Abstract
Forkhead box class O family member proteins (FoxOs) are evolutionarily conserved transcription factors for their highly conserved DNA-binding domain. In mammalian species, all the four FoxO members, FoxO1, FoxO3, FoxO4, and FoxO6, are expressed in different organs. In bone, the first three members are extensively expressed and more studied. Bone development, remodeling, and homeostasis are all regulated by multiple cell lineages, including osteoprogenitor cells, chondrocytes, osteoblasts, osteocytes, osteoclast progenitors, osteoclasts, and the intercellular signaling among these bone cells. The disordered FoxOs function in these bone cells contribute to osteoarthritis, osteoporosis, or other bone diseases. Here, we review the current literature of FoxOs for their roles in bone cells, focusing on helping researchers to develop new therapeutic approaches and prevent or treat the related bone diseases.
Keywords:
FoxOs; bone cells; antioxidant; osteogenesis; osteoclastogenesis; chondrogenesis; bone diseases 1. Introduction
Forkhead box class O family member proteins (FoxOs) are evolutionarily conserved transcription factors [1]. FoxOs translate environmental signaling into gene expression [2], regulating many cellular processes, including cell survival, proliferation, differentiation, apoptosis, oxidative stress resistance, metabolism, inflammation, and aging [3,4,5,6,7,8]. In bone, internal and external environmental changes cause an imbalance of the bone dynamic, resulting in loss of bone mass and strength, and increased risk for fractures and morbidity [9,10]. Recent studies with murine models of cell-specific loss- or gain-of-function of FoxOs, have revealed that FoxOs regulate bone cell functions and their intercellular signaling. These controls affect bone development, alter bone mass, and conduce several bone diseases, such as osteoarthritis, age-related or estrogen-deficiency osteoporosis, and so on [10].
Hence, we will firstly introduce the structure and function of the FoxO family. Then, we will review the recent developments about FoxOs in osteoprogenitor cells, osteoblasts, hematopoietic stem cells, osteoclast progenitors, osteoclasts, and chondrocytes, as well as the intercellular signaling among these bone cells. At the same time, we will also describe the FoxO actions in bone cell biology, including cell survival, self-renewal, cell cycle, proliferation, osteogenesis, osteoclastogenesis, chondrogenesis, autophagy, and apoptosis. All of these will focus on their emerging molecular mechanism, pathological implications, and therapeutic potential for related bone diseases.
2. The FoxO Family
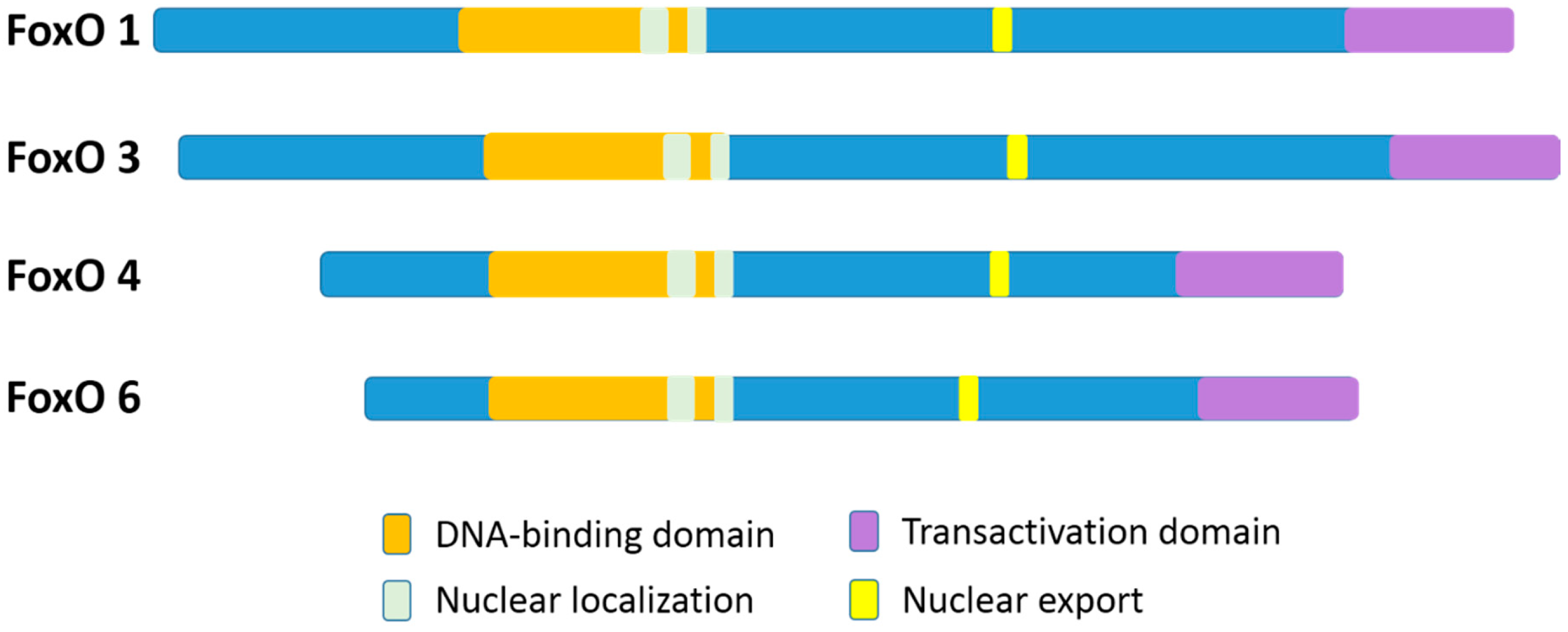
In mammalian species, there are four members of the FoxO family: FoxO1 (FKHR), FoxO3 (FKHRL1), FoxO4 (AFX), and FoxO6 [11]. FoxO1 and FoxO3 proteins are greater than 650 amino acids in size, which are larger than FoxO4 and FoxO6 (nearly 500 amino acids). Although, the FoxO6 gene exhibits major structural differences as compared to the other three family gene members [11]. The four FoxO protein members share obvious sequence homology and possess four clearly different functional motifs, which include a forkhead domain, nuclear localization, nuclear export, and transactivation domains (Figure 1) [12,13]. All the four FoxO members are ubiquitously expressed [14], including FoxO6 that was previously reported to exist basically in the brain but has been more recently discovered in other organs as well [4,15]. In particular, FoxO1, FoxO4, and FoxO6 are more detected in the adipose, musculoskeletal, and nervous tissues, respectively, while FoxO3 is more expressed in the stomach, spleen, kidney, intestine, and cardiac tissues [13]. Nevertheless, FoxO1, FoxO3, and FoxO4 are all expressed in bone cells [16].
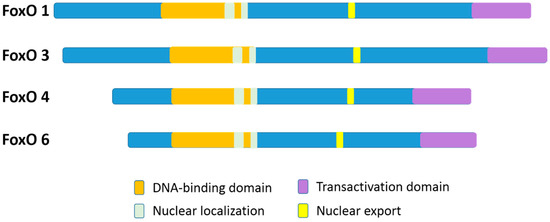
Figure 1.
A schematic diagram of the four different functional motifs of the mammalian FoxO family members (FoxOs). For humans, FoxO1, FoxO3, FoxO4, and FoxO6 proteins are 655 aa, 673 aa, 505 aa, and 492 aa in size, respectively. The functional domains are the forkhead domain, nuclear localization, nuclear export, and a transactivation domain.
In FoxO proteins, the forkhead domain located in the N-terminal portion [17] can recognize two response elements: the insulin-responsive element (5’-(C/A) (A/C)AAA(C/T)AA) and Daf-16 (namely FoxO) family member binding element (5´-GTAAA(T/C)AA) [12,13,14,15,16,18]. Although FoxO1 recognizes both the insulin response element and the Daf-16 family member binding element, it has a higher affinity for the latter [19]. The transactivation domain, located in the C-terminal region of the FoxOs, combines with the cis-regulatory sites of FoxO-target genes [17]. In addition, the nuclear localization sequence and the nuclear export sequence are within the C-terminal DNA binding domain of FoxOs, indispensable for maintaining the FoxO proteins in the nucleus or in the cytosol, respectively [12]. Nuclear FoxO proteins, predominantly as transcription factors, interact with DNA and partner proteins to modulate the transcription of numerously specific target genes [17].
Responding to a variety of external or internal stimuli (including starvation, insulin, growth factors, hormones, cytokines, and oxidative stress), FoxOs act as transcription factors and rarely as co-regulators, either activated or inactivated by post-translational modification (like phosphorylation, ubiquitination, acetylation, and methylation) [9,20]. Further, they are also regulated by lipopolysaccharide (LPS), tumor necrosis factor α (TNF-α), and the interaction with protein partners [21]. These modifications alter the nuclear import and export steps of FoxOs, modify the DNA binding affinity of FoxOs, and alter transcriptional activity for FoxOs’ specific target gene. Therefore, FoxOs take part in regulatory networks that ensure tight and timely transcriptional control of proteins involved in proliferation (FoxO1, FoxO3, and FoxO4), cellular differentiation (FoxO1, FoxO3, and FoxO4), apoptosis (FoxO1, FoxO3, and FoxO4), oxidative stress resistance (FoxO1 and FoxO3), metabolism (FoxO1 and FoxO3), inflammation (FoxO1, FoxO3, and FoxO4) and aging (FoxO1, FoxO3, and FoxO4) in mammals [3,4,5,6,7,8]. What is more, a unifying hypothesis has also highlighted the role of FoxOs as signaling integrators for the maintenance of cell and tissue homeostasis over time and in response to environmental challenges, including metabolic stress, oxidative stress, and growth factor deprivation [9].
3. FoxOs and Bone Cells
As compared to other organs in the body, bone is constantly degraded and replaced throughout one’s life. The relatively dynamic bone modeling and remodeling are governed specifically by osteoprogenitor cells, osteoblasts, osteocytes, hematopoietic stem cells, osteoclast progenitors, osteoclasts, and chondrocytes, as well as the complex intercellular signaling among these bone cells. Except for osteocytes, all bone cells were found expressing FoxO1, 3, and 4, as in several other mammalian cell types. Functions of FoxOs in these cells are pivotal for bone development, remodeling, and homeostasis manipulation under physiological and pathological conditions.
3.1. Bidirectional Regulation of FoxOs in Osteoprogenitor Cells
FoxOs play important roles in the osteogenesis of osteoprogenitor cells containing mesenchymal stem cells (MSCs), the early precursors of osteoblasts. MSCs are multipotent stromal cells. They give rise to chondrocytes, muscle cells, and adipocytes. Moreover, osteogenic differentiation of MSCs into osteoblasts is managed by an array of specific transcription factors (such as transcription factor TCF, runt-related transcription factor 2, and activating transcription factor 4) and genes (such as the alkaline phosphatase gene, runt-related transcription factor 2 gene, and osteocalcin gene), leading to a succession of osteoblast phenotypic markers [16]. Therefore, it is not difficult to envision the bidirectional regulation role of FoxOs in the entire complex and sensitive osteogenic process (Table 1).

