Sledgehammer to Scalpel: Broad Challenges to the Heart and Other Tissues Yield Specific Cellular Responses via Transcriptional Regulation of the ER-Stress Master Regulator ATF6α
Abstract
1. Introduction
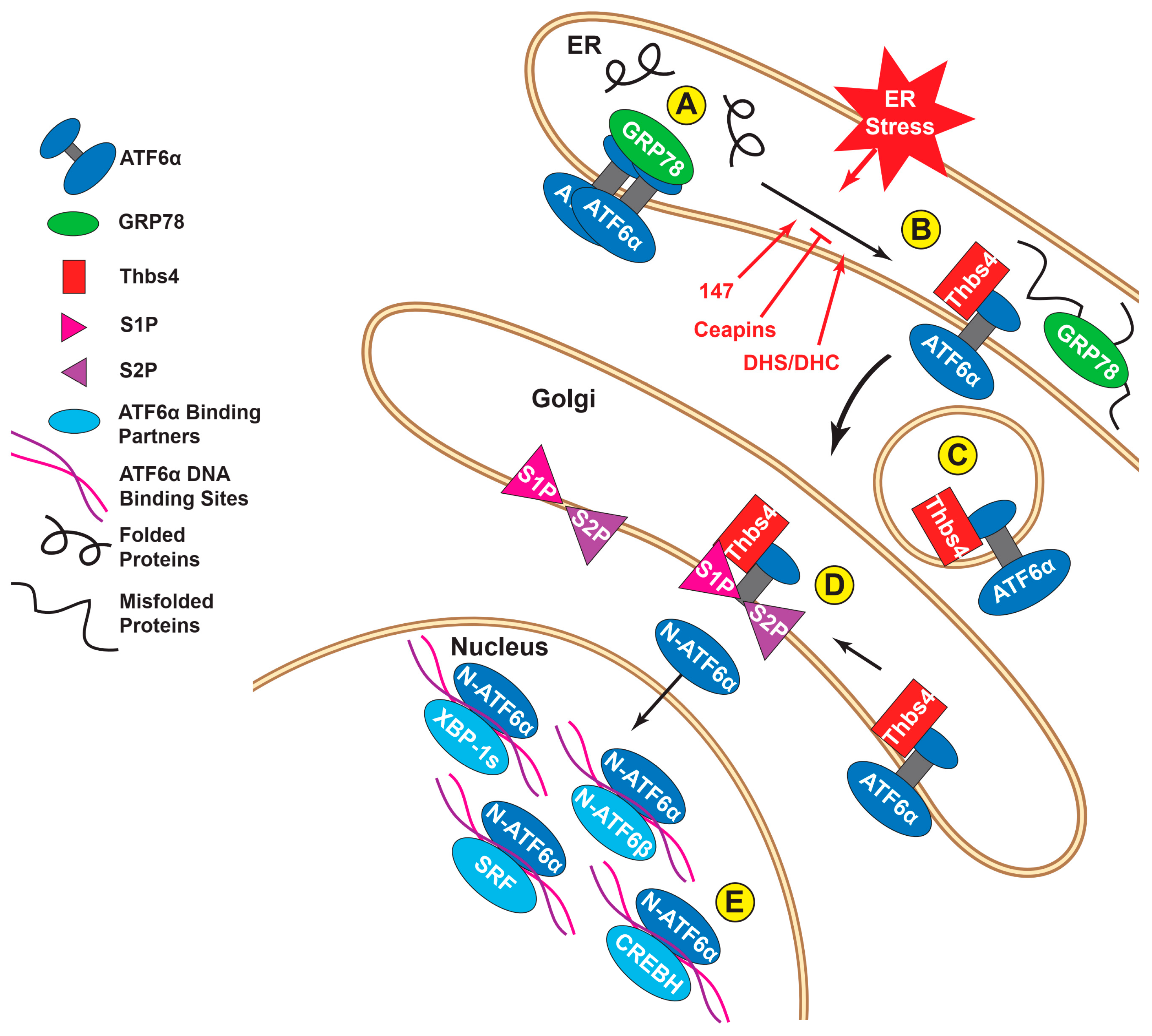
2. ATF6α Overview
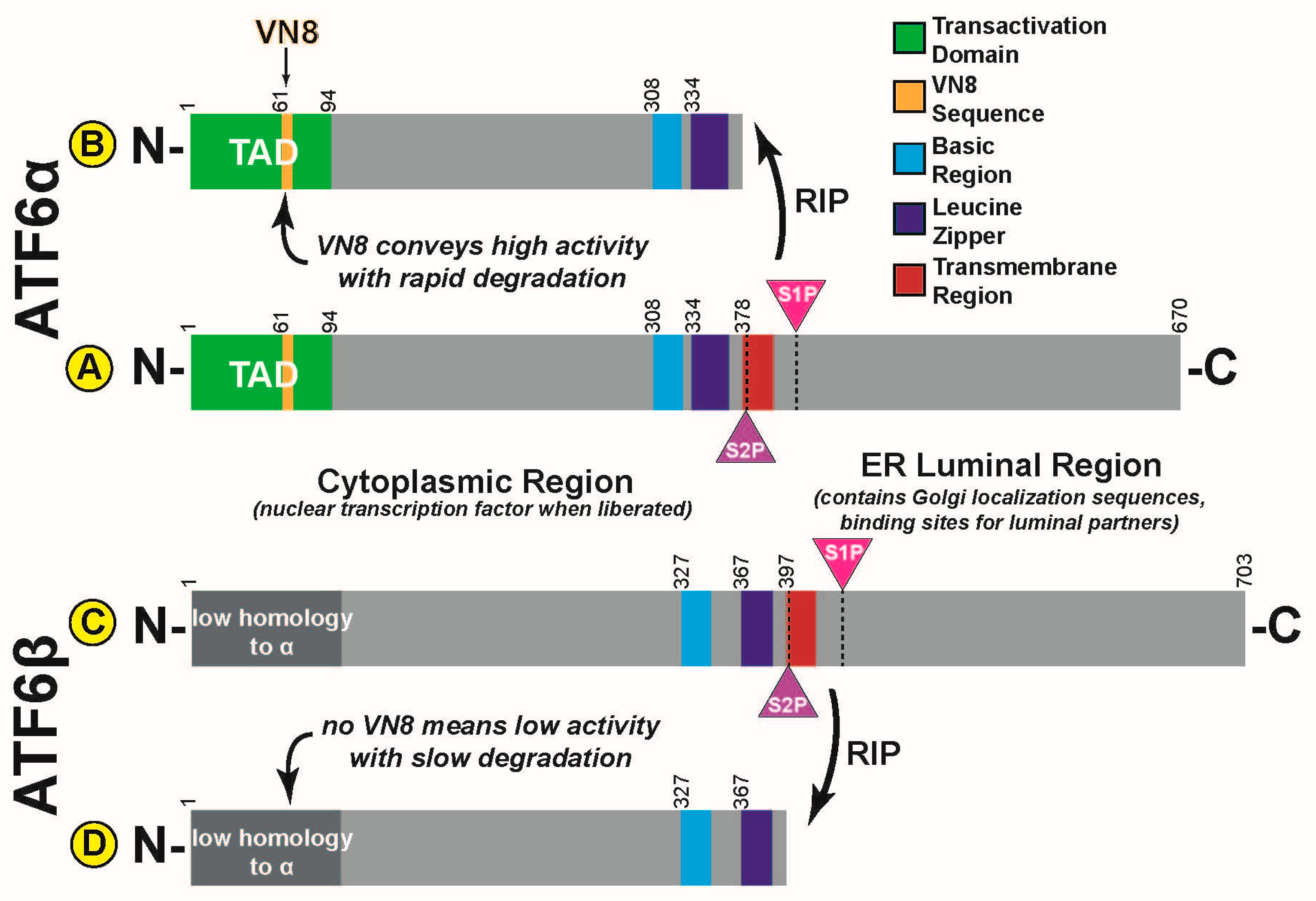
3. ATF6α Activation
4. ATF6α Transcriptional Activity and Degradation
5. ATF6α Dimerization and Nuclear Binding Partners
6. ATF6α Promoter Elements
7. ATF6α Transcriptional Programs
8. Stimulus-Specific ATF6α Transcriptional Programs
9. ATF6α in Disease
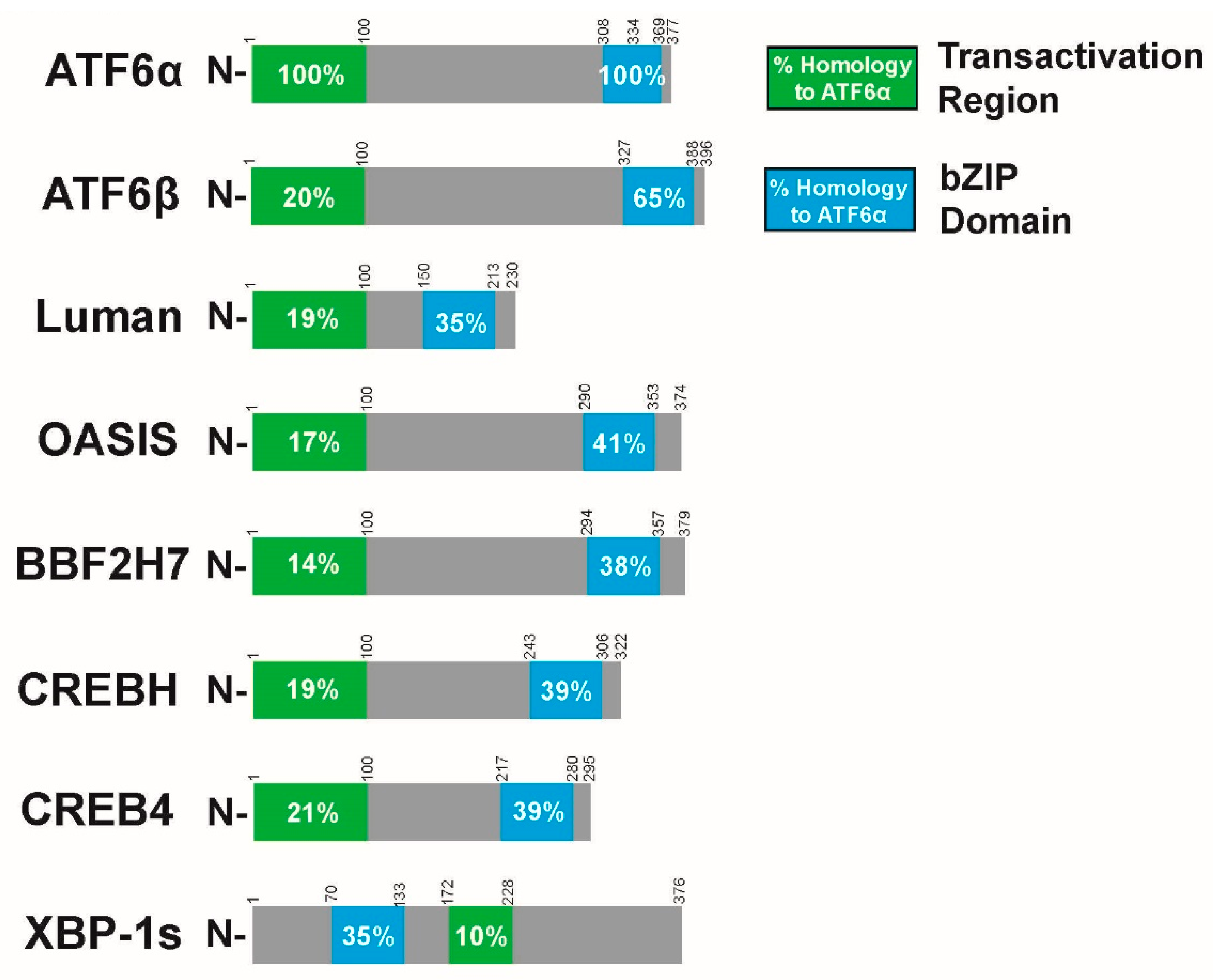
10. ATF6α Relatives
11. ATF6β
12. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ER | endoplasmic reticulum |
| ATF6α | activating transcription factor 6 alpha |
| bZIP | basic leucine zipper |
| HIF1α | hypoxia inducible factor 1 alpha |
| NRF2 | nuclear factor erythroid 2-related factor 2 |
| SRF | serum response factor |
| UPR | unfolded protein response |
| PERK | protein kinase R-like ER kinase |
| IRE1 | inositol-requiring protein 1 |
| eIF2α | eukaryotic initiation factor 2 alpha |
| XBP-1 | x-box-binding protein 1 |
| ATF/CREB | activating transcription factor/cAMP response element binding |
| GRP78 | glucose-regulated protein 78 |
| BiP | binding immunoglobulin protein |
| RIP | regulated intramembrane proteolysis |
| S1P | site-1 protease |
| S2P | site-2 protease |
| Thbs4 | thrombospondin 4 |
| PDI | protein disulfide isomerase |
| PDIA5 | protein disulfide isomerase A5 |
| ERp18 | endoplasmic reticulum protein 18 |
| AEBSF | 4-(2-aminoethyl)benzenesulfonyl fluoride |
| SREBP | sterol regulatory element binding protein |
| TAD | transcriptional activation domain |
| SUG-1 | protease regulatory subunit 8 homolog |
| SRE | serum response elements |
| NF-Y | nuclear transcription factor Y |
| YY-1 | yin-yang 1 |
| XBP-1s | X-box binding protein 1 spliced |
| PGC-1α | Peroxisome proliferator-activated receptor gamma coactivator 1 alpha |
| ERRγ | estrogen related receptor gamma |
| CREBH | cAMP response element binding H |
| COPII | coat protein complex II |
| ERSE | ER stress response elements |
| GRP94 | glucose-regulated protein 94 |
| PDIA6 | protein disulfide isomerase A6 |
| ERAD | ER-associated degradation |
| HRD1 | HMG-CoA reductase degradation protein 1 |
| Derlin3 | degradation in ER protein 3 |
| VIMP | VCP interacting membrane protein |
| CHOP | CCAAT-enhancer-binding protein homologous protein |
| ROS | reactive oxygen species |
| Rheb | Ras homolog enriched in brain |
| mTORC1 | mammalian target of rapamycin complex 1 |
| DHS | dihydrosphingosine |
| DHC | dihydroceramide |
| TAC | transaortic constriction |
| CREB3 | cAMP response element binding 3 |
| OASIS | old astrocyte specifically induced substance |
| BBF2H7 | box B-binding factor 2 human homolog on chromosome 7 |
| CREB4 | cAMP response element binding 4 |
| UPRE | unfolded protein response element |
| CRE | cAMP response element |
References
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 2019, 20, 207–220. [Google Scholar] [CrossRef]
- Spitz, F.; Furlong, E.E. Transcription factors: From enhancer binding to developmental control. Nat. Rev. Genet. 2012, 13, 613–626. [Google Scholar] [CrossRef]
