An Earliest Endosymbiont, Wolbachia massiliensis sp. nov., Strain PL13 from the Bed Bug (Cimex hemipterus), Type Strain of a New Supergroup T
Abstract
:1. Introduction
2. Results
2.1. Isolation, Culture, and Description of the Bacterium
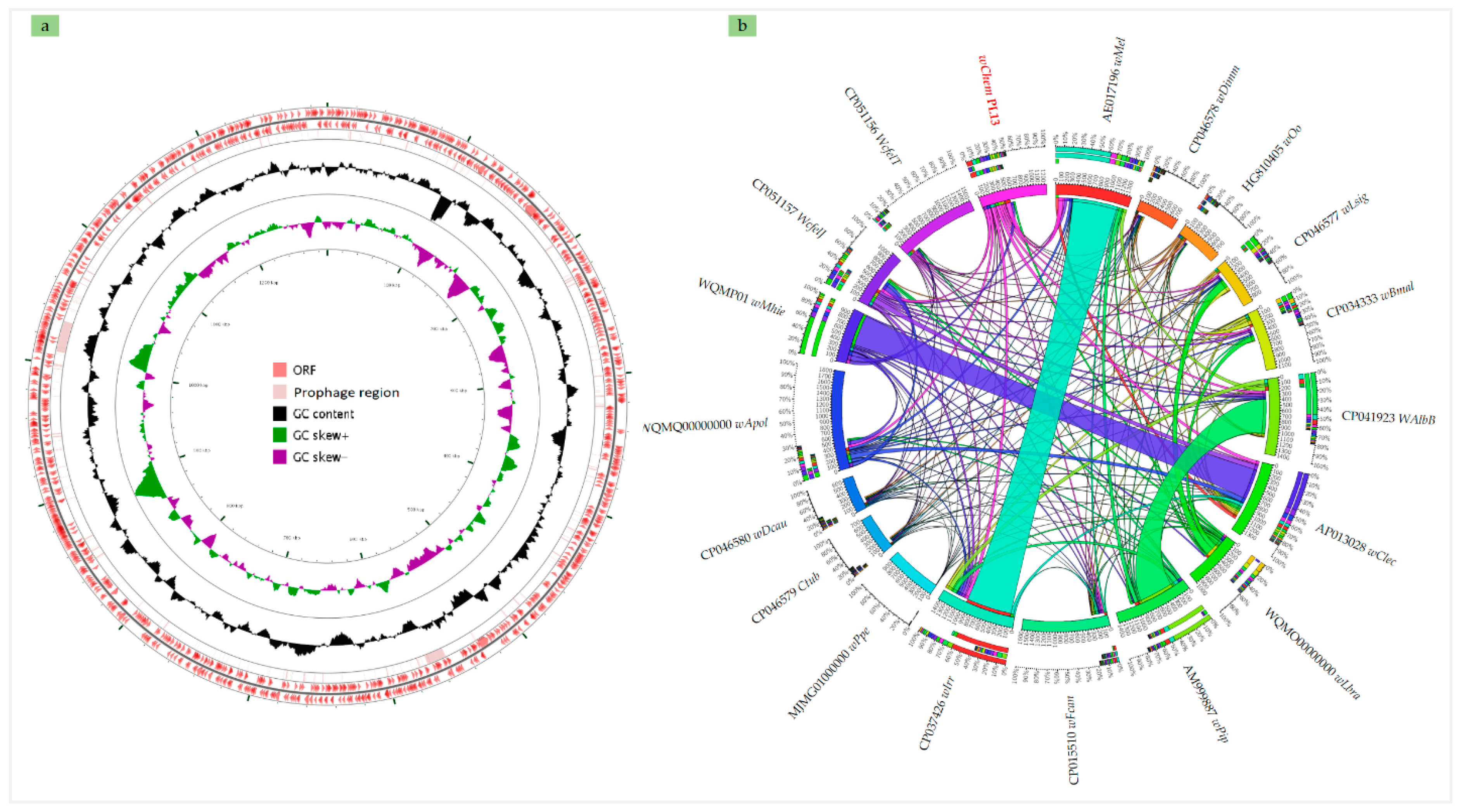
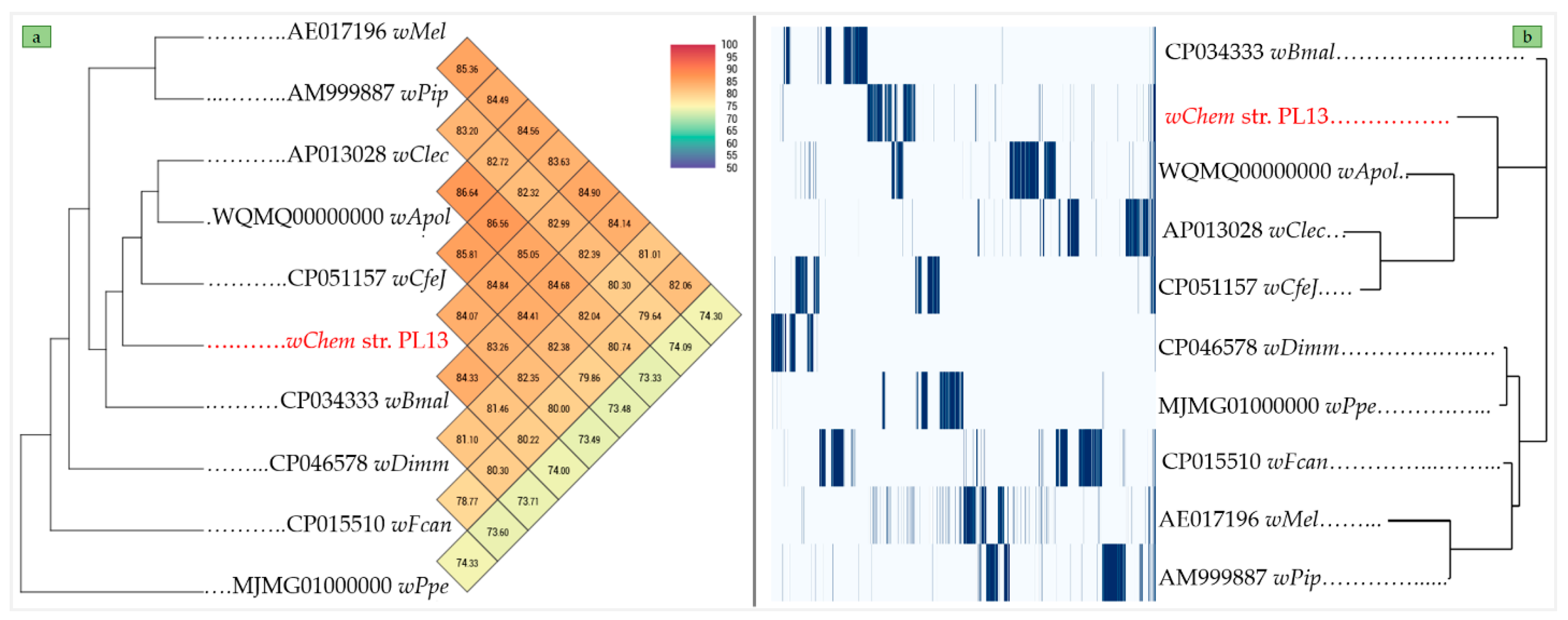
2.2. Genome Sequencing, Annotation and Genomic Comparison
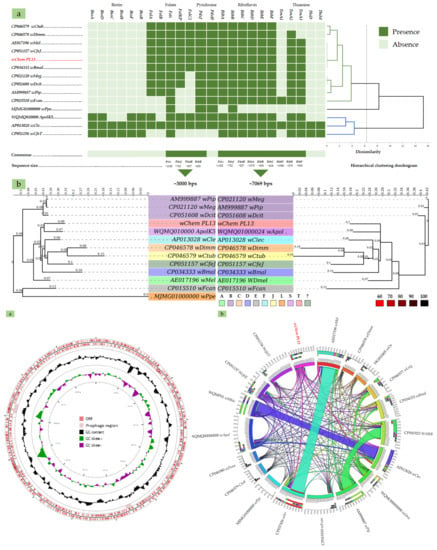
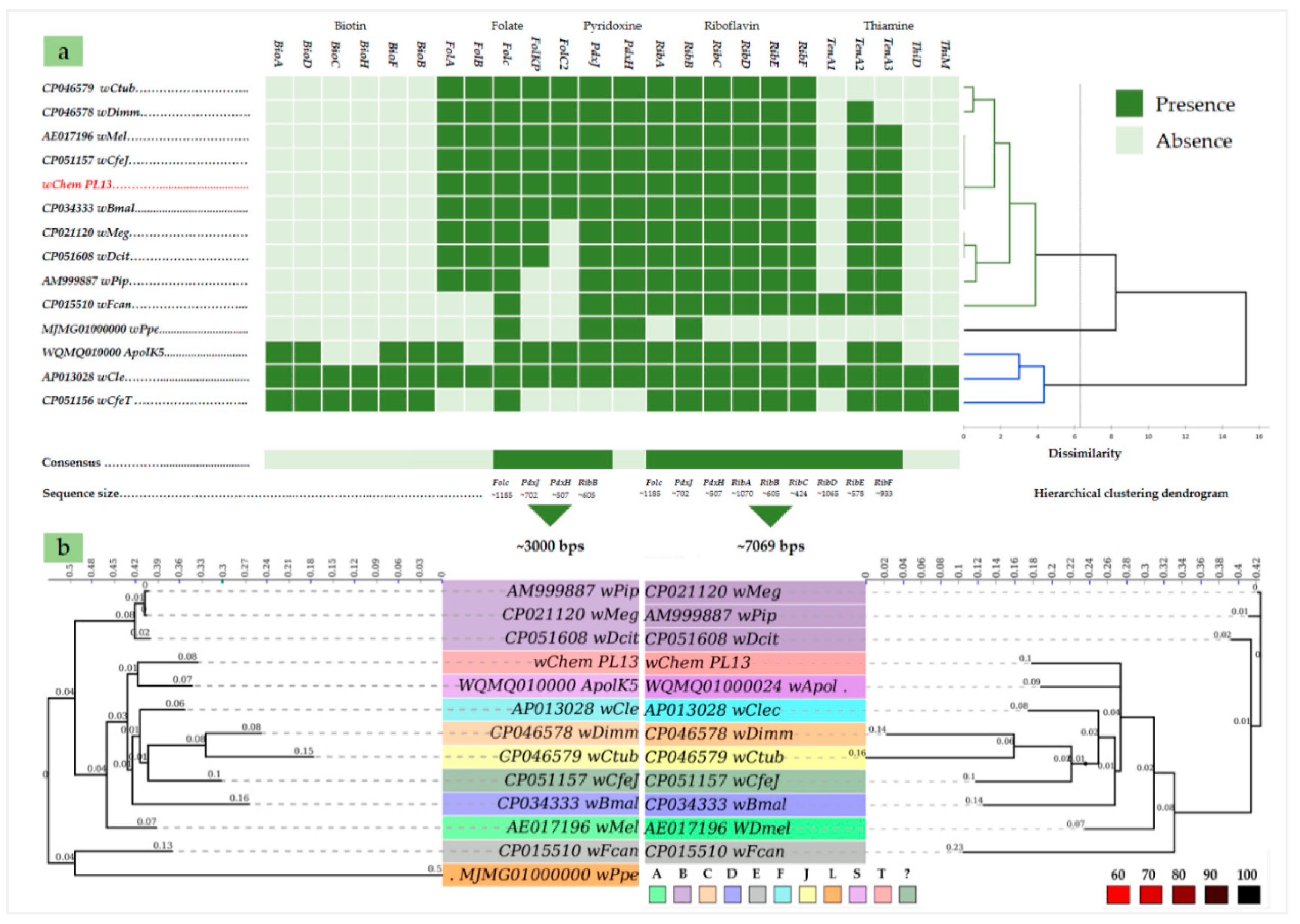
2.3. B-Vitamin Synthesis Patterns in the wChem PL13 and Other Wolbachia Genomes
2.4. Comparative Phylogenies and Placement of Wolbachia sp. Strain wChem PL13 in the Wolbachia T Supergroup
2.5. Description of Wolbachia massiliensis sp. nov.
3. Discussion
4. Materials and Methods
4.1. Source of the Bacterium, Inoculum Preparation and Isolation
4.2. Morphological Characterization and Scanning Electron Microscopy
4.3. Cell Co-Culture Standardization and Wolbachia Production
4.4. Purification of the Bacterium
4.5. Genome Sequencing and De Novo Assembly
4.6. Comparative Genomic Analyses and Annotation
4.7. B-Vitamins Biosynthesis Pathway
4.8. Comparative Phylogenies and Taxonomy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hertig, M. The Rickettsia,Wolbachia pipientis (gen. et sp.n.) and Associated Inclusions of the Mosquito, Culex pipiens. Parasitology 1936, 28, 453–486. [Google Scholar] [CrossRef]
- Zhou, W.; Rousset, F.; O’Neill, S. Phylogeny and PCR–based classification of Wolbachia strains using wsp gene sequences. Proc. R. Soc. B Boil. Sci. 1998, 265, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Khoo, J.J.; Kurtti, T.J.; Husin, N.A.; Beliavskaia, A.; Lim, F.S.; Zulkifli, M.M.S.; Al-Khafaji, A.M.; Hartley, C.; Darby, A.C.; Hughes, G.L.; et al. Isolation and Propagation of Laboratory Strains and a Novel Flea-Derived Field Strain of Wolbachia in Tick Cell Lines. Microorganisms 2020, 8, 988. [Google Scholar] [CrossRef]
- Baldo, L.; Hotopp, J.C.D.; Jolley, K.A.; Bordenstein, S.R.; Biber, S.A.; Choudhury, R.R.; Hayashi, C.; Maiden, M.C.J.; Tettelin, H.; Werren, J.H. Multilocus Sequence Typing System for the Endosymbiont Wolbachia pipientis. Appl. Environ. Microbiol. 2006, 72, 7098–7110. [Google Scholar] [CrossRef] [Green Version]
- Comandatore, F.; Cordaux, R.; Bandi, C.; Blaxter, M.; Darby, A.; Makepeace, B.L.; Montagna, M.; Sassera, D. Supergroup C Wolbachia, mutualist symbionts of filarial nematodes, have a distinct genome structure. Open Biol. 2015, 5, 150099. [Google Scholar] [CrossRef] [Green Version]
- Kampfraath, A.A.; Klasson, L.; Anvar, S.Y.; Vossen, R.H.A.M.; Roelofs, D.; Kraaijeveld, K.; Ellers, J. Genome expansion of an obligate parthenogenesis-associated Wolbachia poses an exception to the symbiont reduction model. BMC Genom. 2019, 20, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Lefoulon, E.; Clark, T.; Borveto, F.; Perriat-Sanguinet, M.; Moulia, C.; Slatko, B.E.; Gavotte, L. Pseudoscorpion Wolbachia symbionts: Diversity and evidence for a new supergroup S. BMC Microbiol. 2020, 20, 1–15. [Google Scholar] [CrossRef]
