Long-Range and Directional Allostery of Actin Filaments Plays Important Roles in Various Cellular Activities
Abstract
1. Introduction
2. Cooperative Binding of ABPs
2.1. Side-Binding ABPs
2.1.1. Myosin II and V
2.1.2. Tropomyosin
2.1.3. Cofilin
2.1.4. Drebrin
2.2. Cross-Linking ABPs
2.2.1. Fimbrin
2.2.2. α-Actinin
2.2.3. Filamin
2.3. End-Binding ABPs
2.3.1. Gelsolin
2.3.2. Formin
3. Allosteric Interaction between ABPs and Actin Filaments Involved in the Formation and Function Mechanism of the Actin-Based Machinery
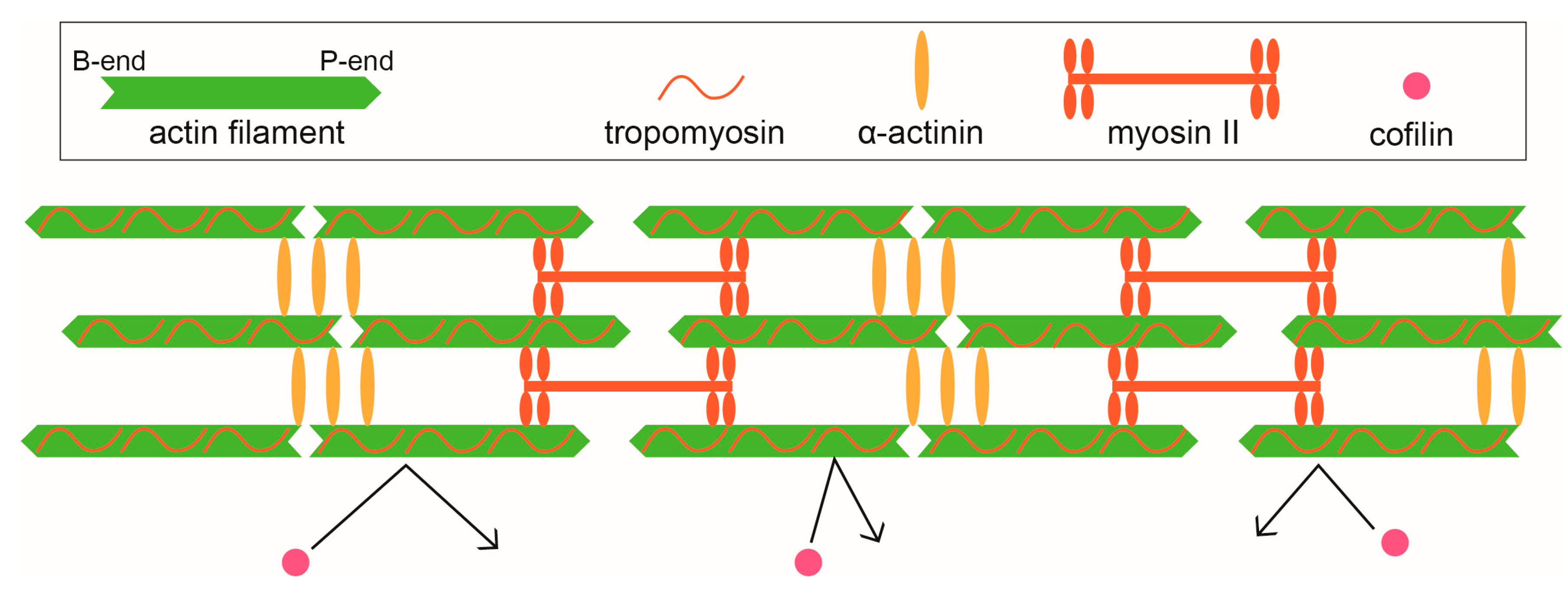
3.1. Stress Fibers
3.2. Lamellipodia
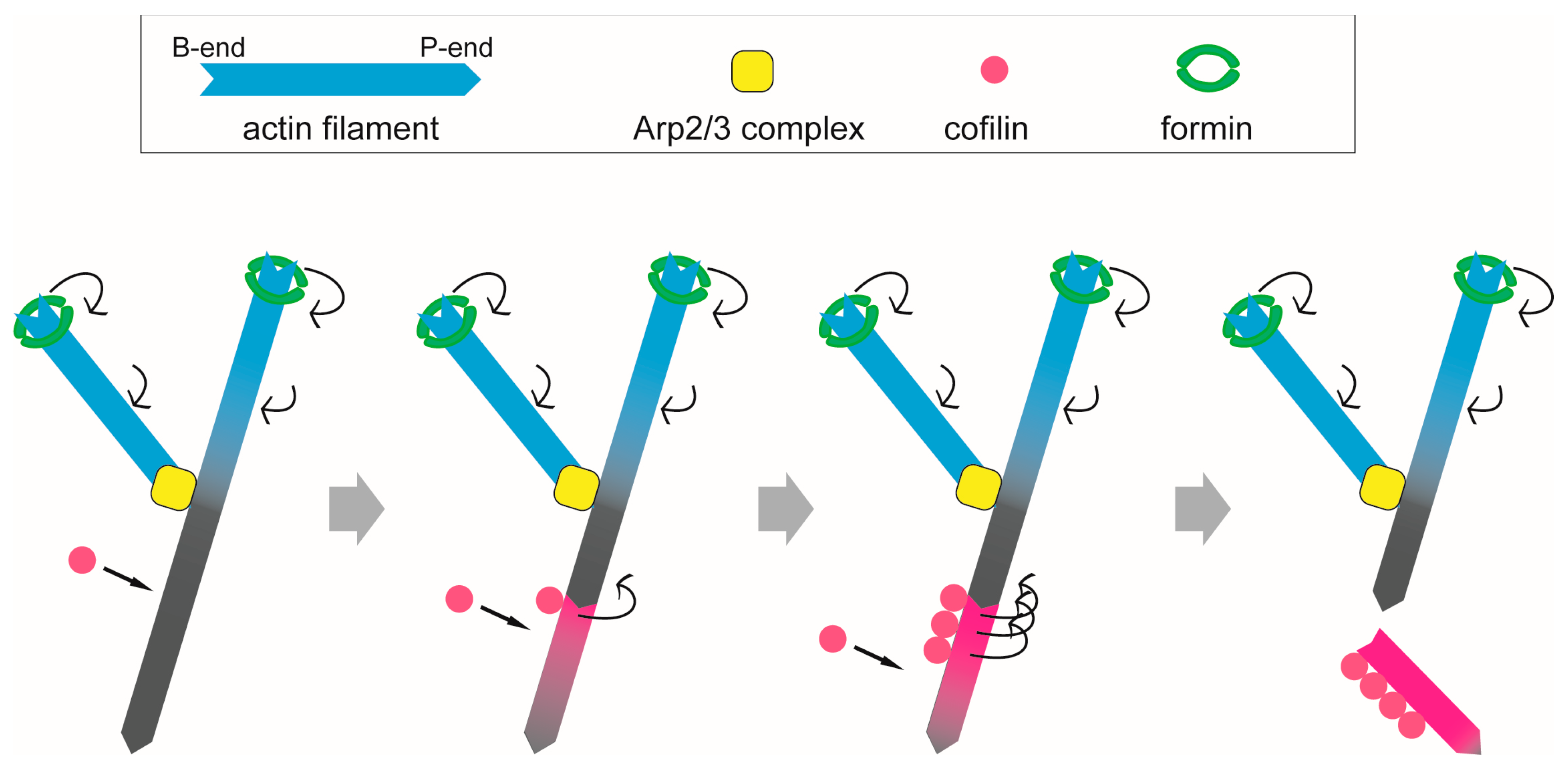
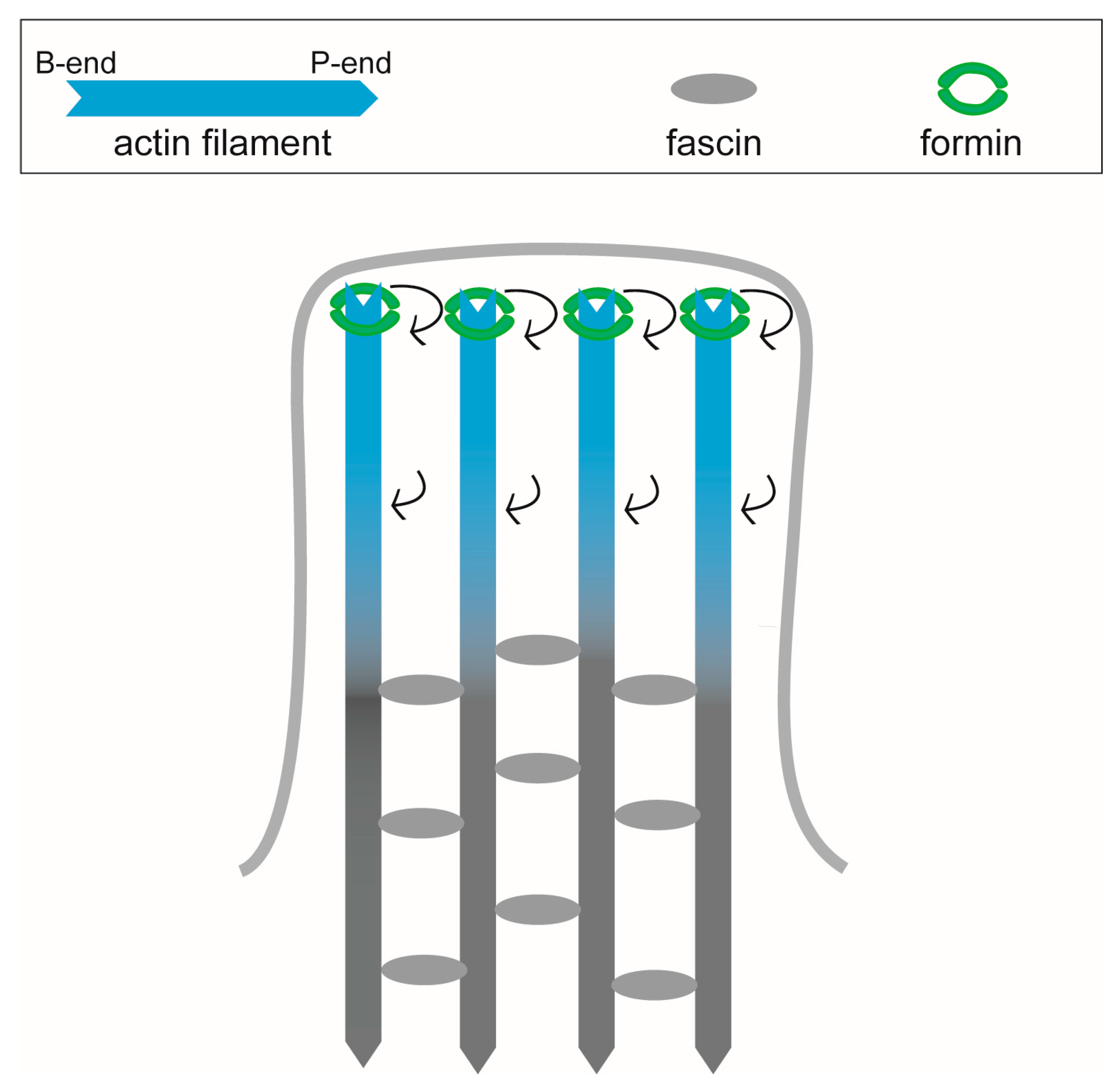
3.3. Filopodia
3.4. Other Actin-Based Machineries
4. Significance of Propagation Distance and Directionality of Long-Range Allostery
5. Future Perspectives
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABD | actin-binding domain |
| ABP | actin-binding protein |
| ADF | actin-depolymerizing factor |
| CH | calponin-homology |
| FRET | fluorescence resonance energy transfer |
| HMM | heavy meromyosin |
| HMM-GFP | GFP-fused HMM |
| HS-AFM | high-speed atomic force microscopy |
References
- Moores, S.L.; Sabry, J.H.; Spudich, J.A. Myosin dynamics in live Dictyostelium cells. Proc. Natl. Acad. Sci. USA 1996, 93, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Yumura, S.; Mori, H.; Fukui, Y. Localization of actin and myosin for the study of ameboid movement in Dictyostelium using improved immunofluorescence. J. Cell Biol. 1984, 99, 894–899. [Google Scholar] [CrossRef] [PubMed]
- Aizawa, H.; Fukui, Y.; Yahara, I. Live dynamics of Dictyostelium cofilin suggests a role in remodeling actin latticework into bundles. J. Cell Sci. 1997, 110, 2333–2344. [Google Scholar] [PubMed]
- Ngo, K.X.; Umeki, N.; Kijima, S.T.; Kodera, N.; Ueno, H.; Furutani-Umezu, N.; Nakajima, J.; Noguchi, T.Q.P.; Nagasaki, A.; Tokuraku, K.; et al. Allosteric regulation by cooperative conformational changes of actin filaments drives mutually exclusive binding with cofilin and myosin. Sci. Rep. 2016, 6, 35449. [Google Scholar] [CrossRef] [PubMed]
- Galkin, V.E.; Orlova, A.; Schroder, G.F.; Egelman, E.H. Structural polymorphism in F-actin. Nat. Struct. Mol. Biol. 2010, 17, 1318–1323. [Google Scholar] [CrossRef] [PubMed]
- Kozuka, J.; Yokota, H.; Arai, Y.; Ishii, Y.; Yanagida, T. Dynamic polymorphism of single actin molecules in the actin filament. Nat. Chem. Biol. 2006, 2, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Uyeda, T.Q.P.; Ngo, K.X.; Kodera, N.; Tokuraku, K. Uni-directional Propagation of Structural Changes in Actin Filaments. In The Role of Water in ATP Hydrolysis Energy Transduction by Protein Machinery; Suzuki, M., Ed.; Springer: Singapore, 2018; pp. 157–177. [Google Scholar]
- Oosawa, F. Macromolecular assembly of actin. In Muscle and Non-muscle Motility; Stracher, A., Ed.; Academic Press: New York, NY, USA, 1983; pp. 151–216. [Google Scholar]
- Egelman, E.H.; Orlova, A. New insights into actin filament dynamics. Curr. Opin. Struct. Biol. 1995, 5, 172–180. [Google Scholar] [CrossRef]
- Oosawa, F.; Fujime, S.; Ishiwata, S.; Mihashi, K. Dynamic property of F-actin and thin filament. Cold Spring Harbor Symp. Quant. Biol. 1972, 37, 277–285. [Google Scholar] [CrossRef]
- Miki, M.; Wahl, P.; Auchet, J.C. Fluorescence anisotropy of labeled F-actin: Influence of divalent cations on the interaction between F-actin and myosin heads. Biochemistry 1982, 21, 3661–3665. [Google Scholar] [CrossRef]
