Developmental Roles of FUSE Binding Protein 1 (Fubp1) in Tooth Morphogenesis
Abstract
1. Introduction
2. Results
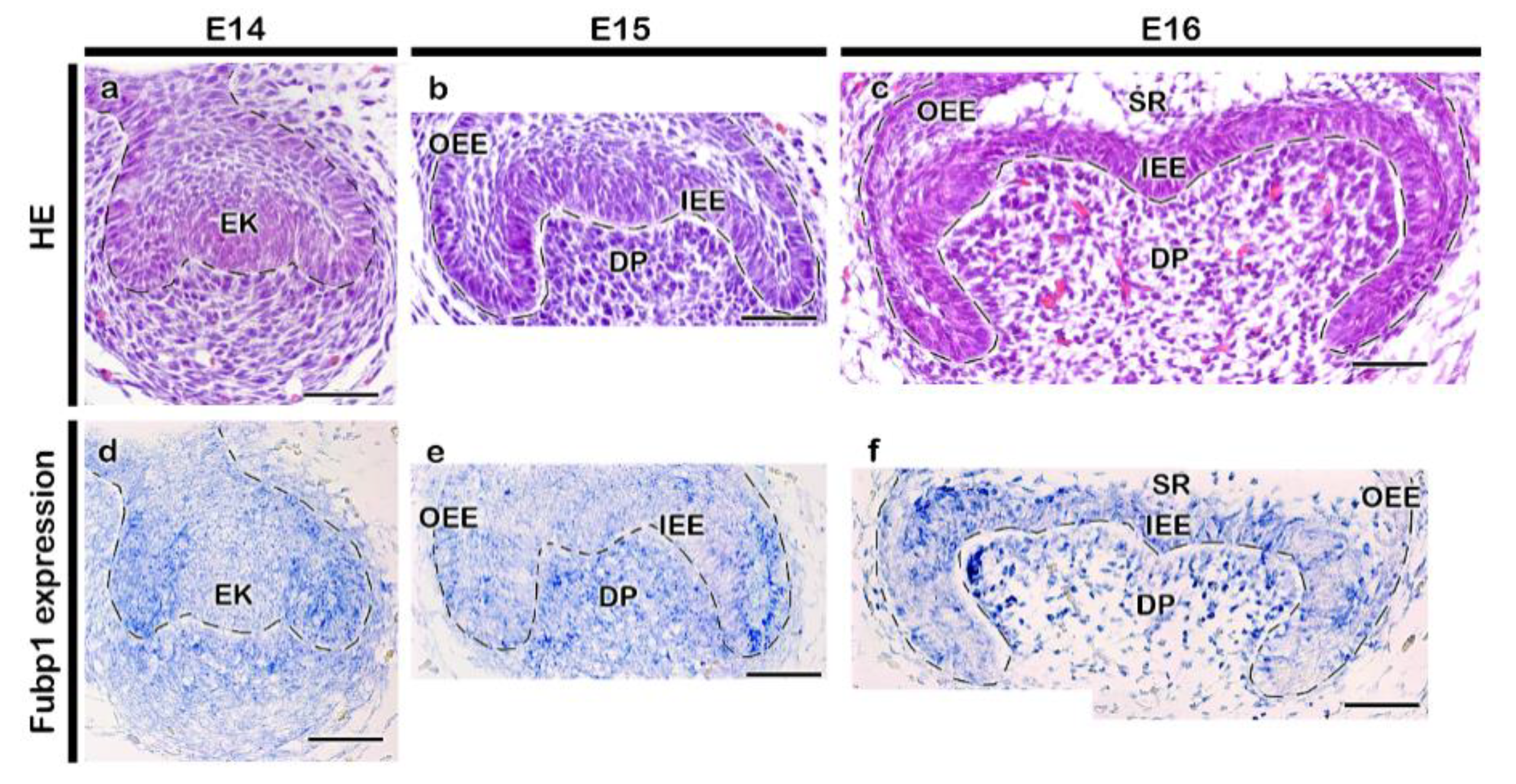
2.1. Expression Pattern and Functional Evaluation of Fubp1
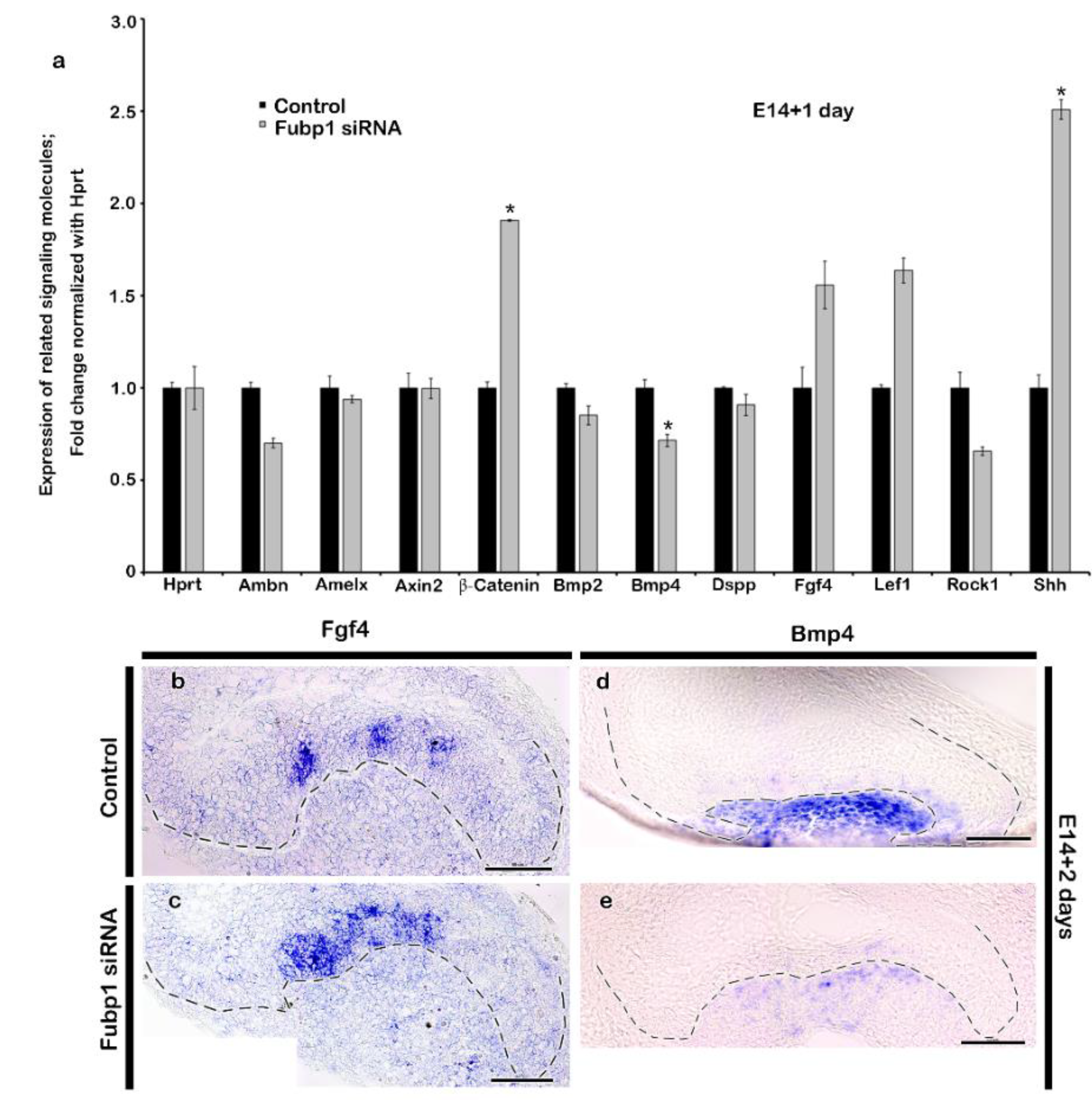
2.2. Altered Expression Patterns of Signalling Molecules by Fubp1 Knock-Down
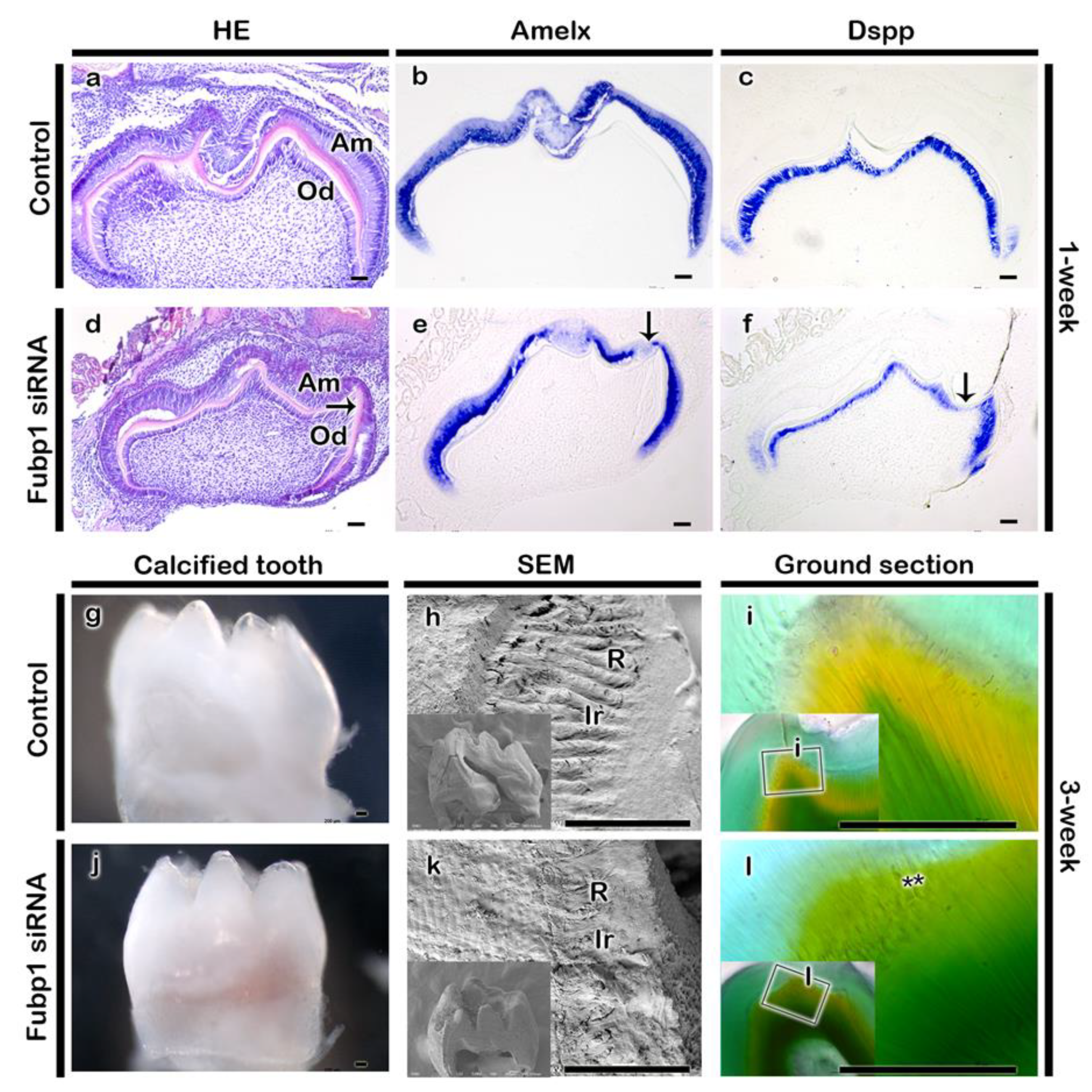
2.3. Fubp1 in Tooth Morphogenesis
3. Discussion
4. Material and Methods
4.1. Animals
4.2. In Situ Hybridization
4.3. siRNA Transfection, In Vitro Organ Cultivation and Kidney Transplantation
4.4. Histology, Immunohistochemistry and Western Blotting
4.5. Quantitative PCR (qPCR)
4.6. Ground Section and Scanning Electron Microscopy (SEM)
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gilbert, S.F.; Barresi, M.J.F. Developmental Biology, 11th Edition 2016. Am. J. Med. Genet. Part A 2017, 173, 1430. [Google Scholar] [CrossRef]
- Kuure, S.; Vuolteenaho, R.; Vainio, S. Kidney morphogenesis: Cellular and molecular regulation. Mech. Dev. 2000, 92, 31–45. [Google Scholar] [CrossRef]
- Pispa, J.; Thesleff, I. Mechanisms of ectodermal organogenesis. Dev. Biol. 2003, 262, 195–205. [Google Scholar] [CrossRef]
- Hogan, B.L.M.; Yingling, J.M. Epithelial/mesenchymal interactions and branching morphogenesis of the lung. Curr. Opin. Genet. Dev. 1998, 8, 481–486. [Google Scholar] [CrossRef]