Table 1.
The bidirectional regulation role of FoxO transcription factors (FoxOs) in osteoprogenitor cells.
3.1.1. FoxOs Promote Osteogenesis
With experiments in vitro, ex vivo, and in vivo, Teixeira et al. have demonstrated FoxO1 as an early positive regulator in the osteogenic differentiation of mesenchymal cells [22]. In mouse embryonic mesenchymal cells (C3H10T1/2 cells), osteogenic stimulants enhance the activity and expression of FoxO1. Similarly, overexpressing FoxO1 significantly increases the expression of osteogenic markers such as runt-related transcription factor 2 (Runx2), alkaline phosphatase (Alp), and osteocalcin (Ocn). Conversely, silencing FoxO1 inhibits the expression of these osteogenic markers, decreases calcification, impairs skeletogenesis and craniofacial development, and especially reduces the size of the embryos as well as bone size in the craniofacial area. In addition, knocking down FoxO1 in mouse embryonic tibia ex vivo has been reported to have similar results as those obtained in vivo, which led to shorter and less mineralized bone [22]. Furthermore, in pre-osteoblastic cells (e.g., MC3T3-E1 cells), the upregulated binding activity of FoxO1 to the Runx2 gene promoter increases the mRNA level of Runx2 and alkaline phosphatase (ALP) activity during the formation of mineralizing nodules. In addition, Runx2 directs pluripotent MSCs to the osteoblast lineage and triggers the expression of major bone matrix protein genes (like type I collagen or Alp) in early progenitors. Then, FoxO1 interacts with Runx2 and could promote osteoblast differentiation [16,23]. Therefore, to some extent, FoxO1 promotes osteoblast differentiation by interacting with Runx2 or its gene promoter (Figure 2). Besides, FoxO1 could also bind to the ALP gene promoter [24]. BMP-2-induced FoxO1 transcription increases the reporter activity of an ALP gene promoter construct [25]. Thus, FoxO1 may also promote osteoblast differentiation by interacting with the ALP gene promoter (Figure 2).
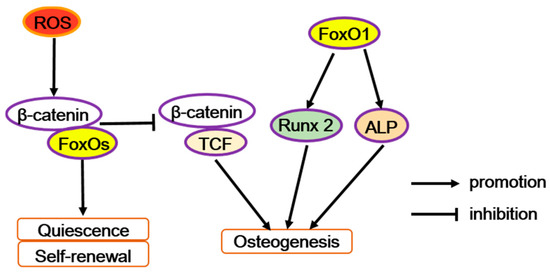
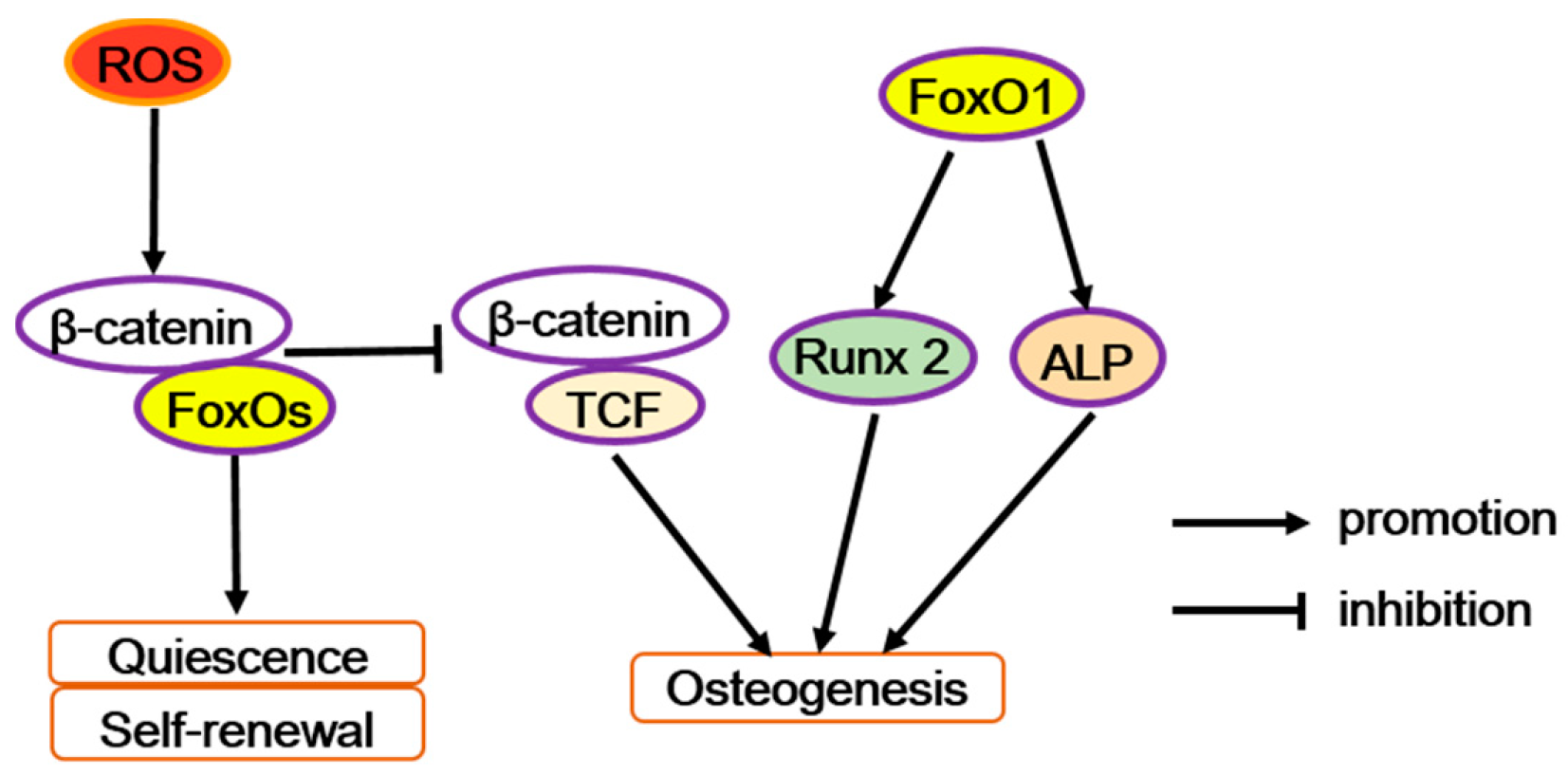
Figure 2.
The diverse function of FoxOs in osteoprogenitor cells. Reactive oxygen species (ROS) can trigger FoxOs mediated transcription and enhances the binding of FoxOs to β-catenin, thus diverting the limited β-catenin pool from transcription factor TCF to FoxO-mediated transcription and thereby decreases osteoblastogenesis. But FoxO1 promotes the transcription of runt-related transcription factor 2 (Runx2) or alkaline phosphatase (Alp), increasing osteoblastogenesis.
Similarly, it has been reported that global deletion of FoxO1, 3, and 4 in mice leads to a reduction in the expression of Runx2, Osterix, and alkaline phosphatase in osteoblastic progenitors in vitro [26], and the mice also display a significant decrease in the number of cell colony-forming unit (CFU) fibroblasts and CFU osteoblasts presented in the bone marrow [21]. That means FoxOs could regulate osteoblast differentiation and proliferation.
On the other hand, FoxO1, as a transcriptional trans-repressor, inhibits the actions of peroxisome proliferator-activated receptor γ (PPARγ) [27], which activates adipogenesis and suppresses osteoblastogenesis. The deletion of FoxOs increases PPARγ expression [21]. Consistent with these findings, FoxO1 restrains adipogenesis by repressing the activity of the PPARγ promoter and by antagonizing PPARγ’s ability by directly binding to PPAR-response elements (PPREs), the promoters of PPARγ target genes in pre-adipocytes [28,29]. As a result, FoxOs improve bone formation, by increasing osteoblast differentiation at the expense of adipocyte differentiation from their common mesenchymal progenitors.
3.1.2. FoxOs Repress Osteogenesis
In contrast to the above findings, some researchers also indicate FoxOs repress the osteogenic actions by suppressing Wnt signaling in osteoblast progenitors. Conditional knocked-out mice of FoxO1, 3, and 4 in bipotential progenitors of osteoblast and adipocytes showed that osteoblast number and bone mass were increased, but unexpectedly, there were no changes in the redox balance. The increased bone mass rooted in the increased proliferation of osteoprogenitor cells and bone formation, which accounted for the upregulation of Wnt/β-catenin signaling and cyclin D1 expression [18]. In addition, overexpression of FoxO1 reduced the MC3T3-E1 cell number and the number of proliferating cell nuclear antigen (PCNA)-positive cells with little effect on apoptosis, indicating that FoxO1 suppressed preosteoblast proliferation [25].
Moreover, in osteoblast progenitors, FoxOs acts as defense molecules in oxidative stress to regulate osteogenic actions. Excessive accumulation of reactive oxygen species (ROS) generally causes oxidative stress and destroys proteins, lipids, and DNA, even consequentially leading to cell death [30]. As a result of the conditional deletion of FoxO1, FoxO3, and FoxO4 in 3-month-old mice, oxidative stress and osteoblast apoptosis increased in bone, and the number of osteoblasts, the rate of bone formation, and bone mass were all decreased at cancellous or cortical sites [26]. So, we can infer that FoxOs can protect osteogenesis against oxidative stress. However, in osteoblast precursors, FoxOs are induced by the oxidative stress (exemplified by H2O2) to bind with β-catenin to co-activate the target genes’ transcriptions of FoxOs at the expense of Wnt/TCF-mediated transcription and osteoblast differentiation (Figure 2) [31]. Moreover, in aging mice, the expression of β-catenin/TCF-target genes decreases whereas FoxO-target genes increases in bone, along with an increase in markers of oxidative stress and a decrease in bone formation [31,32]. In line with this idea, fatty acid-binding protein 4 (FABP4)-Wnt10b mice, which express the Wnt10b transgene in marrow, and the mice with a mutation (G171V) in the low-density lipoprotein receptor related protein 5 (LRP5), both reveal enhanced bone mass and no evidence of age-related loss of bone mass or strength [21,33,34].
Similarly, the lipid oxidation may also suppress Wnt signaling by the same mechanism, and result in the decline in osteoblast number and bone formation that occurs with aging. It has been shown that lipoxygenases oxidize polyunsaturated fatty acids to form products, which bind to and activate PPARγ and generate pro-oxidants like 4-hydroxynonenal (4-HNE) [35]. The Almeida group demonstrated that lipid oxidation increases with age in bone [21]. Similar to H2O2, the 4-HNE also activates FoxOs that in turn attenuate β-catenin/TCF-mediated transcription [36]. Hence, it also enhances PPARγ levels for the suppression of TCF-mediated transcription to PPARγ expression [37,38]. On the other hand, the PPARγ bind to β-catenin and induce β-catenin degradation [36,39], thereby further diminishing β-catenin/TCF-mediated transcription. Therefore, at least in part, it is reasonable to understand that the bone loss with aging might arise from the repressive effect of ROS on Wnt/β-catenin signaling via FoxO activation.