- Tawada, K. Physicochemical studies of F-actin-heavy meromyosin solutions. Biochim. Biophys. Acta 1969, 172, 311–318. [Google Scholar] [CrossRef]
- Fujime, S.; Ishiwata, S. Dynamic study of F-actin by quasielastic scattering of laser light. J. Mol. Biol. 1971, 62, 251–265. [Google Scholar] [CrossRef]
- Loscalzo, J.; Reed, G.H.; Weber, A. Conformational change and cooperativity in actin filaments free of tropomyosin. Proc. Natl. Acad. Sci. USA 1975, 72, 3412–3415. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.D.; Seidel, J.C.; Gergely, J. Rotational dynamics of spin-labeled F-actin in the sub-millisecond time range. J. Mol. Biol. 1979, 132, 257–273. [Google Scholar] [CrossRef]
- Craig, R.; Szent-Gyorgyi, A.G.; Beese, L.; Flicker, P.; Vibert, P.; Cohen, C. Electron microscopy of thin filaments decorated with a Ca2+-regulated myosin. J. Mol. Biol. 1980, 140, 35–55. [Google Scholar] [CrossRef]
- Frado, L.L.; Craig, R. Electron microscopy of the actin-myosin head complex in the presence of ATP. J. Mol. Biol. 1992, 223, 391–397. [Google Scholar] [CrossRef]
- Walker, M.; White, H.; Belknap, B.; Trinick, J. Electron cryomicroscopy of acto-myosin-S1 during steady-state ATP hydrolysis. Biophys. J. 1994, 66, 1563–1572. [Google Scholar] [CrossRef][Green Version]
- Woodrum, D.T.; Rich, S.A.; Pollard, T.D. Evidence for biased bidirectional polymerization of actin filaments using heavy meromyosin prepared by an improved method. J. Cell Biol. 1975, 67, 231–237. [Google Scholar] [CrossRef]
- Orlova, A.; Egelman, E.H. Cooperative rigor binding of myosin to actin is a function of F-actin structure. J. Mol. Biol. 1997, 265, 469–474. [Google Scholar] [CrossRef]
- Tokuraku, K.; Kurogi, R.; Toya, R.; Uyeda, T.Q.P. Novel mode of cooperative binding between myosin and Mg2+-actin filaments in the presence of low concentrations of ATP. J. Mol. Biol. 2009, 386, 149–162. [Google Scholar] [CrossRef]
- Kitazawa, T.; Shuman, H.; Somlyo, A.P. Calcium and magnesium binding to thin and thick filaments in skinned muscle fibres: Electron probe analysis. J. Muscle Res. Cell Motil. 1982, 3, 437–454. [Google Scholar] [CrossRef]
- Hirakawa, R.; Nishikawa, Y.; Uyeda, T.Q.P.; Tokuraku, K. Unidirectional growth of heavy meromyosin clusters along actin filaments revealed by real-time fluorescence microscopy. Cytoskeleton (Hoboken) 2017, 74, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Butters, C.A.; Willadsen, K.A.; Tobacman, L.S. Cooperative interactions between adjacent troponin-tropomyosin complexes may be transmitted through the actin filament. J. Biol. Chem. 1993, 268, 15565–15570. [Google Scholar] [PubMed]
- Khaitlina, S.; Tsaplina, O.; Hinssen, H. Cooperative effects of tropomyosin on the dynamics of the actin filament. FEBS Lett. 2017, 591, 1884–1891. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.R.; Hocky, G.M.; Homa, K.E.; Morganthaler, A.N.; Hitchcock-DeGregori, S.E.; Voth, G.A.; Kovar, D.R. Competition between Tropomyosin, Fimbrin, and ADF/Cofilin drives their sorting to distinct actin filament networks. Elife 2017, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Gateva, G.; Kremneva, E.; Reindl, T.; Kotila, T.; Kogan, K.; Gressin, L.; Gunning, P.W.; Manstein, D.J.; Michelot, A.; Lappalainen, P. Tropomyosin Isoforms Specify Functionally Distinct Actin Filament Populations In Vitro. Curr. Biol. 2017, 27, 705–713. [Google Scholar] [CrossRef] [PubMed]
