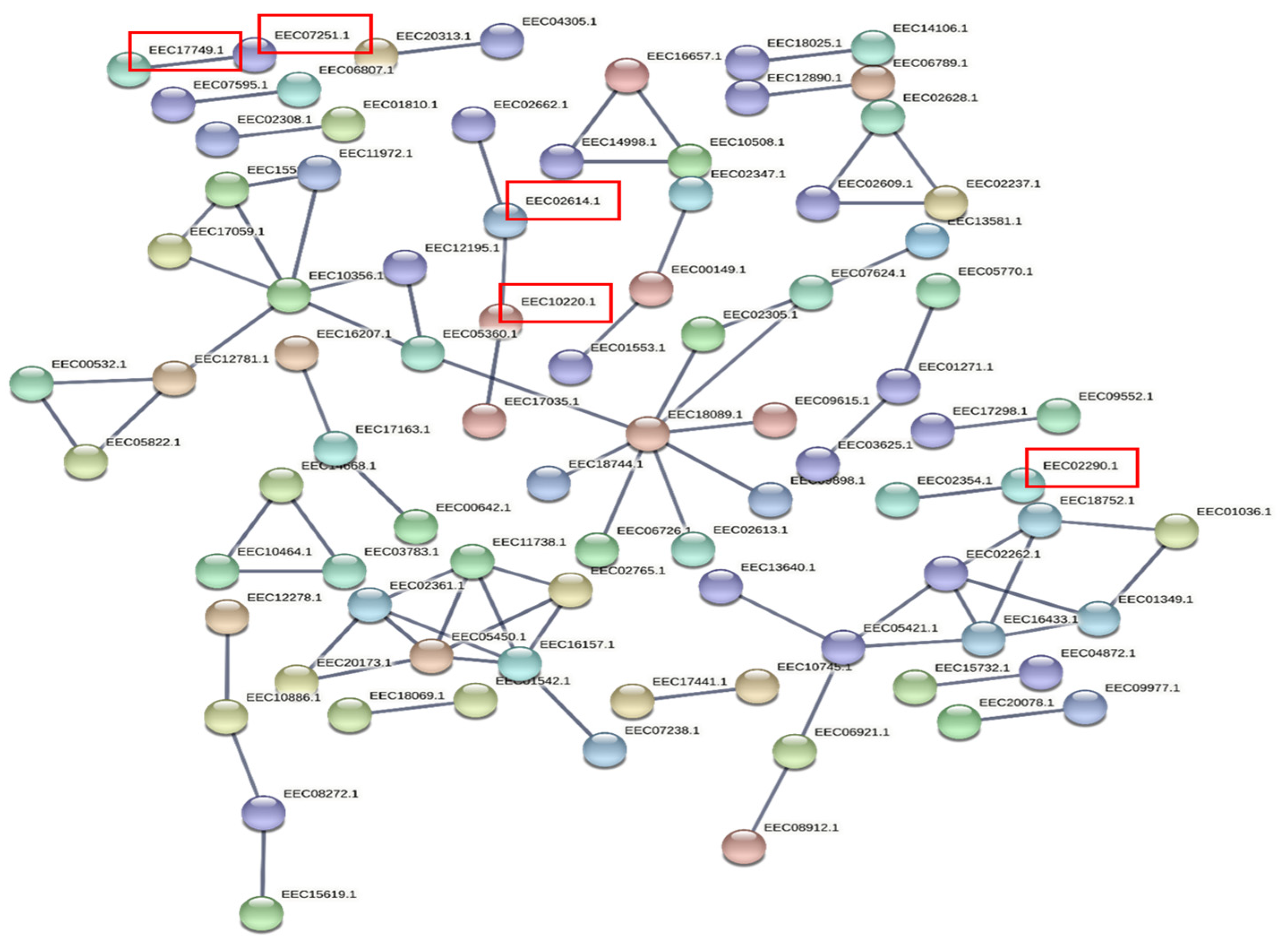
3.5. Prediction of Target Genes and Gene Ontology (GO) and Functional Enrichment Analyses of the Target Network
3.6. Validation of DE miRNAs by qRT-PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell. 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrington, J.C.; Ambros, V. Role of microRNAs in plant and animal development. Science 2003, 301, 336–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for MicroRNA Genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef] [Green Version]
- Lai, E.C. Two decades of miRNA biology: Lessons and Challenges. RNA 2015, 21, 675–677. [Google Scholar] [CrossRef]
- Bartel, D.P.; Chen, C.Z. Micromanagers of gene expression: The Potentially Widespread Influence of Metazoan MicroRNAs. Nat. Rev. Genet. 2004, 5, 396–400. [Google Scholar] [CrossRef]
- Alvarez-Garcia, I.; Miska, E.A. MicroRNA functions in animal development and human disease. Development 2005, 132, 4653–4662. [Google Scholar] [CrossRef] [Green Version]
- Miesen, P.; Ivens, A.; Buck, A.H.; van Rij, R.P. Small RNA Profiling in Dengue Virus 2-Infected Aedes Mosquito Cells Reveals Viral piRNAs and Novel Host miRNAs. PLoS Negl Trop Dis. 2016, 10, e0004452. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Bala, S. miRNA regulation of innate immunity. J. Leukoc. Biol. 2018, 103, 1205–1217. [Google Scholar] [CrossRef]
- Saldaña, M.; Etebari, K.; Hart, C.E.; Widen, S.G.; Wood, T.G.; Thangamani, S.; Asgari, S.; Hughes, G.L. Zika virus alters the microRNA expression profile and elicits an RNAi response in Aedes aegypti mosquitoes. PLoS Negl. Trop. Dis. 2017, 11, e0005760. [Google Scholar] [CrossRef]
- Rigoutsos, I. New tricks for animal microRNAS: Targeting of Amino Acid Coding Regions at Conserved and Nonconserved sites. Cancer Res. 2009, 69, 3245–3248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnall-Levin, M.; Zhao, Y.; Perrimon, N.; Berger, B. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′ UTRs. Proc. Natl. Acad. Sci. USA 2010, 107, 15751–15756. [Google Scholar] [CrossRef] [Green Version]
- Asgari, S. MicroRNAs as regulators of insect host–pathogen interactions and immunity. Adv. Insect Phys. 2018, 55, 19–45. [Google Scholar]
- Flynt, A.S.; Greimann, J.C.; Chung, W.J.; Lima, C.D.; Lai, E.C. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol. Cell. 2010, 38, 900–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, J.M.; Sonenshine, D.E.; Valenzuela, J.G. Exploring the mialome of ticks: An Annotated Catalogue of Midgut Transcripts from the Hard Tick, Dermacentor Variabilis (Acari: Ixodidae). BMC Genom. 2008, 9, 552. [Google Scholar] [CrossRef] [Green Version]
- Antunes, S.; Couto, J.; Ferrolho, J.; Sanches, G.S.; Charrez, J.O.M.; Hernández, N.D.L.C.; Mazuz, M.; Villar, M.; Shkap, V.; De La Fuente, J.; et al. Transcriptome and Proteome Response of Rhipicephalus annulatus Tick Vector to Babesia bigemina Infection. Front. Physiol. 2019, 10, 318. [Google Scholar] [CrossRef]
- Popara, M.; Villar, M.; de la Fuente, J. Proteomics characterization of tick-host-pathogen interactions. Methods Mol. Biol. 2015, 1247, 513–527. [Google Scholar] [CrossRef] [Green Version]