- Bandi, C.; Anderson, T.J.C.; Genchi, C.; Blaxter, M.L. Phylogeny of Wolbachia in filarial nematodes. Proc. R. Soc. B Boil. Sci. 1998, 265, 2407–2413. [Google Scholar] [CrossRef] [Green Version]
- Casiraghi, M.; Bain, O.; Guerrero, R.; Martin, C.; Pocacqua, V.; Gardner, S.L.; Franceschi, A.; Bandi, C. Mapping the presence of Wolbachia pipientis on the phylogeny of filarial nematodes: Evidence for symbiont loss during evolution. Int. J. Parasitol. 2004, 34, 191–203. [Google Scholar] [CrossRef] [Green Version]
- Lefoulon, E.; Bain, O.; Makepeace, B.L.; D’Haese, C.; Uni, S.; Martin, C.; Gavotte, L. Breakdown of coevolution between symbiotic bacteria Wolbachia and their filarial hosts. PeerJ 2016, 4, e1840. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.M.V.; Wasala, S.K.; Howe, D.K.; Peetz, A.B.; Zasada, I.A.; Denver, D.R. Genomic evidence for plant-parasitic nematodes as the earliest Wolbachia hosts. Sci. Rep. 2016, 6, 34955. [Google Scholar] [CrossRef]
- Haegeman, A.; Vanholme, B.; Jacob, J.; Vandekerckhove, T.T.; Claeys, M.; Borgonie, G.; Gheysen, G. An endosymbiotic bacterium in a plant-parasitic nematode: Member of a new Wolbachia supergroup. Int. J. Parasitol. 2009, 39, 1045–1054. [Google Scholar] [CrossRef]
- Ferri, E.; Bain, O.; Barbuto, M.; Martin, C.; Lo, N.; Uni, S.; Landmann, F.; Baccei, S.G.; Guerrero, R.; Lima, S.D.S.; et al. New Insights into the Evolution of Wolbachia Infections in Filarial Nematodes Inferred from a Large Range of Screened Species. PLoS ONE 2011, 6, e20843. [Google Scholar] [CrossRef] [Green Version]
- Lefoulon, E.; Gavotte, L.; Junker, K.; Barbuto, M.; Uni, S.; Landmann, F.; Laaksonen, S.; Saari, S.; Nikander, S.; Lima, S.D.S.; et al. A new type F Wolbachia from Splendidofilariinae (Onchocercidae) supports the recent emergence of this supergroup. Int. J. Parasitol. 2012, 42, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Fischer, P.; Schmetz, C.; Bandi, C.; Bonow, I.; Mand, S.; Fischer, K.; Büttner, D.W. Tunga penetrans: Molecular identification of Wolbachia endobacteria and their recognition by antibodies against proteins of endobacteria from filarial parasites. Exp. Parasitol. 2002, 102, 201–211. [Google Scholar] [CrossRef]
- Gorham, C.H.; Fang, Q.Q.; Durden, L.A. Wolbachia endosymbionts in fleas (Siphonaptera). J. Parasitol. 2003, 89, 283–289. [Google Scholar] [CrossRef]
- Sicard, M.; Bonneau, M.; Weill, M. Wolbachia prevalence, diversity, and ability to induce cytoplasmic incompatibility in mosquitoes. Curr. Opin. Insect Sci. 2019, 34, 12–20. [Google Scholar] [CrossRef]
- Driscoll, T.P.; Verhoeve, V.I.; Gillespie, J.J.; Johnston, J.S.; Guillotte, M.L.; Rennoll-Bankert, K.E.; Rahman, M.S.; Hagen, D.; Elsik, C.G.; Macaluso, K.R.; et al. A chromosome-level assembly of the cat flea genome uncovers rampant gene duplication and genome size plasticity. BMC Biol. 2020, 18, 1–19. [Google Scholar] [CrossRef]
- Dittmar, K.; Whiting, M.F. New Wolbachia endosymbionts from nearctic and neotropical fleas (Siphonaptera). J. Parasitol. 2004, 90, 953–957. [Google Scholar] [CrossRef]
