Redox Components: Key Regulators of Epigenetic Modifications in Plants
Abstract
:1. Introduction
2. Epigenetic Modifications in Plants
2.1. DNA Methylation
2.2. Histone Methylation and Acetylation
2.3. Chromatin Remodeling
3. Redox Components
3.1. ROS and NO
3.2. Enzymatic and Nonenzymatic Antioxidants
4. Redox Regulation of Epigenetic Modifications
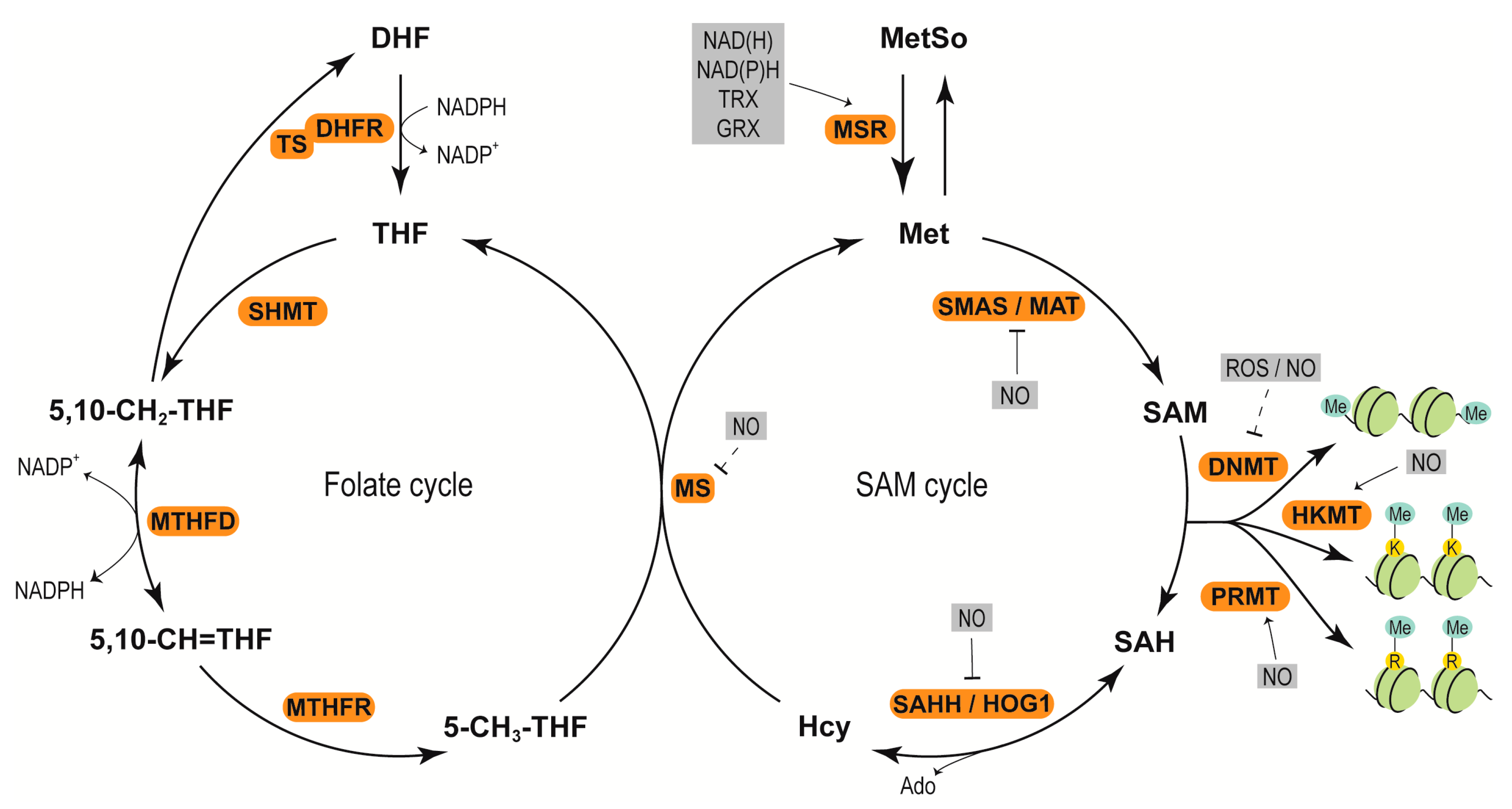
4.1. Redox Regulation of DNA Methylation
4.2. Redox Adjustment of Histone Methylation
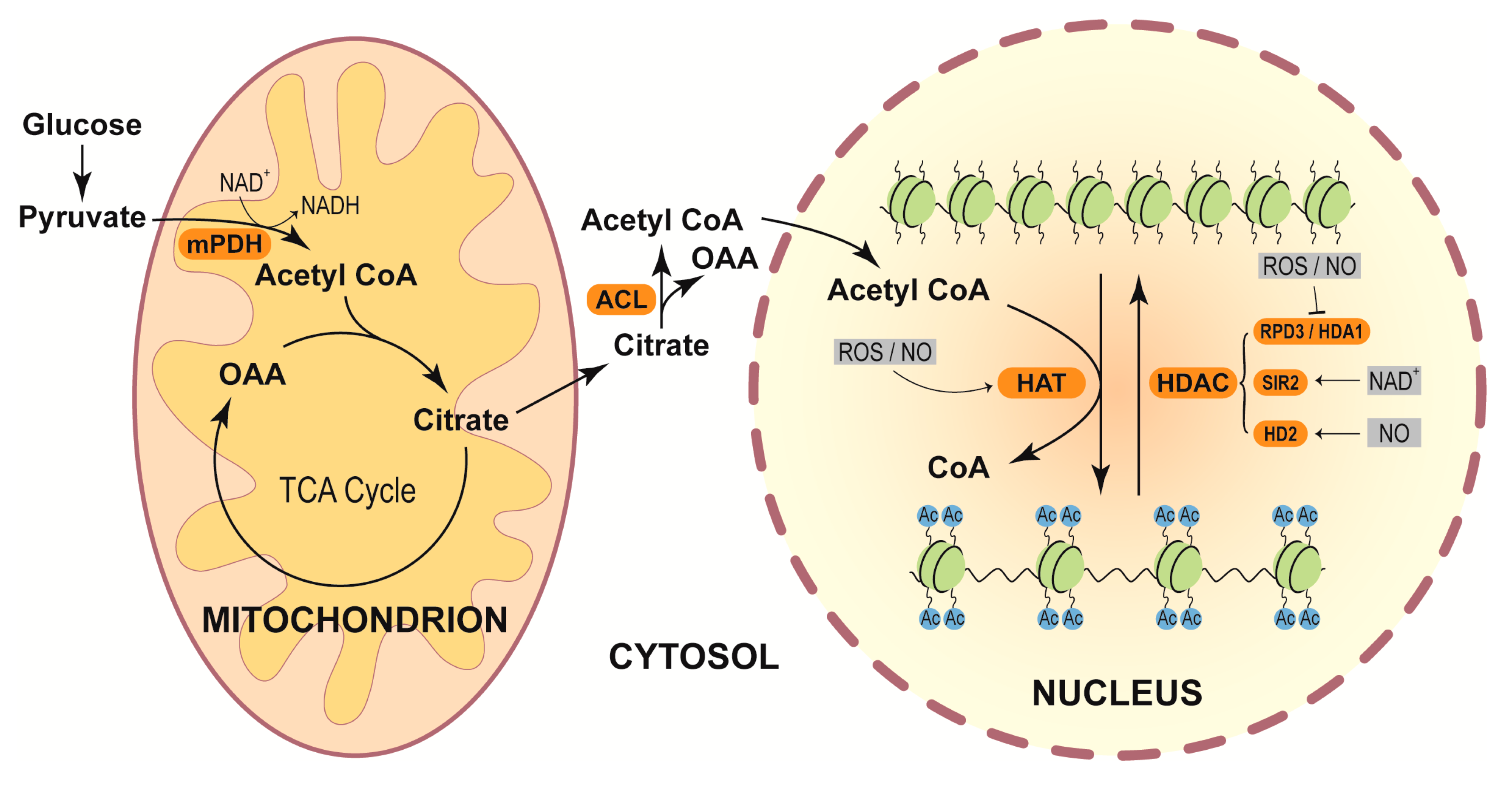
4.3. Redox Regulation of Histone Acetylation
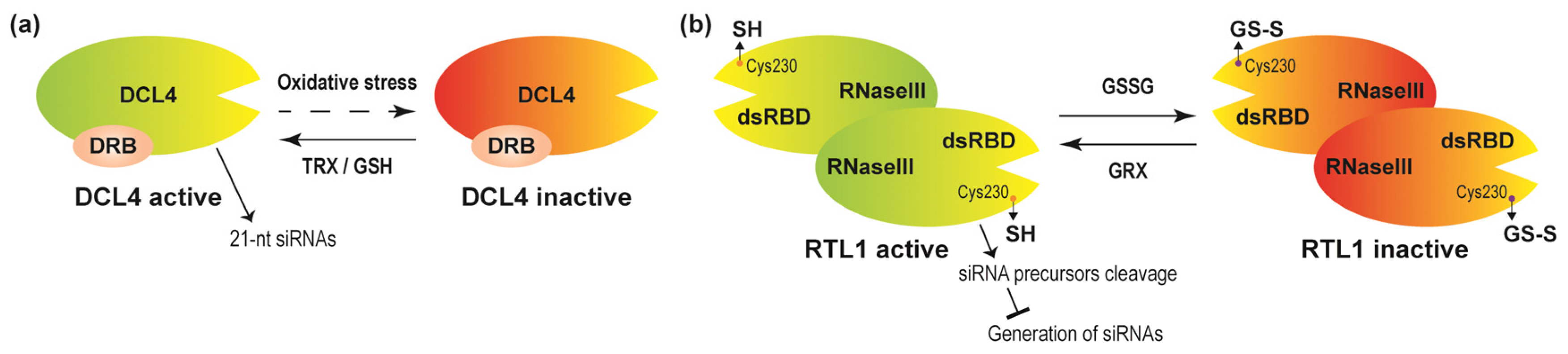
4.4. Redox Affecting Chromatin Remodelers and Other Chromatin-Associated Factors
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ACL | ATP-citrate lyase |
| APX | Ascorbate peroxidase |
| ASC | Ascorbate |
| CAT | Catalase |
| CHD | Chromodomain helicase DNA-binding |
| CMT | Chromomethylase |
| DCL | Dicer-like |
| DDM1 | DNA methylation 1 |
| DHF | Dihydrofolate |
| DHFR | Dihydrofolate reductase |
| DME | Demeter |
| DML | Demeter-like |
| DNMT | DNA methyltransferase |
| DRB | dsRNA-binding protein |
| DRM | Domain rearranged methyltransferase |
| dsRBD | dsRNA binding domains |
| GCN5 | General Control Nondepressible 5 |
| GPX | Glutathione peroxidase |
| GR | Glutathione reductase |
| GRX | Glutaredoxin |
| GSH | Glutathione |
| GSNO | S-Nitrosoglutathione |
| GSSG | Oxidized glutathione |
| H2O2 | Hydrogen peroxide |
| HAT | Histone acetyltransferase |
| Hcy | Homocysteine |
| HD2 | Histone deacetylase 2 |
| HDAC | Histone deacetylase |
| HDM | Histone demethylase |
| HKMT | Histone lysine methyltransferase |
| HMT | Histone methyltransferase |
| HDT2 | Histone deacetylase 2 |
| JMJ | Jumonji C domain-containing protein |
| LSD1 | Lysine-specific demethylase 1 |
| Met | Methionine |
| MET1 | DNA methyltransferase 1 |
| MetSo | Methionine sulfoxide |
| MS | Methionine synthase |
| MSR | Methionine sulfoxide reductase |
| MTHFD1 | Methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase1 |
| MTHFR | Methylenetetrahyrofolate reductase |
| NAD(P)H | Pyridine nucleotides |
| NO | Nitric oxide |
| OAA | Oxaloacetate |
| 1O2 | Singlet oxygen |
| O2•− | Superoxide radical |
| PCD | Programmed cell death |
| PDH | Pyruvate dehydrogenase |
| PR | Pathogensis-related |
| PRMT | Protein arginine methyltransferase |
| RBOH | Respiratory burst oxidase |
| RdDM | RNA directed DNA methylation |
| RDP3/HDA1 | Reduced potassium dependency 3/histone deacetylase 1 |
| ROS | Reactive oxygen species |
| ROS1 | Repressor of silencing 1 |
| RTL | RNAse III-like |
| SA | Salicylic acid |
| SAM | S-Adenosyl-L-methionine |
| SAMS/MAT | S-Adenosyl methionine synthase/methionine adenosyl transferases |
| SAHH/HOG1 | S-Adenosylhomocysteine hydrolase/homologous gene gilencing 1 |
| SHMT | Serine hydroxylmethyl transferase |
| siRNA | Small interfering RNA |
| SIR2 | Silent information regulator 2 |
| SNP | Sodium nitroprusside |
| SOD | Superoxide dismutase |
| stn7 | State transition 7 |
| SWI/SNF | Switching defective/sucrose non-fermenting |
| THF | Tetrahydrofolate |
| Topo VI | Topoisomerase VI |
| TRX | Thioredoxin |
| TS | Thymidylate synthase |
References