- Villar, M.; Ayllón, N.; Alberdi, P.; Moreno, A.; Moreno, M.; Tobes, R.; Mateos-Hernández, L.; Weisheit, S.; Bell-Sakyi, L.; de la Fuente, J. Integrated Metabolomics, Transcriptomics and Proteomics Identifies Metabolic Pathways Affected by Anaplasma phagocytophilum Infection in Tick Cells. Mol. Cell Proteom. 2015, 14, 3154–3172. [Google Scholar] [CrossRef] [Green Version]
- Bechsgaard, J.; Vanthournout, B.; Funch, P.; Vestbo, S.; Gibbs, R.A.; Richards, S.; Sanggaard, K.W.; Enghild, J.J.; Bilde, T. Comparative genomic study of arachnid immune systems indicates loss of beta-1,3-glucanase-related proteins and the immune deficiency pathway. J. Evol. Biol. 2016, 29, 277–291. [Google Scholar] [CrossRef]
- Gulia-Nuss, M.; Nuss, A.; Meyer, J.M.; Sonenshine, D.E.; Roe, R.M.; Waterhouse, R.M.; Sattelle, D.B.; De La Fuente, J.; Ribeiro, J.; Megy, K.; et al. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun. 2016, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Rosa, R.D.; Capelli-Peixoto, J.; Mesquita, R.D.; Kalil, S.P.; Pohl, P.C.; Braz, G.R.; Fogaça, A.C.; Daffre, S. Exploring the immune signalling pathway-related genes of the cattle tick Rhipicephalus microplus: From Molecular Characterization to Transcriptional Profile upon Microbial Challenge. Dev. Comp. Immunol. 2016, 59, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Pal, U.; Kitsou, C.; Drecktrah, D.; Yaş, Ö.B.; Fikrig, E. Interactions Between Ticks and Lyme Disease Spirochetes. Curr. Issues Mol. Biol. 2021, 42, 113–144. [Google Scholar] [CrossRef] [PubMed]
- Bensaci, M.; Bhattacharya, D.; Clark, R.; Hu, L.T. Oral vaccination with vaccinia virus expressing the tick antigen subolesin inhibits tick feeding and transmission of Borrelia burgdorferi. Vaccine 2012, 30, 6040–6046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narasimhan, S.; Kurokawa, C.; Diktas, H.; Strank, N.O.; Černý, J.; Murfin, K.; Cao, Y.; Lynn, G.; Trentleman, J.; Wu, M.-J.; et al. Ixodes scapularis saliva components that elicit responses associated with acquired tick-resistance. Ticks Tick Borne Dis. 2020, 11, 101369. [Google Scholar] [CrossRef]
- Pal, U.; Fikrig, E. Dynamics of Borrelia burgdorferi transmission by nymphal Ixodes dammini ticks. J. Infect. Dis. 2003, 167, 1082–1085. [Google Scholar] [CrossRef]
- Piesman, J.; Schneider, B.S.; Zeidner, N.S. Use of quantitative PCR to measure density of Borrelia burgdorferi in the midgut and salivary glands of feeding tick vectors. J. Clin. Microbiol. 2001, 39, 4145–4148. [Google Scholar] [CrossRef] [Green Version]
- Pal, U.; Fikrig, E. Adaptation of Borrelia burgdorferi in the vector and vertebrate host. Microbes Infect. 2003, 5, 659–666. [Google Scholar] [CrossRef]
- Fikrig, E.; Narasimhan, S. Borrelia burgdorferi—Traveling incognito? Microbes Infect 2006, 8, 1390–1399. [Google Scholar] [CrossRef]
- Ueti, M.W.; Reagan, J.O., Jr.; Knowles, D.P., Jr.; Scoles, G.A.; Shkap, V.; Palmer, G.H. Identification of midgut and salivary glands as specific and distinct barriers to efficient tick-borne transmission of Anaplasma marginale. Infect. Immun 2007, 75, 2959–2964. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, J.M.; Alarcon-Chaidez, F.; Francischetti, I.M.B.; Mans, B.; Mather, T.N.; Valenzuela, J.G.; Wikel, S.K. An annotated catalog of salivary gland transcripts from Ixodes scapularis ticks. Insect Biochem. Mol. Biol. 2006, 36, 111–129. [Google Scholar] [CrossRef]