- Espino, C.I.; Gómez, T.; González, G.; Santos, M.F.B.D.; Solano, J.; Sousa, O.; Moreno, N.; Windsor, D.; Ying, A.; Vilchez, S.; et al. Detection of Wolbachia Bacteria in Multiple Organs and Feces of the Triatomine Insect Rhodnius pallescens (Hemiptera, Reduviidae). Appl. Environ. Microbiol. 2008, 75, 547–550. [Google Scholar] [CrossRef] [Green Version]
- Crainey, J.L.; Wilson, M.D.; Post, R.J. Phylogenetically distinct Wolbachia gene and pseudogene sequences obtained from the African onchocerciasis vector Simulium squamosum. Int. J. Parasitol. 2010, 40, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, T.; Koga, R.; Kikuchi, Y.; Meng, X.-Y.; Fukatsu, T. Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc. Natl. Acad. Sci. USA 2009, 107, 769–774. [Google Scholar] [CrossRef] [Green Version]
- Doudoumis, V.; Alam, U.; Aksoy, E.; Abd-Alla, A.M.; Tsiamis, G.; Brelsfoard, C.; Aksoy, S.; Bourtzis, K. Tsetse-Wolbachia symbiosis: Comes of age and has great potential for pest and disease control. J. Invertebr. Pathol. 2013, 112, S94–S103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tay, S.T. Wolbachia endosymbionts, Rickettsia felis and Bartonella species, in Ctenocephalides felis fleas in a tropical region. J. Vector Ecol. 2013, 38, 200–202. [Google Scholar] [CrossRef]
- Chaisiri, K.; McGarry, J.W.; Morand, S.; Makepeace, B.L. Symbiosis in an overlooked microcosm: A systematic review of the bacterial flora of mites. Parasitology 2015, 142, 1152–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onder, Z.; Ciloglu, A.; Duzlu, O.; Yildirim, A.; Okur, M.; Yetismis, G.; Inci, A. Molecular detection and identification of Wolbachia endosymbiont in fleas (Insecta: Siphonaptera). Folia Microbiol. 2019, 64, 789–796. [Google Scholar] [CrossRef]
- Rosselló-Mora, R.; Amann, R. The species concept for prokaryotes. FEMS Microbiol. Rev. 2001, 25, 39–67. [Google Scholar] [CrossRef]
- Ramírez-Puebla, S.T.; Servín-Garcidueñas, L.E.; Ormeño-Orrillo, E.; De León, A.V.-P.; Rosenblueth, M.; Delaye, L.; Martinez, J.; Martínez-Romero, E. Species in Wolbachia? Proposal for the designation of ‘Candidatus Wolbachia bourtzisii’, ‘Candidatus Wolbachia onchocercicola’, ‘Candidatus Wolbachia blaxteri’, ‘Candidatus Wolbachia brugii’, ‘Candidatus Wolbachia taylori’, ‘Candidatus Wolbachia collembolicola’ and ‘Candidatus Wolbachia multihospitum’ for the different species within Wolbachia supergroups. Syst. Appl. Microbiol. 2015, 38, 390–399. [Google Scholar] [CrossRef]
- Lindsey, A.R.I.; Bordenstein, S.R.; Newton, I.L.; Rasgon, J.L. Wolbachia pipientis should not be split into multiple species: A response to Ramírez-Puebla et al., “Species in Wolbachia? Proposal for the designation of ‘Candidatus Wolbachia bourtzisii’, ‘Candidatus Wolbachia onchocercicola’, ‘Candidatus Wolbachia blaxteri’, ‘Candidatus Wolbachia brugii’, ‘Candidatus Wolbachia taylori’, ‘Candidatus Wolbachia collembolicola’ and ‘Candidatus Wolbachia multihospitum’ for the different species within Wolbachia supergroups”. Syst. Appl. Microbiol. 2016, 39, 220–222. [Google Scholar] [CrossRef] [Green Version]
