Actin Mutations and Their Role in Disease
Abstract
1. Introduction
2. Disease-causing Mutations in the Six Actin Genes, an Overview
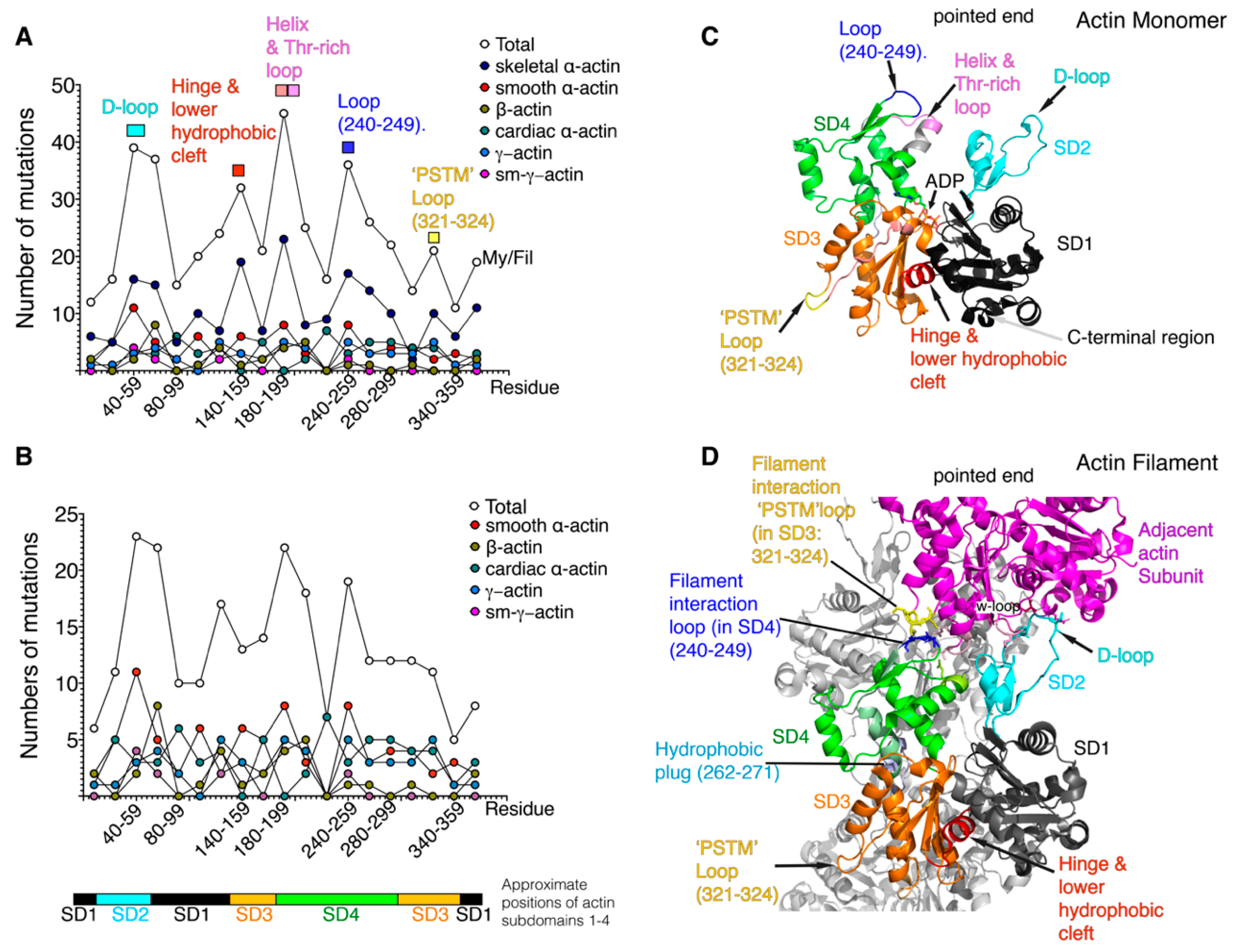
3. Positional Analysis of Disease-Causing Missense Actin Mutations
4. Mutations in Cardiac α-actin that Affect Myosin Interaction with Actin, Directly or Indirectly
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pollard, T.D. Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 2016, 8. [Google Scholar] [CrossRef]
- Varland, S.; Vandekerckhove, J.; Drazic, A. Actin post-translational modifications: The Cinderella of cytoskeletal control. Trends Biochem. Sci. 2019, 44, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, R.; Holmes, K.C. Actin structure and function. Annu. Rev. Biophys. 2011, 40, 169–186. [Google Scholar] [CrossRef] [PubMed]
- Behrmann, E.; Muller, M.; Penczek, P.A.; Mannherz, H.G.; Manstein, D.J.; Raunser, S. Structure of the rigor actin-tropomyosin-myosin complex. Cell 2012, 150, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Iwane, A.H.; Yanagida, T.; Namba, K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature 2010, 467, 724–728. [Google Scholar] [CrossRef]
- Gurel, P.S.; Kim, L.Y.; Ruijgrok, P.V.; Omabegho, T.; Bryant, Z.; Alushin, G.M. Cryo-EM structures reveal specialization at the myosin VI-actin interface and a mechanism of force sensitivity. Elife 2017, 6. [Google Scholar]
- Murakami, K.; Yasunaga, T.; Noguchi, T.Q.; Gomibuchi, Y.; Ngo, K.X.; Uyeda, T.Q.; Wakabayashi, T. Structural basis for actin assembly, activation of ATP hydrolysis, and delayed phosphate release. Cell 2010, 143, 275–287. [Google Scholar] [CrossRef]
- von der Ecken, J.; Muller, M.; Lehman, W.; Manstein, D.J.; Penczek, P.A.; Raunser, S. Structure of the F-actin-tropomyosin complex. Nature 2015, 519, 114–117. [Google Scholar] [CrossRef]
- Huehn, A.R.; Bibeau, J.P.; Schramm, A.C.; Cao, W.; De La Cruz, E.M.; Sindelar, C.V. Structures of cofilin-induced structural changes reveal local and asymmetric perturbations of actin filaments. Proc. Natl. Acad. Sci. USA 2020, 117, 1478–1484. [Google Scholar] [CrossRef]
- Rao, J.N.; Madasu, Y.; Dominguez, R. Mechanism of actin filament pointed-end capping by tropomodulin. Science 2014, 345, 463–467. [Google Scholar] [CrossRef]
- Nowak, K.J.; Ravenscroft, G.; Laing, N.G. Skeletal muscle alpha-actin diseases (actinopathies): Pathology and mechanisms. Acta Neuropathol. 2013, 125, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Wallgren-Pettersson, C.; Sewry, C.A.; Nowak, K.J.; Laing, N.G. Nemaline myopathies. Semin. Pediatr. Neurol. 2011, 18, 230–238. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.C.; Pannu, H.; Tran-Fadulu, V.; Papke, C.L.; Yu, R.K.; Avidan, N.; Bourgeois, S.; Estrera, A.L.; Safi, H.J.; Sparks, E.; et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat. Genet. 2007, 39, 1488–1493. [Google Scholar] [CrossRef] [PubMed]
- Cuoco, J.A.; Busch, C.M.; Klein, B.J.; Benko, M.J.; Stein, R.; Nicholson, A.D.; Marvin, E.A. ACTA2 cerebral arteriopathy: Not just a puff of smoke. Cerebrovasc. Dis. 2018, 46, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Whiffin, N.; Minikel, E.; Walsh, R.; O’Donnell-Luria, A.H.; Karczewski, K.; Ing, A.Y.; Barton, P.J.R.; Funke, B.; Cook, S.A.; MacArthur, D.; et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet. Med. 2017, 19, 1151–1158. [Google Scholar] [CrossRef]