- Patrick, C.D.; Hair, J.A. Laboratory rearing procedures and equipment for multi-host ticks (Acarina: Ixodidae). J. Med. Entomol. 1975, 12, 389–390. [Google Scholar] [CrossRef] [PubMed]
- Karim, S.; Ramakrishnan, V.G.; Tucker, J.S.; Essenberg, R.C.; Sauer, J.R. Amblyomma americanum salivary glands: Double-Stranded RNA-Mediated Gene Silencing of Synaptobrevin Homologue and Inhibition of PGE2 Stimulated Protein Secretion. Insect Biochem. Mol. Biol. 2004, 34, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Budachetri, K.; Meyers, V.C.; Karim, S. Assessment of tick antioxidant responses to exogenous oxidative stressors and insight into the role of catalase in the reproductive fitness of the Gulf Coast tick, Amblyomma maculatum. Insect Mol. Biol. 2016, 25, 283–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Embers, M.E.; Grasperge, B.J.; Jacobs, M.B.; Philipp, M.T. Feeding of ticks on animals for transmission and xenodiagnosis in Lyme disease research. J. Vis. Exp. 2013, 78, e50617. [Google Scholar] [CrossRef]
- Karim, S.; Essenberg, R.C.; Dillwith, J.W.; Tucker, J.S.; Bowman, A.S.; Sauer, J.R. Identification of SNARE and cell trafficking regulatory proteins in the salivary glands of the lone star tick, Amblyomma americanum (L.). Insect Biochem. Mol. Biol. 2002, 32, 1711–1721. [Google Scholar] [CrossRef]
- Kumar, D.; Embers, M.; Mather, T.N.; Karim, S. Is selenoprotein K required for Borrelia burgdorferi infection within the tick vector Ixodes scapularis? Parasites Vectors 2019, 12, 289. [Google Scholar] [CrossRef]
- Zia, M.F.; Flynt, A.S. Detection and Verification of Mammalian Mirtrons by Northern Blotting. Methods Mol. Biol. 2018, 1823, 209–219. [Google Scholar] [CrossRef]
- Stone, B.L.; Russart, N.M.; Gaultney, R.A.; Floden, A.M.; Vaughan, J.A.; Brissette, C.A. The Western progression of lyme disease: Infectious and Nonclonal Borrelia Burgdorferi Sensu Lato Populations in Grand Forks County, North Dakota. Appl. Environ. Microbiol. 2015, 81, 48–58. [Google Scholar] [CrossRef] [Green Version]
- Friedländer, M.R.; Chen, W.; Adamidi, C.; Maaskola, J.; Einspanier, R.; Knespel, S.; Rajewsky, N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008, 26, 407–415. [Google Scholar] [CrossRef]
- Friedlander, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Munderloh, U.G.; Kurtti, T.J. Formulation of medium for tick cell culture. Exp. Appl. Acarol. 1989, 7, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Obonyo, M.; Munderloh, U.G.; Fingerle, V.; Wilske, B.; Kurtti, T.J. Borrelia burgdorferi in tick cell culture modulates expression of outer surface proteins A and C in response to temperature. J. Clin. Microbiol. 1999, 37, 2137–2141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Andrade, J. DEApp: An Interactive Web Interface for Differential Expression Analysis of Next Generation Sequence Data. Source Code Biol. Med. 2017, 12, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sturm, M.; Hackenberg, M.; Langenberger, D.; Frishman, D. TargetSpy: A Supervised Machine Learning Approach for MicroRNA Target Prediction. BMC Bioinform. 2010, 11, 292. [Google Scholar] [CrossRef] [Green Version]