- Lodish, H.F.; Berk, A.; Kaiser, C.A.; Krieger, M.; Bretscher, A.; Ploegh, H.; Amon, A.; Martin, K.C. Molecular Cell Biology, 8th ed.; Macmillan: New York, NY, USA, 2008; pp. xl, 1170, G-1128, I-1136. [Google Scholar]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 175, 598–599. [Google Scholar] [CrossRef]
- Johnson, P.F.; McKnight, S.L. Eukaryotic transcriptional regulatory proteins. Annu. Rev. Biochem. 1989, 58, 799–839. [Google Scholar] [CrossRef]
- Hill, C.S.; Treisman, R. Transcriptional regulation by extracellular signals: Mechanisms and specificity. Cell 1995, 80, 199–211. [Google Scholar] [CrossRef]
- Prabhakar, N.R.; Semenza, G.L. Oxygen Sensing and Homeostasis. Physiology 2015, 30, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Shaw, P.; Chattopadhyay, A. Nrf2-ARE signaling in cellular protection: Mechanism of action and the regulatory mechanisms. J. Cell. Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Treisman, R.; Ammerer, G. The SRF and MCM1 transcription factors. Curr. Opin. Genet. Dev. 1992, 2, 221–226. [Google Scholar] [CrossRef]
- Kaufman, R.J. Orchestrating the unfolded protein response in health and disease. J. Clin. Investig. 2002, 110, 1389–1398. [Google Scholar] [CrossRef]
- Glembotski, C.C.; Rosarda, J.D.; Wiseman, R.L. Proteostasis and Beyond: ATF6 in Ischemic Disease. Trends Mol. Med. 2019, 25, 538–550. [Google Scholar] [CrossRef]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Zhang, Y.; Ron, D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 1999, 397, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Tirasophon, W.; Shen, X.; Michalak, M.; Prywes, R.; Okada, T.; Yoshida, H.; Mori, K.; Kaufman, R.J. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002, 16, 452–466. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Prywes, R. ER stress signaling by regulated proteolysis of ATF6. Methods 2005, 35, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Hai, T.; Hartman, M.G. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: Activating transcription factor proteins and homeostasis. Gene 2001, 273, 1–11. [Google Scholar] [CrossRef]
- Amoutzias, G.D.; Veron, A.S.; Weiner, J., 3rd; Robinson-Rechavi, M.; Bornberg-Bauer, E.; Oliver, S.G.; Robertson, D.L. One billion years of bZIP transcription factor evolution: Conservation and change in dimerization and DNA-binding site specificity. Mol. Biol. Evol. 2007, 24, 827–835. [Google Scholar] [CrossRef]
- Thuerauf, D.J.; Marcinko, M.; Belmont, P.J.; Glembotski, C.C. Effects of the isoform-specific characteristics of ATF6 alpha and ATF6 beta on endoplasmic reticulum stress response gene expression and cell viability. J. Biol. Chem. 2007, 282, 22865–22878. [Google Scholar] [CrossRef]
- Haze, K.; Okada, T.; Yoshida, H.; Yanagi, H.; Yura, T.; Negishi, M.; Mori, K. Identification of the G13 (cAMP-response-element-binding protein-related protein) gene product related to activating transcription factor 6 as a transcriptional activator of the mammalian unfolded protein response. Biochem. J. 2001, 355, 19–28. [Google Scholar] [CrossRef]
- Shen, J.; Chen, X.; Hendershot, L.; Prywes, R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 2002, 3, 99–111. [Google Scholar] [CrossRef]
- Nadanaka, S.; Yoshida, H.; Mori, K. Reduction of disulfide bridges in the lumenal domain of ATF6 in response to glucose starvation. Cell Struct. Funct. 2006, 31, 127–134. [Google Scholar] [CrossRef]