- Pikaard, C.S.; Mittelsten Scheid, O. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 2014, 6, a019315. [Google Scholar] [CrossRef] [PubMed]
- Allis, C.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.M.; Lang, Z.B.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Lebedeva, M.A.; Tvorogova, V.E.; Tikhodeyev, O.N. Epigenetic mechanisms and their role in plant development. Russ. J. Genet. 2017, 53, 1057–1071. [Google Scholar] [CrossRef]
- Lee, K.; Seo, P.J. Dynamic epigenetic changes during plant regeneration. Trends Plant Sci. 2018, 23, 235–247. [Google Scholar] [CrossRef]
- Ramirez-Prado, J.S.; Abulfaraj, A.A.; Rayapuram, N.; Benhamed, M.; Hirt, H. Plant immunity: From signaling to epigenetic control of defense. Trends Plant Sci. 2018, 23, 833–844. [Google Scholar] [CrossRef]
- Chang, Y.N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.K.; Duan, C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant. Biol. 2019. [Google Scholar] [CrossRef]
- Vriet, C.; Hennig, L.; Laloi, C. Stress-induced chromatin changes in plants: Of memories, metabolites and crop improvement. Cell. Mol. Life Sci. 2015, 72, 1261–1273. [Google Scholar] [CrossRef]
- Yamamuro, C.; Zhu, J.K.; Yang, Z. Epigenetic modifications and plant hormone action. Mol. Plant 2016, 9, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Issakidis-Bourguet, E.; Zhou, D.X. Perspectives on the interactions between metabolism, redox, and epigenetics in plants. J. Exp. Bot. 2016, 67, 5291–5300. [Google Scholar] [CrossRef]
- Locato, V.; Cimini, S.; De Gara, L. ROS and redox balance as multifaceted players of cross-tolerance: Epigenetic and retrograde control of gene expression. J. Exp. Bot. 2018, 69, 3373–3391. [Google Scholar] [CrossRef] [PubMed]
- Ageeva-Kieferle, A.; Rudolf, E.E.; Lindermayr, C. Redox-dependent chromatin remodeling: A new function of nitric oxide as architect of chromatin structure in plants. Front. Plant Sci. 2019, 10, 625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Rio, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, D.K.; Palma, J.M.; Corpas, F.J. Antioxidants and Antioxidant Enzymes in Higher Plants., 1st Ed. ed; Springer International Publishing AG: Gewerbestrasse, Switzerland, 2018; pp. 1–162. [Google Scholar]
- He, H.M.; Van Breusegem, F.; Mhamdi, A. Redox-dependent control of nuclear transcription in plants. J. Exp. Bot. 2018, 69, 3359–3372. [Google Scholar] [CrossRef]
- Farnese, F.S.; Menezes-Silva, P.E.; Gusman, G.S.; Oliveira, J.A. When bad guys become good ones: The key role of reactive oxygen species and nitric oxide in the plant response to abiotic stress. Front. Plant Sci. 2016, 7, 471. [Google Scholar] [CrossRef] [Green Version]
- Chan, Z.; Yokawa, K.; Kim, W.Y.; Song, C.P. ROS regulation during plant abiotic stress responses. Front. Plant Sci. 2016, 7, 1536. [Google Scholar] [CrossRef]
- Waszczak, C.; Carmody, M.; Kangasjarvi, J. Reactive oxygen species in plant signaling. Annu. Rev. Plant Biol. 2018, 69, 209–236. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Wei, W.; Zhou, D.X. Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 2015, 20, 614–621. [Google Scholar] [CrossRef]
- Hu, J.L.; Yang, H.J.; Mu, J.Y.; Lu, T.C.; Peng, J.L.; Deng, X.; Kong, Z.S.; Bao, S.L.; Cao, X.F.; Zuo, J.R. Nitric oxide regulates protein methylation during stress responses in plants. Mol. Cell 2017, 67, 702–710. [Google Scholar] [CrossRef]
