Overexpression of Grapevine VvIAA18 Gene Enhanced Salt Tolerance in Tobacco
Abstract
1. Introduction
2. Results
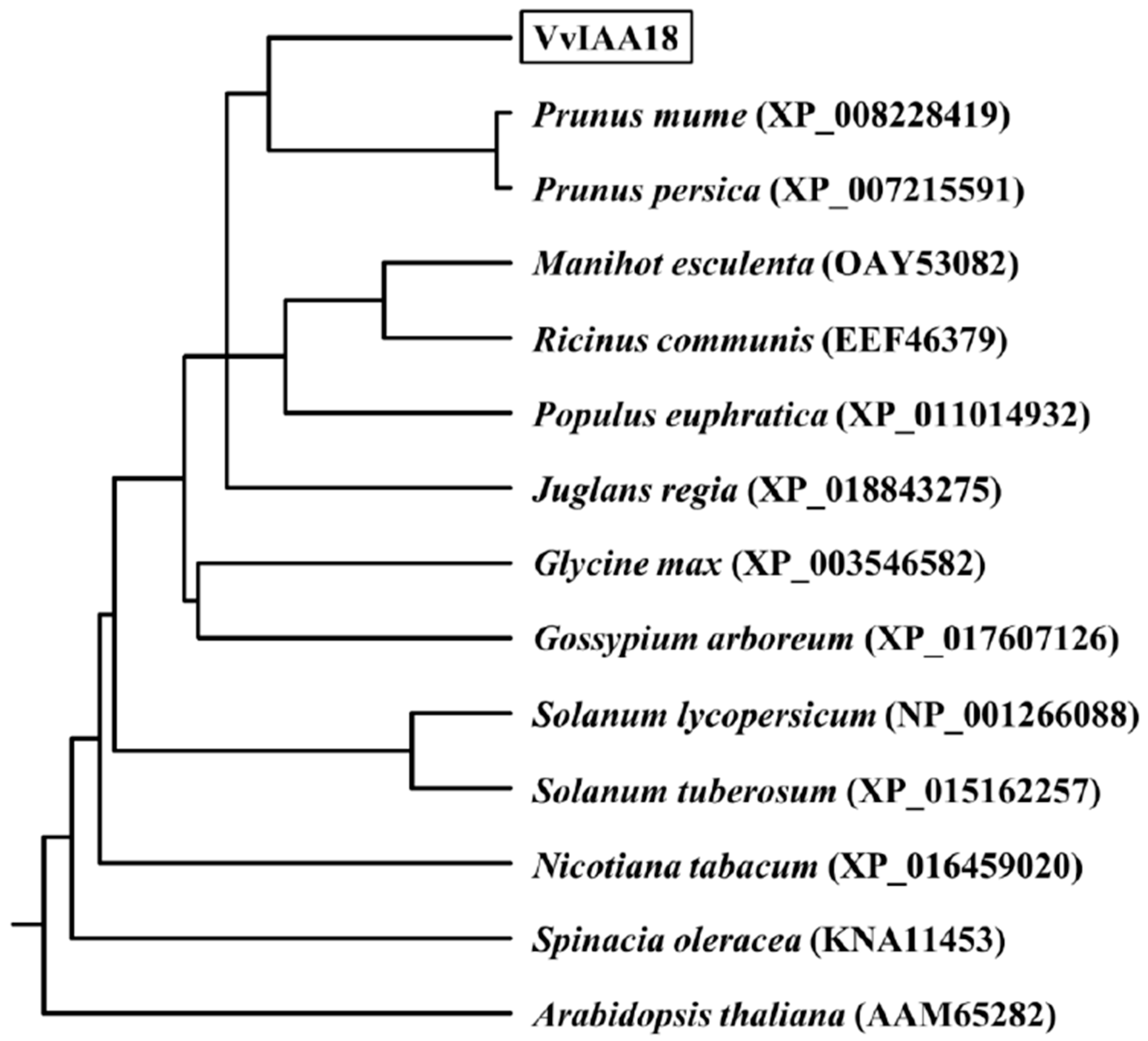
2.1. Cloning and Sequence Analysis of VvIAA18
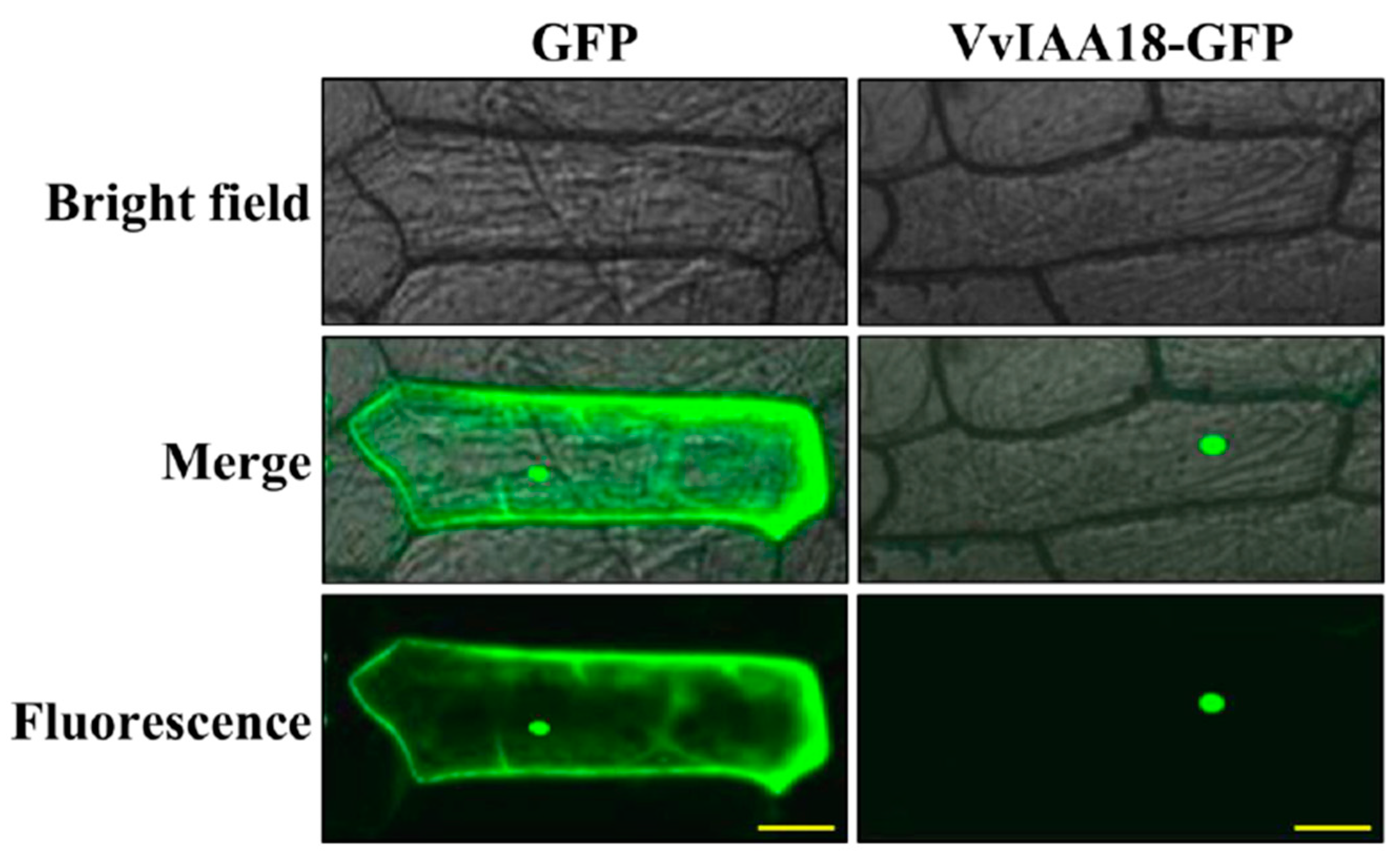
2.2. Nuclear Localization of VvIAA18
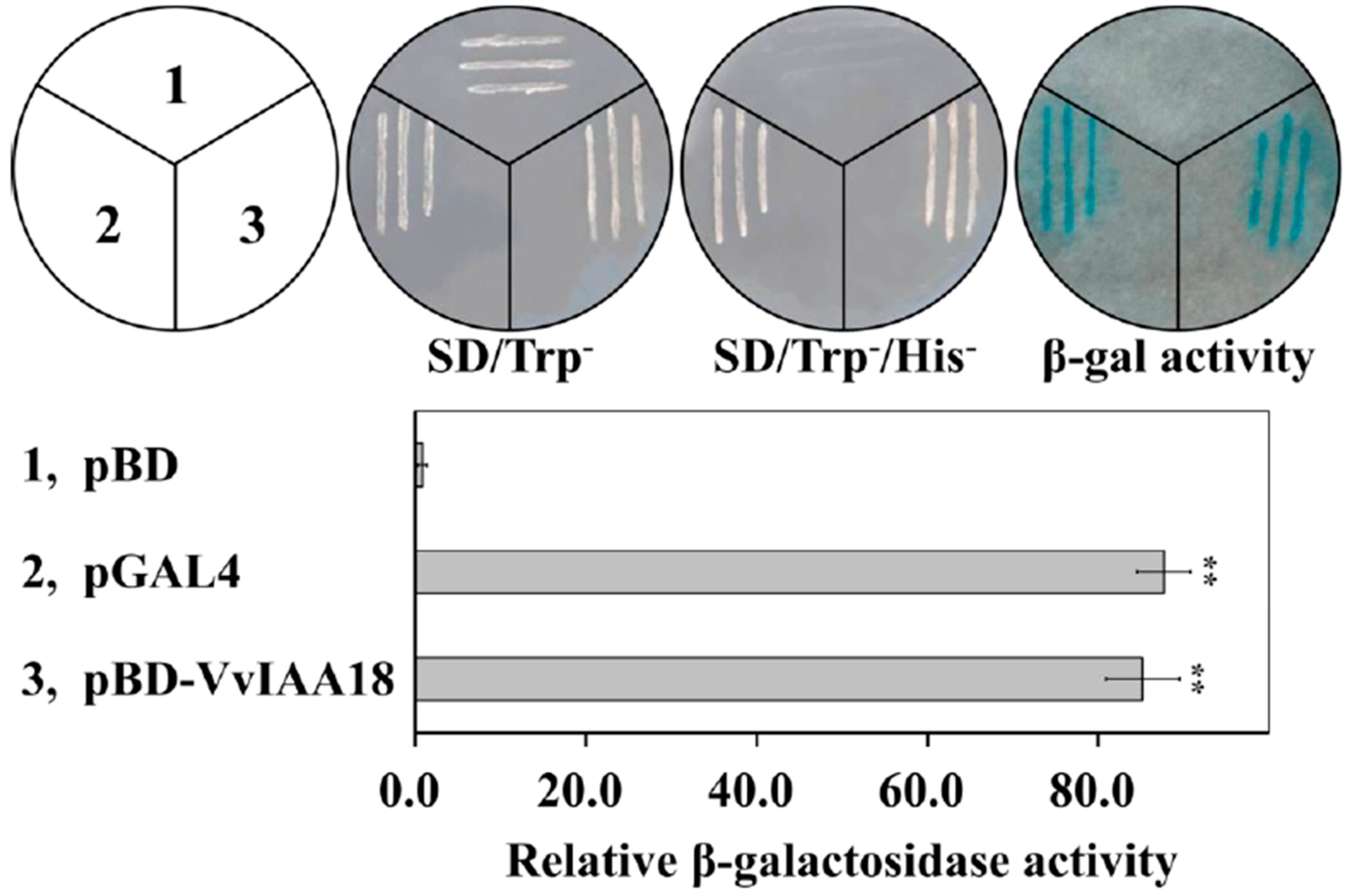
2.3. Transcriptional Activation of VvIAA18
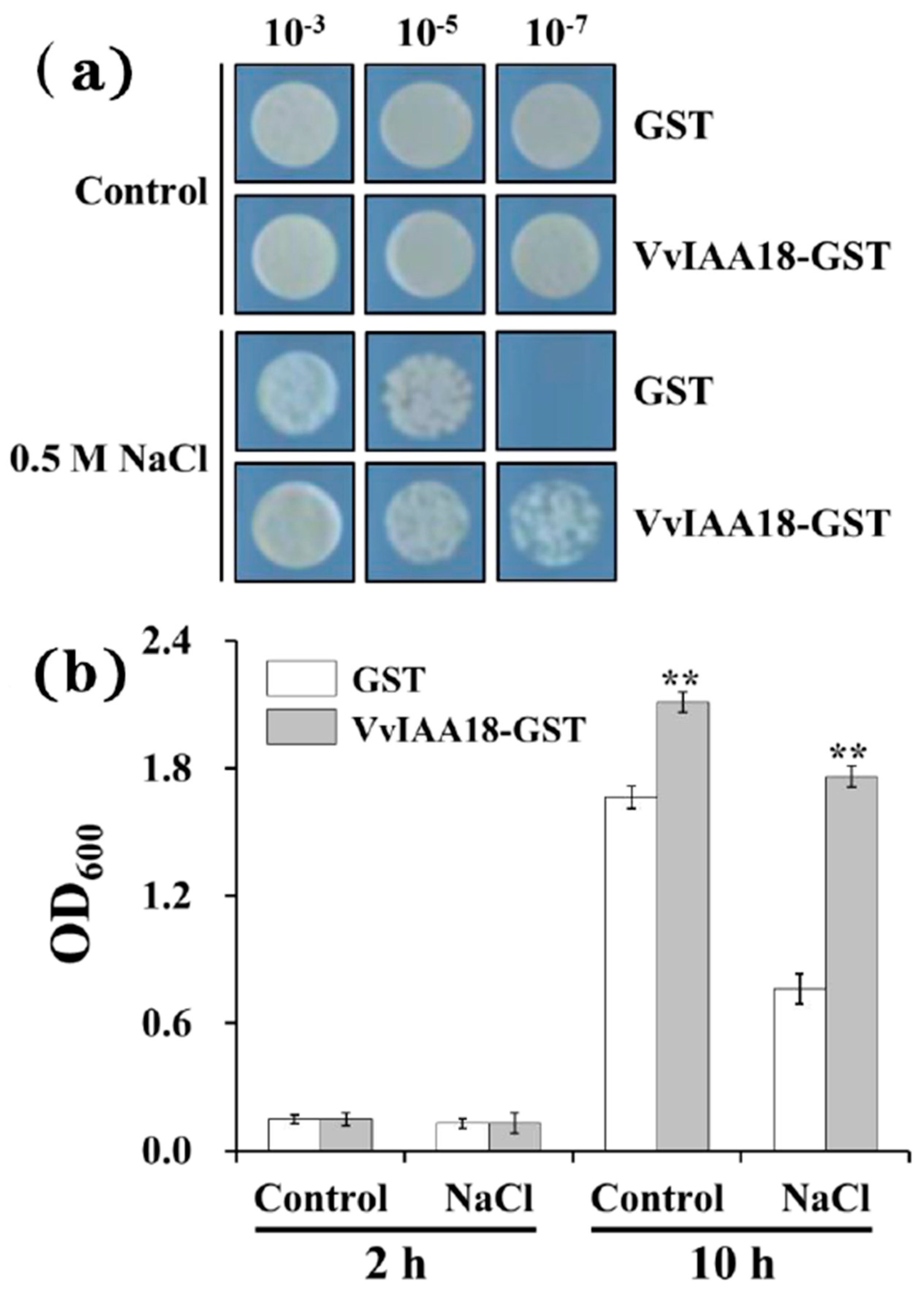
2.4. Improved Salt Tolerance in Escherichia coli
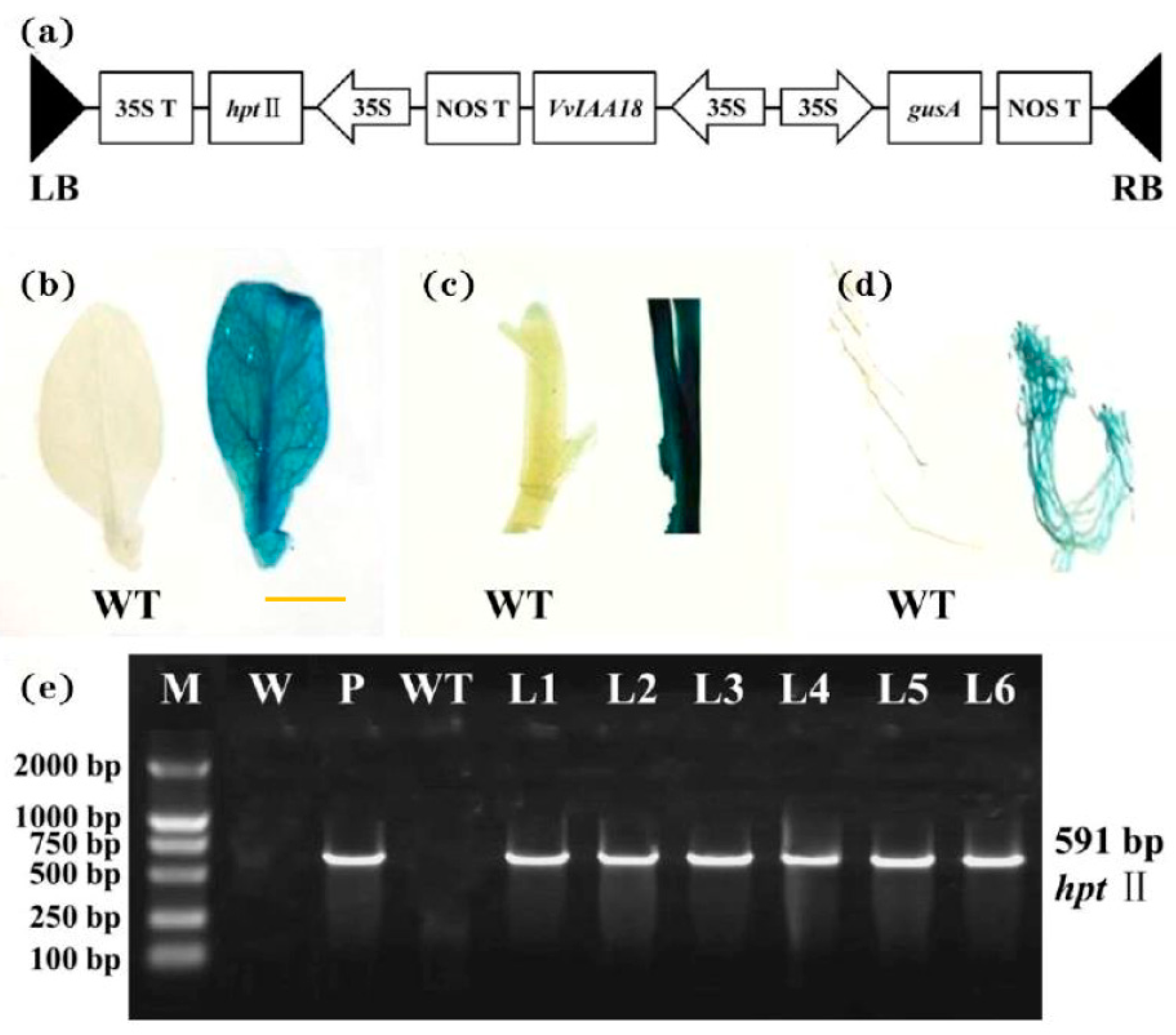
2.5. Regeneration and Identification of Transgenic Tobacco with VvIAA18 Gene
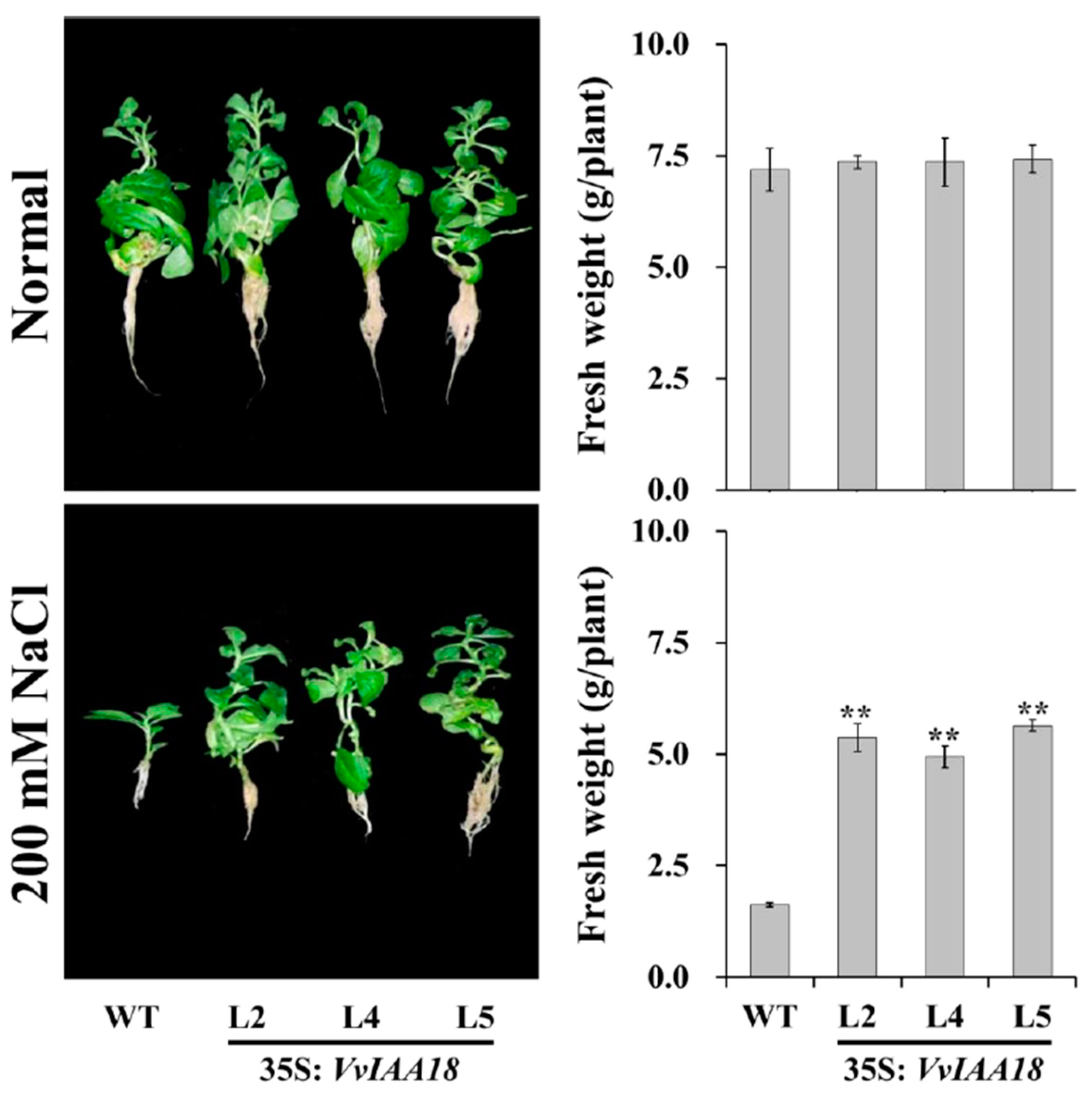
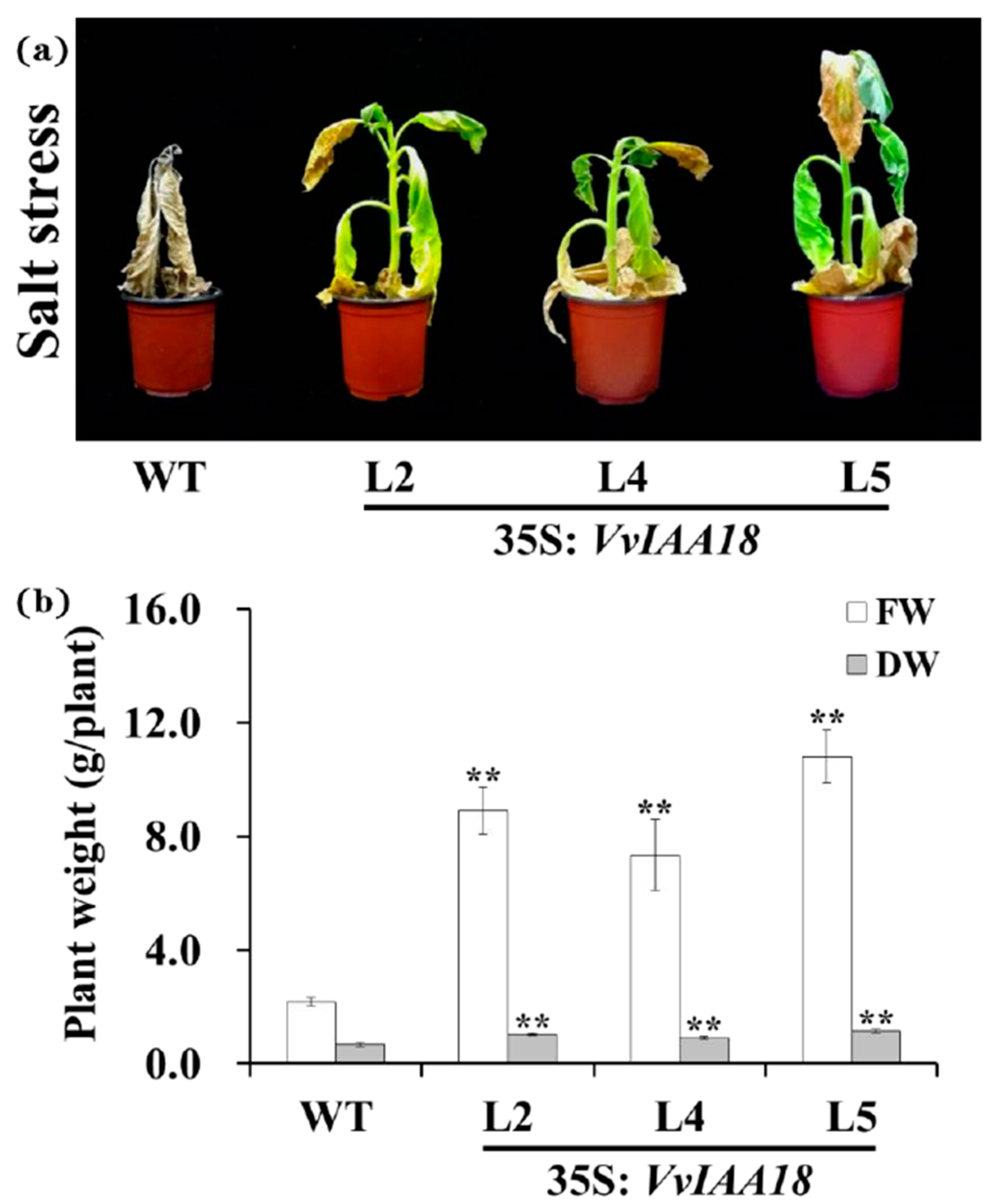
2.6. Enhanced Salt Tolerance in Tobacco Expressing VvIAA18
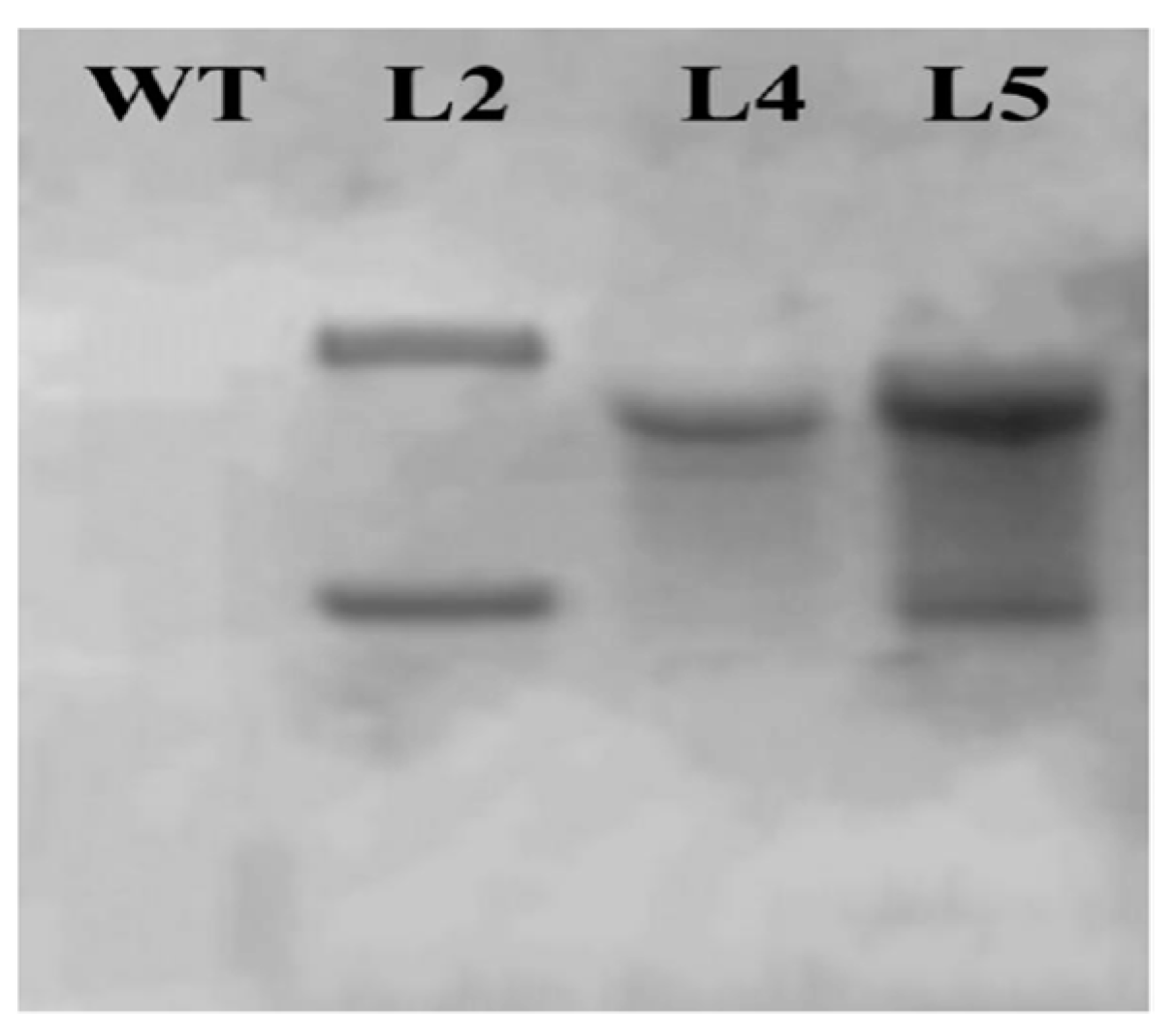
2.7. Southern Blot Analysis of Transgenic Plants
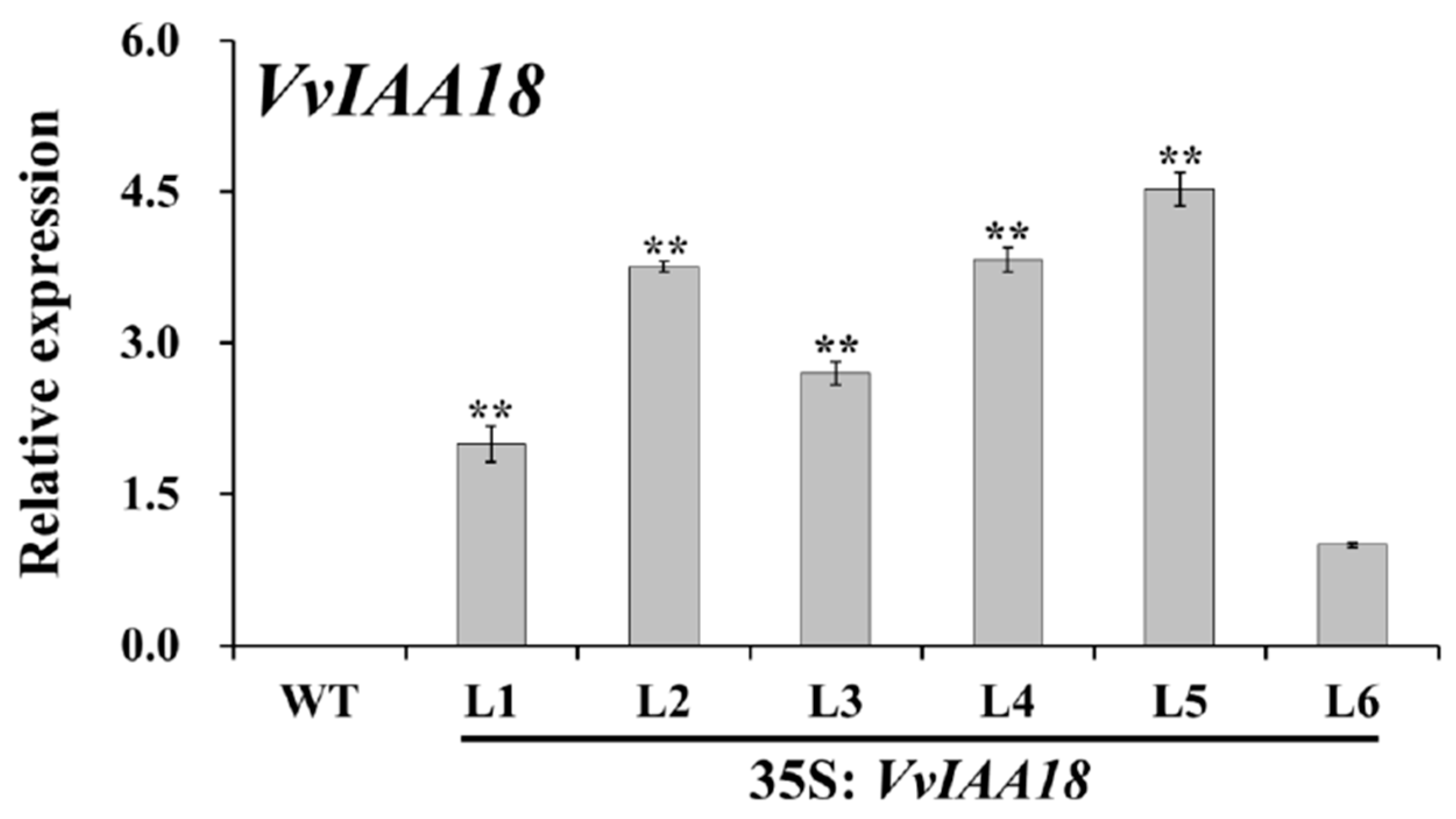
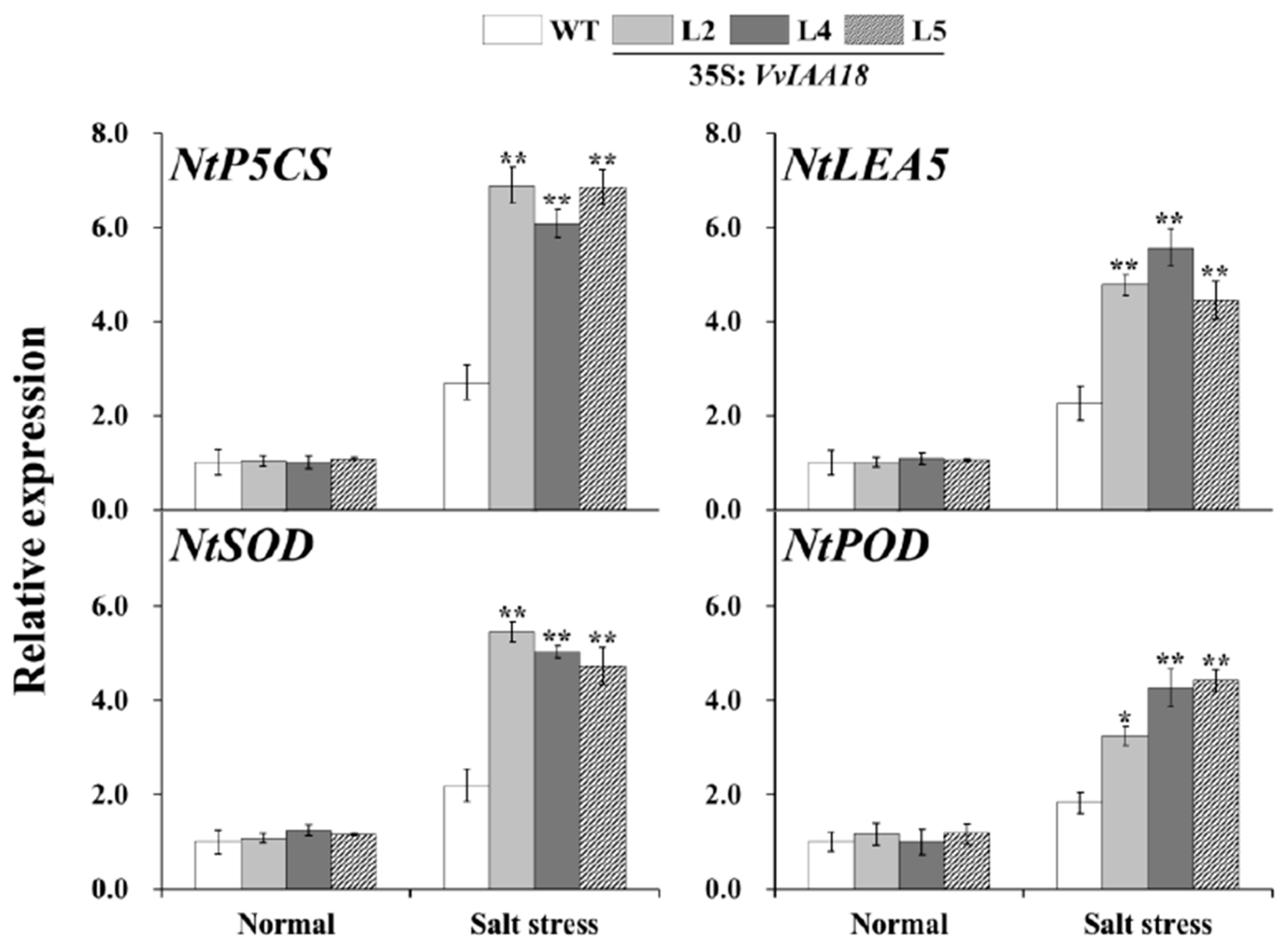
2.8. Increased Expression of the Salt Stress-Responsive Genes in Transgenic Plants
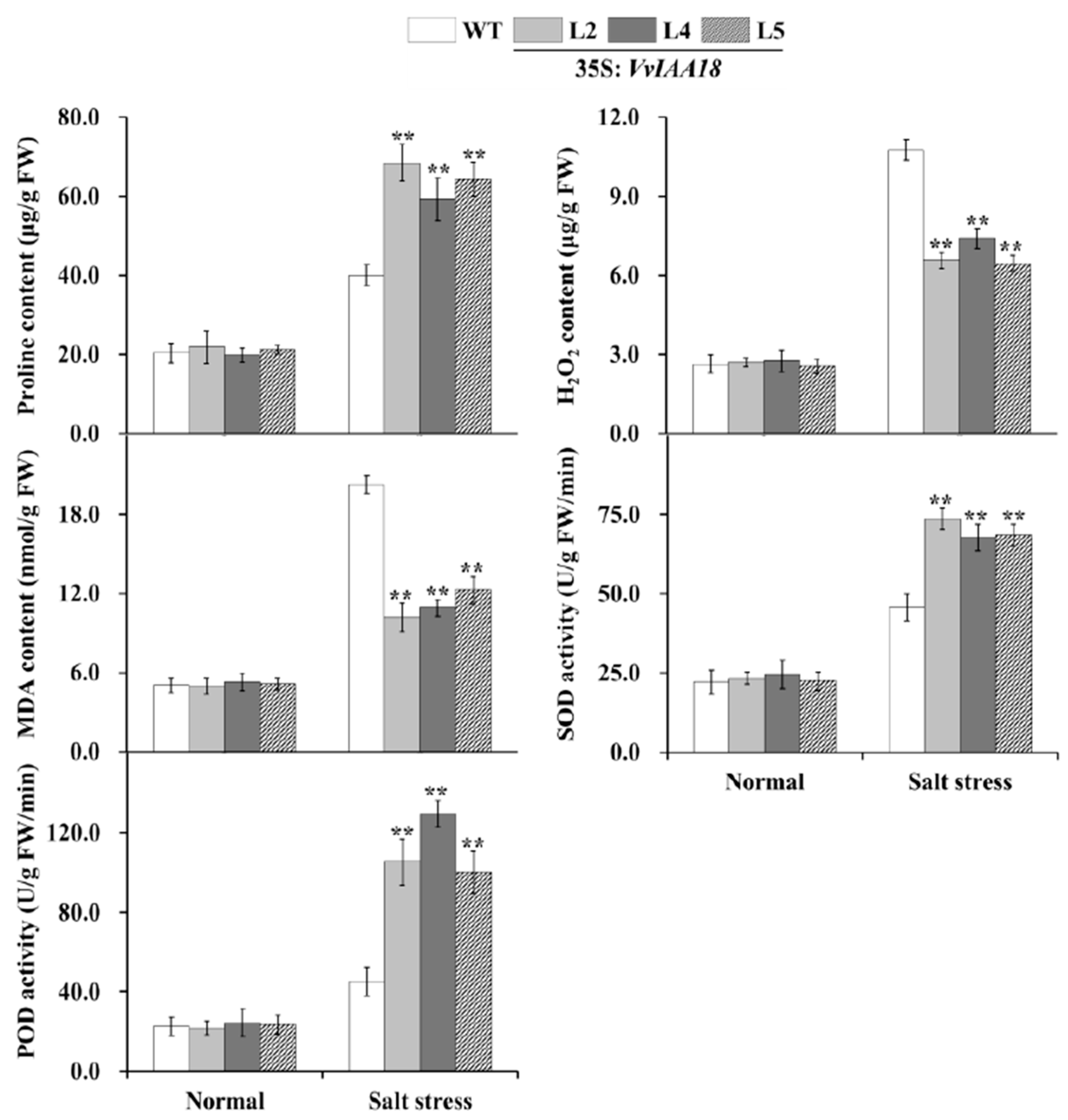
2.9. Promoted Stress Response Physiological Traits in Transgenic Plants
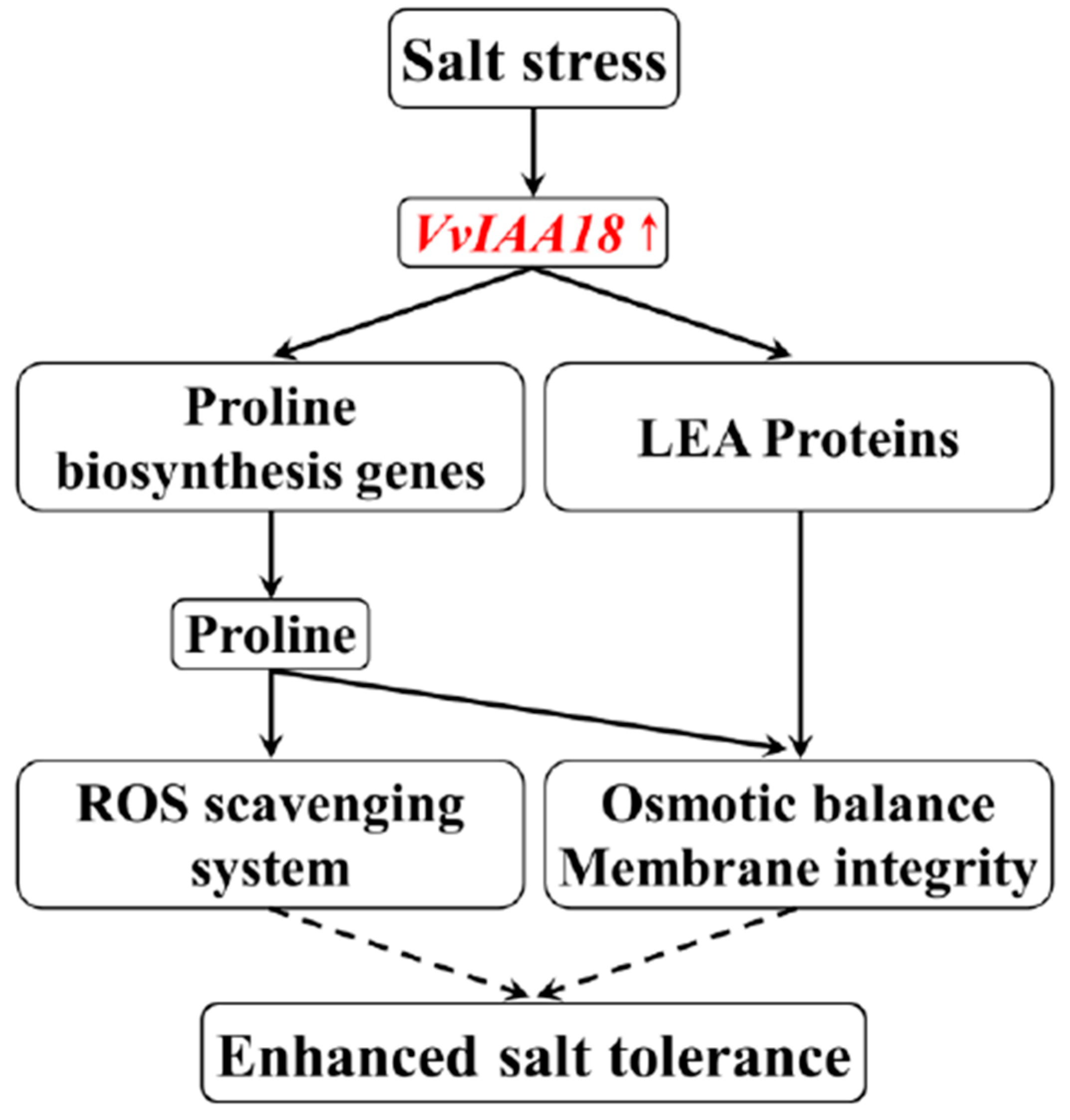
3. Discussion
4. Materials and Methods
4.1. Plant Mterials
4.2. Cloning of Grapevine VvIAA18 Gene
4.3. Sequence Analysis of VvIAA18 Gene
4.4. Subcellular Localization and Transactivation Assay of VvIAA18
4.5. Protein Expression and Bacterial Growth Analysis
4.6. Transformation of Tobacco with VvIAA18
4.7. Molecular Confirmation of Transgenic Plants
4.8. Assay for Salt Tolerance