Responding to oxidative stress, FoxOs also restrain other main osteogenic signaling pathways (Hedgehog signaling and Mitogen-activated protein kinases signaling) while boosting adipogenic signaling pathways [40]. Meanwhile, they also promote expression of antioxidants, which also increases adipogenic differentiation [40]. It is known that adipogenic differentiation is antagonistic to the osteogenic differentiation. Thus, FoxOs may also accelerate bone loss with aging by enhancing adipogenic differentiation. Obviously, in response to oxidative stress, FoxOs play an essential role to maintain skeletal homeostasis in osteoprogenitor cells.
Taken together, in osteoblast progenitor cells, FoxOs have complicated and contradictory features (Figure 2). There is no doubt that FoxOs are important in defense during oxidative stress and prevent cell death triggered by ROS, thus promoting osteogenesis. On one hand, MSCs have relatively high expression of antioxidant-like glycolysis and low ROS levels to keep quiescent [40], but a high ROS level may act as an intracellular signal to drive MSCs to exit from quiescence and result in the activation of a genetic program triggering osteoblast precursors commitment [16]. So, at the initial stage of differentiation, the switching from FoxO-mediated transcription to Wnt/TCF-mediated transcription may benefit osteogenesis. On the other hand, with the progress of osteogenic differentiation, FoxO1 may directly participate in osteoblastogenesis promotion by binding to the ALP and Runx2 gene promoters or directly interacting with Runx2, as well as maintaining a low level of ROS to protect cells from oxidative stress. Besides, FoxOs decrease ROS levels to restrain adipogenic differentiation. Therefore, the specific role of FoxOs may depend on the differentiation stage and ROS level of osteoprogenitor cells. Further studies are still needed to fully reveal the multiple functions of FoxOs in osteoprogenitor cells at a subtle stage during osteogenic differentiation.
3.2. FoxOs Improve Osteoblasts Function
The effects of FoxOs on osteoblasts are also complicated and likely to depend upon specific conditions (Table 2). Nevertheless, most of the literature support the positive effect of FoxOs in osteoblasts. On one hand, FoxOs regulate osteoblast numbers significantly by improving antioxidant defense (Figure 3). Conditionally overexpressed FoxO3 in osteoblasts decreases ROS and further inhibits osteoblast apoptosis with reducing phosphorylation of p66Shc. Consistent with this, conditional deletion of all three FoxO members (FoxO1/3/4) in 3-month-old mice decrease osteoblast numbers due to osteoblast apoptosis with arising oxidative stress. FoxOs inactivation also inhibits osteoblastogenesis [26]. Moreover, osteoblast-specific deletion of FoxO1 leads to increased oxidative stress, which decreases osteoblast numbers and bone mass [41]. More precisely, the FoxO1 inactivation weakens its interaction with activating transcription factor 4 (ATF4), reducing the target gene binding activity of FoxO1 and impeding amino acid import and protein synthesis through ATF4 (Figure 3). Therefore, as transcriptional regulation genes of FoxO1, antioxidant gene (such as Sod2 and catalase) expression decreases, increasing the ROS level and exerting harmful effects on osteoblast survival. And with FoxO1 deletion, the blocked protein synthesis breaks the redox balance by improving the expression of p19ARF (an alternate reading frame protein product of the CDKN2A locus in mice) and p16 and downstream activation of their target protein p53, repressing the cell cycle of the osteoblasts and finally decreasing osteoblast numbers. Additionally, the blocked protein synthesis also suppresses glutathione accumulation, which induces oxidative stress [41]. Taken together, FoxOs enhance the activity of osteoblasts and protect these cells through the induction of antioxidants (Figure 3).

Table 2.
Bone metabolism regulatory functions of FoxO transcription factors in osteoblast.
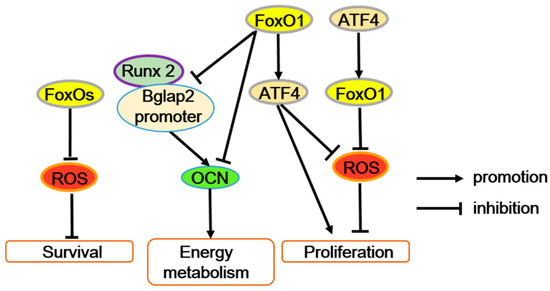
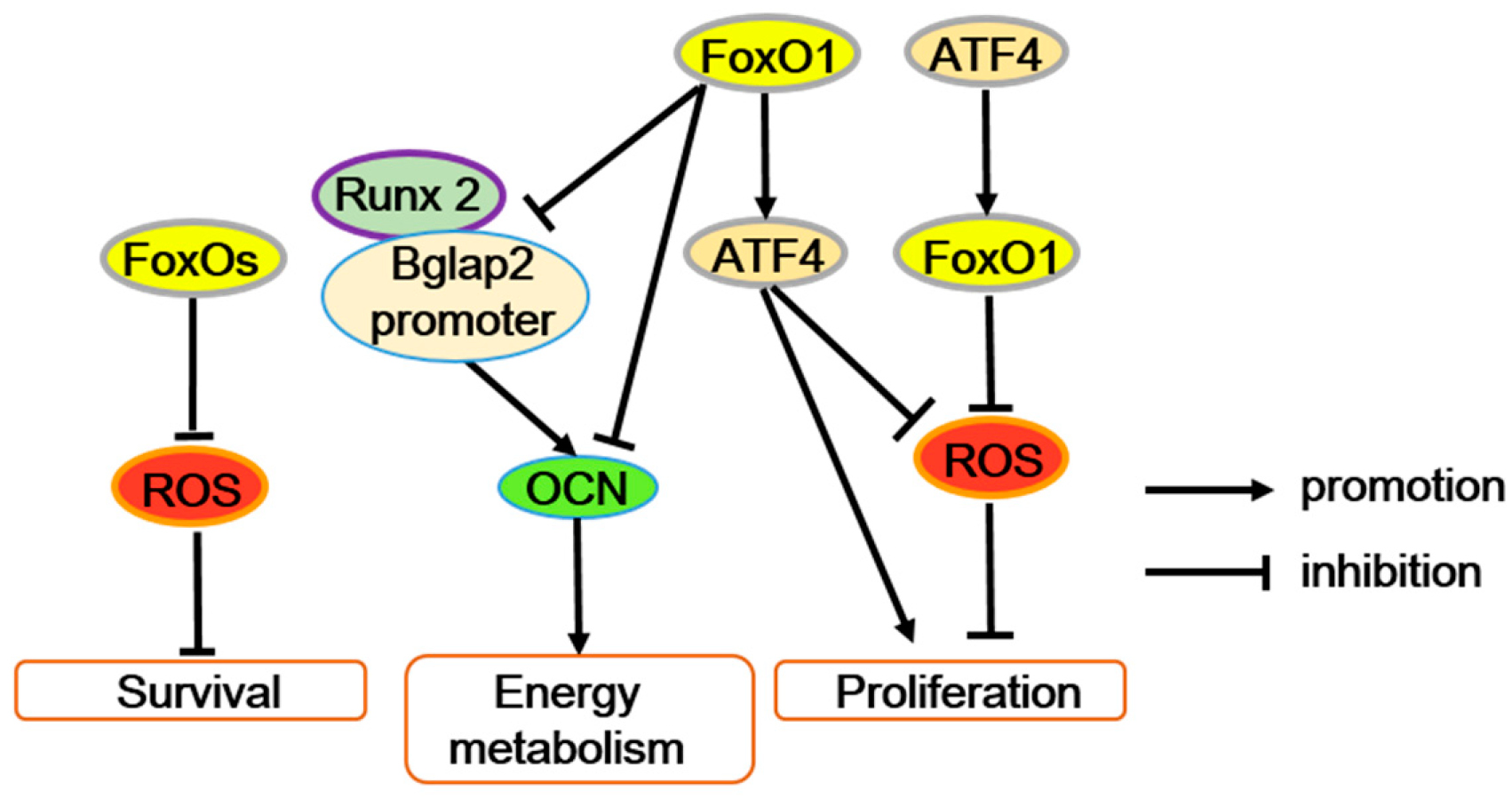
Figure 3.
FoxOs’ function in osteoblasts. FoxOs’ transcriptional activity reduces ROS levels and improves osteoblast survival. The interaction between FoxO1 and activating transcription factor 4 (ATF4) maintains osteoblast normal proliferation by preventing ROS or enhancing protein synthesis. In addition, FoxO1 suppresses the interaction between the Runx2 and Bglap2 (osteocalcin gene) promoters, directly bind to the Bglap2 promote r, or both to inhibit osteocalcin (OCN) expression.
However, FoxO1 can restrain the energy metabolism function of osteoblasts through inhibiting osteocalcin (OCN) expression. Furthermore, FoxO1 can suppress the interaction between the Runx2 and Bglap2 (OCN gene) promoters, directly bind to the Bglap2 promoter, or both to reduce the transcriptional activity of Bglap2 [16,42,43,44]. Generally, OCN also improves bone mineralization, calcium ion homeostasis, and prevents excessive mineralization. Thus, it is possible that FoxO1 mediates the dual roles of OCN in osteogenesis. But, obviously the effect of FoxO1 on Runx2 activity in osteoblasts is opposite in mesenchymal progenitors, and the mechanism is not well known. In addition, accompanied by immune response, FoxO1 also potentially contributes to immune-mediated inhibition of bone formation through accelerating osteoblast apoptosis [45].
Therefore, among the three FoxO proteins, FoxO1 is mainly required for redox balance and proliferation of osteoblasts and thereby to regulate bone formation.
3.3. FoxOs Positively Regulate Hematopoietic Stem Cell Activities
Hematopoietic stem cell (HSC) is an osteoclast progenitor. FoxOs maintain quiescence and self-renewal of HSCs mainly through transcriptional regulation of cell cycle arrest and oxidative stress resistance (Figure 4a). The loss of FoxO3 in HSCs not only leads to oxidative DNA damage in HSCs by interfering in the base excision repair pathway [46], but also damages the HSC pool with regulating the cell cycle through ROS-independent modulations of the tumor suppressor protein ataxia telangiectasia mutated (ATM) and p16INK4a and ROS-mediated activation of p19ARF/p53/p21CIP1/WAF1/Sdi1 tumor suppressor pathways [47]. Likewise, conditional deletion of FoxO1, FoxO3, and FoxO4 in the adult mice hematopoietic system decreases the expression of antioxidant enzyme genes, including catalase, glutathione peroxidase 1 (Gpx-1), and superoxide dismutase Sod1, Sod2, and Sod3, resulting in ROS over-accumulation. The following physiologic oxidative stress increases HSCs’ apoptosis and drives HSCs out of quiescence into the cell cycle. Meanwhile, HSCs repopulating activity have been long-term impaired. All of these imply that FoxOs deficiency enforces HSCs into terminal differentiation at the expense of self-renewal. Finally, combined FoxOs’ deletion markedly decreases HSCs’ population and also expanses myeloid lineage [48]. Therefore, FoxOs are likely to serve as protectors of HSCs, decreasing the number of osteoclast progenitor cells to some degree (Table 3).