- Nadanaka, S.; Okada, T.; Yoshida, H.; Mori, K. Role of disulfide bridges formed in the luminal domain of ATF6 in sensing endoplasmic reticulum stress. Mol. Cell. Biol. 2007, 27, 1027–1043. [Google Scholar] [CrossRef] [PubMed]
- Doroudgar, S.; Thuerauf, D.J.; Marcinko, M.C.; Belmont, P.J.; Glembotski, C.C. Ischemia activates the ATF6 branch of the endoplasmic reticulum stress response. J. Biol. Chem. 2009, 284, 29735–29745. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Snapp, E.L.; Lippincott-Schwartz, J.; Prywes, R. Stable binding of ATF6 to BiP in the endoplasmic reticulum stress response. Mol. Cell. Biol. 2005, 25, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Rawson, R.B.; Komuro, R.; Chen, X.; Dave, U.P.; Prywes, R.; Brown, M.S.; Goldstein, J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 2000, 6, 1355–1364. [Google Scholar] [CrossRef]
- Bertolotti, A.; Zhang, Y.; Hendershot, L.M.; Harding, H.P.; Ron, D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000, 2, 326–332. [Google Scholar] [CrossRef]
- Ma, K.; Vattem, K.M.; Wek, R.C. Dimerization and release of molecular chaperone inhibition facilitate activation of eukaryotic initiation factor-2 kinase in response to endoplasmic reticulum stress. J. Biol. Chem. 2002, 277, 18728–18735. [Google Scholar] [CrossRef]
- Lynch, J.M.; Maillet, M.; Vanhoutte, D.; Schloemer, A.; Sargent, M.A.; Blair, N.S.; Lynch, K.A.; Okada, T.; Aronow, B.J.; Osinska, H.; et al. A thrombospondin-dependent pathway for a protective ER stress response. Cell 2012, 149, 1257–1268. [Google Scholar] [CrossRef]
- Higa, A.; Taouji, S.; Lhomond, S.; Jensen, D.; Fernandez-Zapico, M.E.; Simpson, J.C.; Pasquet, J.M.; Schekman, R.; Chevet, E. Endoplasmic reticulum stress-activated transcription factor ATF6alpha requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol. Cell. Biol. 2014, 34, 1839–1849. [Google Scholar] [CrossRef]
- Oka, O.B.; van Lith, M.; Rudolf, J.; Tungkum, W.; Pringle, M.A.; Bulleid, N.J. ERp18 regulates activation of ATF6alpha during unfolded protein response. EMBO J. 2019, 38, e100990. [Google Scholar] [CrossRef]
- Gallagher, C.M.; Garri, C.; Cain, E.L.; Ang, K.K.; Wilson, C.G.; Chen, S.; Hearn, B.R.; Jaishankar, P.; Aranda-Diaz, A.; Arkin, M.R.; et al. Ceapins are a new class of unfolded protein response inhibitors, selectively targeting the ATF6alpha branch. Elife 2016, 5. [Google Scholar] [CrossRef]
- Gallagher, C.M.; Walter, P. Ceapins inhibit ATF6alpha signaling by selectively preventing transport of ATF6alpha to the Golgi apparatus during ER stress. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Haze, K.; Nadanaka, S.; Yoshida, H.; Seidah, N.G.; Hirano, Y.; Sato, R.; Negishi, M.; Mori, K. A serine protease inhibitor prevents endoplasmic reticulum stress-induced cleavage but not transport of the membrane-bound transcription factor ATF6. J. Biol. Chem. 2003, 278, 31024–31032. [Google Scholar] [CrossRef] [PubMed]
- Pasquato, A.; Rochat, C.; Burri, D.J.; Pasqual, G.; de la Torre, J.C.; Kunz, S. Evaluation of the anti-arenaviral activity of the subtilisin kexin isozyme-1/site-1 protease inhibitor PF-429242. Virology 2012, 423, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Plate, L.; Cooley, C.B.; Chen, J.J.; Paxman, R.J.; Gallagher, C.M.; Madoux, F.; Genereux, J.C.; Dobbs, W.; Garza, D.; Spicer, T.P.; et al. Small molecule proteostasis regulators that reprogram the ER to reduce extracellular protein aggregation. Elife 2016, 5. [Google Scholar] [CrossRef]