4.9. Southern Blot Analysis
4.10. Expression Analysis of VvIAA18 and the Related Genes
4.11. Analyses of Proline, H2O2, and MDA Content and SOD and POD Activities
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.B.; Kong, W.L.; Wong, G.; Fu, L.F.; Peng, R.H.; Li, Z.J.; Yao, Q.H. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genom. 2016, 291, 1545–1559. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.B.; Tong, W.J.; Zhu, H.; Kong, W.L.; Peng, R.H.; Liu, Q.C.; Yao, Q.H. A novel Cys2/His2 zinc finger protein gene from sweetpotato, IbZFP1, is involved in salt and drought tolerance in transgenic Arabidopsis. Planta 2016, 243, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.B.; Zhu, H.; Chen, D.H.; Li, Z.J.; Peng, R.H.; Yao, Q.H. A grape bHLH transcription factor gene, VvbHLH1, increases the accumulation of flavonoids and enhances salt and drought tolerance in transgenic Arabidopsis thaliana. Plant Cell Tiss. Org. 2016, 125, 387–398. [Google Scholar] [CrossRef]
- Wang, F.B.; Zhu, H.; Kong, W.L.; Peng, R.H.; Liu, Q.C.; Yao, Q.H. The Antirrhinum AmDEL gene enhances flavonoids accumulation and salt and drought tolerance in transgenic Arabidopsis. Planta 2016, 244, 59–73. [Google Scholar] [CrossRef]
- Kang, C.; He, S.; Zhai, H.; Li, R.; Zhao, N.; Liu, Q. A sweetpotato auxin response factor gene (IbARF5) is involved in carotenoid biosynthesis and salt and drought tolerance in transgenic Arabidopsis. Front Plant Sci. 2018, 9, 1307. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhang, H.; Wang, T.; Chen, S.X.; Dai, S.J. Proteomics-based investigation of salt-responsive mechanisms in plant roots. J. Proteom. 2013, 82, 230–253. [Google Scholar] [CrossRef]