- Mogensen, J.; Klausen, I.C.; Pedersen, A.K.; Egeblad, H.; Bross, P.; Kruse, T.A.; Gregersen, N.; Hansen, P.S.; Baandrup, U.; Borglum, A.D. Alpha-cardiac actin is a novel disease gene in familial hypertrophic cardiomyopathy. J. Clin. Invest. 1999, 103, R39–43. [Google Scholar] [CrossRef]
- Olson, T.M.; Michels, V.V.; Thibodeau, S.N.; Tai, Y.S.; Keating, M.T. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science 1998, 280, 750–752. [Google Scholar] [CrossRef]
- Maron, B.J.; Shirani, J.; Poliac, L.C.; Mathenge, R.; Roberts, W.C.; Mueller, F.O. Sudden death in young competitive athletes. Clinical, demographic, and pathological profiles. Jama 1996, 276, 199–204. [Google Scholar] [CrossRef]
- Maron, B.J. Hypertrophic cardiomyopathy: A systematic review. Jama 2002, 287, 1308–1320. [Google Scholar] [CrossRef]
- Chang, A.N.; Potter, J.D. Sarcomeric protein mutations in dilated cardiomyopathy. Heart Fail. Rev. 2005, 10, 225–235. [Google Scholar] [CrossRef]
- Seidman, J.G.; Seidman, C. The genetic basis for cardiomyopathy: From mutation identification to mechanistic paradigms. Cell 2001, 104, 557–567. [Google Scholar] [CrossRef]
- Riviere, J.B.; van Bon, B.W.; Hoischen, A.; Kholmanskikh, S.S.; O’Roak, B.J.; Gilissen, C.; Gijsen, S.; Sullivan, C.T.; Christian, S.L.; Abdul-Rahman, O.A.; et al. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser-Winter syndrome. Nat. Genet. 2012, 44, 440–444. [Google Scholar] [CrossRef] [PubMed]
- Verloes, A.; Di Donato, N.; Masliah-Planchon, J.; Jongmans, M.; Abdul-Raman, O.A.; Albrecht, B.; Allanson, J.; Brunner, H.; Bertola, D.; Chassaing, N.; et al. Baraitser-Winter cerebrofrontofacial syndrome: Delineation of the spectrum in 42 cases. Eur. J. Hum. Genet. 2015, 23, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Yates, T.M.; Turner, C.L.; Firth, H.V.; Berg, J.; Pilz, D.T. Baraitser-Winter cerebrofrontofacial syndrome. Clin. Genet. 2017, 92, 3–9. [Google Scholar] [CrossRef]
- Cuvertino, S.; Stuart, H.M.; Chandler, K.E.; Roberts, N.A.; Armstrong, R.; Bernardini, L.; Bhaskar, S.; Callewaert, B.; Clayton-Smith, J.; Davalillo, C.H.; et al. ACTB Loss-of-Function Mutations Result in a Pleiotropic Developmental Disorder. Am. J. Hum. Genet. 2017, 101, 1021–1033. [Google Scholar] [CrossRef]
- Latham, S.L.; Ehmke, N.; Reinke, P.Y.A.; Taft, M.H.; Eicke, D.; Reindl, T.; Stenzel, W.; Lyons, M.J.; Friez, M.J.; Lee, J.A.; et al. Variants in exons 5 and 6 of ACTB cause syndromic thrombocytopenia. Nat. Commun. 2018, 9, 4250. [Google Scholar] [CrossRef]
- Zhu, M.; Yang, T.; Wei, S.; DeWan, A.T.; Morell, R.J.; Elfenbein, J.L.; Fisher, R.A.; Leal, S.M.; Smith, R.J.; Friderici, K.H. Mutations in the gamma-actin gene (ACTG1) are associated with dominant progressive deafness (DFNA20/26). Am. J. Hum. Genet. 2003, 73, 1082–1091. [Google Scholar] [CrossRef]
- Belyantseva, I.A.; Perrin, B.J.; Sonnemann, K.J.; Zhu, M.; Stepanyan, R.; McGee, J.; Frolenkov, G.I.; Walsh, E.J.; Friderici, K.H.; Friedman, T.B.; et al. Gamma-actin is required for cytoskeletal maintenance but not development. Proc. Natl. Acad. Sci. USA 2009, 106, 9703–9708. [Google Scholar] [CrossRef]