- De La Cruz, E.M. Cofilin binding to muscle and non-muscle actin filaments: Isoform-dependent cooperative interactions. J. Mol. Biol. 2005, 346, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, M.; Pope, B.; Maciver, S.K.; Weeds, A.G. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry 1993, 32, 9985–9993. [Google Scholar] [CrossRef]
- Hayakawa, K.; Sakakibara, S.; Sokabe, M.; Tatsumi, H. Single-molecule imaging and kinetic analysis of cooperative cofilin-actin filament interactions. Proc. Natl. Acad. Sci. USA 2014, 111, 9810–9815. [Google Scholar] [CrossRef]
- Hayden, S.M.; Miller, P.S.; Brauweiler, A.; Bamburg, J.R. Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry 1993, 32, 9994–10004. [Google Scholar] [CrossRef]
- Ngo, K.X.; Kodera, N.; Katayama, E.; Ando, T.; Uyeda, T.Q.P. Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. Elife 2015, 4, 04806. [Google Scholar] [CrossRef]
- Galkin, V.E.; Orlova, A.; Kudryashov, D.S.; Solodukhin, A.; Reisler, E.; Schroder, G.F.; Egelman, E.H. Remodeling of actin filaments by ADF/cofilin proteins. Proc. Natl. Acad. Sci. USA 2011, 108, 20568–20572. [Google Scholar] [CrossRef] [PubMed]
- Galkin, V.E.; Orlova, A.; Lukoyanova, N.; Wriggers, W.; Egelman, E.H. Actin depolymerizing factor stabilizes an existing state of F-actin and can change the tilt of F-actin subunits. J. Cell Biol. 2001, 153, 75–86. [Google Scholar] [CrossRef] [PubMed]
- McGough, A.; Pope, B.; Chiu, W.; Weeds, A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J. Cell Biol. 1997, 138, 771–781. [Google Scholar] [CrossRef] [PubMed]
- Suarez, C.; Roland, J.; Boujemaa-Paterski, R.; Kang, H.; McCullough, B.R.; Reymann, A.C.; Guerin, C.; Martiel, J.L.; De la Cruz, E.M.; Blanchoin, L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr. Biol. 2011, 21, 862–868. [Google Scholar] [CrossRef] [PubMed]
- Umeki, N.; Hirose, K.; Uyeda, T.Q.P. Cofilin-induced cooperative conformational changes of actin subunits revealed using cofilin-actin fusion protein. Sci. Rep. 2016, 6, 20406. [Google Scholar] [CrossRef] [PubMed]
- Wioland, H.; Guichard, B.; Senju, Y.; Myram, S.; Lappalainen, P.; Jegou, A.; Romet-Lemonne, G. ADF/Cofilin Accelerates Actin Dynamics by Severing Filaments and Promoting Their Depolymerization at Both Ends. Curr. Biol. 2017, 27, 1956–1967 e1957. [Google Scholar] [CrossRef]
- Noguchi, T.Q.P.; Toya, R.; Ueno, H.; Tokuraku, K.; Uyeda, T.Q.P. Screening of novel dominant negative mutant actins using glycine targeted scanning identifies G146V actin that cooperatively inhibits cofilin binding. Biochem. Biophys. Res. Commun. 2010, 396, 1006–1011. [Google Scholar] [CrossRef]
- Huehn, A.R.; Bibeau, J.P.; Schramm, A.C.; Cao, W.; De La Cruz, E.M.; Sindelar, C.V. Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments. Proc. Natl. Acad. Sci. USA 2020, 117, 1478–1484. [Google Scholar] [CrossRef]
- Umeki, N.; Shibata, K.; Noguchi, T.Q.P.; Hirose, K.; Sako, Y.; Uyeda, T.Q.P. K336I mutant actin alters the structure of neighbouring protomers in filaments and reduces affinity for actin-binding proteins. Sci. Rep. 2019, 9, 5353. [Google Scholar] [CrossRef]
- Sharma, S.; Grintsevich, E.E.; Hsueh, C.; Reisler, E.; Gimzewski, J.K. Molecular cooperativity of drebrin1-300 binding and structural remodeling of F-actin. Biophys. J. 2012, 103, 275–283. [Google Scholar] [CrossRef]