- Yang, S.J.; Vanderbeld, B.; Wan, J.X.; Huang, Y.F. Narrowing down the targets: Towards successful genetic engineering of drought-tolerant crops. Mol. Plant 2010, 3, 469–490. [Google Scholar] [CrossRef]
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to environmental stresses. Plant Cell 1995, 7, 1099–1111. [Google Scholar] [CrossRef]
- Zhu, J.K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Friml, J. Auxin transport-shaping the plant. Curr. Opin. Plant Biol. 2003, 6, 7–12. [Google Scholar] [CrossRef]
- Song, Y.; Wang, L.; Xiong, L. Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 2009, 229, 577–591. [Google Scholar] [CrossRef]
- Vogler, H.; Kuhlemeier, C. Simple hormones but complex signalling. Curr. Opin. Plant Biol. 2003, 6, 51–56. [Google Scholar] [CrossRef]
- Hagen, G.; Guilfoyle, T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol. Biol. 2002, 49, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Liscum, E.; Reed, J.W. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002, 49, 387–400. [Google Scholar] [CrossRef]
- Berleth, T.; Krogan, N.T.; Scarpella, E. Auxin signals-turning genes on and turning cells around. Curr. Opin. Plant Biol. 2004, 7, 553–563. [Google Scholar] [CrossRef]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971. [Google Scholar]