- John, B.; Enright, A.J.; Aravin, A.; Tuschl, T.; Sander, C.; Marks, D.S. Human MicroRNA targets. PLoS Biol. 2004, 2, e363. [Google Scholar] [CrossRef] [Green Version]
- Kertesz, M.; Iovino, N.; Unnerstall, U.; Gaul, U.; Segal, E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007, 39, 1278–1284. [Google Scholar] [CrossRef]
- Aparicio-Puerta, E.; Lebrón, R.; Rueda, A.; Gómez-Martín, C.; Giannoukakos, S.; Jáspez, D.; Medina, J.M.; Zubković, A.; Jurak, I.; Fromm, B.; et al. sRNAbench and sRNAtoolbox 2019: Intuitive Fast Small RNA Profiling and Differential Expression. Nucleic Acids Res. 2019, 47, W530–W535. [Google Scholar] [CrossRef] [Green Version]
- Min, H.; Yoon, S. Got target?: Computational Methods for MicroRNA Target Prediction and Their Extension. Exp. Mol. Med. 2010, 42, 233–244. [Google Scholar] [CrossRef] [Green Version]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-Protein Interaction Networks, with Increased Coverage and Integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef] [Green Version]
- Toronen, P.; Medlar, A.; Holm, L. PANNZER2: A Rapid Functional Annotation Web Server. Nucleic Acids Res. 2018, 46, W84–W88. [Google Scholar] [CrossRef]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A.; et al. WEGO 2.0: A Web Tool for Analyzing and Plotting GO Annotations, 2018 Update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; He, B.; He, J.; Mao, X. Upregulation of miR-153 promotes cell proliferation via downregulation of the PTEN tumor suppressor gene in human prostate cancer. Prostate 2013, 73, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Xu, D.; Zhang, X. Characterization of host microRNAs that respond to DNA virus infection in a crustacean. BMC Genomics 2012, 13, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, T.; Zhang, X. Functional analysis of a crustacean microRNA in host-virus interactions. J. Virol. 2012, 86, 12997–13004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hussain, M.; Torres, S.; Schnettler, E.; Funk, A.; Grundhoff, A.; Pijlman, G.P.; Khromykh, A.; Asgari, S. West Nile virus encodes a microRNA-like small RNA in the 3′ untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res. 2012, 40, 2210–2223. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Jiang, Y.; Xu, X.; Su, X.; Liu, Y.; Ma, Y.; Zhao, Y.; Shen, Z.; Huang, B.; Cau, X. Inducible degradation of lncRNA Sros1 promotes IFN-gamma-mediated activation of innate immune responses by stabilizing Stat1 mRNA. Nat. Immunol. 2019, 20, 1621–1630. [Google Scholar] [CrossRef]
- Wang, F.; Gong, H.; Zhang, H.; Zhou, Y.; Cao, J.; Zhou, J. Lipopolysaccharide-Induced Differential Expression of miRNAs in Male and Female Rhipicephalus haemaphysaloides Ticks. PLoS ONE. 2015, 10, e0139241. [Google Scholar] [CrossRef]
- Bond, D.; Foley, E. A quantitative RNAi screen for JNK modifiers identifies Pvr as a novel regulator of Drosophila immune signaling. PLoS Pathog. 2009, 5, e1000655. [Google Scholar] [CrossRef] [Green Version]
- Fullaondo, A.; Lee, S.Y. Identification of putative miRNA involved in Drosophila melanogaster immune response. Dev. Comp. Immunol. 2012, 36, 267–273. [Google Scholar] [CrossRef]