- Di Donato, N.; Kuechler, A.; Vergano, S.; Heinritz, W.; Bodurtha, J.; Merchant, S.R.; Breningstall, G.; Ladda, R.; Sell, S.; Altmuller, J.; et al. Update on the ACTG1-associated Baraitser-Winter cerebrofrontofacial syndrome. Am. J. Med. Genet. A 2016, 170, 2644–2651. [Google Scholar] [CrossRef]
- Wangler, M.F.; Gonzaga-Jauregui, C.; Gambin, T.; Penney, S.; Moss, T.; Chopra, A.; Probst, F.J.; Xia, F.; Yang, Y.; Werlin, S.; et al. Heterozygous de novo and inherited mutations in the smooth muscle actin (ACTG2) gene underlie megacystis-microcolon-intestinal hypoperistalsis syndrome. PLoS Genet. 2014, 10, e1004258. [Google Scholar] [CrossRef]
- Ravenscroft, G.; Pannell, S.; O’Grady, G.; Ong, R.; Ee, H.C.; Faiz, F.; Marns, L.; Goel, H.; Kumarasinghe, P.; Sollis, E.; et al. Variants in ACTG2 underlie a substantial number of Australasian patients with primary chronic intestinal pseudo-obstruction. Neurogastroenterol. Motil. 2018, 30, e13371. [Google Scholar] [CrossRef] [PubMed]
- Collins, R.R.J.; Barth, B.; Megison, S.; Pfeifer, C.M.; Rice, L.M.; Harris, S.; Timmons, C.F.; Rakheja, D. ACTG2-associated visceral myopathy with chronic intestinal pseudoobstruction, intestinal malrotation, hypertrophic pyloric stenosis, choledochal cyst, and a novel missense mutation. Int. J. Surg. Pathol. 2019, 27, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W.; Mannherz, H.G.; Suck, D.; Pai, E.F.; Holmes, K.C. Atomic structure of the actin:DNase I complex. Nature 1990, 347, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Huehn, A.; Cao, W.; Elam, W.A.; Liu, X.; De La Cruz, E.M.; Sindelar, C.V. The actin filament twist changes abruptly at boundaries between bare and cofilin-decorated segments. J. Biol. Chem. 2018, 293, 5377–5383. [Google Scholar] [CrossRef]
- von der Ecken, J.; Heissler, S.M.; Pathan-Chhatbar, S.; Manstein, D.J.; Raunser, S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature 2016, 534, 724–728. [Google Scholar] [CrossRef]
- Hegyi, G.; Premecz, G.; Sain, B.; Muhlrad, A. Selective carbethoxylation of the histidine residues of actin by diethylpyrocarbonate. Eur. J. Biochem. 1974, 44, 7–12. [Google Scholar] [CrossRef]
- Normand, E.A.; Braxton, A.; Nassef, S.; Ward, P.A.; Vetrini, F.; He, W.; Patel, V.; Qu, C.; Westerfield, L.E.; Stover, S.; et al. Clinical exome sequencing for fetuses with ultrasound abnormalities and a suspected Mendelian disorder. Genome Med. 2018, 10, 74. [Google Scholar] [CrossRef]
- Posey, J.E.; Harel, T.; Liu, P.; Rosenfeld, J.A.; James, R.A.; Coban Akdemir, Z.H.; Walkiewicz, M.; Bi, W.; Xiao, R.; Ding, Y.; et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N. Engl. J. Med. 2017, 376, 21–31. [Google Scholar] [CrossRef]
- Walsh, R.; Thomson, K.L.; Ware, J.S.; Funke, B.H.; Woodley, J.; McGuire, K.J.; Mazzarotto, F.; Blair, E.; Seller, A.; Taylor, J.C.; et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet. Med. 2017, 19, 192–203. [Google Scholar] [CrossRef]
- Proost, D.; Vandeweyer, G.; Meester, J.A.; Salemink, S.; Kempers, M.; Ingram, C.; Peeters, N.; Saenen, J.; Vrints, C.; Lacro, R.V.; et al. Performant mutation identification using targeted next-generation sequencing of 14 thoracic aortic aneurysm genes. Hum. Mutat. 2015, 36, 808–814. [Google Scholar] [CrossRef]