- Sharma, S.; Grintsevich, E.E.; Phillips, M.L.; Reisler, E.; Gimzewski, J.K. Atomic force microscopy reveals drebrin induced remodeling of f-actin with subnanometer resolution. Nano Lett 2011, 11, 825–827. [Google Scholar] [CrossRef] [PubMed]
- Galkin, V.E.; Orlova, A.; Cherepanova, O.; Lebart, M.C.; Egelman, E.H. High-resolution cryo-EM structure of the F-actin-fimbrin/plastin ABD2 complex. Proc. Natl. Acad. Sci. USA 2008, 105, 1494–1498. [Google Scholar] [CrossRef] [PubMed]
- Hanein, D.; Matsudaira, P.; DeRosier, D.J. Evidence for a conformational change in actin induced by fimbrin (N375) binding. J. Cell Biol. 1997, 139, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Skau, C.T.; Kovar, D.R. Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr. Biol. 2010, 20, 1415–1422. [Google Scholar] [CrossRef]
- Craig-Schmidt, M.C.; Robson, R.M.; Goll, D.E.; Stromer, M.H. Effect of alpha-actinin on actin structure. Release of bound nucleotide. Biochim. Biophys. Acta 1981, 670, 9–16. [Google Scholar] [CrossRef]
- Galkin, V.E.; Orlova, A.; Salmazo, A.; Djinovic-Carugo, K.; Egelman, E.H. Opening of tandem calponin homology domains regulates their affinity for F-actin. Nat. Struct. Mol. Biol. 2010, 17, 614–616. [Google Scholar] [CrossRef]
- Shibata, K.; Nagasaki, A.; Adachi, H.; Uyeda, T.Q.P. Actin binding domain of filamin distinguishes posterior from anterior actin filaments in migrating Dictyostelium cells. Biophys Physicobiol 2016, 13, 321–331. [Google Scholar] [CrossRef]
- Hansen, S.D.; Kwiatkowski, A.V.; Ouyang, C.Y.; Liu, H.; Pokutta, S.; Watkins, S.C.; Volkmann, N.; Hanein, D.; Weis, W.I.; Mullins, R.D.; et al. Alpha E-catenin actin-binding domain alters actin filament conformation and regulates binding of nucleation and disassembly factors. Mol. Biol. Cell 2013, 24, 3710–3720. [Google Scholar] [CrossRef]
- Orlova, A.; Prochniewicz, E.; Egelman, E.H. Structural dynamics of F-actin: II. Cooperativity in structural transitions. J. Mol. Biol. 1995, 245, 598–607. [Google Scholar] [CrossRef]
- Selden, L.A.; Kinosian, H.J.; Newman, J.; Lincoln, B.; Hurwitz, C.; Gershman, L.C.; Estes, J.E. Severing of F-actin by the amino-terminal half of gelsolin suggests internal cooperativity in gelsolin. Biophys. J. 1998, 75, 3092–3100. [Google Scholar] [CrossRef]
- Papp, G.; Bugyi, B.; Ujfalusi, Z.; Barko, S.; Hild, G.; Somogyi, B.; Nyitrai, M. Conformational changes in actin filaments induced by formin binding to the barbed end. Biophys. J. 2006, 91, 2564–2572. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huxley, H.E. The mechanism of muscular contraction. Science 1969, 164, 1356–1365. [Google Scholar] [CrossRef] [PubMed]
- Bailey, K. Tropomyosin: A new asymmetric protein component of muscle. Nature 1946, 157, 368. [Google Scholar] [CrossRef] [PubMed]
- Ebashi, S.; Endo, M. Calcium ion and muscle contraction. Prog. Biophys. Mol. Biol. 1968, 18, 123–183. [Google Scholar] [CrossRef]
- Weber, A.; Murray, J.M. Molecular control mechanisms in muscle contraction. Physiol. Rev. 1973, 53, 612–673. [Google Scholar] [CrossRef]
- Nishida, E.; Maekawa, S.; Sakai, H. Cofilin, a protein in porcine brain that binds to actin filaments and inhibits their interactions with myosin and tropomyosin. Biochemistry 1984, 23, 5307–5313. [Google Scholar] [CrossRef]
- Yonezawa, N.; Nishida, E.; Sakai, H. pH control of actin polymerization by cofilin. J Biol Chem 1985, 260, 14410–14412. [Google Scholar]