- Artigas-Jerónimo, S.; Alberdi, P.; Villar Rayo, M.; Cabezas-Cruz, A.; Prados, P.J.E.; Mateos-Hernández, L.; de la Fuente, J. Anaplasma phagocytophilum modifies tick cell microRNA expression and upregulates isc-mir-79 to facilitate infection by targeting the Roundabout protein 2 pathway. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Freitak, D.; Knorr, E.; Vogel, H.; Vilcinskas, A. Gender- and stressor-specific microRNA expression in Tribolium castaneum. Biol. Lett. 2012, 8, 860–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zheng, Y.; Jagadeeswaran, G.; Ren, R.; Sunkar, R.; Jiang, H. Identification of conserved and novel microRNAs in Manduca sexta and their possible roles in the expression regulation of immunity-related genes. Insect. Biochem. Mol. Biol. 2014, 47, 12–22. [Google Scholar] [CrossRef] [Green Version]
- Hackenberg, M.; Langenberger, D.; Schwarz, A.; Erhart, J.; Kotsyfakis, M. In silico target network analysis of de novo-discovered, tick saliva-specific microRNAs reveals important combinatorial effects in their interference with vertebrate host physiology. RNA 2017, 23, 1259–1269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Huang, Y.; Zhang, Q.; Zhou, H.; Jin, P.; Ma, F. The miR-317 functions as a negative regulator of Toll immune response and influences Drosophila survival. Dev. Comp. Immunol. 2019, 95, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Pushpavalli, S.N.; Sarkar, A.; Bag, I.; Hunt, C.R.; Ramaiah, M.J.; Pandita, T.K.; Bhadra, U.; Pal-Bhadra, M. Argonaute-1 functions as a mitotic regulator by controlling Cyclin B during DDrosophila early embryogenesis. FASEB J. 2014. [CrossRef]
- Fricke, C.; Green, D.; Smith, D.; Dalmay, T.; Chapman, T. MicroRNAs influence reproductive responses by females to male sex peptide in drosophila melanogaster. Genetics 2014, 198, 1603–1619. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; He, Y.; Wang, C.; Zhang, X. Characterization of miRNAs from hydrothermal vent 20 Rimicaris exoculata. Mar. Genomics. 2015, 24, 371–378. [Google Scholar] [CrossRef]
- Hermance, M.E.; Widen, S.G.; Wood, T.G.; Thangamani, S. Ixodes scapularis salivary gland microRNAs are differentially expressed during Powassan virus transmission. Sci. Rep. 2019, 9, 1–17. [Google Scholar] [CrossRef]
- Boulias, K.; Horvitz, H.R. The C. elegans MicroRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab. 2012, 15, 439–450. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, K.; Vilcinskas, A. Development and immunity-related microRNAs of the lepidopteran model host Galleria mellonella. BMC Genom. 2014, 15, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular Pathways and Physiological Roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Sun, Y.; Zhang, X. Noncoding miRNAs bridge virus infection and host autophagy in shrimp in vivo. FASEB J. 2017, 31, 2854–2868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Avila-Bonilla, R.G.; Yocupicio-Monroy, M.; Marchat, L.A.; De Nova-Ocampo, M.A.; Del Angel, R.M.; Salas-Benito, J.S. Analysis of the miRNA profile in C6/36 cells persistently infected with dengue virus type 2. Virus Res. 2017, 232, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, Y.; Wu, J.; Zheng, P.; Li, Y.; Zheng, X.; Puthiyakunnon, S.; Tu, Z.; Chen, X.