- Fremont, S.; Romet-Lemonne, G.; Houdusse, A.; Echard, A. Emerging roles of MICAL family proteins - From actin oxidation to membrane trafficking during cytokinesis. J. Cell Sci. 2017, 130, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Grintsevich, E.E.; Yesilyurt, H.G.; Rich, S.K.; Hung, R.J.; Terman, J.R.; Reisler, E. F-actin dismantling through a redox-driven synergy between Mical and cofilin. Nat. Cell Biol. 2016, 18, 876–885. [Google Scholar] [CrossRef] [PubMed]
- Alto, L.T.; Terman, J.R. MICALs. Curr Biol 2018, 28, R538–541. [Google Scholar] [CrossRef] [PubMed]
- Hesketh, S.; Srisawat, K.; Sutherland, H.; Jarvis, J.; Burniston, J. On the rate of synthesis of individual proteins within and between different striated muscles of the rat. Proteomes 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Wang, J.; Liu, X.; Wang, Y.; Chen, Y.; Sun, K.; Gao, S.; Zhang, C.; Wang, Z.; Zhang, Y.; et al. Multiple gene mutations, not the type of mutation, are the modifier of left ventricle hypertrophy in patients with hypertrophic cardiomyopathy. Mol. Biol. Rep. 2013, 40, 3969–3976. [Google Scholar] [CrossRef] [PubMed]
- Fischer, J.; Weide, T.; Barnekow, A. The MICAL proteins and rab1: A possible link to the cytoskeleton? Biochem. Biophys. Res. Commun. 2005, 328, 415–423. [Google Scholar] [CrossRef]
- Laing, N.G.; Dye, D.E.; Wallgren-Pettersson, C.; Richard, G.; Monnier, N.; Lillis, S.; Winder, T.L.; Lochmuller, H.; Graziano, C.; Mitrani-Rosenbaum, S.; et al. Mutations and polymorphisms of the skeletal muscle alpha-actin gene (ACTA1). Hum. Mutat. 2009, 30, 1267–1277. [Google Scholar] [CrossRef]
- Beuchle, D.; Schwarz, H.; Langegger, M.; Koch, I.; Aberle, H. Drosophila MICAL regulates myofilament organization and synaptic structure. Mech. Dev. 2007, 124, 390–406. [Google Scholar] [CrossRef]
- Hoffjan, S.; Waldmuller, S.; Blankenfeldt, W.; Kotting, J.; Gehle, P.; Binner, P.; Epplen, J.T.; Scheffold, T. Three novel mutations in the ACTA2 gene in German patients with thoracic aortic aneurysms and dissections. Eur J. Hum. Genet. 2011, 19, 520–524. [Google Scholar] [CrossRef][Green Version]
- Disabella, E.; Grasso, M.; Gambarin, F.I.; Narula, N.; Dore, R.; Favalli, V.; Serio, A.; Antoniazzi, E.; Mosconi, M.; Pasotti, M.; et al. Risk of dissection in thoracic aneurysms associated with mutations of smooth muscle alpha-actin 2 (ACTA2). Heart 2011, 97, 321–326. [Google Scholar] [CrossRef]
- Morin, M.; Bryan, K.E.; Mayo-Merino, F.; Goodyear, R.; Mencia, A.; Modamio-Hoybjor, S.; del Castillo, I.; Cabalka, J.M.; Richardson, G.; Moreno, F.; et al. In vivo and in vitro effects of two novel gamma-actin (ACTG1) mutations that cause DFNA20/26 hearing impairment. Hum. Mol. Genet. 2009, 18, 3075–3089. [Google Scholar] [CrossRef] [PubMed]
- Sparrow, J.C.; Nowak, K.J.; Durling, H.J.; Beggs, A.H.; Wallgren-Pettersson, C.; Romero, N.; Nonaka, I.; Laing, N.G. Muscle disease caused by mutations in the skeletal muscle alpha-actin gene (ACTA1). NMD 2003, 13, 519–531. [Google Scholar] [CrossRef]
- Bhoj, E.J.; Haye, D.; Toutain, A.; Bonneau, D.; Nielsen, I.K.; Lund, I.B.; Bogaard, P.; Leenskjold, S.; Karaer, K.; Wild, K.T.; et al. Phenotypic spectrum associated with SPECC1L pathogenic variants: New families and critical review of the nosology of Teebi, Opitz GBBB, and Baraitser-Winter syndromes. Eur. J. Med. Genet. 2019, 62, 103588. [Google Scholar] [CrossRef] [PubMed]