- Paxman, R.; Plate, L.; Blackwood, E.A.; Glembotski, C.; Powers, E.T.; Wiseman, R.L.; Kelly, J.W. Pharmacologic ATF6 activating compounds are metabolically activated to selectively modify endoplasmic reticulum proteins. Elife 2018, 7. [Google Scholar] [CrossRef]
- Tam, A.B.; Roberts, L.S.; Chandra, V.; Rivera, I.G.; Nomura, D.K.; Forbes, D.J.; Niwa, M. The UPR Activator ATF6 Responds to Proteotoxic and Lipotoxic Stress by Distinct Mechanisms. Dev. Cell 2018, 46, 327–343.e327. [Google Scholar] [CrossRef]
- Hai, T.W.; Liu, F.; Coukos, W.J.; Green, M.R. Transcription factor ATF cDNA clones: An extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989, 3, 2083–2090. [Google Scholar] [CrossRef]
- Thuerauf, D.J.; Morrison, L.E.; Hoover, H.; Glembotski, C.C. Coordination of ATF6-mediated transcription and ATF6 degradation by a domain that is shared with the viral transcription factor, VP16. J. Biol. Chem. 2002, 277, 20734–20739. [Google Scholar] [CrossRef]
- Desterro, J.M.; Rodriguez, M.S.; Hay, R.T. Regulation of transcription factors by protein degradation. Cell Mol. Life Sci. 2000, 57, 1207–1219. [Google Scholar] [CrossRef]
- Thuerauf, D.J.; Morrison, L.; Glembotski, C.C. Opposing roles for ATF6alpha and ATF6beta in endoplasmic reticulum stress response gene induction. J. Biol. Chem. 2004, 279, 21078–21084. [Google Scholar] [CrossRef]
- Fonseca, S.G.; Ishigaki, S.; Oslowski, C.M.; Lu, S.; Lipson, K.L.; Ghosh, R.; Hayashi, E.; Ishihara, H.; Oka, Y.; Permutt, M.A.; et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J. Clin. Investig. 2010, 120, 744–755. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Li, M.; Mao, C.; Lee, A.S. Endoplasmic reticulum stress triggers an acute proteasome-dependent degradation of ATF6. J. Cell. Biochem. 2004, 92, 723–732. [Google Scholar] [CrossRef]
- Zhu, Q.; Yao, J.; Wani, G.; Chen, J.; Wang, Q.E.; Wani, A.A. The ubiquitin-proteasome pathway is required for the function of the viral VP16 transcriptional activation domain. FEBS Lett. 2004, 556, 19–25. [Google Scholar] [CrossRef]
- Hong, M.; Luo, S.; Baumeister, P.; Huang, J.M.; Gogia, R.K.; Li, M.; Lee, A.S. Underglycosylation of ATF6 as a novel sensing mechanism for activation of the unfolded protein response. J. Biol. Chem. 2004, 279, 11354–11363. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Yang, Z.; Zhang, K.; Fang, D.; Sun, F. SUMOylation represses the transcriptional activity of the Unfolded Protein Response transducer ATF6. Biochem. Biophys Res. Commun. 2017, 494, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Geng, F.; Wenzel, S.; Tansey, W.P. Ubiquitin and proteasomes in transcription. Annu. Rev. Biochem. 2012, 81, 177–201. [Google Scholar] [CrossRef]
- Freedman, D.A.; Levine, A.J. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol. Cell. Biol. 1998, 18, 7288–7293. [Google Scholar] [CrossRef]
- Chen, L.; Madura, K. Yeast importin-alpha (Srp1) performs distinct roles in the import of nuclear proteins and in targeting proteasomes to the nucleus. J. Biol. Chem. 2014, 289, 32339–32352. [Google Scholar] [CrossRef]