- Hu, Y.; Lu, Y.; Zhao, Y.; Zhou, D.X. Histone acetylation dynamics integrates metabolic activity to regulate plant response to stress. Front. Plant Sci. 2019, 10, 1236. [Google Scholar] [CrossRef]
- Zhao, T.; Zhan, Z.; Jiang, D. Histone modifications and their regulatory roles in plant development and environmental memory. J. Genet. Genom. 2019. [Google Scholar] [CrossRef] [PubMed]
- Law, J.A.; Jacobsen, S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010, 11, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Bartels, A.; Han, Q.; Nair, P.; Stacey, L.; Gaynier, H.; Mosley, M.; Huang, Q.Q.; Pearson, J.K.; Hsieh, T.F.; An, Y.Q.C.; et al. Dynamic DNA methylation in plant growth and development. Int. J. Mol. Sci. 2018, 19, 2144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bewick, A.J.; Schmitz, R.J. Gene body DNA methylation in plants. Curr. Opin. Plant Biol. 2017, 36, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef]
- Cuerda-Gil, D.; Slotkin, R.K. Non-canonical RNA-directed DNA methylation. Nat. Plants 2016, 2, 16163. [Google Scholar] [CrossRef]
- Parrilla-Doblas, J.T.; Roldan-Arjona, T.; Ariza, R.R.; Cordoba-Canero, D. Active DNA demethylation in plants. Int. J. Mol. Sci. 2019, 20, 4683. [Google Scholar] [CrossRef] [Green Version]
- Luger, K.; Mader, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 1997, 389, 251–260. [Google Scholar]
- Ueda, M.; Seki, M. Histone modifications form epigenetic regulatory networks to regulate abiotic stress response. Plant Physiol. 2020, 182, 15–26. [Google Scholar] [CrossRef] [Green Version]
- He, S.B.; Yan, S.H.; Wang, P.; Zhu, W.; Wang, X.W.; Shen, Y.; Shao, K.J.; Xin, H.P.; Li, S.H.; Li, L.J. Comparative analysis of genome-wide chromosomal histone modification patterns in maize cultivars and their wild relatives. PLoS ONE 2014, 9, e97364. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.; He, S.; Yan, S.; Li, Y.; Li, H.; Zhang, H.; Zhao, L.; Li, L. Exogenous EDDS modifies copper-induced various toxic responses in rice. Protoplasma 2014, 251, 1213–1221. [Google Scholar] [CrossRef]
- Liu, C.Y.; Lu, F.L.; Cui, X.; Cao, X.F. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Chen, X. Dynamics of histone H3 lysine 27 trimethylation in plant development. Curr. Opin. Plant Biol. 2011, 14, 123–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berr, A.; Shafiq, S.; Shen, W.H. Histone modifications in transcriptional activation during plant development. BBA-Gene Regul. Mech. 2011, 1809, 567–576. [Google Scholar]
- Xiao, J.; Lee, U.S.; Wagner, D. Tug of war: Adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 2016, 34, 41–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marmorstein, R.; Zhou, M.M. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harbor Perspect. Biol. 2014, 6, a018762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.J.; Hou, X.M.; Tan, L.M.; Shao, C.R.; Huang, H.W.; Li, Y.Q.; Li, L.; Cai, T.; Chen, S.; He, X.J. The Arabidopsis acetylated histone-binding protein BRAT1 forms a complex with BRP1 and prevents transcriptional silencing. Nat. Commun. 2016, 7, 11715. [Google Scholar] [CrossRef]