- Bobkov, A.A.; Muhlrad, A.; Pavlov, D.A.; Kokabi, K.; Yilmaz, A.; Reisler, E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J. Mol. Biol. 2006, 356, 325–334. [Google Scholar] [CrossRef]
- Shirao, T.; Kojima, N.; Kato, Y.; Obata, K. Molecular cloning of a cDNA for the developmentally regulated brain protein, drebrin. Brain Res. 1988, 464, 71–74. [Google Scholar] [CrossRef]
- Lin, Y.C.; Koleske, A.J. Mechanisms of synapse and dendrite maintenance and their disruption in psychiatric and neurodegenerative disorders. Annu. Rev. Neurosci. 2010, 33, 349–378. [Google Scholar] [CrossRef]
- Bretscher, A.; Weber, K. Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J. Cell Biol. 1980, 86, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil, R.R. Structure and evolution of the actin crosslinking proteins. Bioessays 1991, 13, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, J.H.; Kwiatkowski, D.J. Actin-binding proteins. Curr. Opin. Cell Biol. 1991, 3, 87–97. [Google Scholar] [CrossRef]
- Matsudaira, P. Modular organization of actin crosslinking proteins. Trends Biochem. Sci. 1991, 16, 87–92. [Google Scholar] [CrossRef]
- Otto, J.J. Actin-bundling proteins. Curr. Opin. Cell Biol. 1994, 6, 105–109. [Google Scholar] [CrossRef]
- Maruyama, K.; Ebashi, S. Alpha-actinin, a new structural protein from striated muscle. II. Action on actin. J. Biochem. 1965, 58, 13–19. [Google Scholar] [CrossRef]
- Wang, K.; Ash, J.F.; Singer, S.J. Filamin, a new high-molecular-weight protein found in smooth muscle and non-muscle cells. Proc. Natl. Acad. Sci. USA 1975, 72, 4483–4486. [Google Scholar] [CrossRef]
- Stossel, T.P.; Condeelis, J.; Cooley, L.; Hartwig, J.H.; Noegel, A.; Schleicher, M.; Shapiro, S.S. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001, 2, 138–145. [Google Scholar] [CrossRef]
- Byfield, F.J.; Wen, Q.; Levental, I.; Nordstrom, K.; Arratia, P.E.; Miller, R.T.; Janmey, P.A. Absence of filamin A prevents cells from responding to stiffness gradients on gels coated with collagen but not fibronectin. Biophys. J. 2009, 96, 5095–5102. [Google Scholar] [CrossRef]
- Lynch, C.D.; Gauthier, N.C.; Biais, N.; Lazar, A.M.; Roca-Cusachs, P.; Yu, C.H.; Sheetz, M.P. Filamin depletion blocks endoplasmic spreading and destabilizes force-bearing adhesions. Mol. Biol. Cell 2011, 22, 1263–1273. [Google Scholar] [CrossRef]
- Luo, T.; Mohan, K.; Iglesias, P.A.; Robinson, D.N. Molecular mechanisms of cellular mechanosensing. Nat. Mater. 2013, 12, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Forster, C.; Nakamura, F.; Glogauer, M. Filamin-A regulates neutrophil uropod retraction through RhoA during chemotaxis. PLoS ONE 2013, 8, e79009. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.L.; Stossel, T.P. Control of cytoplasmic actin gel-sol transformation by gelsolin, a calcium-dependent regulatory protein. Nature 1979, 281, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Mass, R.L.; Zeller, R.; Woychik, R.P.; Vogt, T.F.; Leder, P. Disruption of formin-encoding transcripts in two mutant limb deformity alleles. Nature 1990, 346, 853–855. [Google Scholar] [CrossRef] [PubMed]
- Courtemanche, N. Mechanisms of formin-mediated actin assembly and dynamics. Biophys. Rev. 2018, 10, 1553–1569. [Google Scholar] [CrossRef] [PubMed]
- Ridley, A.J. Life at the leading edge. Cell 2011, 145, 1012–1022. [Google Scholar] [CrossRef]