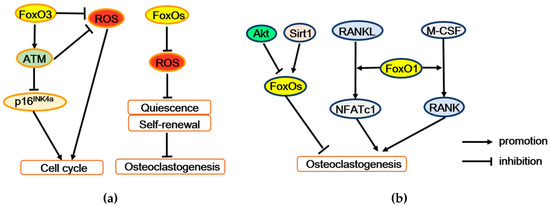
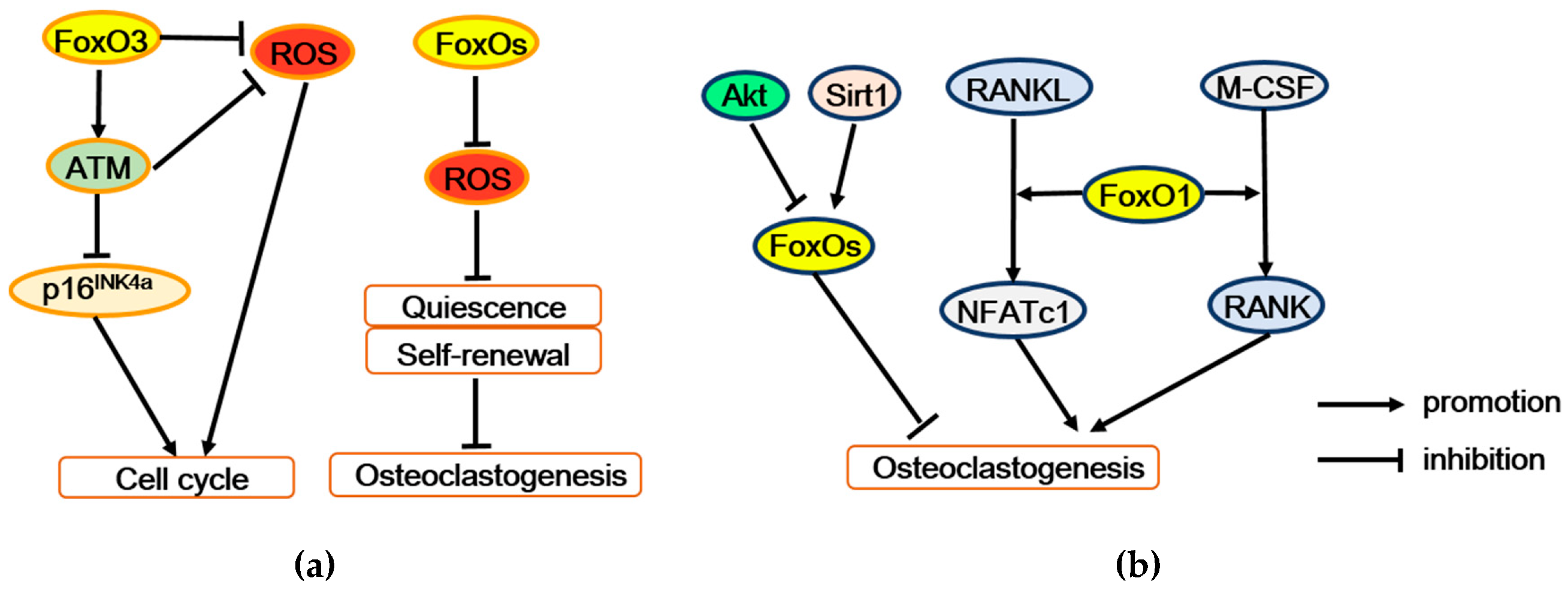
Figure 4.
FoxOs regulate osteoclastogenesis and bone resorption. (a) FoxOs promote survival and self-renewal of hematopoietic stem cells by enhancing the expression of antioxidant enzymes. Moreover, FoxO3 also regulates expression of the ATM tumor suppressor to decrease ROS level and p16INK4a expression, thereby maintaining cell cycling and self-renewal. (b) FoxO1 stimulates osteoclastogenesis may by mediating the effect of M-CSF or RANKL on osteoclast precursors. But conversely, FoxOs restrain osteoclastogenesis, which mediated the regulation of Akt or Sirt1. ATM = ataxia telangiectasia mutated, p16INK4a = p16 (also known as multiple tumor suppressor 1), Akt = Protein kinase B, Sirt1 = Sirtuin 1, RANK = receptor activator of the NF-κB, RANKL = receptor activator of the NF-κB ligand, M-CSF = macrophage colony-stimulating factor 1, NFATc1 = nuclear factor of activated T cells 1.

Table 3.
Bone metabolism regulatory functions of FoxO transcription factors in hematopoietic stem cell.
3.4. FoxOs Regulate Osteoclastogenesis
3.4.1. FoxOs Activate Osteoclastogenesis
Complex osteoclastogenesis is governed by macrophage colony-stimulating factor 1 (M-CSF) and receptor activator of the NF-κB (RANK) ligand (RANKL), as well as other cytokines secreted by osteoblasts and osteocytes that control various steps of the osteoclast differentiation process, including precursor proliferation, commitment, differentiation, and maturation [49]. In osteoclast precursors, M-CSF can stimulate expression of RANK, which promotes osteoclast differentiation [50]. FoxO1 deletion in bone marrow macrophages (BMMs) decreases M-CSF-induced RANK expression and migration of osteoclast precursors [51]. RANKL indirectly induces nuclear factor of activated T cells 1 (NFATc1) of osteoclast precursors, dramatically facilitating osteoclast differentiation [52]. FoxO1 deletion in osteoclast precursors reduces expression and nuclear localization of NFATc1 [51]. In line with this, conditional deletion of FoxO1, FoxO3, and FoxO4 in 3-month-old mice reduces the expression of osteoclastogenesis related factors like the calcitonin receptor, tartrate-resistant acid phosphatase (TRAP), and cathepsin K [26]. Therefore, FoxOs should activate osteoclastogenesis by mediating the effect of M-CSF or RANKL on osteoclast precursors (Figure 4b).
3.4.2. FoxOs Suppress Osteoclastogenesis
Other researchers have demonstrated that FoxOs negatively mediate RANKL-induced osteoclast formation. Overexpressing FoxO3 in monocyte/macrophage lineage cells decreases several resorption markers and increases bone mass [21]. The phosphoinositide 3-kinase (PI3K)/Akt pathway is the main negative regulator of FoxOs activity [6]. In osteoclast precursors, RANKL activates Akt signaling (like Akt/PI3K signaling) to promote proliferation, differentiation, and also attenuate apoptosis [50,53,54]. Along with these evidence, the deletion of FoxO1, FoxO3, and FoxO4 or overexpression of FoxO3 or mitochondria-targeted catalase in osteoclast precursors further elucidate that FoxOs upregulate the H2O2-inactivating enzyme catalase, and attenuates H2O2 accumulation, partly restraining the RANKL–Akt mediated osteoclastogenesis action [55]. Besides, Tan et al. have indicated that FoxO1 antagonizes ROS generation and thereby represses Akt, mitogen-activated protein kinases (MAPKs), and nuclear factor kappa-B (NF-κB), including the activated pathways of RANKL-induced osteoclastogenesis [56]. What is more, the inhibitory effect of FoxO1 on osteoclastogenesis is partially mediated by suppression of transcription factor MYC [57], which has been shown to reduce ROS production by inhibiting mitochondrial function [4,58,59]. Taken together, phosphorylation and inhibition of FoxO activity might partially mediate the positive effects of RANKL-induced Akt in osteoclast precursors (Figure 4b).
Beyond that, RANKL also prevents FoxO activity via acetylation. But Sirtuin 1 (Sirt1) deacetylates FoxO1, FoxO3, and FoxO4, and thereby stimulates FoxO-mediated transcription of catalase and hemeoxygenase-1 (HO-1). In osteoclast precursors, HO-1 attenuates mitochondrial oxidative phosphorylation and ATP production [60] and, along with catalase, reduce H2O2 levels. Both low ATP and H2O2 levels repress osteoclastogenesis [61]. Therefore, Sirt1 contributes to the anti-osteoclastogenic effects of FoxOs (Figure 4b). Similarly, mammalian sterile 20-like (Mst) kinase 1 (Mst1) is also a FoxO activating kinase and might facilitate osteoclast apoptosis via activation of FoxO-mediated transcription. When osteoclasts apoptosis is induced by serum withdrawal, staurosporine or bisphosphonates, Mst1 is identified as a key intermediate [62]. In detail, these apoptosis-inducing treatments promote the caspase cleavage of Mst1 into a positive 34 kDa species that keeps the catalytic domain and conduces to phosphorylate FoxOs [63]. However, it remains unknown whether FoxOs mediate the actions of Mst1 on osteoclast generation and survival.
3.4.3. FoxOs, Expressed in Osteoblasts, Indirectly Repress Osteoclastogenesis
Unexpectedly, FoxOs also repress osteoclastogenesis indirectly via osteoblasts. In mice, conditional deletion of FoxO1 or overexpression of FoxO3 in osteoblasts results in increased or reduced osteoclast numbers, respectively [26]. Hence, activated FoxOs in osteoblastic cells also reduce osteoclast numbers. Osteoprotegerin (OPG), largely produced by osteoblasts, can bind and neutralize RANKL and block osteoclast formation in vitro and bone resorption in vivo [64]. Targeted deletion of FoxO1 in osteoblasts in mice diminishes OPG expression in bone. And FoxO1 activated or not in osteoblasts, in vitro, increases or decreases OPG, respectively [42]. Therefore, FoxO1 may control osteoclast numbers by increasing the OPG expression of osteoblasts. However, mice with overexpressed FoxO3 in osteoblasts does not exhibit any altering OPG levels in bone, hence indicating that FoxO3 might regulate osteoclast numbers independent of OPG [26].
Above all, in osteoclastogenesis, a relatively high ROS level increases HSC apoptosis and drives HSCs out of quiescence into the cell cycle and further terminal differentiation at the expense of self-renewal. But FoxOs fight against oxidative stress and thus protect HSCs from osteoclast differentiation. Moreover, in osteoclast precursor cells, relatively high ROS level promotes osteoclastogenesis [65]. But FoxOs also attenuate osteoclastogenesis or osteoclast activity largely for their oxidative resistance role, which is activated by Sirt1 or Mst1 and suppressed by Akt. In contrast, some researchers have suggested that FoxO1 stimulates osteoclastogenesis by mediating the effects of MSC-F or RANKL on osteoclast precursors. Nevertheless, ROS level is important for FoxOs for controlling osteoclastogenesis; the majority of the literature demonstrates that FoxOs negatively regulate osteoclastogenesis (Figure 4; Table 3 and Table 4).