- Nie, W.F.; Lei, M.; Zhang, M.; Tang, K.; Huang, H.; Zhang, C.; Miki, D.; Liu, P.; Yang, Y.; Wang, X.; et al. Histone acetylation recruits the SWR1 complex to regulate active DNA demethylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2019, 116, 16641–16650. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Yue, M.; Zheng, X.; Gautam, M.; He, S.; Li, L. The role of promoter-associated histone acetylation of Haem Oxygenase-1 (HO-1) and Giberellic Acid-Stimulated Like-1 (GSL-1) genes in heat-induced lateral root primordium inhibition in maize. Front. Plant Sci. 2018, 9, 1520. [Google Scholar] [CrossRef] [Green Version]
- Pandey, R.; Muller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Ding, A.B.; Zhong, X. Functions and mechanisms of plant histone deacetylases. Sci. China Life Sci. 2020, 63, 206–216. [Google Scholar] [CrossRef] [Green Version]
- Ojolo, S.P.; Cao, S.; Priyadarshani, S.V.G.N.; Li, W.; Yan, M.; Aslam, M.; Zhao, H.; Qin, Y. Regulation of plant growth and development: A review from a chromatin remodeling perspective. Front. Plant Sci. 2018, 9, 1232. [Google Scholar] [CrossRef]
- Han, S.K.; Wu, M.F.; Cui, S.; Wagner, D. Roles and activities of chromatin remodeling ATPases in plants. Plant J. 2015, 83, 62–77. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Miao, Y.; Song, C.P. Behind the scenes: The roles of reactive oxygen species in guard cells. New Phytol. 2014, 201, 1121–1140. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Song, C.P.; Wang, B.; Zhou, J.; Kangasjärvi, J.; Zhu, J.K.; Gong, Z. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 2018, 60, 67–88. [Google Scholar]
- Marino, D.; Dunand, C.; Puppo, A.; Pauly, N. A burst of plant NADPH oxidases. Trends Plant Sci. 2012, 17, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.Y.; Zhang, H.; Sun, L.R.; Jiao, Y.H.; Zhang, G.Z.; Miao, C.; Hao, F.S. NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 2012, 63, 305–317. [Google Scholar] [CrossRef]
- Jiao, Y.H.; Sun, L.R.; Song, Y.L.; Wang, L.M.; Liu, L.P.; Zhang, L.Y.; Liu, B.; Li, N.; Miao, C.; Hao, F.S. AtrbohD and AtrbohF positively regulate abscisic acid-inhibited primary root growth by affecting Ca2+ signalling and auxin response of roots in Arabidopsis. J. Exp. Bot. 2013, 64, 4183–4192. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Sun, L.R.; Zhang, L.Y.; Song, Y.L.; Hu, P.P.; Li, C.; Hao, F.S. AtrbohD and AtrbohF negatively regulate lateral root development by changing the localized accumulation of superoxide in primary roots of Arabidopsis. Planta 2015, 241, 591–602. [Google Scholar] [CrossRef]
- Chen, Q.; Yang, G. Signal function studies of ROS, especially RBOH dependent ROS, in plant growth, development and environmental stress. J. Plant Growth Regul. 2019. [Google Scholar] [CrossRef]
- Sun, L.; Zhao, Z.J.; Hao, F.S. NADPH oxidases, essential players of hormone signalings in plant development and response to stresses. Plant Signal. Behav. 2019, 14, 1657343. [Google Scholar] [CrossRef]
- Czarnocka, W.; Karpinski, S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef]
- Kolbert, Z.; Barroso, J.B.; Brouquisse, R.; Corpas, F.J.; Gupta, K.J.; Lindermayr, C.; Loake, G.J.; Palma, J.M.; Petřivalský, M.; Wendehenne, D.; et al. A forty year journey: The generation and roles of NO in plants. Nitric Oxide. 2019. [Google Scholar] [CrossRef] [Green Version]
