The Position and Complex Genomic Architecture of Plant T-DNA Insertions Revealed by 4SEE
Abstract
1. Introduction
2. Results
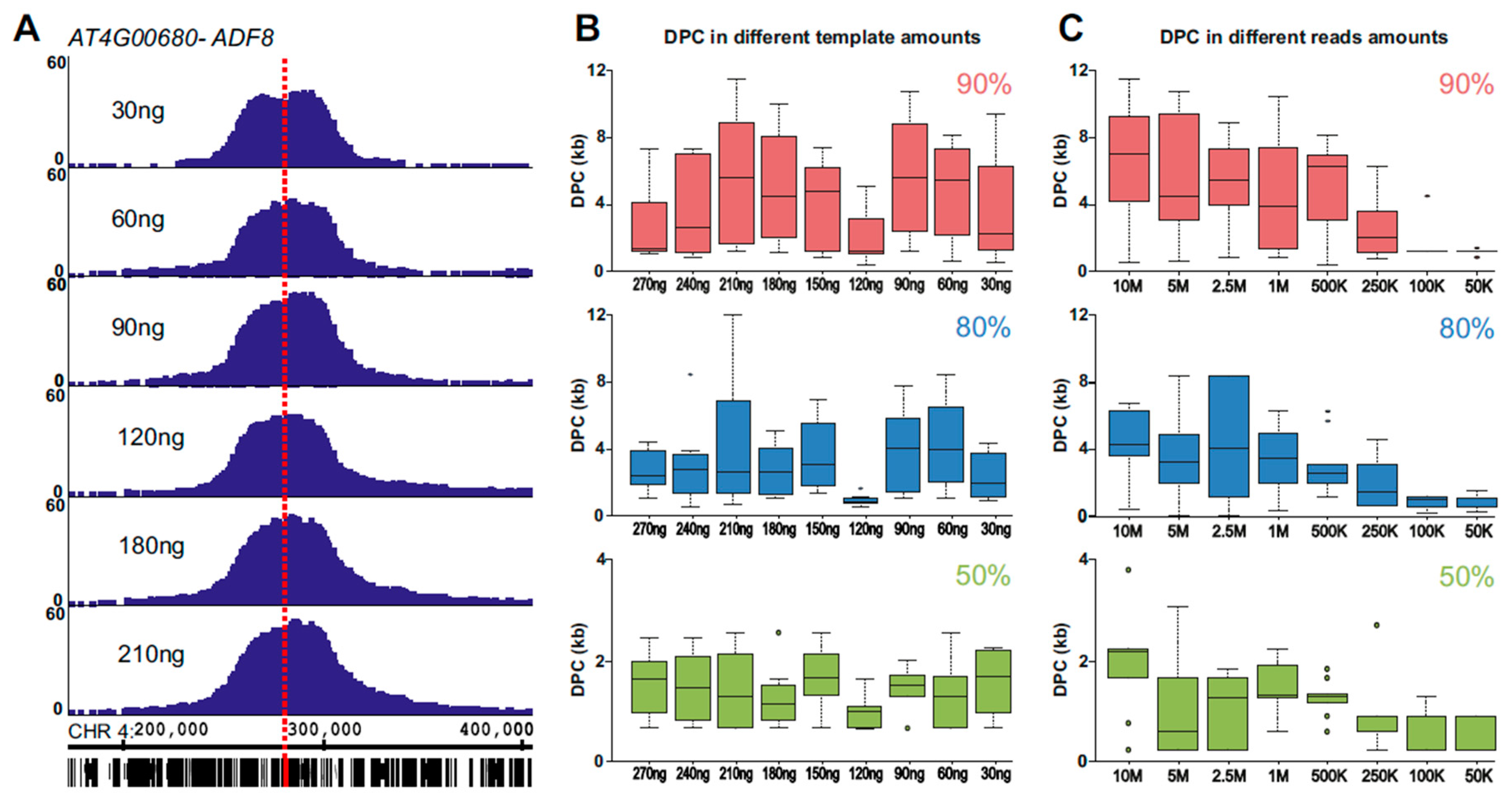
2.1. 4C Signal As A Pointer for Genomic Location
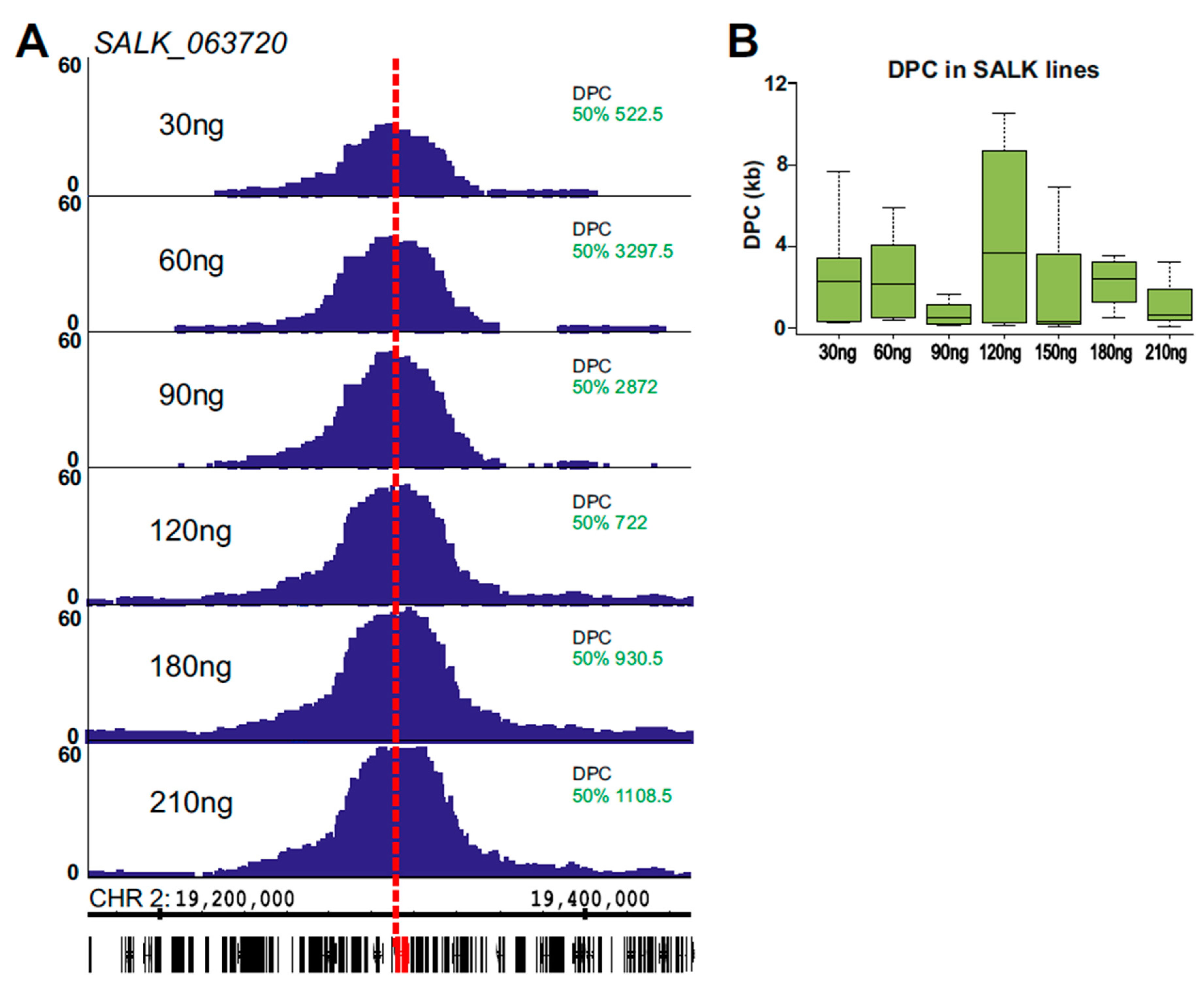
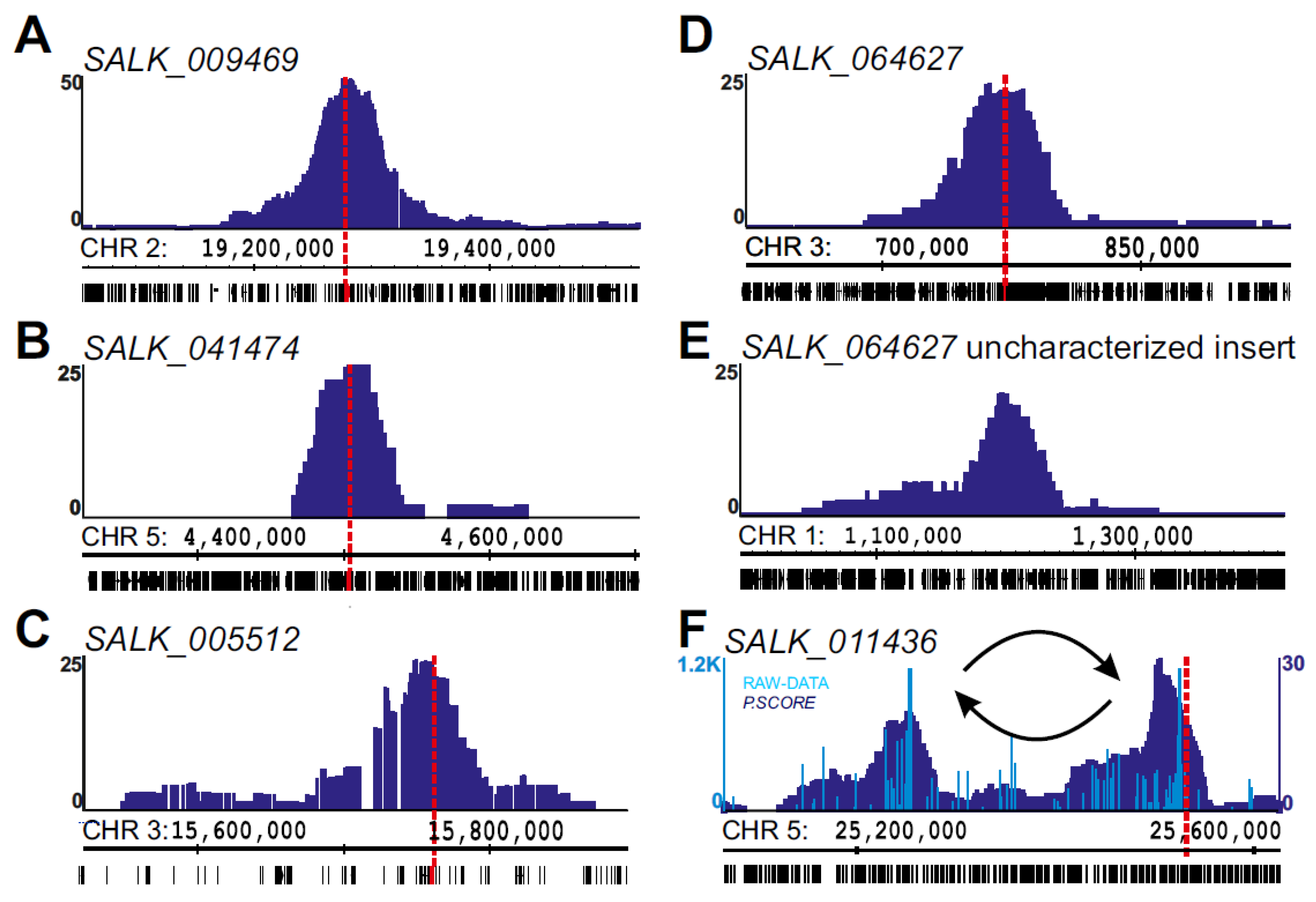
2.2. Using 4SEE As A Pointer for T-DNA Insertion Sites
2.3. Chromosomal Rearrangements Uncovered by 4SEE
2.4. INTACT ADF8p:NTF/ACT2p:BirA Transgene Position and Chromatin Composition
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Circular Chromosome Conformation Capture (4C)
4.2.1. Nucleus isolation and fixation
4.2.2. Enzymatic Digestion, Reverse Crosslinking, and Ligation
4.2.3. 4C Template Purification and Amplification
4.3. 4SEE Analysis of 4C-seq Data
4.4. Mapping T-DNA Insertion Sites and Chromosomal Translocations from Chromatin Accessibility Data
4.5. DNA Fluorescence In Situ Hybridization (FISH)
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 4C | Circular chromosome confirmation capture |
| bp | Base pair |
| DPC | Distance from peak center |
| NGS | Next-generation sequencing |
| RE | Restriction enzyme |
| WT | Wild type |
References
- Sardesai, N.; Subramanyam, S. Agrobacterium: A Genome-Editing Tool-Delivery System. Curr. Top. Microbiol. Immunol. 2018, 418, 463–488. [Google Scholar]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Rosso, M.G.; Li, Y.; Strizhov, N.; Reiss, B.; Dekker, K.; Weisshaar, B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 2003, 53, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Eggenberger, A.L.; Banakar, R.; McCaw, M.E.; Zhu, H.; Main, M.; Kang, M.; Gelvin, S.B.; Wang, K. CRISPR/Cas9-mediated targeted T-DNA integration in rice. Plant Mol. Biol. 2019, 99, 317–328. [Google Scholar] [CrossRef]
- Nacry, P.; Camilleri, C.; Courtial, B.; Caboche, M.; Bouchez, D. Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics 1998, 149, 641–650. [Google Scholar]