-G. The expression profile of Aedes albopictus miRNAs is altered by dengue virus serotype-2 infection. Cell Biosci. 2015, 5, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ramasamy, E.; Taank, V.; Anderson, J.F.; Sultana, H.; Neelakanta, G. Repression of tick microRNA-133 induces organic anion transporting polypeptide expression critical for Anaplasma phagocytophilum survival in the vector and transmission to the vertebrate host. PLoS Genet. 2020, 16, e1008856. [Google Scholar] [CrossRef]
- Khanal, S.; Sultana, H.; Catravas, J.D.; Carlyon, J.A.; Neelakanta, G. Anaplasma phagocytophilum infection modulates expression of megakaryocyte cell cycle genes through phosphatidylinositol-3-kinase signaling. PLoS ONE. 2017, 12, e0182898. [Google Scholar] [CrossRef] [Green Version]
- Sultana, H.; Neelakanta, G.; Kantor, F.S.; Malawista, S.E.; Fish, D.; Montgomery, R.R.; Fikrig, E. Anaplasma phagocytophilum induces actin phosphorylation to selectively regulate gene transcription in Ixodes scapularis ticks. J. Exp. Med. 2010, 207, 1727–1743. [Google Scholar] [CrossRef] [Green Version]
- Taank, V.; Dutta, S.; Dasgupta, A.; Steeves, T.K.; Fish, D.; Anderson, J.F.; Sultana, H.; Neelakanta, G. Human rickettsial pathogen modulates arthropod organic anion transporting polypeptide and tryptophan pathway for its survival in ticks. Sci. Rep. 2017, 7, 13256. [Google Scholar] [CrossRef]
- Turck, J.W.; Taank, V.; Neelakanta, G.; Sultana, H. Ixodes scapularis Src tyrosine kinase facilitates Anaplasma phagocytophilum survival in its arthropod vector. Ticks Tick Borne Dis. 2019, 10, 838–847. [Google Scholar] [CrossRef]
- Neelakanta, G.; Sultana, H.; Fish, D.; Anderson, J.F.; Fikrig, E. Anaplasma phagocytophilum induces Ixodes scapularis ticks to express an antifreeze glycoprotein gene that enhances their survival in the cold. J. Clin. Invest. 2010, 120, 3179–3190. [Google Scholar] [CrossRef] [Green Version]
- Barrero, R.A.; Keeble-Gagnère, G.; Zhang, B.; Moolhuijzen, P.; Ikeo, K.; Tateno, Y.; Gojobori, T.; Guerrero, F.D.; Lew-Tabor, A.; Bellgrad, M. Evolutionary conserved microRNAs are ubiquitously expressed compared to tick-specific miRNAs in the cattle tick Rhipicephalus (Boophilus) microplus. BMC Genom. 2011, 12, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawaz, M.; Malik, M.I.; Zhang, H.; Gebremedhin, M.B.; Cao, J.; Zhou, Y.; Zhou, J. miRNA profile of extracellular vesicles isolated from saliva of Haemaphysalis longicornis tick. Acta Trop. 2020, 212, 105718. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell. Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Coakley, G.; Maizels, R.M.; Buck, A.H. Exosomes and Other Extracellular Vesicles: The New Communicators in Parasite Infections. Trends Parasitol. 2015, 31, 477–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambertz, U.; Oviedo Ovando, M.E.; Vasconcelos, E.J.; Unrau, P.J.; Myler, P.J.; Reiner, N.E. Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both old and new world Leishmania providing evidence for conserved exosomal RNA Packaging. BMC Genomics. 2015, 16, 151. [Google Scholar] [CrossRef] [Green Version]