- Tiwari, S.B.; Hagen, G.; Guilfoyle, T. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 2003, 15, 533–543. [Google Scholar] [CrossRef]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. ARF1, a transcription factor that binds to auxin response elements. Science 1997, 276, 1865–1868. [Google Scholar] [CrossRef]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Activation and repression of transcription by auxin-response factors. Proc. Natl. Acad. Sci. USA 1999, 96, 5844–5849. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.T. Further characterization of auxin-regulated mRNAs in hypocotyl sections of mung bean [Vigna radiata (L.) Wilczek]: Sequence homology to genes for fatty-acid desaturases and atypical late-embryogenesis-abundant protein, and the mode of expression of the mRNAs. Planta 1994, 192, 359–364. [Google Scholar] [PubMed]
- Reed, J.W. Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci. 2001, 6, 420–425. [Google Scholar] [CrossRef]
- Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. OsIAA1, an Aux/IAA cDNA from rice, and changes in its expression as influenced by auxin and light. DNA Res. 2001, 8, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, A.; Umemura, I.; Gomi, K.; Hasegawa, Y.; Kitano, H.; Sazuka, T.; Matsuoka, M. Production and characterization of auxininsensitive rice by overexpression of a mutagenized rice IAA protein. Plant J. 2006, 46, 297–306. [Google Scholar] [CrossRef]