- Kreis, T.E.; Birchmeier, W. Stress fiber sarcomeres of fibroblasts are contractile. Cell 1980, 22, 555–561. [Google Scholar] [CrossRef]
- Tojkander, S.; Gateva, G.; Lappalainen, P. Actin stress fibers—Assembly, dynamics and biological roles. J. Cell Sci. 2012, 125, 1855–1864. [Google Scholar] [CrossRef]
- Tsaturyan, A.K.; Koubassova, N.; Ferenczi, M.A.; Narayanan, T.; Roessle, M.; Bershitsky, S.Y. Strong binding of myosin heads stretches and twists the actin helix. Biophys. J. 2005, 88, 1902–1910. [Google Scholar] [CrossRef]
- Holmes, K.C.; Angert, I.; Kull, F.J.; Jahn, W.; Schroder, R.R. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature 2003, 425, 423–427. [Google Scholar] [CrossRef]
- Hayakawa, K.; Tatsumi, H.; Sokabe, M. Actin filaments function as a tension sensor by tension-dependent binding of cofilin to the filament. J. Cell Biol. 2011, 195, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Chrzanowska-Wodnicka, M.; Burridge, K. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 1996, 133, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Ono, S.; Abe, H.; Obinata, T. Stimulus-dependent disorganization of actin filaments induced by overexpression of cofilin in C2 myoblasts. Cell Struct. Funct. 1996, 21, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.J.; Sen, S.; Sweeney, H.L.; Discher, D.E. Matrix elasticity directs stem cell lineage specification. Cell 2006, 126, 677–689. [Google Scholar] [CrossRef]
- Lazarides, E. Tropomyosin antibody: The specific localization of tropomyosin in nonmuscle cells. J. Cell Biol. 1975, 65, 549–561. [Google Scholar] [CrossRef]
- Lazarides, E.; Burridge, K. Alpha-actinin: Immunofluorescent localization of a muscle structural protein in nonmuscle cells. Cell 1975, 6, 289–298. [Google Scholar] [CrossRef]
- Tojkander, S.; Gateva, G.; Schevzov, G.; Hotulainen, P.; Naumanen, P.; Martin, C.; Gunning, P.W.; Lappalainen, P. A molecular pathway for myosin II recruitment to stress fibers. Curr. Biol. 2011, 21, 539–550. [Google Scholar] [CrossRef]
- Tang, N.; Ostap, E.M. Motor domain-dependent localization of myo1b (myr-1). Curr. Biol. 2001, 11, 1131–1135. [Google Scholar] [CrossRef]
- Kuragano, M.; Uyeda, T.Q.P.; Kamijo, K.; Murakami, Y.; Takahashi, M. Different contributions of nonmuscle myosin IIA and IIB to the organization of stress fiber subtypes in fibroblasts. Mol. Biol. Cell 2018, 29, 911–922. [Google Scholar] [CrossRef]
- Uyeda, T.Q.P.; Iwadate, Y.; Umeki, N.; Nagasaki, A.; Yumura, S. Stretching actin filaments within cells enhances their affinity for the myosin II motor domain. PLoS ONE 2011, 6, e26200. [Google Scholar] [CrossRef]
- Krause, M.; Gautreau, A. Steering cell migration: Lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol. 2014, 15, 577–590. [Google Scholar] [CrossRef] [PubMed]
- Van Rheenen, J.; Condeelis, J.; Glogauer, M. A common cofilin activity cycle in invasive tumor cells and inflammatory cells. J. Cell Sci. 2009, 122, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Tanaka, K.; Yamashiro, S.; Narita, A.; Watanabe, N. Helical rotation of the diaphanous-related formin mDia1 generates actin filaments resistant to cofilin. Proc. Natl. Acad. Sci. 2018, 115, E5000–E5007. [Google Scholar] [CrossRef]