Table 4.
Bone metabolism regulatory functions of FoxO transcription factors in osteoclast and progenitor cells.
3.5. FoxOs Facilitate Chondrogenesis
In human and mice models, FoxO1 and FoxO3 expressions are dramatically decreased in the superficial zone of cartilage regions during joint aging. There are obvious FoxO phosphorylation and cytoplasmic localization in chondrocyte clusters of osteoarthritis (OA) cartilage [66]. Furthermore, combined deletion of FoxO1, 3, and 4 in mice growth plate chondrocytes (Col2Cre-FoxO1, 3, and 4 triple knockout; Col2Cre-TKO) and Col2Cre-FoxO1 single knockout results in the overall body and tail length increasing, the articular cartilage becoming thicker in young mice, and severe skeletal deformities occurring at older stages, such as hyperkyphosis or OA-like changes in cartilage, synovium, and subchondral bone. These abnormalities are associated with the distinctly elongated hypertrophic zone of the growth plate or the increased height of the proliferative zone of the proximal tibial growth plate [67,68]. Noticeably, FoxOs are indispensable for the growth, development, and health of cartilage (Table 5).

Table 5.
Bone metabolism regulatory functions of FoxO transcription factors in chondrocytes.
Chondrogenesis begins with the mesenchymal cells differentiating to chondrocytes, which then proliferate, hypertrophically differentiate, and terminally be apoptotic. FoxO3A expression is upregulated during chondrogenesis of human primary MSCs, blocking the hypertrophic differentiation and promoting apoptosis [69]. In human chondrocytes, downregulation of FoxO transcription factors with siFoxO1 and siFoxO3 reduces cell viability in response to ROS. The reduced antioxidant proteins, such as the ROS scavengers, GPX-1, and catalase, lead to an increase in intracellular oxidative stress. And the simultaneously raised autophagy-related proteins, such as microtubule-associated protein 1A/1B-light chain 3 (LC3) and Beclin1, give rise to ROS-induced apoptosis [70]. Furthermore, similar to the strongly reduced sirtuin 1 (SIRT-1) expression in OA cartilage [71], FoxO1 or FoxO1+3 (FoxO1 and FoxO3) downregulation also suppresses SIRT-1; this regulates oxidative stress and autophagy by modulating the signaling of FoxO and p53 [72], improves human chondrocytes survival via reducing proapoptotic protein tyrosine phosphatase 1B (PTP1B) [73], and also manages autophagy directly via deacetylation of key components (such as ATG5, 7, and 8) in the autophagy induction network [74]. Besides, the decreased SIRT-1, in response to siFoxO, brought about reduced FoxO transcriptional activity [70]. Collectively, in response to ROS in chondrocytes, FoxOs mediate antioxidant and autophagy proteins through altering the SIRT-1 level to some extent (Figure 5).
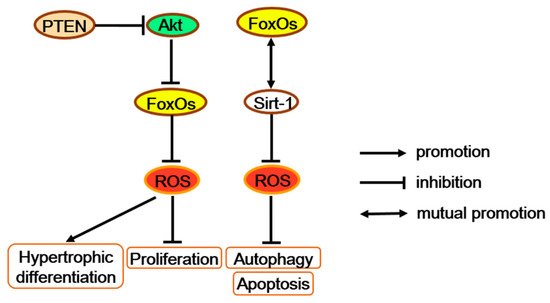
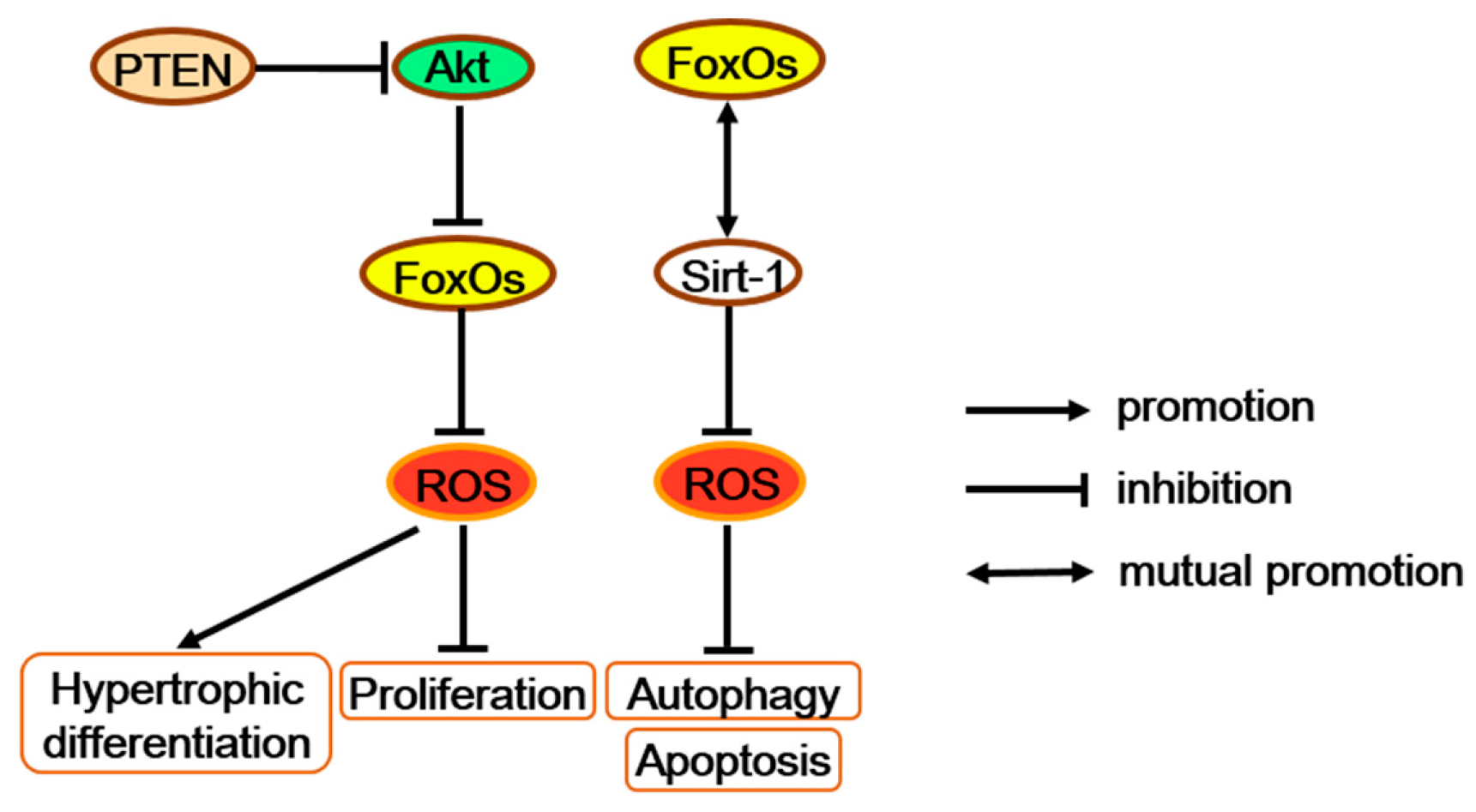
Figure 5.
FoxOs increase antioxidants and further regulate multiple functions of chondrocytes. FoxOs possibly promote proliferation and block hypertrophic differentiation along with the PTEN/Akt/FoxO signaling. FoxOs also repress apoptosis and autophagy with altering the SIRT-1 protein level. PTEN = phosphatase and tensin homolog deleted on chromosome ten.
Combined deletion of FoxO1, 3, and 4 in mice growth plate chondrocytes (chondrocyte triple knock-out; CTKO) significantly reduces expression levels of oxidative stress defense genes, such as Gpx3, Sepp1, and Sesn3, in the primary chondrocytes, which may elevate the levels of ROS and promote more proliferated chondrocytes turning to pre-hypertrophic and hypertrophic chondrocytes [67,75]. Interestingly, chondrocyte-specific inactivation of the tumor suppressor gene phosphatase and tensin homolog deleted on chromosome ten (PTEN) also generated a disorganized neonatal growth plate, hyperkyphosis, and an increased total body length, which is similar to the phenotype of FoxOs CTKO mice [76]. Besides, PTEN can act as the upstream of FoxOs by regulating Akt activation [24]. Moreover, the Akt/FoxO pathway enhances chondrocyte proliferation but decreases chondrocyte maturation and cartilage matrix production [77]. Therefore, as antioxidant factors in chondrocytes, FoxOs may promote proliferation and block hypertrophic differentiation through the signaling along the PTEN/Akt/FoxO axis (Figure 5).
In addition, FoxO1 downregulation in chondrocytes also dramatically upregulates the expression of the inflammatory factor, chemerin, which increases chondrocyte apoptosis to some degree [70]. Through transcription factor FoxO1, pro-inflammatory cytokines TNF-α activates chondrocyte apoptosis and accelerates the loss of cartilage in diabetic fracture healing [78,79]. Moreover, TNF-α promote the chemokine expression of chondrocytes in diabetic fracture healing [80]. Hence, FoxO1 may protect diabetic fracture healing from an inflammation effect through TNF-α/chemokine.
In short, by decreasing ROS accumulation during chondrogenesis, FoxOs promote proliferation and block hypertrophic differentiation along with the PTEN/Akt/FoxO signaling, as well as repress apoptosis and autophagy by altering the Sirt1 protein level. All of these demonstrate that FoxOs contribute to relieving osteoarthritis and improve the growth, development, and health of cartilage.
4. Conclusion and Perspective
In summary, through FoxO deletion or overexpression in early osteoblast progenitors, osteoblasts, hematopoietic stem cells, osteoclast precursors, or chondrocytes, these sufficient and seminal papers have revealed the complicated and prominent roles of FoxO transcription factors not only in regulating bone cells function, but also in controlling the bone modeling or remodeling and preserving skeletal homeostasis.