- Del Castello, F.; Nejamkin, A.; Cassia, R.; Correa-Aragunde, N.; Fernández, B.; Foresi, N.; Lombardo, C.; Ramirez, L.; Lamattina, L. The era of nitric oxide in plant biology: Twenty years tying up loose ends. Nitric Oxide. 2019, 85, 17–27. [Google Scholar] [CrossRef]
- Sun, L.R.; Yue, C.M.; Hao, F.S. Update on roles of nitric oxide in regulating stomatal closure. Plant Signal. Behav. 2019, 14, e1649569. [Google Scholar] [CrossRef] [PubMed]
- Mengel, A.; Chaki, M.; Shekariesfahlan, A.; Lindermayr, C. Effect of nitric oxide on gene transcription—S-nitrosylation of nuclear proteins. Front. Plant Sci. 2013, 4, 293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, L.; Trujillo-Hernandez, J.A.; Reichheld, J.P. Thiol based redox signaling in plant nucleus. Front. Plant Sci. 2018, 9, 705. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox signaling in plants. Antioxid. Redox Signal. 2013, 18, 2087–2090. [Google Scholar] [CrossRef]
- Soares, C.; Carvalho, M.E.A.; Azevedo, R.A.; Fidalgo, F. Plants facing oxidative challenges-A little help from the antioxidant networks. Environ. Exp. Bot. 2019, 161, 4–25. [Google Scholar] [CrossRef]
- Foyer, C.H.; Halliwell, B. The presence of glutathione and glutathione reductase in chloroplasts: A proposed role in ascorbic acid metabolism. Planta 1976, 133, 21–25. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Rossi, R.; Colombo, G.; Giustarini, D.; Milzani, A. Protein S-glutathionylation: A regulatory device from bacteria to humans. Trends Biochem. Sci. 2009, 34, 85–96. [Google Scholar] [CrossRef]
- Zaffagnini, M.; Fermani, S.; Marchand, C.H.; Costa, A.; Sparla, F.; Rouhier, N.; Geigenberger, P.; Lemaire, S.D.; Trost, P. Redox homeostasis in photosynthetic organisms: Novel and established thiol-based molecular mechanisms. Antioxid. Redox Signal. 2019, 31, 155–210. [Google Scholar] [CrossRef]
- Choi, C.S.; Sano, H. Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase-like protein in tobacco plants. Mol. Genet. Genom. 2007, 277, 589–600. [Google Scholar] [CrossRef]
- Poborilova, Z.; Ohlsson, A.B.; Berglund, T.; Vildova, A.; Provaznik, I.; Babula, P. DNA hypomethylation concomitant with the overproduction of ROS induced by naphthoquinone juglone on tobacco BY-2 suspension cells. Environ. Exp. Bot. 2015, 113, 28–39. [Google Scholar] [CrossRef]
- Berglund, T.; Wallstrom, A.; Nguyen, T.V.; Laurell, C.; Ohlsson, A.B. Nicotinamide; antioxidative and DNA hypomethylation effects in plant cells. Plant Physiol. Biochem. 2017, 118, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.F.; Zhuang, T.T.; Yin, W.C.; Miao, Y.L.; Wang, B.; Zhang, Y.H.; Lin, X.Y.; Xu, C.M.; von Wettstein, D.; Rustgi, S.; et al. DNA methylation changes induced in rice by exposure to high concentrations of the nitric oxide modulator, sodium nitroprusside. Plant Mol. Biol. Rep. 2015, 33, 1428–1440. [Google Scholar] [CrossRef]
- Rai, K.K.; Rai, N.; Rai, S.P. Salicylic acid and nitric oxide alleviate high temperature induced oxidative damage in Lablab purpureus L plants by regulating bio-physical processes and DNA methylation. Plant Physiol. Biochem. 2018, 128, 72–88. [Google Scholar] [CrossRef] [PubMed]