- Weeks, A.R.; Breeuwer, J.A.J. Wolbachia–induced parthenogenesis in a genus of phytophagous mites. Proc. R. Soc. B Boil. Sci. 2001, 268, 2245–2251. [Google Scholar] [CrossRef] [Green Version]
- Hornett, E.A.; Duplouy, A.M.R.; Davies, N.; Roderick, G.K.; Wedell, N.; Hurst, G.D.D.; Charlat, S. You can’t keep a good parasite down: Evolution of a male-killer suppressor uncovers cytoplasmic incompatibility. Evol. Int. J. Org. Evol. 2008, 62, 1258–1263. [Google Scholar] [CrossRef] [PubMed]
- Werren, J.H.; Baldo, L.; Clark, M.E. Wolbachia: Master manipulators of invertebrate biology. Nat. Rev. Genet. 2008, 6, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Kern, P.; Cook, J.M.; Kageyama, D.; Riegler, M. Double trouble: Combined action of meiotic drive and Wolbachia feminization in Eurema butterflies. Biol. Lett. 2015, 11, 20150095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fenn, K.; Blaxter, M. Wolbachia genomes: Revealing the biology of parasitism and mutualism. Trends Parasitol. 2006, 22, 60–65. [Google Scholar] [CrossRef]
- Pike, N.; Kingcombe, R. Antibiotic treatment leads to the elimination of Wolbachia endosymbionts and sterility in the diplodiploid collembolan Folsomia candida. BMC Biol. 2009, 7, 54. [Google Scholar] [CrossRef] [Green Version]
- Bourtzis, K.; Dobson, S.L.; Xi, Z.; Rasgon, J.L.; Calvitti, M.; Moreira, L.A.; Bossin, H.C.; Moretti, R.; Anthony, L.; Hughes, G.L.; et al. Acta Tropica Harnessing mosquito—Wolbachia symbiosis for vector and disease control. Acta Trop. 2014, 132, S150–S163. [Google Scholar] [CrossRef]
- Bouchery, T.; Lefoulon, E.; Karadjian, G.; Nieguitsila, A.; Martin, C. The symbiotic role of Wolbachia in Onchocercidae and its impact on filariasis. Clin. Microbiol. Infect. 2013, 19, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Kamtchum-Tatuene, J.; Makepeace, B.L.; Benjamin, L.; Baylis, M.; Solomon, T. The potential role of Wolbachia in controlling the transmission of emerging human arboviral infections. Curr. Opin. Infect. Dis. 2017, 30, 108–116. [Google Scholar] [CrossRef] [Green Version]
- Hughes, G.L.; Rasgon, J.L. Transinfection: A method to investigate Wolbachia-host interactions and control arthropod-borne disease. Insect Mol. Biol. 2013, 23, 141–151. [Google Scholar] [CrossRef] [Green Version]
- Fisher, M.L.; Watson, D.W.; Osborne, J.A.; Mochizuki, H.; Breen, M.; Schal, C. Growth kinetics of endosymbiont Wolbachia in the common bed bug, Cimex lectularius. Sci. Rep. 2018, 8, 11444. [Google Scholar] [CrossRef]
- Meriweather, M.; Matthews, S.; Rio, R.; Baucom, R.S. A 454 Survey Reveals the Community Composition and Core Microbiome of the Common Bed Bug (Cimex lectularius) across an Urban Landscape. PLoS ONE 2013, 8, e61465. [Google Scholar] [CrossRef] [Green Version]