- Mousavi, S.J.; Hamdy Doweidar, M. Role of mechanical cues in cell differentiation and proliferation: A 3D numerical model. PLoS ONE 2015, 10, e0124529. [Google Scholar] [CrossRef] [PubMed]
- Thesleff, I. The genetic basis of tooth development and dental defects. Am. J. Med. Genet. Part A 2006, 140A, 2530–2535. [Google Scholar] [CrossRef]
- Du, W.; Hu, J.K.-H.; Du, W.; Klein, O.D. Lineage tracing of epithelial cells in developing teeth reveals two strategies for building signalling centers. J. Biol. Chem. 2017, 292, 15062–15069. [Google Scholar] [CrossRef]
- Nirvani, M.; Khuu, C.; Utheim, T.P.; Hollingen, H.S.; Amundsen, S.F.; Sand, L.P.; Sehic, A. Circadian rhythms and gene expression during mouse molar tooth development. Acta Odontol. Scand. 2017, 75, 144–153. [Google Scholar] [CrossRef]
- Aurrekoetxea, M.; Irastorza, I.; García-Gallastegui, P.; Jiménez-Rojo, L.; Nakamura, T.; Yamada, Y.; Ibarretxe, G.; Unda, F.J. Wnt/β-Catenin Regulates the Activity of Epiprofin/Sp6, SHH, FGF, and BMP to Coordinate the Stages of Odontogenesis. Front. Cell Dev. Biol. 2016, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.-J.; Liu, J.; Dundr, M.; Nie, Z.; Sanford, S.; Levens, D. FBPs are calibrated molecular tools to adjust gene expression. Mol. Cell. Biol. 2006, 26, 6584–6597. [Google Scholar] [CrossRef]