- Forsbach, A.; Schubert, D.; Lechtenberg, B.; Gils, M.; Schmidt, R. A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol. Biol. 2003, 52, 161–176. [Google Scholar] [CrossRef]
- Clark, K.A.; Krysan, P.J. Chromosomal translocations are a common phenomenon in Arabidopsis thaliana T-DNA insertion lines. Plant J. 2010, 64, 990–1001. [Google Scholar] [CrossRef]
- Kleinboelting, N.; Huep, G.; Appelhagen, I.; Viehoever, P.; Li, Y.; Weisshaar, B. The Structural Features of Thousands of T-DNA Insertion Sites Are Consistent with a Double-Strand Break Repair-Based Insertion Mechanism. Mol. Plant 2015, 8, 1651–1664. [Google Scholar] [CrossRef]
- Cockram, J.; Mackay, I. Genetic Mapping Populations for Conducting High-Resolution Trait Mapping in Plants. Adv. Biochem. Eng. Biotechnol. 2018, 164, 109–138. [Google Scholar]
- Ochman, H.; Gerber, A.S.; Hartl, D.L. Genetic applications of an inverse polymerase chain reaction. Genetics 1988, 120, 621–623. [Google Scholar] [PubMed]
- Liu, Y.G.; Mitsukawa, N.; Oosumi, T.; Whittier, R.F. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 1995, 8, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Braam, J. Restriction Site Extension PCR: A Novel Method for High-Throughput Characterization of Tagged DNA Fragments and Genome Walking. PLoS ONE 2010, 5, e10577. [Google Scholar] [CrossRef] [PubMed]
- Lepage, É.; Zampini, É.; Boyle, B.; Brisson, N. Time- and Cost-Efficient Identification of T-DNA Insertion Sites through Targeted Genomic Sequencing. PLoS ONE 2013, 8, e70912. [Google Scholar] [CrossRef]
- Jupe, F.; Rivkin, A.C.; Michael, T.P.; Zander, M.; Motley, S.T.; Sandoval, J.P.; Slotkin, R.K.; Chen, H.; Castanon, R.; Nery, J.R.; et al. The complex architecture and epigenomic impact of plant T-DNA insertions. PLoS Genet. 2019, 15, e1007819. [Google Scholar] [CrossRef]