- Akhoundi, M.; Cannet, A.; Loubatier, C.; Berenger, J.-M.; Izri, A.; Marty, P.; Delaunay, P. Molecular characterization of Wolbachia infection in bed bugs (Cimex lectularius) collected from several localities in France. Parasite 2016, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriyama, M.; Nikoh, N.; Hosokawa, T.; Fukatsu, T. Riboflavin provisioning underlies Wolbachia’s fitness contribution to its insect host. MBio 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, M. Estimation of evolutionary distances between homologous nucleotide sequences. Proc. Natl. Acad. Sci. USA 1981, 78, 454–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waddell, P.J.; Steel, M. General Time-Reversible Distances with Unequal Rates across Sites: Mixing Γ and Inverse Gaussian Distributions with Invariant Sites. Mol. Phylogenet. Evol. 1997, 8, 398–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bordenstein, S.R.; Rosengaus, R.B. Discovery of a Novel Wolbachia Supergroup in Isoptera. Curr. Microbiol. 2005, 51, 393–398. [Google Scholar] [CrossRef]
- Glowska, E.; Dragun-Damian, A.; Dabert, M.; Gerth, M. New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae). Infect. Genet. Evol. 2015, 30, 140–146. [Google Scholar] [CrossRef]
- Stackebrandt, E. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 33, 152–155. [Google Scholar]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- O’Neill, S.L.; Pettigrew, M.M.; Sinkins, S.P.; Braig, H.R.; Andreadis, T.G.; Tesh, R.B. In vitro cultivation of Wolbachia pipientis in an Aedes albopictus cell line. Insect Mol. Biol. 1997, 6, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Fenollar, F.; La Scola, B.; Inokuma, H.; Dumler, J.S.; Taylor, M.J.; Raoult, D. Culture and Phenotypic Characterization of a Wolbachia pipientis Isolate. J. Clin. Microbiol. 2003, 41, 5434–5441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hague, M.T.J.; Caldwell, C.N.; Cooper, B.S. Divergent effects of Wolbachia on host temperature preference. bioRxiv 2020, 59812. [Google Scholar] [CrossRef]
- Lefoulon, E.; Vaisman, N.; Frydman, H.M.; Sun, L.; Voland, L.; Foster, J.M.; Slatko, B.E. Large Enriched Fragment Targeted Sequencing (LEFT-SEQ) Applied to Capture of Wolbachia Genomes. Sci. Rep. 2019, 9, 5939. [Google Scholar] [CrossRef] [PubMed]
- Lefoulon, E.; Clark, T.; Guerrero, R.; Cañizales, I.; Cardenas-Callirgos, J.M.; Junker, K.; Vallarino-Lhermitte, N.; Makepeace, B.L.; Darby, A.C.; Foster, J.M.; et al. Diminutive, degraded but dissimilar: Wolbachia genomes from filarial nematodes do not conform to a single paradigm. bioRxiv 2020, 3, 1–45. [Google Scholar] [CrossRef]
- Lindsey, A.R.I.; Werren, J.H.; Richards, S.; Stouthamer, R. Comparative Genomics of a Parthenogenesis-Inducing Wolbachia Symbiont. G3 Genes Genomes Genet. 2016, 6, 2113–2123. [Google Scholar] [CrossRef]
- Foster, J.; Ganatra, M.; Kamal, I.; Ware, J.; Makarova, K.; Ivanova, N.; Bhattacharyya, A.; Kapatral, V.; Kumar, S.; Posfai, J.; et al. The Wolbachia Genome of Brugia malayi: Endosymbiont Evolution within a Human Pathogenic Nematode. PLoS Biol. 2005, 3, e121. [Google Scholar] [CrossRef]
- Wang, N.; Jia, S.; Xu, H.; Liu, Y.; Huang, D.W. Multiple Horizontal Transfers of Bacteriophage WO and Host Wolbachia in Fig Wasps in a Closed Community. Front. Microbiol. 2016, 7, 136. [Google Scholar] [CrossRef]