- Goley, E.D.; Welch, M.D. The ARP2/3 complex: An actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006, 7, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Carlier, M.F.; Laurent, V.; Santolini, J.; Melki, R.; Didry, D.; Xia, G.X.; Hong, Y.; Chua, N.H.; Pantaloni, D. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: Implication in actin-based motility. J. Cell Biol. 1997, 136, 1307–1322. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D.; Borisy, G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell 2003, 112, 453–465. [Google Scholar] [CrossRef]
- Mattila, P.K.; Lappalainen, P. Filopodia: Molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 2008, 9, 446–454. [Google Scholar] [CrossRef]
- Machesky, L.M.; Li, A. Fascin: Invasive filopodia promoting metastasis. Commun. Integr. Biol. 2010, 3, 263–270. [Google Scholar] [CrossRef]
- Adams, J.C. Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 2004, 16, 590–596. [Google Scholar] [CrossRef]
- Nakagawa, H.; Terasaki, A.G.; Suzuki, H.; Ohashi, K.; Miyamoto, S. Short-term retention of actin filament binding proteins on lamellipodial actin bundles. FEBS Lett. 2006, 580, 3223–3228. [Google Scholar] [CrossRef]
- Cohan, C.S.; Welnhofer, E.A.; Zhao, L.; Matsumura, F.; Yamashiro, S. Role of the actin bundling protein fascin in growth cone morphogenesis: Localization in filopodia and lamellipodia. Cell Motil. Cytoskeleton 2001, 48, 109–120. [Google Scholar] [CrossRef]
- Shin, H.; Purdy Drew, K.R.; Bartles, J.R.; Wong, G.C.; Grason, G.M. Cooperativity and frustration in protein-mediated parallel actin bundles. Phys. Rev. Lett. 2009, 103, 238102. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.; Kole, T.P.; Lee, J.S.; Fedorov, E.; Almo, S.C.; Schafer, B.W.; Wirtz, D. How actin crosslinking and bundling proteins cooperate to generate an enhanced cell mechanical response. Biochem. Biophys. Res. Commun. 2005, 334, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Nagy, S.; Rock, R.S. Structured post-IQ domain governs selectivity of myosin X for fascin-actin bundles. J. Biol. Chem. 2010, 285, 26608–26617. [Google Scholar] [CrossRef] [PubMed]
- Gordon, A.M.; Homsher, E.; Regnier, M. Regulation of contraction in striated muscle. Physiol. Rev. 2000, 80, 853–924. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K. Connectin, an elastic protein from myofibrils. J. Biochem. 1976, 80, 405–407. [Google Scholar] [CrossRef]
- Wang, K.; McClure, J.; Tu, A. Titin: Major myofibrillar components of striated muscle. Proc. Natl. Acad. Sci. USA 1979, 76, 3698–3702. [Google Scholar] [CrossRef]
- Wang, K.; Ramirez-Mitchell, R. A network of transverse and longitudinal intermediate filaments is associated with sarcomeres of adult vertebrate skeletal muscle. J. Cell Biol. 1983, 96, 562–570. [Google Scholar] [CrossRef]
- Fowler, V.M. Tropomodulin: A cytoskeletal protein that binds to the end of erythrocyte tropomyosin and inhibits tropomyosin binding to actin. J. Cell Biol. 1990, 111, 471–481. [Google Scholar] [CrossRef]
- Taniguchi, K.; Takeya, R.; Suetsugu, S.; Kan, O.M.; Narusawa, M.; Shiose, A.; Tominaga, R.; Sumimoto, H. Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J. Biol. Chem. 2009, 284, 29873–29881. [Google Scholar] [CrossRef]
- Alberts, B. Molecular Biology of the Cell, 6th ed.; Garland Science, Taylor and Francis Group: New York, NY, USA, 2015; p. 1. [Google Scholar]