- Wu, L.; Di, D.-W.; Zhang, D.; Song, B.; Luo, P.; Guo, G.-Q. Frequent problems and their resolutions by using thermal asymmetric interlaced PCR (TAIL-PCR) to clone genes in Arabidopsis T-DNA tagged mutants. Biotechnol. Biotechnol. Equip. 2015, 29, 260–267. [Google Scholar] [CrossRef]
- Krysan, P.J.; Young, J.C.; Sussman, M.R. T-DNA as an Insertional Mutagen in Arabidopsis. Plant Cell 1999, 11, 2283–2290. [Google Scholar] [CrossRef]
- Wu, E.; Lenderts, B.; Glassman, K.; Berezowska-Kaniewska, M.; Christensen, H.; Asmus, T.; Zhen, S.; Chu, U.; Cho, M.-J.; Zhao, Z.-Y. Optimized Agrobacterium-mediated sorghum transformation protocol and molecular data of transgenic sorghum plants. Vitr. Cell. Dev. Biol. Plant 2014, 50, 9–18. [Google Scholar] [CrossRef]
- Bartlett, J.; Smedley, M.; Harwood, W. Analysis of T-DNA/Host-Plant DNA Junction Sequences in Single-Copy Transgenic Barley Lines. Biology 2014, 3, 39–55. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, Z.; Zhuang, C.; Huang, J. Cascade of chromosomal rearrangements caused by a heterogeneous T-DNA integration supports the double-stranded break repair model for T-DNA integration. Plant J. 2017, 90, 954–965. [Google Scholar] [CrossRef]
- Inagaki, S.; Henry, I.M.; Lieberman, M.C.; Comai, L. High-Throughput Analysis of T-DNA Location and Structure Using Sequence Capture. PLoS ONE 2015, 10, e0139672. [Google Scholar] [CrossRef] [PubMed]
- De Vree, P.J.P.; de Wit, E.; Yilmaz, M.; van de Heijning, M.; Klous, P.; Verstegen, M.J.A.M.; Wan, Y.; Teunissen, H.; Krijger, P.H.L.; Geeven, G.; et al. Targeted sequencing by proximity ligation for comprehensive variant detection and local haplotyping. Nat. Biotechnol. 2014, 32, 1019–1025. [Google Scholar] [CrossRef] [PubMed]
- Hakim, O.; Misteli, T. SnapShot: Chromosome Conformation Capture. Cell 2012, 148, 1068.e1–2. [Google Scholar] [CrossRef] [PubMed]