- Çakir, B.; Kiliçkaya, O.; Olcay, A.C. Genome-wide analysis of Aux/IAA genes in Vitis vinifera: Cloning and expression profiling of a grape Aux/IAA gene in response to phytohormone and abiotic stresses. Acta Physiol. Plant. 2013, 35, 365–377. [Google Scholar] [CrossRef]
- Remington, D.L.; Vision, T.J.; Guilfoyle, T.J.; Reed, J.W. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol. 2004, 135, 1738–1752. [Google Scholar] [CrossRef]
- Jain, M.; Kaur, N.; Garg, R.; Thakur, J.K.; Tyagi, A.K.; Khurana, J.P. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct. Integr. Genom. 2006, 6, 47–59. [Google Scholar] [CrossRef]
- Wang, S.; Bai, Y.; Shen, C.; Wu, Y.; Zhang, S.; Jiang, D.; Guilfoyle, T.J.; Chen, M.; Qi, Y. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genom. 2010, 10, 533–546. [Google Scholar] [CrossRef]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors: Recent advances in auxin biology. J. Plant Growth Regul. 2001, 10, 281–291. [Google Scholar] [CrossRef]
- Kepinski, S.; Leyser, O. Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc. Natl. Acad. Sci. USA 2004, 101, 12381–12386. [Google Scholar] [CrossRef] [PubMed]
- Thakur, J.K.; Jain, M.; Tyagi, A.K.; Khurana, J.P. Exogenous auxin enhances the degradation of a light down-regulated and nuclear-localized OsiIAA1, an Aux/IAA protein from rice, via proteasome. Bba-Gene Struct. Expr. 2005, 1730, 196–205. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.L.; Xu, Z.F. Ectopic overexpression of an AUXIN/INDOLE-3-ACETIC ACID (Aux/IAA) gene OsIAA4 in rice induces morphological changes and reduces responsiveness to auxin. Int. J. Mol. Sci. 2013, 14, 13645–13656. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Wang, F.B.; Si, Z.Z.; Huo, J.X.; Xing, L.; An, Y.Y.; He, S.Z.; Liu, Q.C. A myo-inositol-1-phosphate synthase gene, IbMIPS1, enhances salt and drought tolerance and stem nematode resistance in transgenic sweetpotato. Plant Biotechnol. J. 2016, 14, 592–602. [Google Scholar] [CrossRef]
- Liu, D.G.; He, S.Z.; Zhai, H.; Wang, L.J.; Zhao, Y.; Wang, B.; Li, R.J.; Liu, Q.C. Overexpression of IbP5CR enhances salt tolerance in transgenic sweetpotato. Plant Cell Tiss. Org. 2014, 117, 1–16. [Google Scholar] [CrossRef]
- Verbruggen, N.; Hermans, C. Proline Accumulation in Plants: A Review. Amino Acids 2008, 35, 753–759. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Bioch. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Yin, Z.; Jiang, J.; Zhang, M.; Guo, X.; Ye, Z.; Zhao, Y.; Xiong, H.; Zhang, Z.; et al. OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signalling in rice. Plant Biotechnol. J. 2017, 15, 183–196. [Google Scholar] [CrossRef]
- Zhang, H.; Han, B.; Wang, T.; Chen, S.; Li, H.; Zhang, Y.; Dai, S. Mechanisms of plant salt response: Insights from proteomics. J. Proteome Res. 2012, 11, 49–67. [Google Scholar] [CrossRef]
- Liu, D.G.; He, S.Z.; Song, X.J.; Zhai, H.; Liu, N.; Zhang, D.D.; Ren, Z.T.; Liu, Q.C. IbSIMT1, a novel salt-induced methyltransferase gene from Ipomoea batatas, is involved in salt tolerance. Plant Cell Tiss. Org. 2015, 120, 701–715. [Google Scholar] [CrossRef]