- He, L.; Liu, J.; Collins, I.; Sanford, S.; O’Connell, B.; Benham, C.J.; Levens, D. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 2000, 19, 1034–1044. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, Q.M. Far upstream element binding protein 1: A commander of transcription, translation and beyond. Oncogene 2013, 32, 2907–2916. [Google Scholar] [CrossRef] [PubMed]
- Nie, Z.; Hu, G.; Wei, G.; Cui, K.; Yamane, A.; Resch, W.; Wang, R.; Green, D.R.; Tessarollo, L.; Casellas, R.; et al. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012, 151, 68–79. [Google Scholar] [CrossRef]
- Dang, C.V. c-Myc Target Genes Involved in Cell Growth, Apoptosis, and Metabolism. Mol. Cell. Biol. 1999, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Davis-Smyth, T.; Duncan, R.C.; Zheng, T.; Michelotti, G.; Levens, D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J. Biol. Chem. 1996, 271, 31679–31687. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Levens, D. Minireview Molecules and c-myc Expression: Keep the Noise Down! Mol. Cells 2005, 20, 157–166. [Google Scholar] [PubMed]
- Duan, J.; Bao, X.; Ma, X.; Zhang, Y.; Ni, D.; Wang, H.; Zhang, F.; Du, Q.; Fan, Y.; Chen, J.; et al. Upregulation of far upstream element-binding protein 1 (FUBP1) promotes tumor proliferation and tumorigenesis of clear cell renal cell carcinoma. PLoS ONE 2017, 12, e0169852. [Google Scholar] [CrossRef]
- Hwang, I.; Cao, D.; Na, Y.; Kim, D.-Y.; Zhang, T.; Yao, J.; Oh, H.; Hu, J.; Zheng, H.; Yao, Y.; et al. Far Upstream Element-Binding Protein 1 Regulates LSD1 Alternative Splicing to Promote Terminal Differentiation of Neural Progenitors. Stem Cell Rep. 2018, 10, 1208–1221. [Google Scholar] [CrossRef]
- Zhou, W.; Chung, Y.J.; Parrilla Castellar, E.R.; Zheng, Y.; Chung, H.J.; Bandle, R.; Liu, J.; Tessarollo, L.; Batchelor, E.; Aplan, P.D.; et al. Far upstream element binding protein plays a crucial role in embryonic development, hematopoiesis, and stabilizing myc expression levels. Am. J. Pathol. 2016, 186, 701–715. [Google Scholar] [CrossRef]
- Adhikari, N.; Neupane, S.; Gwon, G.-J.; Kim, J.-Y.; An, C.-H.; Lee, S.; Sohn, W.-J.; Lee, Y.; Kim, J.-Y. Grhl3 modulates epithelial structure formation of the circumvallate papilla during mouse development. Histochem. Cell Biol. 2017, 147, 5–16. [Google Scholar] [CrossRef]
- Neupane, S.; Sohn, W.-J.; Rijal, G.; Lee, Y.-J.; Lee, S.; Yamamoto, H.; An, C.-H.; Cho, S.-W.; Lee, Y.; Shin, H.