In particular, FoxOs regulate bone cell survival, cell cycle, proliferation, differentiation, autophagy, even apoptosis, and also participate in network control among different kinds of bone cells (Figure 6). They are intimately involved in many bone physiological and pathological activities, such as bone development, bone remodeling, energy metabolism, aging, osteoarthritis, low bone mass in type 1 diabetes, postmenopausal osteoporosis, age-related bone loss, and myeloid leukemia. For example, FoxOs improve expression of antioxidant enzymes and maintain relatively low ROS levels to protect MSCs/HSCs from osteogenesis/osteoclastogenesis. During differentiation, ROS levels are increased due to enhanced cell metabolism. FoxOs promote osteogenesis and inhibit adipogenesis or osteoclastogenesis by decreasing ROS levels. Moreover, age-related mechanisms intrinsic to bone and oxidative stress are more proven as the protagonist of osteoporosis (including postmenopausal osteoporosis) [81]. Therefore, in bone, all the three FoxO members (FoxO1, 3, and 4) jointly mediate the redox balance of these bone cells, thereby regulating bone metabolism and also involved in osteoporosis pathology. Above all, these roles of FoxOs may help in the development of pharmacological agents, targeting FoxOs to further elucidate the treatment of bone metabolism diseases.
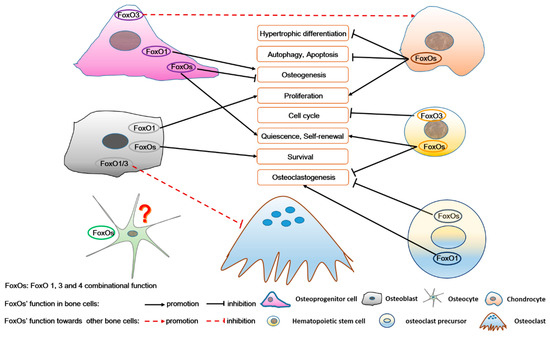
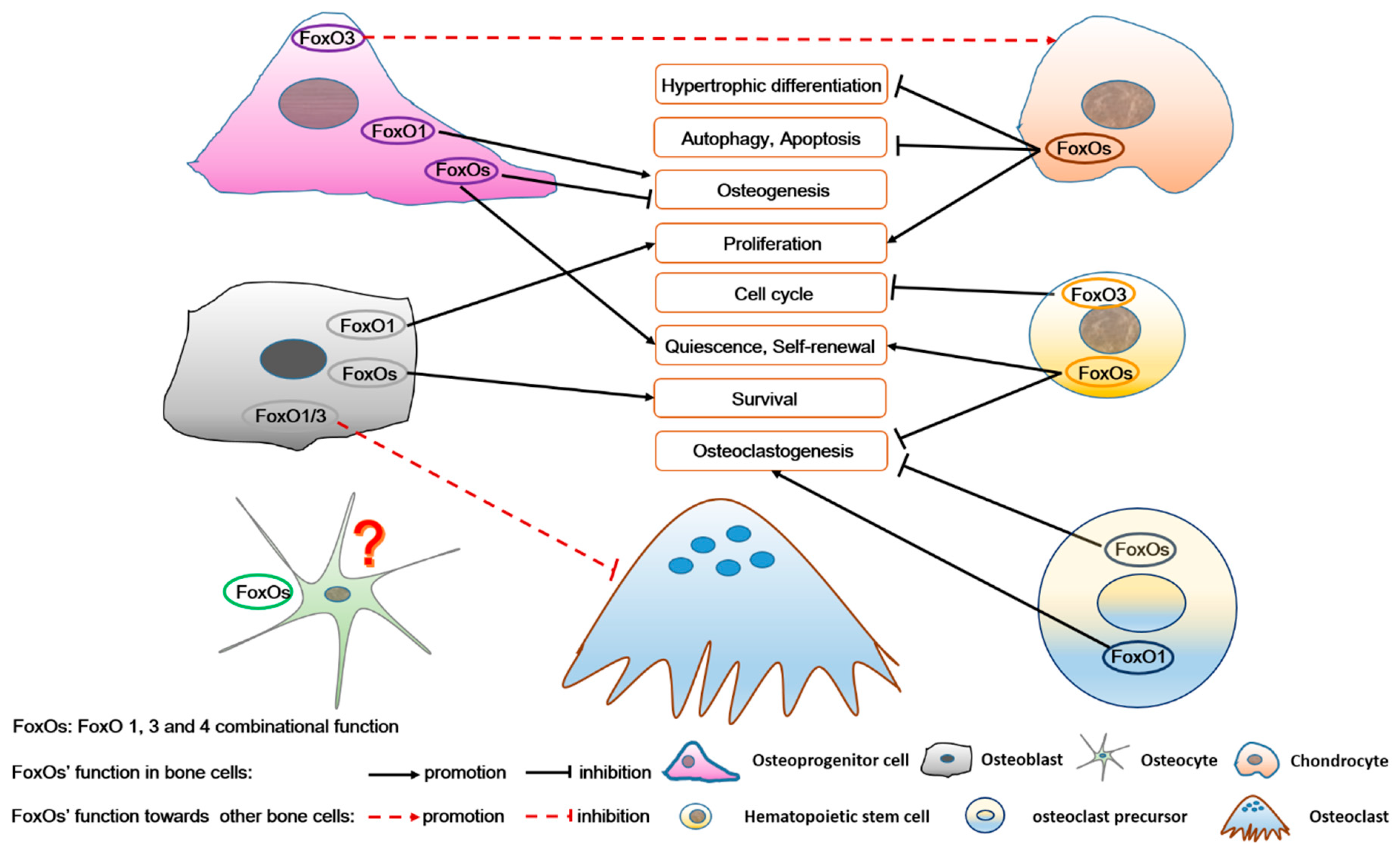
Figure 6.
The role of FoxOs in osteoprogenitor cells, chondrocytes, osteoblasts, osteocytes, hematopoietic stem cells, and osteoclast precursors, as well as in the complex intercellular signaling among these bone cells. FoxOs not only regulate survival, self-renewal, cell cycle, proliferation, differentiation, autophagy, and apoptosis of bone cells, but also participate in the interaction among these bone cells. FoxO’s function in osteocytes is unknown.
Before that, several questions should be paid attention to, including but not limited to the following: Firstly, how FoxO factors balance the positive and negative regulation of bone formation? On one hand, FoxO1 facilitates bone formation by interacting with Runx2 in osteoprogenitor cells or with ATF4 in osteoblasts, while diminishing bone formation by mediating the function of M-CSF/RANKL in osteoclasts. Here, FoxO1 should be an important factor for bone formation, but not involved in redox regulation. On the other hand, FoxOs mediate the redox balance in or among bone cells, thereby regulating bone metabolism and homeostasis. Therefore, the balance role of FoxOs may depend on the bone metabolism stage and bone redox homeostasis. Secondly, what are the complete and precise mechanisms of FoxOs performing functions among the different kinds of bone cells? For example, FoxOs regulate redox balance involved in the activities of all kinds of bone cells. Then, how the FoxOs mediate ROS signaling in or among bone cells? Thirdly, how FoxOs regulate bone remodeling and preserve skeletal homeostasis during aging in bone? For example, with aging, excessive osteocyte apoptosis correlates with oxidative stress and favors osteoclastogenesis, which leads to increased bone remodeling imbalance and bone loss. This results in the occurrence of several bone diseases, especially osteoporosis. Then, does FoxOs inhibit osteocyte apoptosis by regulating an anti-oxidation system in osteocytes? And what is the mechanism involved therein? Is it effective to treat and prevent bone loss? In light of this evidence, more research is necessary to further illuminate the precise role of FoxOs, especially for the development of new therapeutic approaches.
Author Contributions
X.M. and Y.T. conceived the concept of this manuscript and wrote the first draft version of it, which was then equally developed by P.S., C.Y., X.L., X.W., Y.G., S.P., A.R.W., A.Q. (Abdul Qadir) and A.Q. (Airong Qian) revised and proofread the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (nos. No.81801871) and China Postdoctoral Science Foundation (nos. No. 2017M613210).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| FoxOs | Forkhead box class O family member proteins |
| LPS | Lipopolysaccharide |
| TNF-α | Tumor necrosis factor α |
| MSCs | Mesenchymal stem cells |
| Runx2 | Runt-related transcription factor 2 |
| ALP | Alkaline phosphatase |
| OCN | Osteocalcin |
| CFU | Colony forming unit |
| PPARγ | Peroxisome proliferator-activated receptor γ |
| PPREs | PPAR-response elements |
| PCNA | Proliferating cell nuclear antigen |
| ROS | Reactive oxygen species |
| FABP4 | Fatty acid-binding protein 4 |
| 4-HNE | 4-hydroxynonenal |
| ATF4 | Activating transcription factor 4 |
| BFR | Bone formation rate |
| HSC | Hematopoietic stem cell |
| Gpx-1 | Glutathione peroxidase 1 |
| M-CSF | Macrophage-colony stimulating factor |
| RANK | Receptor activator of NF-κ B |
| RANKL | Receptor activator of NF-κ B ligand |
| BMMCs | Bone marrow mononuclear cells |
| NFATc1 | Nuclear factor of activated T cells 1 |
| Akt | Protein kinase B |
| SIRT-1 | Sirtuin 1 |
| TRAP | Tartrate-resistant acid phosphatase |
| MAPKs | Mitogen-activated protein kinases |
| NF-κB | Nuclear factor kappa-B |
| HO-1 | Hemeoxygenase-1 |
| Mst 1 | Mammalian Sterile 20-like kinase 1 |
| OPG | Osteoprotegerin |
| AP-1 | Activator protein 1 |
| PI3K | phosphoinositide 3-kinase |
| OA | Osteoarthritis |
| PTP1B | Protein tyrosine phosphatase 1B |
| CTKO | Chondrocyte triple knock-out |
| PTEN | Phosphatase and tensin homolog deleted on chromosome ten |
References
- Carter, M.E.; Brunet, A. FOXO transcription factors. Curr. Biol. 2007, 17, R113–R114. [Google Scholar] [CrossRef] [PubMed]
- Avila-Flores, A.; Arranz-Nicolas, J.; Merida, I. Transcriptional Activity of FOXO Transcription Factors Measured by Luciferase Assays. In FOXO Transcription Factors; Springer: Berlin, Germany, 2019; pp. 91–102. [Google Scholar]
- Genin, E.C.; Caron, N.; Vandenbosch, R.; Nguyen, L.; Malgrange, B. Concise review: Forkhead pathway in the control of adult neurogenesis. Stem Cells 2014, 32, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.S.; Binderup, T.; Jensen, K.T.; Therkelsen, I.; Borup, R.; Nilsson, E.; Multhaupt, H.; Bouchard, C.; Quistorff, B.; Kjær, A. FoxO3A promotes metabolic adaptation to hypoxia by antagonizing Myc function. EMBO J. 2011, 30, 4554–4570. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, G.; Kaestner, K.H. SnapShot: Forkhead transcription factors I. Cell 2007, 130, 1160. [Google Scholar] [CrossRef]
- Calnan, D.; Brunet, A. The foxo code. Oncogene 2008, 27, 2276. [Google Scholar] [CrossRef]
- Henderson, S.T.; Johnson, T.E. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr. Biol. 2001, 11, 1975–1980. [Google Scholar] [CrossRef]
- Webb, A.E.; Brunet, A. FOXO transcription factors: Key regulators of cellular quality control. Trends Biochem. Sci. 2014, 39, 159–169. [Google Scholar] [CrossRef]
- Eijkelenboom, A.; Burgering, B.M. FOXOs: Signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 2013, 14, 83. [Google Scholar] [CrossRef]
- Kim, H.-N.; Iyer, S.; Ring, R.; Almeida, M. The role of FoxOs in bone health and disease. In Current Topics in Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2018; Volume 127, pp. 149–163. [Google Scholar]
- Bullock, M. FOXO factors and breast cancer: Outfoxing endocrine resistance. Endocr. Relat. Cancer 2016, 23, R113–R130. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, Y.; Graves, D.T. FOXO transcription factors: Their clinical significance and regulation. BioMed Res. Int. 2014, 2014, 925350. [Google Scholar] [CrossRef]
- De Brachène, A.C.; Demoulin, J.-B. FOXO transcription factors in cancer development and therapy. Cell. Mol. Life Sci. 2016, 73, 1159–1172. [Google Scholar] [CrossRef] [PubMed]