- Bao, A.K.; Wang, S.M.; Wu, G.Q.; Xi, J.J.; Zhang, J.L.; Wang, C.M. Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.). Plant Sci. 2009, 176, 232–240. [Google Scholar] [CrossRef]
- Zou, J.; Liu, C.F.; Liu, A.; Zou, D.; Chen, X.B. Overexpression of OsHsp17.0 and OsHsp23.7 enhances drought and salt tolerance in rice. J. Plant Physiol. 2012, 169, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Jia, F.; Qi, S.; Li, H.; Liu, P.; Li, P.; Wu, C.; Zheng, C.; Huang, J. Overexpression of late embryogenesis abundant 14 enhances Arabidopsis salt stress tolerance. Biochem. Biophys. Res. Commun. 2014, 454, 505–511. [Google Scholar] [CrossRef]
- Banerjee, A.; Roychoudhury, A. Group II late embryogenesis abundant (LEA) proteins: Structural and functional aspects in plant abiotic stress. Plant Growth Regul. 2015, 79, 1–17. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, C.; Lü, P.; Jiang, G.; Liu, X.; Dai, F.; Gao, J. RhNAC3, a stress-associated NAC transcription factor, has a role in dehydration tolerance through regulating osmotic stress-related genes in rose petals. Plant Biotechnol. J. 2014, 12, 38–48. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. Embo J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Lou, X.M.; Yao, Q.H.; Zhang, Z.; Peng, R.H.; Xiong, A.S.; Wang, K.K. Expression of human hepatitis B virus large surface antigen gene in transgenic tomato. Clin. Vaccine Immunol. 2007, 14, 464–469. [Google Scholar] [CrossRef]