- Ramachandra, R.N.; Wikel, S.K. Effects of Dermacentor andersoni (Acari: Ixodidae) salivary gland extracts on Bos indicus and B. taurus lymphocytes and macrophages: In vitro cytokine elaboration and lymphocyte blastogenesis. J. Med. Entomol. 1995, 32, 338–345. [Google Scholar] [CrossRef]
- Urioste, S.; Hall, L.R.; Telford, S.R., 3rd; Titus, R.G. Saliva of the Lyme disease vector, Ixodes dammini, blocks cell activation by a nonprostaglandin E2-dependent mechanism. J. Exp. Med. 1994, 180, 1077–1085. [Google Scholar] [CrossRef] [Green Version]
- Kung, F.; Anguita, J.; Pal, U. Borrelia burgdorferi and tick proteins supporting pathogen persistence in the vector. Future Microbiol. 2013, 8, 41–56. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Sturrock, A.; Weis, J.J. Intracellular localization of Borrelia burgdorferi within human endothelial cells. Infect. Immun. 1991, 59, 671–678. [Google Scholar] [CrossRef] [Green Version]
- Georgilis, K.; Peacocke, M.; Klempner, M.S. Fibroblasts protect the Lyme disease spirochete, Borrelia burgdorferi, from ceftriaxone in vitro. J. Infect. Dis. 1992, 166, 440–444. [Google Scholar] [CrossRef] [Green Version]
- Klempner, M.S.; Noring, R.; Rogers, R.A. Invasion of human skin fibroblasts by the Lyme disease spirochete, Borrelia burgdorferi. J. Infect. Dis. 1993, 167, 1074–1081. [Google Scholar] [CrossRef] [PubMed]
- Wawrzeniak, K.; Gaur, G.; Sapi, E.; Senejani, A.G. Effect of Borrelia burgdorferi Outer Membrane Vesicles on Host Oxidative Stress Response. Antibiotics 2020, 9, 275. [Google Scholar] [CrossRef] [PubMed]
- Gaur, G.; Sawant, J.Y.; Chavan, A.S.; Khatri, V.A.; Liu, Y.-H.; Zhang, M.; Sapi, E. Effect of Invasion of Borrelia burgdorferi in Normal and Neoplastic Mammary Epithelial Cells. Antibiotics 2021, 10, 1295. [Google Scholar] [CrossRef] [PubMed]
- Kaneda, K.; Masuzawa, T.; Yasugami, K.; Suzuki, T.; Suzuki, Y.; Yanagihara, Y. Glycosphingolipid-binding protein of Borrelia burgdorferi sensu lato. Infect. Immun. 1997, 65, 3180–3185. [Google Scholar] [CrossRef] [Green Version]
- O’Neal, A.J.; Butler, L.R.; Rolandelli, A.; Gilk, S.D.; Pedra, J.H. Lipid hijacking: A unifying theme in vector-borne diseases. Elife 2020, 9, e61675. [Google Scholar] [CrossRef]
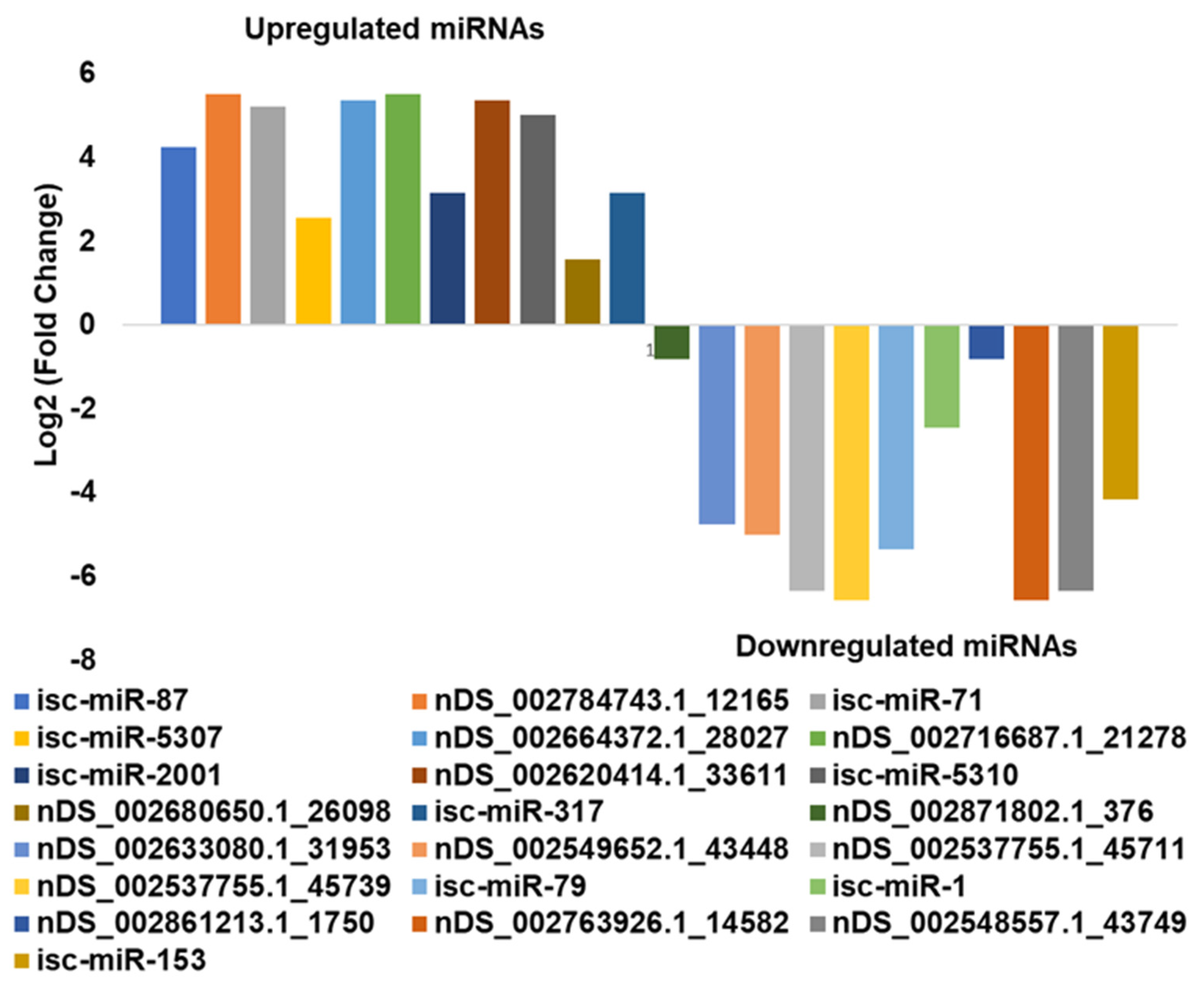
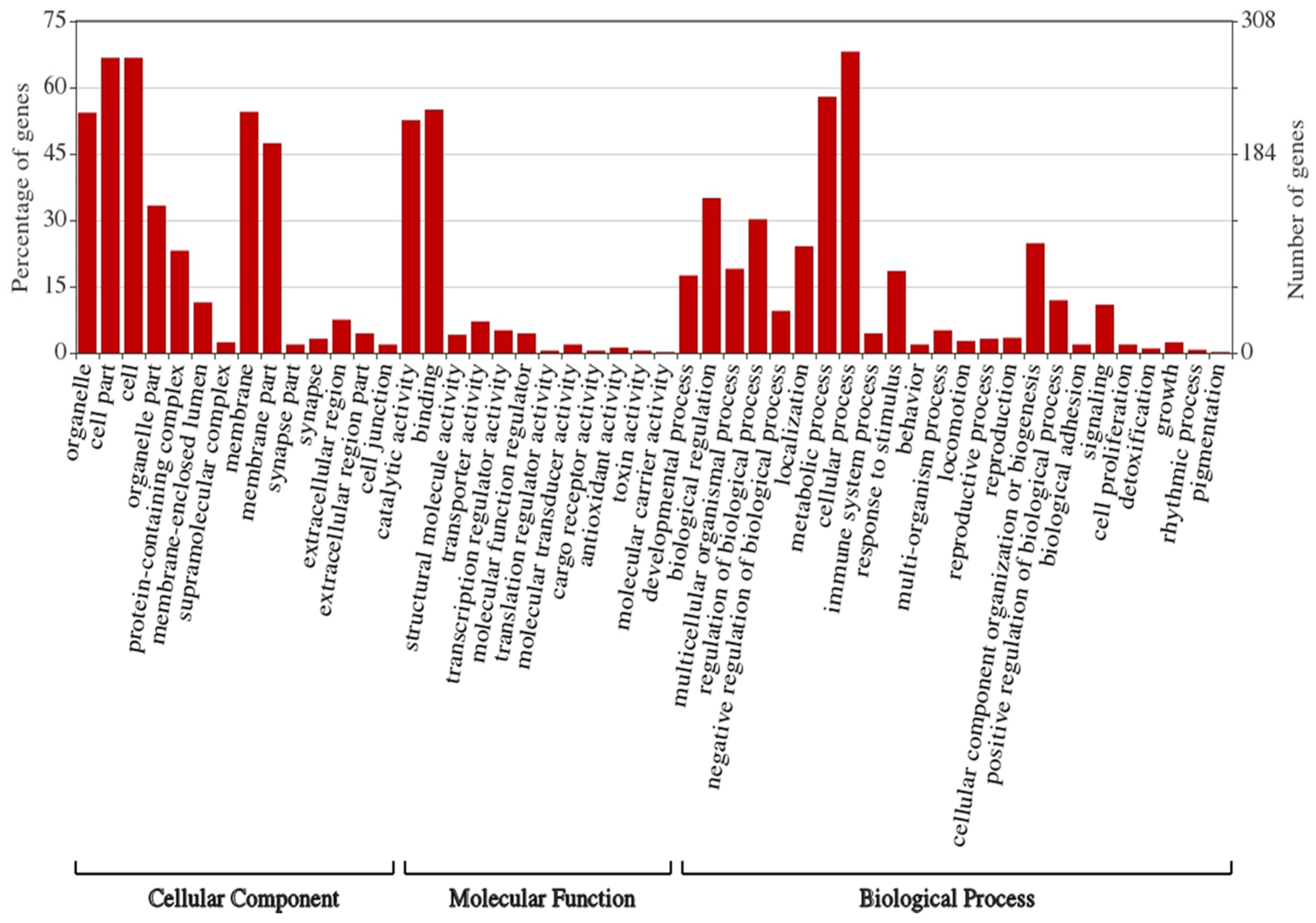
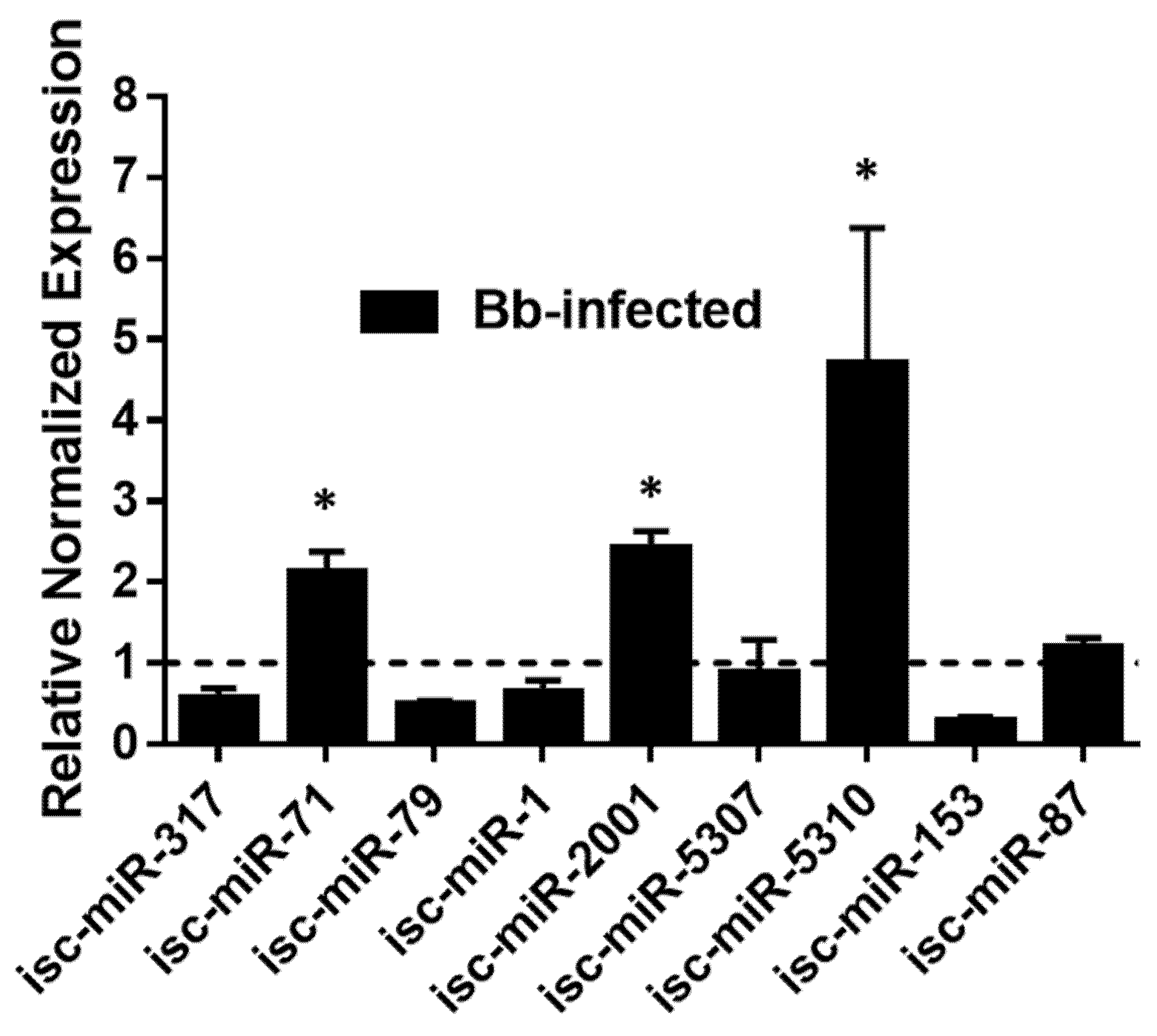
| Differentially Expressed miRNA * | Log2FC | logCPM | LR | p-Value | FDR |
|---|---|---|---|---|---|
| isc-miR-87 | 4.245604 | 11.83289 | 33.20241 | 8.30 × 10−9 | 4.65 × 10−8 |
| nDS_002784743.1_12165 | 5.511035 | 11.48887 | 29.7299 | 4.97 × 10−8 | 1.55 × 10−7 |
| isc-miR-71 | 5.226443 | 11.95849 | 48.94389 | 2.63 × 10−12 | 3.69 × 10−11 |
| isc-miR-5307 | 2.570363 | 12.34313 | 24.71399 | 6.65 × 10−7 | 1.55 × 10−6 |
| nDS_002664372.1_28027 | 5.362184 | 11.43227 | 26.80077 | 2.26 × 10−7 | 5.84 × 10−7 |
| nDS_002716687.1_21278 | 5.511153 | 11.48887 | 29.72059 | 4.99 × 10−8 | 1.55 × 10−7 |
| isc-miR-2001 | 3.146271 | 11.78845 | 19.75379 | 8.81 × 10−6 | 1.64 × 10−5 |
| nDS_002620414.1_33611 | 5.361301 | 11.43227 | 26.76848 | 2.29 × 10−7 | 5.84 × 10−7 |
| isc-miR-5310 | 5.009416 | 11.31195 | 20.86089 | 4.94 × 10−6 | 1.06 × 10−5 |