- Whittington, J.R.; Poole, A.T.; Dutta, E.H.; Munn, M.B. A novel mutation in ACTG2 gene in mother with chronic intestinal pseudoobstruction and fetus with megacystis microcolon intestinal hypoperistalsis syndrome. Case Rep. Genet. 2017, 2017, 9146507. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.C.; Papke, C.L.; Tran-Fadulu, V.; Regalado, E.S.; Avidan, N.; Johnson, R.J.; Kim, D.H.; Pannu, H.; Willing, M.C.; Sparks, E.; et al. Mutations in smooth muscle alpha-actin (ACTA2) cause coronary artery disease, stroke, and Moyamoya disease, along with thoracic aortic disease. Am. J. Hum. Genet. 2009, 84, 617–627. [Google Scholar] [CrossRef]
- Meng, L.; Pammi, M.; Saronwala, A.; Magoulas, P.; Ghazi, A.R.; Vetrini, F.; Zhang, J.; He, W.; Dharmadhikari, A.V.; Qu, C.; et al. Use of exome sequencing for infants in intensive care units: Ascertainment of severe single-gene disorders and effect on medical management. JAMA Pediatr. 2017, 171, e173438. [Google Scholar] [CrossRef]
- Batchelor, M.; Wolny, M.; Dougan, L.; Paci, E.; Knight, P.J.; Peckham, M. Myosin tails and single alpha-helical domains. Biochem. Soc. Trans. 2015, 43, 58–63. [Google Scholar] [CrossRef]
- Iglesias, A.; Anyane-Yeboa, K.; Wynn, J.; Wilson, A.; Truitt Cho, M.; Guzman, E.; Sisson, R.; Egan, C.; Chung, W.K. The usefulness of whole-exome sequencing in routine clinical practice. Genet. Med. 2014, 16, 922–931. [Google Scholar] [CrossRef]
- Weerakkody, R.; Ross, D.; Parry, D.A.; Ziganshin, B.; Vandrovcova, J.; Gampawar, P.; Abdullah, A.; Biggs, J.; Dumfarth, J.; Ibrahim, Y.; et al. Targeted genetic analysis in a large cohort of familial and sporadic cases of aneurysm or dissection of the thoracic aorta. Genet. Med. 2018, 20, 1414–1422. [Google Scholar] [CrossRef]
- Reza, N.; Garg, A.; Merrill, S.L.; Chowns, J.L.; Rao, S.; Owens, A.T. ACTA1 novel likely pathogenic variant in a family with dilated cardiomyopathy. Circ. Genom. Precis. Med. 2018, 11, e002243. [Google Scholar] [CrossRef]
- Rainger, J.; Williamson, K.A.; Soares, D.C.; Truch, J.; Kurian, D.; Gillessen-Kaesbach, G.; Seawright, A.; Prendergast, J.; Halachev, M.; Wheeler, A.; et al. A recurrent de novo mutation in ACTG1 causes isolated ocular coloboma. Hum. Mutat. 2017, 38, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Sandestig, A.; Green, A.; Jonasson, J.; Vogt, H.; Wahlstrom, J.; Pepler, A.; Ellnebo, K.; Biskup, S.; Stefanova, M. Could dissimilar phenotypic effects of ACTB missense mutations reflect the actin conformational change? Two novel mutations and literature review. Mol. Syndromol. 2019, 9, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Ohlsson, M.; Tajsharghi, H.; Darin, N.; Kyllerman, M.; Oldfors, A. Follow-up of nemaline myopathy in two patients with novel mutations in the skeletal muscle alpha-actin gene (ACTA1). NMD 2004, 14, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Regalado, E.S.; Guo, D.C.; Prakash, S.; Bensend, T.A.; Flynn, K.; Estrera, A.; Safi, H.; Liang, D.; Hyland, J.; Child, A.; et al. Aortic disease presentation and outcome associated with ACTA2 mutations. Circ. Cardiovasc. Genet. 2015, 8, 457–864. [Google Scholar] [CrossRef] [PubMed]