- Jiang, T.; Zhai, H.; Wang, F.B.; Yang, N.K.; Wang, B.; He, S.Z.; Liu, Q.C. Cloning and characterization of a carbohydrate metabolism-associated gene IbSnRK1 from sweetpotato. Sci. Hortic. 2013, 158, 22–32. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 1985, 5, 69–76. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Dang, C.; Ye, Y.; Wang, Z.; Hu, L.; Zhang, F.; Zhang, Y.; Qian, X.; Shi, J.; Guo, Y.; et al. Overexpression of Grapevine VvIAA18 Gene Enhanced Salt Tolerance in Tobacco. Int. J. Mol. Sci. 2020, 21, 1323. https://doi.org/10.3390/ijms21041323
Li W, Dang C, Ye Y, Wang Z, Hu L, Zhang F, Zhang Y, Qian X, Shi J, Guo Y, et al. Overexpression of Grapevine VvIAA18 Gene Enhanced Salt Tolerance in Tobacco. International Journal of Molecular Sciences. 2020; 21(4):1323. https://doi.org/10.3390/ijms21041323
Chicago/Turabian StyleLi, Wei, Changxi Dang, Yuxiu Ye, Zunxin Wang, Laibao Hu, Fan Zhang, Yang Zhang, Xingzhi Qian, Jiabin Shi, Yanyun Guo, and et al. 2020. "Overexpression of Grapevine VvIAA18 Gene Enhanced Salt Tolerance in Tobacco" International Journal of Molecular Sciences 21, no. 4: 1323. https://doi.org/10.3390/ijms21041323
APA StyleLi, W., Dang, C., Ye, Y., Wang, Z., Hu, L., Zhang, F., Zhang, Y., Qian, X., Shi, J., Guo, Y., Zhou, Q., Wang, T., Chen, X., & Wang, F. (2020). Overexpression of Grapevine VvIAA18 Gene Enhanced Salt Tolerance in Tobacco. International Journal of Molecular Sciences, 21(4), 1323. https://doi.org/10.3390/ijms21041323