- Nikoh, N.; Hosokawa, T.; Moriyama, M.; Oshima, K.; Hattori, M.; Fukatsu, T. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc. Natl. Acad. Sci. USA 2014, 111, 10257–10262. [Google Scholar] [CrossRef] [Green Version]
- Balvín, O.; Roth, S.; Talbot, B.; Reinhardt, K. Co-speciation in bedbug Wolbachia parallel the pattern in nematode hosts. Sci. Rep. 2018, 8, 8797. [Google Scholar] [CrossRef]
- Akman, L.; Yamashita, A.; Watanabe, H.; Oshima, K.; Shiba, T.; Hattori, M.; Aksoy, S. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 2002, 32, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Schneider, I. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 1972, 27, 353–365. [Google Scholar] [PubMed]
- Maina, A.N.; Luce-Fedrow, A.; Omulo, S.; Hang, J.; Chan, T.-C.; Ade, F.; Jima, D.D.; Ogola, E.; Ge, H.; Breiman, R.F.; et al. Isolation and characterization of a novel Rickettsia species (Rickettsia asembonensis sp. nov.) obtained from cat fleas (Ctenocephalides felis). Int. J. Syst. Evol. Microbiol. 2016, 66, 4512–4517. [Google Scholar] [CrossRef] [PubMed]
- Luce-Fedrow, A.; Macaluso, K.R.; Richards, A.L. Growth of Rickettsia felis in Drosophila melanogaster S2 Cells. Vector-Borne Zoonotic Dis. 2014, 14, 101–110. [Google Scholar] [CrossRef]
- Mediannikov, O.; Matsumoto, K.; Samoylenko, I.; Drancourt, M.; Roux, V.; Rydkina, E.; Davoust, B.; Tarasevich, I.; Brouqui, P.; Fournier, P.-E. Rickettsia raoultii sp. nov., a spotted fever group rickettsia associated with Dermacentor ticks in Europe and Russia. Int. J. Syst. Evol. Microbiol. 2008, 58, 1635–1639. [Google Scholar] [CrossRef] [Green Version]
- Diop, A.; Barker, S.C.; Eberhard, M.; Barker, D.; Nguyen, T.T.; Di Pinto, F.; Raoult, D.; Mediannikov, O. Rickettsia fournieri sp. nov., a novel spotted fever group rickettsia from Argas lagenoplastis ticks in Australia. Int. J. Syst. Evol. Microbiol. 2018, 68, 3781–3784. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; A Pelletier, D.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Laidoudi, Y.; Davoust, B.; Varloud, M.; Niang, E.H.A.; Fenollar, F.; Mediannikov, O. Development of a multiplex qPCR-based approach for the diagnosis of Dirofilaria immitis, D. repens and Acanthocheilonema reconditum. Parasites Vectors 2020, 13, 319. [Google Scholar] [CrossRef]
- Tanizawa, Y.; Fujisawa, T.; Kaminuma, E.; Nakamura, Y.; Arita, M. DFAST and DAGA: Web-based integrated genome annotation tools and resources. Biosci. Microbiota Food Health 2016, 35, 173–184. [Google Scholar] [CrossRef] [Green Version]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2017, 34, 1037–1039. [Google Scholar] [CrossRef] [Green Version]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Driscoll, T.P.; Verhoeve, V.I.; Gillespie, J.J.; Johnston, J.S.; Guillotte, M.L.; Rennoll-Bankert, K.E.; Rahman, M.S.; Hagen, D.; Elsik, C.G.; Macaluso, K.R.; et al. Cat fleas in flux: Rampant gene duplication, genome size plasticity, and paradoxical Wolbachia infection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.; Kim, Y.O.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; Von Jan, M.; Klenk, H.-P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [Green Version]