- Rahikainen, M.; Alegre, S.; Trotta, A.; Pascual, J.; Kangasjarvi, S. Trans-methylation reactions in plants: Focus on the activated methyl cycle. Physiol. Plant 2018, 162, 162–176. [Google Scholar] [CrossRef]
- Lindermayr, C.; Saalbach, G.; Durner, J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2005, 137, 921–930. [Google Scholar] [CrossRef] [Green Version]
- Puyaubert, J.; Fares, A.; Reze, N.; Peltier, J.B.; Baudouin, E. Identification of endogenously S-nitrosylated proteins in Arabidopsis plantlets: Effect of cold stress on cysteine nitrosylation level. Plant Sci. 2014, 215–216, 150–156. [Google Scholar]
- Chaki, M.; Valderrama, R.; Fernandez-Ocana, A.M.; Carreras, A.; Lopez-Jaramillo, J.; Luque, F.; Palma, J.M.; Pedrajas, J.R.; Begara-Morales, J.C.; Sanchez-Calvo, B.; et al. Protein targets of tyrosine nitration in sunflower (Helianthus annuus L.) hypocotyls. J. Exp. Bot. 2009, 60, 4221–4234. [Google Scholar]
- Lozano-Juste, J.; Colom-Moreno, R.; Leon, J. In vivo protein tyrosine nitration in Arabidopsis thaliana. J. Exp. Bot. 2011, 62, 3501–3517. [Google Scholar] [CrossRef] [Green Version]
- Rouhier, N.; Vieira Dos Santos, C.; Tarrago, L.; Rey, P. Plant methionine sulfoxide reductase A and B multigenic families. Photosynth. Res. 2006, 89, 247–262. [Google Scholar] [CrossRef]
- Petriacq, P.; de Bont, L.; Hager, J.; Didierlaurent, L.; Mauve, C.; Guerard, F.; Noctor, G.; Pelletier, S.; Renou, J.P.; Tcherkez, G.; et al. Inducible NAD overproduction in Arabidopsis alters metabolic pools and gene expression correlated with increased salicylate content and resistance to Pst-AvrRpm1. Plant J. 2012, 70, 650–665. [Google Scholar] [CrossRef]
- Dos Santos, C.V.; Laugier, E.; Tarrago, L.; Massot, V.; Issakidis-Bourguet, E.; Rouhier, N.; Rey, P. Specificity of thioredoxins and glutaredoxins as electron donors to two distinct classes of Arabidopsis plastidial methionine sulfoxide reductases B. FEBS Lett. 2007, 581, 4371–4376. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.M.; Deng, X.Y.; Miki, D.; Cutler, S.; La, H.G.; Hou, Y.J.; Oh, J.; Zhu, J.K. Sulfamethazine suppresses epigenetic silencing in Arabidopsis by impairing folate synthesis. Plant Cell 2012, 24, 1230–1241. [Google Scholar] [CrossRef] [Green Version]
- Gorelova, V.; De Lepeleire, J.; Van Daele, J.; Pluim, D.; Mei, C.; Cuypers, A.; Leroux, O.; Rebeille, F.; Schellens, J.H.M.; Blancquaert, D.; et al. Dihydrofolate reductase/thymidylate synthase fine-tunes the folate status and controls redox homeostasis in plants. Plant Cell 2017, 29, 2831–2853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groth, M.; Moissiard, G.; Wirtz, M.; Wang, H.F.; Garcia-Salinas, C.; Ramos-Parra, P.A.; Bischof, S.; Feng, S.H.; Cokus, S.J.; John, A.; et al. MTHFD1 controls DNA methylation in Arabidopsis. Nat. Commun. 2016, 7, 11640. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Wang, T.; Xu, S.Y.; Xu, S.X.; Wu, L.J.; Wu, Y.J.; Bian, P. Radiation-induced epigenetic bystander effects demonstrated in Arabidopsis Thaliana. Radiat. Res. 2015, 183, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.G.; Wang, X.G.; Tang, K.; Zhang, H.M.; Mangrauthia, S.K.; Lei, M.G.; Hsu, C.C.; Hou, Y.J.; Wang, C.G.; Li, Y.; et al. MET18 connects the cytosolic iron-sulfur cluster assembly pathway to active DNA demethylation in Arabidopsis. PLoS Genet. 2015, 11, e1005559. [Google Scholar] [CrossRef] [Green Version]