- Dang, F.W.; Chen, L.; Madura, K. Catalytically Active Proteasomes Function Predominantly in the Cytosol. J. Biol. Chem. 2016, 291, 18765–18777. [Google Scholar] [CrossRef]
- Rodriguez-Martinez, J.A.; Reinke, A.W.; Bhimsaria, D.; Keating, A.E.; Ansari, A.Z. Combinatorial bZIP dimers display complex DNA-binding specificity landscapes. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Huet, A.; Parlakian, A.; Arnaud, M.C.; Glandieres, J.M.; Valat, P.; Fermandjian, S.; Paulin, D.; Alpert, B.; Zentz, C. Mechanism of binding of serum response factor to serum response element. FEBS J. 2005, 272, 3105–3119. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Johansen, F.E.; Prywes, R. Interaction of ATF6 and serum response factor. Mol. Cell. Biol. 1997, 17, 4957–4966. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Okada, T.; Haze, K.; Yanagi, H.; Yura, T.; Negishi, M.; Mori, K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol. Cell. Biol. 2000, 20, 6755–6767. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Baumeister, P.; Roy, B.; Phan, T.; Foti, D.; Luo, S.; Lee, A.S. ATF6 as a transcription activator of the endoplasmic reticulum stress element: Thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol. Cell. Biol. 2000, 20, 5096–5106. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Sato, T.; Matsui, T.; Sato, M.; Okada, T.; Yoshida, H.; Harada, A.; Mori, K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell 2007, 13, 365–376. [Google Scholar] [CrossRef]
- Wu, J.; Ruas, J.L.; Estall, J.L.; Rasbach, K.A.; Choi, J.H.; Ye, L.; Bostrom, P.; Tyra, H.M.; Crawford, R.W.; Campbell, K.P.; et al. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1alpha/ATF6alpha complex. Cell Metab. 2011, 13, 160–169. [Google Scholar] [CrossRef]
- Misra, J.; Kim, D.K.; Choi, W.; Koo, S.H.; Lee, C.H.; Back, S.H.; Kaufman, R.J.; Choi, H.S. Transcriptional cross talk between orphan nuclear receptor ERRgamma and transmembrane transcription factor ATF6alpha coordinates endoplasmic reticulum stress response. Nucleic Acids Res. 2013, 41, 6960–6974. [Google Scholar] [CrossRef][Green Version]
- Zhang, K.; Shen, X.; Wu, J.; Sakaki, K.; Saunders, T.; Rutkowski, D.T.; Back, S.H.; Kaufman, R.J. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 2006, 124, 587–599. [Google Scholar] [CrossRef]
- Chen, X.; Shen, J.; Prywes, R. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J. Biol. Chem. 2002, 277, 13045–13052. [Google Scholar] [CrossRef]
- Vinson, C.; Acharya, A.; Taparowsky, E.J. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim. Biophys. Acta. 2006, 1759, 4–12. [Google Scholar] [CrossRef]
- Yoshida, H.; Haze, K.; Yanagi, H.; Yura, T.; Mori, K. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem. 1998, 273, 33741–33749. [Google Scholar] [CrossRef] [PubMed]
- Kokame, K.; Kato, H.; Miyata, T. Identification of ERSE-II, a new cis-acting element responsible for the ATF6-dependent mammalian unfolded protein response. J. Biol. Chem. 2001, 276, 9199–9205. [Google Scholar] [CrossRef] [PubMed]
- Belmont, P.J.; Tadimalla, A.; Chen, W.J.; Martindale, J.J.; Thuerauf, D.J.; Marcinko, M.; Gude, N.; Sussman, M.A.; Glembotski, C.C. Coordination of growth and endoplasmic reticulum stress signaling by regulator of calcineurin 1 (RCAN1), a novel ATF6-inducible gene. J. Biol. Chem. 2008, 283, 14012–14021. [Google Scholar] [CrossRef] [PubMed]