- Belton, J.-M.; McCord, R.P.; Gibcus, J.H.; Naumova, N.; Zhan, Y.; Dekker, J. Hi–C: A comprehensive technique to capture the conformation of genomes. Methods 2012, 58, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Burton, J.N.; Adey, A.; Patwardhan, R.P.; Qiu, R.; Kitzman, J.O.; Shendure, J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 2013, 31, 1119–1125. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, N.; Dekker, J. High-throughput genome scaffolding from in vivo DNA interaction frequency. Nat. Biotechnol. 2013, 31, 1143–1147. [Google Scholar] [CrossRef]
- Jiao, W.-B.; Accinelli, G.G.; Hartwig, B.; Kiefer, C.; Baker, D.; Severing, E.; Willing, E.-M.; Piednoel, M.; Woetzel, S.; Madrid-Herrero, E.; et al. Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Res. 2017, 27, 778–786. [Google Scholar] [CrossRef]
- Moll, K.M.; Zhou, P.; Ramaraj, T.; Fajardo, D.; Devitt, N.P.; Sadowsky, M.J.; Stupar, R.M.; Tiffin, P.; Miller, J.R.; Young, N.D.; et al. Strategies for optimizing BioNano and Dovetail explored through a second reference quality assembly for the legume model, Medicago truncatula. BMC Genom. 2017, 18, 578. [Google Scholar] [CrossRef]
- Grob, S.; Grossniklaus, U. Invasive DNA elements modify the nuclear architecture of their insertion site by KNOT-linked silencing in Arabidopsis thaliana. Genome Biol. 2019, 20, 120. [Google Scholar] [CrossRef]
- Cao, J.; Ye, C.; Hao, G.; Dabney-Smith, C.; Hunt, A.G.; Li, Q.Q. Root Hair Single Cell Type Specific Profiles of Gene Expression and Alternative Polyadenylation Under Cadmium Stress. Front. Plant Sci. 2019, 10, 589. [Google Scholar] [CrossRef]