- Ilkovski, B.; Cooper, S.T.; Nowak, K.; Ryan, M.M.; Yang, N.; Schnell, C.; Durling, H.J.; Roddick, L.G.; Wilkinson, I.; Kornberg, A.J.; et al. Nemaline myopathy caused by mutations in the muscle alpha-skeletal-actin gene. Am. J. Hum. Genet. 2001, 68, 1333–1343. [Google Scholar] [CrossRef] [PubMed]
- Weitensteiner, V.; Zhang, R.; Bungenberg, J.; Marks, M.; Gehlen, J.; Ralser, D.J.; Hilger, A.C.; Sharma, A.; Schumacher, J.; Gembruch, U.; et al. Exome sequencing in syndromic brain malformations identifies novel mutations in ACTB, and SLC9A6, and suggests BAZ1A as a new candidate gene. Birth Defects Res. 2018, 110, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Lukoyanova, N.; VanLoock, M.S.; Orlova, A.; Galkin, V.E.; Wang, K.; Egelman, E.H. Each actin subunit has three nebulin binding sites: Implications for steric blocking. Curr. Biol. 2002, 12, 383–388. [Google Scholar] [CrossRef]
- Pappas, C.T.; Bliss, K.T.; Zieseniss, A.; Gregorio, C.C. The Nebulin family: An actin support group. Trends Cell Biol. 2011, 21, 29–37. [Google Scholar] [CrossRef]
- Kiss, B.; Lee, E.J.; Ma, W.; Li, F.W.; Tonino, P.; Mijailovich, S.M.; Irving, T.C.; Granzier, H.L. Nebulin stiffens the thin filament and augments cross-bridge interaction in skeletal muscle. Proc. Natl. Acad. Sci. USA 2018, 115, 10369–10374. [Google Scholar] [CrossRef]
- Hernandez, D.A.; Bennett, C.M.; Dunina-Barkovskaya, L.; Wedig, T.; Capetanaki, Y.; Herrmann, H.; Conover, G.M. Nebulette is a powerful cytolinker organizing desmin and actin in mouse hearts. Mol. Biol. Cell 2016, 27, 3869–3882. [Google Scholar] [CrossRef]
- Bang, M.L.; Chen, J. Roles of nebulin family members in the heart. Circ. J. 2015, 79, 2081–2087. [Google Scholar] [CrossRef] [PubMed]
- Meyer, L.C.; Wright, N.T. Structure of giant muscle proteins. Front. Physiol. 2013, 4, 368. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Saha, S.; Shabalina, S.A.; Kashina, A. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science 2010, 329, 1534–1537. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Mundia, M.M.; Zhang, F.; Demers, R.W.; Korobova, F.; Svitkina, T.; Perieteanu, A.A.; Dawson, J.F.; Kashina, A. Arginylation regulates intracellular actin polymer level by modulating actin properties and binding of capping and severing proteins. Mol. Biol. Cell 2010, 21, 1350–1361. [Google Scholar] [CrossRef] [PubMed]
- Drazic, A.; Aksnes, H.; Marie, M.; Boczkowska, M.; Varland, S.; Timmerman, E.; Foyn, H.; Glomnes, N.; Rebowski, G.; Impens, F.; et al. NAA80 is actin’s N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc. Natl. Acad. Sci. USA 2018, 115, 4399–4404. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Terman, J.R. MICAL redox enzymes and actin remodeling: New links to classical tumorigenic and cancer pathways. Mol. Cell Oncol. 2018, 5, e1384881. [Google Scholar] [CrossRef] [PubMed]
- Milligan, R.A. Protein-protein interactions in the rigor actomyosin complex. Proc. Natl. Acad. Sci. USA 1996, 93, 21–26. [Google Scholar] [CrossRef]
- Milligan, R.A.; Whittaker, M.; Safer, D. Molecular structure of F-actin and location of surface binding sites. Nature 1990, 348, 217–221. [Google Scholar] [CrossRef]
- Marston, S. The molecular mechanisms of mutations in actin and myosin that cause inherited myopathy. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef]