- Blackwood, E.A.; Hofmann, C.; Santo Domingo, M.; Bilal, A.S.; Sarakki, A.; Stauffer, W.; Arrieta, A.; Thuerauf, D.J.; Kolkhorst, F.W.; Muller, O.J.; et al. ATF6 Regulates Cardiac Hypertrophy by Transcriptional Induction of the mTORC1 Activator, Rheb. Circ. Res. 2019, 124, 79–93. [Google Scholar] [CrossRef]
- Haze, K.; Yoshida, H.; Yanagi, H.; Yura, T.; Mori, K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 1999, 10, 3787–3799. [Google Scholar] [CrossRef]
- Vekich, J.A.; Belmont, P.J.; Thuerauf, D.J.; Glembotski, C.C. Protein disulfide isomerase-associated 6 is an ATF6-inducible ER stress response protein that protects cardiac myocytes from ischemia/reperfusion-mediated cell death. J. Mol. Cell. Cardiol. 2012, 53, 259–267. [Google Scholar] [CrossRef]
- Werner, E.D.; Brodsky, J.L.; McCracken, A.A. Proteasome-dependent endoplasmic reticulum-associated protein degradation: An unconventional route to a familiar fate. Proc. Natl. Acad. Sci. USA 1996, 93, 13797–13801. [Google Scholar] [CrossRef]
- Hampton, R.Y.; Gardner, R.G.; Rine, J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell 1996, 7, 2029–2044. [Google Scholar] [CrossRef]
- Nadav, E.; Shmueli, A.; Barr, H.; Gonen, H.; Ciechanover, A.; Reiss, Y. A novel mammalian endoplasmic reticulum ubiquitin ligase homologous to the yeast Hrd1. Biochem. Biophys. Res. Commun. 2003, 303, 91–97. [Google Scholar] [CrossRef]
- Kikkert, M.; Doolman, R.; Dai, M.; Avner, R.; Hassink, G.; van Voorden, S.; Thanedar, S.; Roitelman, J.; Chau, V.; Wiertz, E. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J. Biol. Chem. 2004, 279, 3525–3534. [Google Scholar] [CrossRef] [PubMed]
- Doroudgar, S.; Volkers, M.; Thuerauf, D.J.; Khan, M.; Mohsin, S.; Respress, J.L.; Wang, W.; Gude, N.; Muller, O.J.; Wehrens, X.H.; et al. Hrd1 and ER-Associated Protein Degradation, ERAD, are Critical Elements of the Adaptive ER Stress Response in Cardiac Myocytes. Circ. Res. 2015, 117, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Belmont, P.J.; Chen, W.J.; San Pedro, M.N.; Thuerauf, D.J.; Gellings Lowe, N.; Gude, N.; Hilton, B.; Wolkowicz, R.; Sussman, M.A.; Glembotski, C.C. Roles for endoplasmic reticulum-associated degradation and the novel endoplasmic reticulum stress response gene Derlin-3 in the ischemic heart. Circ. Res. 2010, 106, 307–316. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.K.; Blackwood, E.A.; Azizi, K.; Thuerauf, D.J.; Fahem, A.G.; Hofmann, C.; Kaufman, R.J.; Doroudgar, S.; Glembotski, C.C. ATF6 Decreases Myocardial Ischemia/Reperfusion Damage and Links ER Stress and Oxidative Stress Signaling Pathways in the Heart. Circ. Res. 2017, 120, 862–875. [Google Scholar] [CrossRef] [PubMed]
- Oda, Y.; Okada, T.; Yoshida, H.; Kaufman, R.J.; Nagata, K.; Mori, K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J. Cell. Biol. 2006, 172, 383–393. [Google Scholar] [CrossRef]
- Reeves, M.A.; Hoffmann, P.R. The human selenoproteome: Recent insights into functions and regulation. Cell. Mol. Life Sci. 2009, 66, 2457–2478. [Google Scholar] [CrossRef]
- Glembotski, C.C. Roles for ATF6 and the sarco/endoplasmic reticulum protein quality control system in the heart. J. Mol. Cell. Cardiol. 2014, 71, 11–15. [Google Scholar] [CrossRef]