- Deal, R.B.; Henikoff, S. The INTACT method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat. Protoc. 2011, 6, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Deal, R.B.; Henikoff, S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 2010, 18, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Maher, K.A.; Bajic, M.; Kajala, K.; Reynoso, M.; Pauluzzi, G.; West, D.A.; Zumstein, K.; Woodhouse, M.; Bubb, K.; Dorrity, M.W.; et al. Profiling of Accessible Chromatin Regions across Multiple Plant Species and Cell Types Reveals Common Gene Regulatory Principles and New Control Modules. Plant Cell 2018, 30, 15–36. [Google Scholar] [CrossRef] [PubMed]
- Foley, S.W.; Gosai, S.J.; Wang, D.; Selamoglu, N.; Sollitti, A.C.; Köster, T.; Steffen, A.; Lyons, E.; Daldal, F.; Garcia, B.A.; et al. A Global View of RNA-Protein Interactions Identifies Post-transcriptional Regulators of Root Hair Cell Fate. Dev. Cell 2017, 41, 204–220.e5. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, A.M.; Arsovski, A.A.; Lempe, J.; Bubb, K.L.; Weirauch, M.T.; Sabo, P.J.; Sandstrom, R.; Thurman, R.E.; Neph, S.; Reynolds, A.P.; et al. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep. 2014, 8, 2015–2030. [Google Scholar] [CrossRef]
- Sun, L.; Ge, Y.; Sparks, J.A.; Robinson, Z.T.; Cheng, X.; Wen, J.; Blancaflor, E.B. TDNAscan: A Software to Identify Complete and Truncated T-DNA Insertions. Front. Genet. 2019, 10, 685. [Google Scholar] [CrossRef]
- Gang, H.; Liu, G.; Zhang, M.; Zhao, Y.; Jiang, J.; Chen, S. Comprehensive characterization of T-DNA integration induced chromosomal rearrangement in a birch T-DNA mutant. BMC Genom. 2019, 20, 311. [Google Scholar] [CrossRef]
- McCarty, D.R.; Mark Settles, A.; Suzuki, M.; Tan, B.C.; Latshaw, S.; Porch, T.; Robin, K.; Baier, J.; Avigne, W.; Lai, J.; et al. Steady-state transposon mutagenesis in inbred maize. Plant J. 2005, 44, 52–61. [Google Scholar] [CrossRef]