- Olson, T.M.; Doan, T.P.; Kishimoto, N.Y.; Whitby, F.G.; Ackerman, M.J.; Fananapazir, L. Inherited and de novo mutations in the cardiac actin gene cause hypertrophic cardiomyopathy. J. Mol. Cell Cardiol. 2000, 32, 1687–1694. [Google Scholar] [CrossRef]
- Vang, S.; Corydon, T.J.; Borglum, A.D.; Scott, M.D.; Frydman, J.; Mogensen, J.; Gregersen, N.; Bross, P. Actin mutations in hypertrophic and dilated cardiomyopathy cause inefficient protein folding and perturbed filament formation. Febs J. 2005, 272, 2037–2049. [Google Scholar] [CrossRef] [PubMed]
- Bookwalter, C.S.; Trybus, K.M. Functional consequences of a mutation in an expressed human alpha-cardiac actin at a site implicated in familial hypertrophic cardiomyopathy. J. Biol. Chem. 2006, 281, 16777–16784. [Google Scholar] [CrossRef] [PubMed]
- Wolny, M.; Colegrave, M.; Colman, L.; White, E.; Knight, P.J.; Peckham, M. Cardiomyopathy mutations in the tail of beta-cardiac myosin modify the coiled-coil structure and affect integration into thick filaments in muscle sarcomeres in adult cardiomyocytes. J. Biol. Chem. 2013, 288, 31952–31962. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Ono, K.; Ono, S. Mutations in Caenorhabditis elegans actin, which are equivalent to human cardiomyopathy mutations, cause abnormal actin aggregation in nematode striated muscle. F1000Res. 2019, 8, 279. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Dyer, E.; Stuckey, D.J.; Copeland, O.; Leung, M.C.; Bayliss, C.; Messer, A.; Wilkinson, R.; Tremoleda, J.L.; Schneider, M.D.; et al. Molecular mechanism of the E99K mutation in cardiac actin (ACTC Gene) that causes apical hypertrophy in man and mouse. J. Biol. Chem. 2011, 286, 27582–27593. [Google Scholar] [CrossRef]
- Rommelaere, H.; Waterschoot, D.; Neirynck, K.; Vandekerckhove, J.; Ampe, C. A method for rapidly screening functionality of actin mutants and tagged actins. Biol. Proced. Online 2004, 6, 235–249. [Google Scholar] [CrossRef][Green Version]
- Brault, V.; Sauder, U.; Reedy, M.C.; Aebi, U.; Schoenenberger, C.A. Differential epitope tagging of actin in transformed Drosophila produces distinct effects on myofibril assembly and function of the indirect flight muscle. Mol. Biol. Cell 1999, 10, 135–149. [Google Scholar] [CrossRef]
- Debold, E.P.; Saber, W.; Cheema, Y.; Bookwalter, C.S.; Trybus, K.M.; Warshaw, D.M.; Vanburen, P. Human actin mutations associated with hypertrophic and dilated cardiomyopathies demonstrate distinct thin filament regulatory properties in vitro. J. Mol. Cell Cardiol. 2010, 48, 286–292. [Google Scholar] [CrossRef]
- Barua, B.; Fagnant, P.M.; Winkelmann, D.A.; Trybus, K.M.; Hitchcock-DeGregori, S.E. A periodic pattern of evolutionarily conserved basic and acidic residues constitutes the binding interface of actin-tropomyosin. J. Biol. Chem. 2013, 288, 9602–9609. [Google Scholar] [CrossRef]
- Bai, F.; Caster, H.M.; Rubenstein, P.A.; Dawson, J.F.; Kawai, M. Using baculovirus/insect cell expressed recombinant actin to study the molecular pathogenesis of HCM caused by actin mutation A331P. J. Mol. Cell Cardiol. 2014, 74, 64–75. [Google Scholar] [CrossRef]