- Auch, A.F.; Klenk, H.-P.; Göker, M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand. Genom. Sci. 2010, 2, 142–148. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [Green Version]
- A Benson, D.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Ostell, J.; Pruitt, K.D.; Sayers, E.W. GenBank. Nucleic Acids Res. 2017, 46, D41–D47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39 (Suppl. 2), W29–W37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [Green Version]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol. Biol. Evol. 2009, 27, 221–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milne, I.; Lindner, D.; Bayer, M.; Husmeier, D.; McGuire, G.; Marshall, D.F.; Wright, F. TOPALi v2: A rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 2008, 25, 126–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.; Biosciences, I.; Carlsbad, C. BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2011, 2, 60–61. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Lorenzo-Carballa, M.O.; Torres-Cambas, Y.; Heaton, K.; Hurst, G.; Charlat, S.; Sherratt, T.N.; Van Gossum, H.; Cordero-Rivera, A.; Beatty, C.D. Widespread Wolbachia infection in an insular radiation of damselflies (Odonata, Coenagrionidae). Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Strain Information | wChem PL13 | wClec | wDmel | wPip | wFcan | wApol | wCfeJ | wBmal | wDimm | wPpe |
|---|---|---|---|---|---|---|---|---|---|---|
| Host type | Insects | Nematodes | ||||||||
| Wolbachia host | C. hemipterus | C. lectularius | D. melanogaster | C. quinquefasciatus | F. candida | A. politus | C. felis | B. malayi | D. immitis | P. penetrans |
| Supergroup | New supergroup “T” | F | A | B | E | S | Undescribed | D | C | L |
| Genome features | ||||||||||
| Accession Number | CP061738 | AP013028 | AE017196 | AM999887 | CP015510 | WQMQ00000000 | CP051157 | CP034333 | CP046578 | MJMG01000000 |
| Total length (bp) | 1,291,339 | 1,250,060 | 1,267,782 | 1,482,455 | 1,801,626 | 1,445,964 | 1,201,647 | 1,080,064 | 920,122 | 975,127 |
| No. of contigs | 1 | 1 | 1 | 1 | 1 | 373 | 1 | 1 | 1 | 12 |
| GC content (%) | 35.4 | 36.3 | 35.2 | 34.2 | 34.4 | 35.6 | 35.6 | 34.2 | 32.7 | 32.2 |
| N50 | 1,291,339 | 1,250,060 | 1,267,782 | 1,482,455 | 1,801,626 | 5741 | 1,201,647 | 1,080,064 | 920,122 | 9,555 |
| Gap ratio (%) | 0.326793 | 0.0 | 0.0 | 0.006746 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.13547 |
| No. of CDSs | 1194 | 1226 | 1211 | 1395 | 1591 | 1546 | 1045 | 1017 | 709 | 939 |
| No. of rRNA | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| No. of tRNA | 32 | 34 | 34 | 34 | 35 | 39 | 34 | 34 | 34 | 35 |
| No. of CRISPRS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Coding ratio (%) | 78.5 | 77.3 | 81.6 | 84.4 | 86.9 | 66.1 | 82.3 | 70.1 | 70.7 | 84.8 |