- Couturier, J.; Chibani, K.; Jacquot, J.P.; Rouhier, N. Cysteine-based redox regulation and signaling in plants. Front. Plant Sci. 2013, 4, 105. [Google Scholar] [CrossRef] [Green Version]
- Seta, A.; Tabara, M.; Nishibori, Y.; Hiraguri, A.; Ohkama-Ohtsu, N.; Yokoyama, T.; Hara, S.; Yoshida, K.; Hisabori, T.; Fukudome, A.; et al. Post-translational regulation of the dicing activities of Arabidopsis DICER-LIKE 3 and 4 by inorganic phosphate and the redox state. Plant Cell Physiol. 2017, 58, 485–495. [Google Scholar] [CrossRef]
- Charbonnel, C.; Niazi, A.K.; Elvira-Matelot, E.; Nowak, E.; Zytnicki, M.; de Bures, A.; Jobet, E.; Opsomer, A.; Shamandi, N.; Nowotny, M.; et al. The siRNA suppressor RTL1 is redox-regulated through glutathionylation of a conserved cysteine in the double-stranded-RNA-binding domain. Nucleic Acids Res. 2017, 45, 11891–11907. [Google Scholar] [CrossRef] [Green Version]
- Fukudome, A.; Fukuhara, T. Plant dicer-like proteins: Double-stranded RNA-cleaving enzymes for small RNA biogenesis. J. Plant Res. 2017, 130, 33–44. [Google Scholar] [CrossRef]
- Shamandi, N.; Zytnicki, M.; Charbonnel, C.; Elvira-Matelot, E.; Bochnakian, A.; Comella, P.; Mallory, A.C.; Lepere, G.; Saez-Vasquez, J.; Vaucheret, H. Plants encode a general siRNA suppressor that is induced and suppressed by viruses. PLoS Biol. 2015, 13, e1002326. [Google Scholar] [CrossRef] [Green Version]
- Hussain, A.; Mun, B.G.; Imran, Q.M.; Lee, S.U.; Adamu, T.A.; Shahid, M.; Kim, K.M.; Yun, B.W. Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 975. [Google Scholar] [CrossRef] [Green Version]
- Blanc, R.S.; Richard, S. Arginine methylation: The coming of age. Mol. Cell 2017, 65, 8–24. [Google Scholar] [CrossRef] [Green Version]
- Ojima, Y.; Suryadarma, P.; Tsuchida, K.; Taya, M. Accumulation of pyruvate by changing the redox status in Escherichia coli. Biotechnol. Lett. 2012, 34, 889–893. [Google Scholar] [CrossRef]
- Miernyk, J.A.; Randall, D.D. Some kinetic and regulatory properties of the pea mitochondrial pyruvate dehydrogenase complex. Plant Physiol. 1987, 83, 306–310. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Zhao, L.; Hou, H.L.; Zhang, H.; Huang, Y.; Wang, Y.P.; Li, H.; Gao, F.; Yan, S.H.; Li, L.J. Epigenetic changes are associated with programmed cell death induced by heat stress in seedling leaves of Zea mays. Plant Cell Physiol. 2015, 56, 965–976. [Google Scholar] [CrossRef] [Green Version]
- Dietzel, L.; Glasser, C.; Liebers, M.; Hiekel, S.; Courtois, F.; Czarnecki, O.; Schlicke, H.; Yan, Z.B.; Borner, T.; Mayer, K.; et al. Identification of early nuclear target genes of plastidial redox signals that trigger the long-term response of Arabidopsis to light quality shifts. Mol. Plant 2015, 8, 1237–1252. [Google Scholar] [CrossRef] [Green Version]
- Mhamdi, A.; Hager, J.; Chaouch, S.; Queval, G.; Han, Y.; Taconnat, L.; Saindrenan, P.; Gouia, H.; Issakidis-Bourguet, E.; Renou, J.P.; et al. Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 2010, 153, 1144–1160. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.M.; Song, H.R.; Han, S.K.; Han, M.; Kim, C.Y.; Park, J.; Lee, Y.H.; Jeon, J.S.; Noh, Y.S.; Noh, B. HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 2012, 71, 135–146. [Google Scholar] [CrossRef]