- Tsitsipatis, D.; Klotz, L.-O.; Steinbrenner, H. Multifaceted functions of the forkhead box transcription factors FoxO1 and FoxO3 in skin. Biochim. Biophys. Acta. Gen. Subj. 2017, 1861, 1057–1064. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; da Fontoura, C.S.; Moreno, M.; Holton, N.E.; Sweat, M.; Sweat, Y.; Lee, M.K.; Arbon, J.; Bidlack, F.B.; Thedens, D.R. FoxO6 regulates Hippo signaling and growth of the craniofacial complex. PLoS Genet. 2018, 14, e1007675. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Gong, Y.; Xu, L.; Zhou, M.; Li, J.; Song, J. Bidirectional regulation of osteogenic differentiation by the FOXO subfamily of Forkhead transcription factors in mammalian MSCs. Cell Prolif. 2019, 52, e12540. [Google Scholar] [CrossRef]
- Fu, Z.; Tindall, D. FOXOs, cancer and regulation of apoptosis. Oncogene 2008, 27, 2312. [Google Scholar] [CrossRef]
- Iyer, S.; Ambrogini, E.; Bartell, S.M.; Han, L.; Roberson, P.K.; de Cabo, R.; Jilka, R.L.; Weinstein, R.S.; O’Brien, C.A.; Manolagas, S.C. FOXOs attenuate bone formation by suppressing Wnt signaling. J. Clin. Investig. 2013, 123, 3409–3419. [Google Scholar] [CrossRef]
- Brent, M.M.; Anand, R.; Marmorstein, R. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure 2008, 16, 1407–1416. [Google Scholar] [CrossRef]
- Murtaza, G.; Khan, A.K.; Rashid, R.; Muneer, S.; Hasan, S.M.F.; Chen, J. FOXO transcriptional factors and long-term living. Oxid. Med. Cell. Longev. 2017, 2017, 3494289. [Google Scholar] [CrossRef]
- Almeida, M. Unraveling the role of FoxOs in bone—insights from mouse models. Bone 2011, 49, 319–327. [Google Scholar] [CrossRef]
- Teixeira, C.C.; Liu, Y.; Thant, L.M.; Pang, J.; Palmer, G.; Alikhani, M. Foxo1, a novel regulator of osteoblast differentiation and skeletogenesis. J. Biol. Chem. 2010, 285, 31055–31065. [Google Scholar] [CrossRef]
- Komori, T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010, 339, 189. [Google Scholar] [CrossRef] [PubMed]
- Van der Horst, A.; Burgering, B.M. Stressing the role of FoxO proteins in lifespan and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 440. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, M.F.; Flowers, S.; Bhattacharya, R.; Faibish, D.; Behl, Y.; Kotton, D.N.; Gerstenfeld, L.; Moran, E.; Graves, D.T. FOXO1 modulates osteoblast differentiation. Bone 2011, 48, 1043–1051. [Google Scholar] [CrossRef] [PubMed]
- Ambrogini, E.; Almeida, M.; Martin-Millan, M.; Paik, J.-H.; DePinho, R.A.; Han, L.; Goellner, J.; Weinstein, R.S.; Jilka, R.L.; O’Brien, C.A. FoxO-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metab. 2010, 11, 136–146. [Google Scholar] [CrossRef]
- Klotz, L.-O.; Steinbrenner, H. Cellular adaptation to xenobiotics: Interplay between xenosensors, reactive oxygen species and FOXO transcription factors. Redox Biol. 2017, 13, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Armoni, M.; Harel, C.; Karni, S.; Chen, H.; Bar-Yoseph, F.; Ver, M.R.; Quon, M.J.; Karnieli, E. FOXO1 represses peroxisome proliferator-activated receptor-gamma1 and -gamma2 gene promoters in primary adipocytes. A novel paradigm to increase insulin sensitivity. J. Biol. Chem. 2006, 281, 19881–19891. [Google Scholar] [CrossRef]
- Fan, W.; Imamura, T.; Sonoda, N.; Sears, D.D.; Patsouris, D.; Kim, J.J.; Olefsky, J.M. FOXO1 transrepresses peroxisome proliferator-activated receptor γ transactivation, coordinating an insulin-induced feed-forward response in adipocytes. J. Biol. Chem. 2009, 284, 12188–12197. [Google Scholar] [CrossRef]
- Muller, F.L.; Lustgarten, M.S.; Jang, Y.; Richardson, A.; Van Remmen, H. Trends in oxidative aging theories. Free Radic. Biol. Med. 2007, 43, 477–503. [Google Scholar] [CrossRef]
- Almeida, M.; Han, L.; Martin-Millan, M.; O’Brien, C.A.; Manolagas, S.C. Oxidative stress antagonizes Wnt signaling in osteoblast precursors by diverting β-catenin from T cell factor-to forkhead box O-mediated transcription. J. Biol. Chem. 2007, 282, 27298–27305. [Google Scholar] [CrossRef]
- Almeida, M.; Han, L.; Martin-Millan, M.; Plotkin, L.I.; Stewart, S.A.; Roberson, P.K.; Kousteni, S.; O’Brien, C.A.; Bellido, T.; Parfitt, A.M. Skeletal involution by age-associated oxidative stress and its acceleration by loss of sex steroids. J. Biol. Chem. 2007, 282, 27285–27297. [Google Scholar] [CrossRef]
- Bennett, C.N.; Longo, K.A.; Wright, W.S.; Suva, L.J.; Lane, T.F.; Hankenson, K.D.; MacDougald, O.A. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc. Natl. Acad. Sci. 2005, 102, 3324–3329. [Google Scholar] [CrossRef] [PubMed]
- Babij, P.; Zhao, W.; Small, C.; Kharode, Y.; Yaworsky, P.J.; Bouxsein, M.L.; Reddy, P.S.; Bodine, P.V.; Robinson, J.A.; Bhat, B. High bone mass in mice expressing a mutant LRP5 gene. J. Bone Miner. Res. 2003, 18, 960–974. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; Porter, N.A.; Brash, A.R. Routes to 4-hydroxynonenal: Fundamental issues in the mechanisms of lipid peroxidation. J. Biol. Chem. 2008, 283, 15539–15543. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.; Ambrogini, E.; Han, L.; Manolagas, S.C.; Jilka, R.L. Increased lipid oxidation causes oxidative stress, increased peroxisome proliferator-activated receptor-γ expression, and diminished pro-osteogenic Wnt signaling in the skeleton. J. Biol. Chem. 2009, 284, 27438–27448. [Google Scholar] [CrossRef]
- Kang, S.; Bennett, C.N.; Gerin, I.; Rapp, L.A.; Hankenson, K.D.; MacDougald, O.A. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein α and peroxisome proliferator-activated receptor γ. J. Biol. Chem. 2007, 282, 14515–14524. [Google Scholar] [CrossRef]
- Okamura, M.; Kudo, H.; Wakabayashi, K.-i.; Tanaka, T.; Nonaka, A.; Uchida, A.; Tsutsumi, S.; Sakakibara, I.; Naito, M.; Osborne, T.F. COUP-TFII acts downstream of Wnt/β-catenin signal to silence PPARγ gene expression and repress adipogenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 5819–5824. [Google Scholar] [CrossRef]
- Sharma, C.; Pradeep, A.; Wong, L.; Rana, A.; Rana, B. Peroxisome proliferator-activated receptor γ activation can regulate β-catenin levels via a proteasome-mediated and adenomatous polyposis coli-independent pathway. J. Biol. Chem. 2004, 279, 35583–35594. [Google Scholar] [CrossRef]
- Atashi, F.; Modarressi, A.; Pepper, M.S. The role of reactive oxygen species in mesenchymal stem cell adipogenic and osteogenic differentiation: A review. Stem Cells Dev. 2015, 24, 1150–1163. [Google Scholar] [CrossRef]
- Rached, M.-T.; Kode, A.; Xu, L.; Yoshikawa, Y.; Paik, J.-H.; DePinho, R.A.; Kousteni, S. FoxO1 is a positive regulator of bone formation by favoring protein synthesis and resistance to oxidative stress in osteoblasts. Cell Metab. 2010, 11, 147–160. [Google Scholar] [CrossRef]
- Clemens, T.L.; Karsenty, G. The osteoblast: An insulin target cell controlling glucose homeostasis. J. Bone Miner. Res. 2011, 26, 677–680. [Google Scholar] [CrossRef]
- Yang, S.; Xu, H.; Yu, S.; Cao, H.; Fan, J.; Ge, C.; Fransceschi, R.T.; Dong, H.H.; Xiao, G. Foxo1 mediates insulin-like growth factor 1 (IGF1)/insulin regulation of osteocalcin expression by antagonizing Runx2 in osteoblasts. J. Biol. Chem. 2011, 286, 19149–19158. [Google Scholar] [CrossRef] [PubMed]
- Rached, M.-T.; Kode, A.; Silva, B.C.; Jung, D.Y.; Gray, S.; Ong, H.; Paik, J.-H.; DePinho, R.A.; Kim, J.K.; Karsenty, G. FoxO1 expression in osteoblasts regulates glucose homeostasis through regulation of osteocalcin in mice. J. Clin. Investig. 2010, 120, 357–368. [Google Scholar] [CrossRef]
- Behl, Y.; Siqueira, M.; Ortiz, J.; Li, J.; Desta, T.; Faibish, D.; Graves, D.T. Activation of the acquired immune response reduces coupled bone formation in response to a periodontal pathogen. J. Immunol. 2008, 181, 8711–8718. [Google Scholar] [CrossRef]
- Bigarella, C.L.; Li, J.; Rimmelé, P.; Liang, R.; Sobol, R.W.; Ghaffari, S. FOXO3 transcription factor is essential for protecting hematopoietic stem and progenitor cells from oxidative DNA damage. J. Biol. Chem. 2017, 292, 3005–3015. [Google Scholar] [CrossRef] [PubMed]
- Yalcin, S.; Zhang, X.; Luciano, J.P.; Mungamuri, S.K.; Marinkovic, D.; Vercherat, C.; Sarkar, A.; Grisotto, M.; Taneja, R.; Ghaffari, S. Foxo3 is essential for the regulation of ataxia telangiectasia mutated and oxidative stress-mediated homeostasis of hematopoietic stem cells. J. Biol. Chem. 2008, 283, 25692–25705. [Google Scholar] [CrossRef] [PubMed]