| nDS_002680650.1_26098 | 1.567661 | 15.43032 | 103.3589 | 2.80 × 10−24 | 7.83 × 10−23 |
| isc-miR-317 | 3.146271 | 11.78845 | 19.75379 | 8.81 × 10−6 | 1.64 × 10−5 |
| nDS_002871802.1_376 | −0.80359 | 15.73883 | 19.22785 | 1.16 × 10−5 | 1.91 × 10−5 |
| nDS_002633080.1_31953 | −4.7431 | 11.95812 | 10.11663 | 0.001469 | 0.001959 |
| nDS_002549652.1_43448 | −4.9974 | 12.11011 | 12.13051 | 0.000496 | 0.000731 |
| nDS_002537755.1_45711 | −6.34111 | 13.07343 | 30.24285 | 3.81 × 10−8 | 1.52 × 10−7 |
| nDS_002537755.1_45739 | −6.57147 | 13.2637 | 36.56262 | 1.48 × 10−9 | 1.03 × 10−8 |
| isc-miR-79 dme-miR-9a-5p | −5.35587 | 12.34273 | 15.64359 | 7.65 × 10−5 | 0.000119 |
| isc-miR-1 dme-miR-1-3p | −2.44072 | 12.67077 | 10.50794 | 0.001189 | 0.001664 |
| nDS_002861213.1_1750 | −0.80359 | 15.73883 | 19.22785 | 1.16 × 10−5 | 1.91 × 10−5 |
| nDS_002763926.1_14582 | −6.57147 | 13.2637 | 36.56262 | 1.48 × 10−9 | 1.03 × 10−8 |
| nDS_002548557.1_43749 | −6.34111 | 13.07343 | 30.24285 | 3.81 × 10−8 | 1.52 × 10−7 |
| isc-miR-153 | −4.15091 | 11.64621 | 6.563386 | 0.01041 | 0.013249 |
| microRNA | Role of Ortholog microRNA | Target Genes | Reference |
|---|---|---|---|
| isc-miR-153 | Development, immune response | [52] | |
| isc-miR-1 | Stress response, immunity, development, facilitating infection | HSP60, HSP70, GATA4 | [53,54,55,56] |
| isc-miR-79 | Innate immunity, differentiation, apoptosis | Roundabout protein 2 pathway (robo2), DRAPER, HP2 (Hemolymph protease), P38, pvr, puc | [57,58,59,60,61,62] |
| isc-miR-317 | Negative influencer of Toll pathway, host homeostatic response (targeting Gap junctions, TRP channels), mitotic regulation, Fecundity | Dif-Rc, Thioredoxin peroxidase-2, STAT | [62,63,64,65,66] |
| isc-miR-2001 | Under extreme environmental conditions | [67] | |
| isc-miR-5307 | Propagation of Powassan virus inside vero cells | [68] | |
| isc-miR-71 | Stress response, development, facilitates WSSV infection | cap-1, riok1, Actin1, myD88, IML3 | [62,69,70,71,72] |
| isc-miR-87 | Pathogen survival (disruption of Toll and IMD pathway) | Serine/Threnine Kinase, Toll 1A, Putative TLR 5b, FADD | [62,73,74] |
| isc-miR-5310 | Pathogen survival by modulating signaling pathways, Feeding behavior | [75,76,77,78,79,80,81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, D.; Downs, L.P.; Embers, M.; Flynt, A.S.; Karim, S. Identification of microRNAs in the Lyme Disease Vector Ixodes scapularis. Int. J. Mol. Sci. 2022, 23, 5565. https://doi.org/10.3390/ijms23105565
Kumar D, Downs LP, Embers M, Flynt AS, Karim S. Identification of microRNAs in the Lyme Disease Vector Ixodes scapularis. International Journal of Molecular Sciences. 2022; 23(10):5565. https://doi.org/10.3390/ijms23105565
Chicago/Turabian StyleKumar, Deepak, Latoyia P. Downs, Monica Embers, Alex Sutton Flynt, and Shahid Karim. 2022. "Identification of microRNAs in the Lyme Disease Vector Ixodes scapularis" International Journal of Molecular Sciences 23, no. 10: 5565. https://doi.org/10.3390/ijms23105565
APA StyleKumar, D., Downs, L. P., Embers, M., Flynt, A. S., & Karim, S. (2022). Identification of microRNAs in the Lyme Disease Vector Ixodes scapularis. International Journal of Molecular Sciences, 23(10), 5565. https://doi.org/10.3390/ijms23105565