- Wojtaszek, P. Oxidative burst: An early plant response to pathogen infection. Biochem. J. 1997, 322, 681–692. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Zhang, H.; Yu, B.; Xiong, L.; Xia, Y. Proteomic identification of early salicylate- and flg22-responsive redox-sensitive proteins in Arabidopsis. Sci. Rep. 2015, 5, 8625. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhao, Y.; Zhou, D.X. Rice NAD+-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes. Nucleic Acids Res. 2017, 45, 12241–12255. [Google Scholar] [CrossRef] [Green Version]
- Mengel, A.; Ageeva, A.; Georgii, E.; Bernhardt, J.; Wu, K.; Durner, J.; Lindermayr, C. Nitric oxide modulates histone acetylation at stress genes by inhibition of histone deacetylases. Plant Physiol. 2017, 173, 1434–1452. [Google Scholar] [CrossRef] [Green Version]
- Kuang, J.F.; Chen, J.Y.; Luo, M.; Wu, K.Q.; Sun, W.; Jiang, Y.M.; Lu, W.J. Histone deacetylase HD2 interacts with ERF1 and is involved in longan fruit senescence. J. Exp. Bot. 2012, 63, 441–454. [Google Scholar] [CrossRef] [Green Version]
- Chaki, M.; Shekariesfahlan, A.; Ageeva, A.; Mengel, A.; von Toerne, C.; Durner, J.; Lindermayr, C. Identification of nuclear target proteins for S-nitrosylation in pathogen-treated Arabidopsis thaliana cell cultures. Plant Sci. 2015, 238, 115–126. [Google Scholar] [CrossRef]
- Zemach, A.; Kim, M.Y.; Hsieh, P.H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef] [Green Version]
- Jeddeloh, J.A.; Stokes, T.L.; Richards, E.J. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 1999, 22, 94–97. [Google Scholar] [CrossRef]
- Ho, K.K.; Zhang, H.; Golden, B.L.; Ogas, J. PICKLE is a CHD subfamily II ATP-dependent chromatin remodeling factor. Biochim. Biophys. Acta 2012, 1829, 199–210. [Google Scholar] [CrossRef] [Green Version]
- Simkova, K.; Moreau, F.; Pawlak, P.; Vriet, C.; Baruah, A.; Alexandre, C.; Hennig, L.; Apel, K.; Laloi, C. Integration of stress-related and reactive oxygen species-mediated signals by Topoisomerase VI in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2012, 109, 16360–16365. [Google Scholar] [CrossRef] [Green Version]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
R. M., S.K.; Wang, Y.; Zhang, X.; Cheng, H.; Sun, L.; He, S.; Hao, F. Redox Components: Key Regulators of Epigenetic Modifications in Plants. Int. J. Mol. Sci. 2020, 21, 1419. https://doi.org/10.3390/ijms21041419
R. M. SK, Wang Y, Zhang X, Cheng H, Sun L, He S, Hao F. Redox Components: Key Regulators of Epigenetic Modifications in Plants. International Journal of Molecular Sciences. 2020; 21(4):1419. https://doi.org/10.3390/ijms21041419
Chicago/Turabian StyleR. M., Saravana Kumar, Yibin Wang, Xiaopan Zhang, Hui Cheng, Lirong Sun, Shibin He, and Fushun Hao. 2020. "Redox Components: Key Regulators of Epigenetic Modifications in Plants" International Journal of Molecular Sciences 21, no. 4: 1419. https://doi.org/10.3390/ijms21041419
APA StyleR. M., S. K., Wang, Y., Zhang, X., Cheng, H., Sun, L., He, S., & Hao, F. (2020). Redox Components: Key Regulators of Epigenetic Modifications in Plants. International Journal of Molecular Sciences, 21(4), 1419. https://doi.org/10.3390/ijms21041419