- Schwayer, C.; Sikora, M.; Slovakova, J.; Kardos, R.; Heisenberg, C.P. Actin Rings of Power. Dev. Cell 2016, 37, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Pelham, R.J.; Chang, F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature 2002, 419, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Field, C.M.; Alberts, B.M. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J. Cell Biol. 1995, 131, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Kumar, S.; Mizoguchi, A.; Ide, C.; Kinoshita, A.; Haraguchi, T.; Hiraoka, Y.; Noda, M. Nedd5, a mammalian septin, is a novel cytoskeletal component interacting with actin-based structures. Genes Dev. 1997, 11, 1535–1547. [Google Scholar] [CrossRef] [PubMed]
- Laporte, D.; Ojkic, N.; Vavylonis, D.; Wu, J.Q. alpha-Actinin and fimbrin cooperate with myosin II to organize actomyosin bundles during contractile-ring assembly. Mol. Biol. Cell 2012, 23, 3094–3110. [Google Scholar] [CrossRef]
- Schoumacher, M.; Goldman, R.D.; Louvard, D.; Vignjevic, D.M. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 2010, 189, 541–556. [Google Scholar] [CrossRef]
- Doki, C.; Nishida, K.; Saito, S.; Shiga, M.; Ogara, H.; Kuramoto, A.; Kuragano, M.; Nozumi, M.; Igarashi, M.; Nakagawa, H.; et al. Microtubule elongation along actin filaments induced by microtubule-associated protein 4 contributes to the formation of cellular protrusions. J. Biochem. 2020. [Google Scholar] [CrossRef]
- Muto, E.; Sakai, H.; Kaseda, K. Long-range cooperative binding of kinesin to a microtubule in the presence of ATP. J. Cell Biol. 2005, 168, 691–696. [Google Scholar] [CrossRef]
- Shima, T.; Morikawa, M.; Kaneshiro, J.; Kambara, T.; Kamimura, S.; Yagi, T.; Iwamoto, H.; Uemura, S.; Shigematsu, H.; Shirouzu, M.; et al. Kinesin-binding-triggered conformation switching of microtubules contributes to polarized transport. J. Cell Biol. 2018, 217, 4164–4183. [Google Scholar] [CrossRef]
- Laing, N.G.; Dye, D.E.; Wallgren-Pettersson, C.; Richard, G.; Monnier, N.; Lillis, S.; Winder, T.L.; Lochmuller, H.; Graziano, C.; Mitrani-Rosenbaum, S.; et al. Mutations and polymorphisms of the skeletal muscle alpha-actin gene (ACTA1). Hum. Mutat. 2009, 30, 1267–1277. [Google Scholar] [CrossRef]
- Sparrow, J.C.; Nowak, K.J.; Durling, H.J.; Beggs, A.H.; Wallgren-Pettersson, C.; Romero, N.; Nonaka, I.; Laing, N.G. Muscle disease caused by mutations in the skeletal muscle alpha-actin gene (ACTA1). Neuromuscul. Disord. 2003, 13, 519–531. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tokuraku, K.; Kuragano, M.; Uyeda, T.Q.P. Long-Range and Directional Allostery of Actin Filaments Plays Important Roles in Various Cellular Activities. Int. J. Mol. Sci. 2020, 21, 3209. https://doi.org/10.3390/ijms21093209
Tokuraku K, Kuragano M, Uyeda TQP. Long-Range and Directional Allostery of Actin Filaments Plays Important Roles in Various Cellular Activities. International Journal of Molecular Sciences. 2020; 21(9):3209. https://doi.org/10.3390/ijms21093209
Chicago/Turabian StyleTokuraku, Kiyotaka, Masahiro Kuragano, and Taro Q. P. Uyeda. 2020. "Long-Range and Directional Allostery of Actin Filaments Plays Important Roles in Various Cellular Activities" International Journal of Molecular Sciences 21, no. 9: 3209. https://doi.org/10.3390/ijms21093209
APA StyleTokuraku, K., Kuragano, M., & Uyeda, T. Q. P. (2020). Long-Range and Directional Allostery of Actin Filaments Plays Important Roles in Various Cellular Activities. International Journal of Molecular Sciences, 21(9), 3209. https://doi.org/10.3390/ijms21093209