- Wu, J.; Rutkowski, D.T.; Dubois, M.; Swathirajan, J.; Saunders, T.; Wang, J.; Song, B.; Yau, G.D.; Kaufman, R.J. ATF6alpha optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev. Cell 2007, 13, 351–364. [Google Scholar] [CrossRef]
- Kaufman, R.J. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev. 1999, 13, 1211–1233. [Google Scholar] [CrossRef]
- Marciniak, S.J.; Yun, C.Y.; Oyadomari, S.; Novoa, I.; Zhang, Y.; Jungreis, R.; Nagata, K.; Harding, H.P.; Ron, D. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004, 18, 3066–3077. [Google Scholar] [CrossRef]
- Ubeda, M.; Habener, J.F. CHOP gene expression in response to endoplasmic-reticular stress requires NFY interaction with different domains of a conserved DNA-binding element. Nucleic Acids Res. 2000, 28, 4987–4997. [Google Scholar] [CrossRef] [PubMed]
- Gotoh, T.; Oyadomari, S.; Mori, K.; Mori, M. Nitric oxide-induced apoptosis in RAW 264.7 macrophages is mediated by endoplasmic reticulum stress pathway involving ATF6 and CHOP. J. Biol. Chem. 2002, 277, 12343–12350. [Google Scholar] [CrossRef] [PubMed]
- Oyadomari, S.; Mori, M. Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death. Differ. 2004, 11, 381–389. [Google Scholar] [CrossRef]
- Martindale, J.J.; Fernandez, R.; Thuerauf, D.; Whittaker, R.; Gude, N.; Sussman, M.A.; Glembotski, C.C. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ. Res. 2006, 98, 1186–1193. [Google Scholar] [CrossRef]
- Meex, S.J.; Weissglas-Volkov, D.; van der Kallen, C.J.; Thuerauf, D.J.; van Greevenbroek, M.M.; Schalkwijk, C.G.; Stehouwer, C.D.; Feskens, E.J.; Heldens, L.; Ayoubi, T.A.; et al. The ATF6-Met[67]Val substitution is associated with increased plasma cholesterol levels. Arterioscler. Thromb. Vasc. Biol. 2009, 29, 1322–1327. [Google Scholar] [CrossRef][Green Version]
- Blackwood, E.A.; Azizi, K.; Thuerauf, D.J.; Paxman, R.J.; Plate, L.; Kelly, J.W.; Wiseman, R.L.; Glembotski, C.C. Pharmacologic ATF6 activation confers global protection in widespread disease models by reprograming cellular proteostasis. Nat. Commun. 2019, 10, 187. [Google Scholar] [CrossRef]
- Sharma, R.B.; O’Donnell, A.C.; Stamateris, R.E.; Ha, B.; McCloskey, K.M.; Reynolds, P.R.; Arvan, P.; Alonso, L.C. Insulin demand regulates beta cell number via the unfolded protein response. J. Clin. Investig. 2015, 125, 3831–3846. [Google Scholar] [CrossRef]
- Kohl, S.; Zobor, D.; Chiang, W.C.; Weisschuh, N.; Staller, J.; Gonzalez Menendez, I.; Chang, S.; Beck, S.C.; Garcia Garrido, M.; Sothilingam, V.; et al. Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia. Nat. Genet. 2015, 47, 757–765. [Google Scholar] [CrossRef]
- Bailey, D.; O’Hare, P. Transmembrane bZIP transcription factors in ER stress signaling and the unfolded protein response. Antioxid. Redox Signal. 2007, 9, 2305–2321. [Google Scholar] [CrossRef]
- Chan, C.P.; Kok, K.H.; Jin, D.Y. CREB3 subfamily transcription factors are not created equal: Recent insights from global analyses and animal models. Cell Biosci. 2011, 1, 6. [Google Scholar] [CrossRef]
- Asada, R.; Kanemoto, S.; Kondo, S.; Saito, A.; Imaizumi, K. The signalling from endoplasmic reticulum-resident bZIP transcription factors involved in diverse cellular physiology. J. Biochem. 2011, 149, 507–518. [Google Scholar] [CrossRef] [PubMed]