- Tothova, Z.; Kollipara, R.; Huntly, B.J.; Lee, B.H.; Castrillon, D.H.; Cullen, D.E.; McDowell, E.P.; Lazo-Kallanian, S.; Williams, I.R.; Sears, C. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 2007, 128, 325–339. [Google Scholar] [CrossRef]
- Buck, D.W.; Dumanian, G.A. Bone biology and physiology: Part I. The fundamentals. Plast. Reconstr. Surg. 2012, 129, 1314–1320. [Google Scholar] [CrossRef]
- Boyce, B. Advances in the regulation of osteoclasts and osteoclast functions. J. Dent. Res. 2013, 92, 860–867. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, G.; Jeon, H.H.; Elazizi, M.; La, L.B.; Hameedaldeen, A.; Xiao, E.; Tian, C.; Alsadun, S.; Choi, Y. FOXO1 mediates RANKL-induced osteoclast formation and activity. J. Immunol. 2015, 194, 2878–2887. [Google Scholar] [CrossRef]
- Nakashima, T.; Hayashi, M.; Takayanagi, H. New insights into osteoclastogenic signaling mechanisms. Trends Endocrinol. Metab. 2012, 23, 582–590. [Google Scholar] [CrossRef]
- Sugatani, T.; Hruska, K.A. Akt1/Akt2 and mammalian target of rapamycin/Bim play critical roles in osteoclast differentiation and survival, respectively, whereas Akt is dispensable for cell survival in isolated osteoclast precursors. J. Biol. Chem. 2005, 280, 3583–3589. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, N.; Kugimiya, F.; Oshima, Y.; Ohba, S.; Ikeda, T.; Saito, T.; Shinoda, Y.; Kawasaki, Y.; Ogata, N.; Hoshi, K. Akt1 in osteoblasts and osteoclasts controls bone remodeling. PLoS ONE 2007, 2, e1058. [Google Scholar] [CrossRef]
- Bartell, S.M.; Kim, H.-N.; Ambrogini, E.; Han, L.; Iyer, S.; Ucer, S.S.; Rabinovitch, P.; Jilka, R.L.; Weinstein, R.S.; Zhao, H. FoxO proteins restrain osteoclastogenesis and bone resorption by attenuating H 2 O 2 accumulation. Nat. Commun. 2014, 5, 3773. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.K.; Choi, Y.G.; Baik, J.Y.; Han, S.Y.; Jeong, D.-w.; Bae, Y.S.; Kim, N.; Lee, S.Y. A crucial role for reactive oxygen species in RANKL-induced osteoclast differentiation. Blood 2005, 106, 852–859. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Guan, H.; Xie, L.; Mi, B.; Fang, Z.; Li, J.; Li, F. FOXO1 inhibits osteoclastogenesis partially by antagnozing MYC. Sci. Rep. 2015, 5, 16835. [Google Scholar] [CrossRef]
- Ferber, E.C.; Peck, B.; Delpuech, O.; Bell, G.P.; East, P.; Schulze, A. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ. 2012, 19, 968. [Google Scholar] [CrossRef]
- Peck, B.; Ferber, E.C.; Schulze, A. Antagonism between FOXO and MYC regulates cellular powerhouse. Front. Oncol. 2013, 3, 96. [Google Scholar] [CrossRef]
- Ke, K.; Safder, M.; Sul, O.-J.; Kim, W.-K.; Suh, J.-H.; Joe, Y.; Chung, H.-T.; Choi, H.-S. Hemeoxygenase-1 maintains bone mass via attenuating a redox imbalance in osteoclast. Mol. Cell. Endocrinol. 2015, 409, 11–20. [Google Scholar] [CrossRef]
- Kim, H.-N.; Han, L.; Iyer, S.; de Cabo, R.; Zhao, H.; O’Brien, C.A.; Manolagas, S.C.; Almeida, M. Sirtuin1 suppresses osteoclastogenesis by deacetylating FoxOs. Mol. Endocrinol. 2015, 29, 1498–1509. [Google Scholar] [CrossRef]
- Reszka, A.A.; Halasy-Nagy, J.M.; Masarachia, P.J.; Rodan, G.A. Bisphosphonates act directly on the osteoclast to induce caspase cleavage of mst1 kinase during apoptosis A link between inhibition of the mevalonate pathway and regulation of an apoptosis-promoting kinase. J. Biol. Chem. 1999, 274, 34967–34973. [Google Scholar] [CrossRef]
- Jang, S.-W.; Yang, S.-J.; Srinivasan, S.; Ye, K. Akt phosphorylates MstI and prevents its proteolytic activation, blocking FOXO3 phosphorylation and nuclear translocation. J. Biol. Chem. 2007, 282, 30836–30844. [Google Scholar] [CrossRef] [PubMed]
- Boyle, W.J.; Simonet, W.S.; Lacey, D.L. Osteoclast differentiation and activation. Nature 2003, 423, 337. [Google Scholar] [CrossRef]
- Domazetovic, V.; Marcucci, G.; Iantomasi, T.; Brandi, M.L.; Vincenzini, M.T. Oxidative stress in bone remodeling: Role of antioxidants. Clin. Cases Miner. Bone Metab. 2017, 14, 209. [Google Scholar] [CrossRef] [PubMed]
- Akasaki, Y.; Hasegawa, A.; Saito, M.; Asahara, H.; Iwamoto, Y.; Lotz, M. Dysregulated FOXO transcription factors in articular cartilage in aging and osteoarthritis. Osteoarthr. Cartil. 2014, 22, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Eelen, G.; Verlinden, L.; Maes, C.; Beullens, I.; Gysemans, C.; Paik, J.-H.; DePinho, R.A.; Bouillon, R.; Carmeliet, G.; Verstuyf, A. Forkhead box O transcription factors in chondrocytes regulate endochondral bone formation. J. Steroid Biochem. Mol. Biol. 2016, 164, 337–343. [Google Scholar] [CrossRef]
- Matsuzaki, T.; Alvarez-Garcia, O.; Mokuda, S.; Nagira, K.; Olmer, M.; Gamini, R.; Miyata, K.; Akasaki, Y.; Su, A.I.; Asahara, H. FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci. Transl. Med. 2018, 10, eaan0746. [Google Scholar] [CrossRef]
- Djouad, F.; Bony, C.; Canovas, F.; Fromigue, O.; Reme, T.; Jorgensen, C.; Noel, D. Transcriptomic analysis identifies Foxo3A as a novel transcription factor regulating mesenchymal stem cell chrondrogenic differentiation. Cloning Stem Cells 2009, 11, 407–416. [Google Scholar] [CrossRef]
- Akasaki, Y.; Alvarez-Garcia, O.; Saito, M.; Caramés, B.; Iwamoto, Y.; Lotz, M.K. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 2014, 66, 3349–3358. [Google Scholar] [CrossRef]
- Fujita, N.; Matsushita, T.; Ishida, K.; Kubo, S.; Matsumoto, T.; Takayama, K.; Kurosaka, M.; Kuroda, R. Potential involvement of SIRT1 in the pathogenesis of osteoarthritis through the modulation of chondrocyte gene expressions. J. Orthop. Res. 2011, 29, 511–515. [Google Scholar] [CrossRef]
- Salminen, A.; Kaarniranta, K. SIRT1: Regulation of longevity via autophagy. Cell. Signal. 2009, 21, 1356–1360. [Google Scholar] [CrossRef]
- Gagarina, V.; Gabay, O.; Dvir-Ginzberg, M.; Lee, E.J.; Brady, J.K.; Quon, M.J.; Hall, D.J. SirT1 enhances survival of human osteoarthritic chondrocytes by repressing protein tyrosine phosphatase 1B and activating the insulin-like growth factor receptor pathway. Arthritis Rheum. 2010, 62, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Ng, F.; Tang, B.L. Sirtuins’ modulation of autophagy. J. Cell. Physiol. 2013, 228, 2262–2270. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Miyamoto, T.; Fujita, N.; Kubota, Y.; Ito, K.; Takubo, K.; Miyamoto, K.; Ninomiya, K.; Suzuki, T.; Iwasaki, R. Reactive oxygen species induce chondrocyte hypertrophy in endochondral ossification. J. Exp. Med. 2007, 204, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Ford-Hutchinson, A.F.; Ali, Z.; Lines, S.E.; Hallgrímsson, B.; Boyd, S.K.; Jirik, F.R. Inactivation of Pten in osteo-chondroprogenitor cells leads to epiphyseal growth plate abnormalities and skeletal overgrowth. J. Bone Miner. Res. 2007, 22, 1245–1259. [Google Scholar] [CrossRef] [PubMed]
- Rokutanda, S.; Fujita, T.; Kanatani, N.; Yoshida, C.A.; Komori, H.; Liu, W.; Mizuno, A.; Komori, T. Akt regulates skeletal development through GSK3, mTOR, and FoxOs. Dev. Biol. 2009, 328, 78–93. [Google Scholar] [CrossRef] [PubMed]
- Kayal, R.A.; Siqueira, M.; Alblowi, J.; McLean, J.; Krothapalli, N.; Faibish, D.; Einhorn, T.A.; Gerstenfeld, L.C.; Graves, D.T. TNF-α mediates diabetes-enhanced chondrocyte apoptosis during fracture healing and stimulates chondrocyte apoptosis Through FOXO1. J. Bone Miner. Res. 2010, 25, 1604–1615. [Google Scholar] [CrossRef] [PubMed]
- Alblowi, J.; Kayal, R.A.; Siqueria, M.; McKenzie, E.; Krothapalli, N.; McLean, J.; Conn, J.; Nikolajczyk, B.; Einhorn, T.A.; Gerstenfeld, L. High levels of tumor necrosis factor-α contribute to accelerated loss of cartilage in diabetic fracture healing. Am. J. Pathol. 2009, 175, 1574–1585. [Google Scholar] [CrossRef]
- Alblowi, J.; Tian, C.; Siqueira, M.F.; Kayal, R.A.; McKenzie, E.; Behl, Y.; Gerstenfeld, L.; Einhorn, T.A.; Graves, D.T. Chemokine expression is upregulated in chondrocytes in diabetic fracture healing. Bone 2013, 53, 294–300. [Google Scholar] [CrossRef]
- Manolagas, S.C. From estrogen-centric to aging and oxidative stress: A revised perspective of the pathogenesis of osteoporosis. Endocr. Rev. 2010, 31, 266–300. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).