- Saito, T.; Ariizumi, T.; Okabe, Y.; Asamizu, E.; Hiwasa-Tanase, K.; Fukuda, N.; Mizoguchi, T.; Yamazaki, Y.; Aoki, K.; Ezura, H. TOMATOMA: A Novel Tomato Mutant Database Distributing Micro-Tom Mutant Collections. Plant Cell Physiol. 2011, 52, 283–296. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Raskina, O.; Belyayev, A.; Nevo, E. Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosom. Res. 2004, 12, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Lysak, M.; Fransz, P.; Schubert, I. Cytogenetic analyses of Arabidopsis. Methods Mol. Biol. 2006, 323, 173–186. [Google Scholar] [PubMed]
- Celeda, D.; Aldinger, K.; Haar, F.-M.; Hausmann, M.; Durm, M.; Ludwig, H.; Cremer, C. Rapid fluorescence in situ hybridization with repetitive DNA probes: Quantification by digital image analysis. Cytometry 1994, 17, 13–25. [Google Scholar] [CrossRef] [PubMed]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krispil, R.; Tannenbaum, M.; Sarusi-Portuguez, A.; Loza, O.; Raskina, O.; Hakim, O. The Position and Complex Genomic Architecture of Plant T-DNA Insertions Revealed by 4SEE. Int. J. Mol. Sci. 2020, 21, 2373. https://doi.org/10.3390/ijms21072373
Krispil R, Tannenbaum M, Sarusi-Portuguez A, Loza O, Raskina O, Hakim O. The Position and Complex Genomic Architecture of Plant T-DNA Insertions Revealed by 4SEE. International Journal of Molecular Sciences. 2020; 21(7):2373. https://doi.org/10.3390/ijms21072373
Chicago/Turabian StyleKrispil, Ronen, Miriam Tannenbaum, Avital Sarusi-Portuguez, Olga Loza, Olga Raskina, and Ofir Hakim. 2020. "The Position and Complex Genomic Architecture of Plant T-DNA Insertions Revealed by 4SEE" International Journal of Molecular Sciences 21, no. 7: 2373. https://doi.org/10.3390/ijms21072373
APA StyleKrispil, R., Tannenbaum, M., Sarusi-Portuguez, A., Loza, O., Raskina, O., & Hakim, O. (2020). The Position and Complex Genomic Architecture of Plant T-DNA Insertions Revealed by 4SEE. International Journal of Molecular Sciences, 21(7), 2373. https://doi.org/10.3390/ijms21072373