| Completeness (%) | 98.00 | 98.00 | 98.73 | 99.45 | 97.27 | 95.89 | 98.36 | 99.09 | 98.00 | 93.32 |
| Contamination (%) | 0.36 | 0.36 | 0.00 | 0.00 | 1.55 | 19.67 | 0.36 | 0.00 | 0.00 | 2.73 |
| No. of prophage | 2 | 3 | 3 | 4 | 6 | 2 | 0 | 0 | 0 | 1 |
| Strain | Wolbachia Host | Accession Number | DDH | Distance | Prob. DDH ≥ 70% | G + C Difference | Model C.I. |
|---|---|---|---|---|---|---|---|
| wPpe | P. penetrans | MJMG01000000 | 19.8 | 0.2216 | 0 | 3.21 | [17.6–22.2%] |
| Ctub | C. tuberocauda | CP046579 | 23 | 0.1901 | 0 | 3.09 | [20.7–25.5%] |
| WCfelT | C. felis | CP051156 | 23.6 | 0.1853 | 0 | 0.19 | [21.3–26%] |
| wFcan | F. candida | CP015510 | 24.1 | 0.1809 | 0.01 | 1.02 | [21.8–26.6%] |
| wDimm | D. immitis | CP046578 | 24.8 | 0.1757 | 0.01 | 2.67 | [22.5–27.3%] |
| wCmeg | C. megacephala | CP021120 | 27 | 0.1599 | 0.03 | 1.42 | [24.7–29.5%] |
| wDcit | D. citri | CP051608 | 27 | 0.1604 | 0.03 | 1.38 | [24.6–29.4%] |
| wPip | C. quinquefasciatus | AM999887 | 27.1 | 0.1598 | 0.03 | 1.18 | [24.7–29.5%] |
| wCfeJ | C. felis | CP051157 | 27.8 | 0.155 | 0.04 | 0.2 | [25.4–30.3%] |
| wBmal | B. malayi | CP034333 | 28.4 | 0.1512 | 0.05 | 1.19 | [26–30.9%] |
| wDmel | D. melanogaster | AE017196 | 29.3 | 0.1462 | 0.08 | 0.14 | [26.9–31.8%] |
| wClec | C. lectularius | AP013028 | 29.3 | 0.1462 | 0.08 | 0.88 | [26.9–31.8%] |
| wApolK5 | A. politus | WQMQ00000000 | 30 | 0.1423 | 0.1 | 0.23 | [27.6–32.5%] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laidoudi, Y.; Levasseur, A.; Medkour, H.; Maaloum, M.; Ben Khedher, M.; Sambou, M.; Bassene, H.; Davoust, B.; Fenollar, F.; Raoult, D.; et al. An Earliest Endosymbiont, Wolbachia massiliensis sp. nov., Strain PL13 from the Bed Bug (Cimex hemipterus), Type Strain of a New Supergroup T. Int. J. Mol. Sci. 2020, 21, 8064. https://doi.org/10.3390/ijms21218064
Laidoudi Y, Levasseur A, Medkour H, Maaloum M, Ben Khedher M, Sambou M, Bassene H, Davoust B, Fenollar F, Raoult D, et al. An Earliest Endosymbiont, Wolbachia massiliensis sp. nov., Strain PL13 from the Bed Bug (Cimex hemipterus), Type Strain of a New Supergroup T. International Journal of Molecular Sciences. 2020; 21(21):8064. https://doi.org/10.3390/ijms21218064
Chicago/Turabian StyleLaidoudi, Younes, Anthony Levasseur, Hacène Medkour, Mossaab Maaloum, Mariem Ben Khedher, Masse Sambou, Hubert Bassene, Bernard Davoust, Florence Fenollar, Didier Raoult, and et al. 2020. "An Earliest Endosymbiont, Wolbachia massiliensis sp. nov., Strain PL13 from the Bed Bug (Cimex hemipterus), Type Strain of a New Supergroup T" International Journal of Molecular Sciences 21, no. 21: 8064. https://doi.org/10.3390/ijms21218064
APA StyleLaidoudi, Y., Levasseur, A., Medkour, H., Maaloum, M., Ben Khedher, M., Sambou, M., Bassene, H., Davoust, B., Fenollar, F., Raoult, D., & Mediannikov, O. (2020). An Earliest Endosymbiont, Wolbachia massiliensis sp. nov., Strain PL13 from the Bed Bug (Cimex hemipterus), Type Strain of a New Supergroup T. International Journal of Molecular Sciences, 21(21), 8064. https://doi.org/10.3390/ijms21218064