- Chow, M.L.; Shaffer, J.F.; Harris, S.P.; Dawson, J.F. Altered interactions between cardiac myosin binding protein-C and alpha-cardiac actin variants associated with cardiomyopathies. Arch. Biochem. Biophys. 2014, 550–551, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Dyer, E.; Stuckey, D.; Leung, M.C.; Memo, M.; Mansfield, C.; Ferenczi, M.; Liu, K.; Redwood, C.; Nowak, K.; et al. Investigation of a transgenic mouse model of familial dilated cardiomyopathy. J. Mol. Cell Cardiol. 2010, 49, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Marston, S.; Zamora, J.E. Troponin structure and function: A view of recent progress. J. Muscle Res. Cell Motil. 2020, 41, 71–89. [Google Scholar] [CrossRef] [PubMed]
- Mundia, M.M.; Demers, R.W.; Chow, M.L.; Perieteanu, A.A.; Dawson, J.F. Subdomain location of mutations in cardiac actin correlate with type of functional change. PLoS ONE 2012, 7, e36821. [Google Scholar] [CrossRef] [PubMed]
- Dahari, M.; Dawson, J.F. Do cardiac actin mutations lead to altered actomyosin interactions? Biochem. Cell Biol. 2015, 93, 330–334. [Google Scholar] [CrossRef]
- Muller, M.; Mazur, A.J.; Behrmann, E.; Diensthuber, R.P.; Radke, M.B.; Qu, Z.; Littwitz, C.; Raunser, S.; Schoenenberger, C.A.; Manstein, D.J.; et al. Functional characterization of the human alpha-cardiac actin mutations Y166C and M305L involved in hypertrophic cardiomyopathy. Cell Mol. Life Sci. 2012, 69, 3457–3479. [Google Scholar] [CrossRef]
- Lindqvist, J.; Penisson-Besnier, I.; Iwamoto, H.; Li, M.; Yagi, N.; Ochala, J. A myopathy-related actin mutation increases contractile function. Acta Neuropathol. 2012, 123, 739–746. [Google Scholar] [CrossRef]
- Fan, J.; Chan, C.; McNamara, E.L.; Nowak, K.J.; Iwamoto, H.; Ochala, J. Molecular consequences of the myopathy-related D286G mutation on actin function. Front. Physiol. 2018, 9, 1756. [Google Scholar] [CrossRef]
- Chan, C.; Fan, J.; Messer, A.E.; Marston, S.B.; Iwamoto, H.; Ochala, J. Myopathy-inducing mutation H40Y in ACTA1 hampers actin filament structure and function. Biochim. Biophys. Acta 2016, 1862, 1453–1458. [Google Scholar] [CrossRef]
- Sevdali, M.; Kumar, V.; Peckham, M.; Sparrow, J. Human congenital myopathy actin mutants cause myopathy and alter Z-disc structure in Drosophila flight muscle. Neuromuscul. Disord. 2013, 23, 243–255. [Google Scholar] [CrossRef]
- Anthony Akkari, P.; Nowak, K.J.; Beckman, K.; Walker, K.R.; Schachat, F.; Laing, N.G. Production of human skeletal alpha-actin proteins by the baculovirus expression system. Biochem. Biophys. Res. Commun. 2003, 307, 74–79. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parker, F.; Baboolal, T.G.; Peckham, M. Actin Mutations and Their Role in Disease. Int. J. Mol. Sci. 2020, 21, 3371. https://doi.org/10.3390/ijms21093371
Parker F, Baboolal TG, Peckham M. Actin Mutations and Their Role in Disease. International Journal of Molecular Sciences. 2020; 21(9):3371. https://doi.org/10.3390/ijms21093371
Chicago/Turabian StyleParker, Francine, Thomas G. Baboolal, and Michelle Peckham. 2020. "Actin Mutations and Their Role in Disease" International Journal of Molecular Sciences 21, no. 9: 3371. https://doi.org/10.3390/ijms21093371
APA StyleParker, F., Baboolal, T. G., & Peckham, M. (2020). Actin Mutations and Their Role in Disease. International Journal of Molecular Sciences, 21(9), 3371. https://doi.org/10.3390/ijms21093371