- DenBoer, L.M.; Hardy-Smith, P.W.; Hogan, M.R.; Cockram, G.P.; Audas, T.E.; Lu, R. Luman is capable of binding and activating transcription from the unfolded protein response element. Biochem. Biophys. Res. Commun 2005, 331, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Omori, Y.; Imai, J.; Suzuki, Y.; Watanabe, S.; Tanigami, A.; Sugano, S. OASIS is a transcriptional activator of CREB/ATF family with a transmembrane domain. Biochem. Biophys. Res. Commun 2002, 293, 470–477. [Google Scholar] [CrossRef]
- Horiuchi, K.; Tohmonda, T.; Morioka, H. The unfolded protein response in skeletal development and homeostasis. Cell. Mol. Life Sci. 2016, 73, 2851–2869. [Google Scholar] [CrossRef]
- Kondo, S.; Saito, A.; Hino, S.; Murakami, T.; Ogata, M.; Kanemoto, S.; Nara, S.; Yamashita, A.; Yoshinaga, K.; Hara, H.; et al. BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol. Cell. Biol. 2007, 27, 1716–1729. [Google Scholar] [CrossRef]
- Stirling, J.; O’Hare, P. CREB4, a transmembrane bZip transcription factor and potential new substrate for regulation and cleavage by S1P. Mol. Biol. Cell 2006, 17, 413–426. [Google Scholar] [CrossRef]
- Kondo, S.; Saito, A.; Asada, R.; Kanemoto, S.; Imaizumi, K. Physiological unfolded protein response regulated by OASIS family members, transmembrane bZIP transcription factors. Iubmb Life 2011, 63, 233–239. [Google Scholar] [CrossRef]
- Forouhan, M.; Mori, K.; Boot-Handford, R.P. Paradoxical roles of ATF6alpha and ATF6beta in modulating disease severity caused by mutations in collagen X. Matrix Biol. 2018, 70, 50–71. [Google Scholar] [CrossRef]
- Correll, R.N.; Grimes, K.M.; Prasad, V.; Lynch, J.M.; Khalil, H.; Molkentin, J.D. Overlapping and differential functions of ATF6alpha versus ATF6beta in the mouse heart. Sci. Rep. 2019, 9, 2059. [Google Scholar] [CrossRef]
- Vaquerizas, J.M.; Kummerfeld, S.K.; Teichmann, S.A.; Luscombe, N.M. A census of human transcription factors: Function, expression and evolution. Nat. Rev. Genet. 2009, 10, 252–263. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stauffer, W.T.; Arrieta, A.; Blackwood, E.A.; Glembotski, C.C. Sledgehammer to Scalpel: Broad Challenges to the Heart and Other Tissues Yield Specific Cellular Responses via Transcriptional Regulation of the ER-Stress Master Regulator ATF6α. Int. J. Mol. Sci. 2020, 21, 1134. https://doi.org/10.3390/ijms21031134
Stauffer WT, Arrieta A, Blackwood EA, Glembotski CC. Sledgehammer to Scalpel: Broad Challenges to the Heart and Other Tissues Yield Specific Cellular Responses via Transcriptional Regulation of the ER-Stress Master Regulator ATF6α. International Journal of Molecular Sciences. 2020; 21(3):1134. https://doi.org/10.3390/ijms21031134
Chicago/Turabian StyleStauffer, Winston T., Adrian Arrieta, Erik A. Blackwood, and Christopher C. Glembotski. 2020. "Sledgehammer to Scalpel: Broad Challenges to the Heart and Other Tissues Yield Specific Cellular Responses via Transcriptional Regulation of the ER-Stress Master Regulator ATF6α" International Journal of Molecular Sciences 21, no. 3: 1134. https://doi.org/10.3390/ijms21031134
APA StyleStauffer, W. T., Arrieta, A., Blackwood, E. A., & Glembotski, C. C. (2020). Sledgehammer to Scalpel: Broad Challenges to the Heart and Other Tissues Yield Specific Cellular Responses via Transcriptional Regulation of the ER-Stress Master Regulator ATF6α. International Journal of Molecular Sciences, 21(3), 1134. https://doi.org/10.3390/ijms21031134