-I.; et al. Developmental regulations of Perp in mice molar morphogenesis. Cell Tissue Res. 2014, 358, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Balic, A. Concise Review: Cellular and Molecular Mechanisms Regulation of Tooth Initiation. Stem Cells 2019, 37, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Uchibe, K.; Shimizu, H.; Yokoyama, S.; Kuboki, T.; Asahara, H. Identification of novel transcription-regulating genes expressed during murine molar development. Dev. Dyn. 2012, 241, 1217–1226. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.; Murphy, M.J.; Oskarsson, T.; Kaloulis, K.; Bettess, M.D.; Oser, G.M.; Pasche, A.-C.; Knabenhans, C.; Macdonald, H.R.; Trumpp, A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004, 18, 2747–2763. [Google Scholar] [CrossRef]
- Luo, W.; Chen, J.; Li, L.; Ren, X.; Cheng, T.; Lu, S.; Lawal, R.A.; Nie, Q.; Zhang, X.; Hanotte, O. c-Myc inhibits myoblast differentiation and promotes myoblast proliferation and muscle fibre hypertrophy by regulating the expression of its target genes, miRNAs and lincRNAs. Cell Death Differ. 2019, 26, 426–442. [Google Scholar] [CrossRef]
- Melnik, S.; Werth, N.; Boeuf, S.; Hahn, E.-M.; Gotterbarm, T.; Anton, M.; Richter, W. Impact of c-MYC expression on proliferation, differentiation, and risk of neoplastic transformation of human mesenchymal stromal cells. Stem Cell Res. Ther. 2019, 10, 73. [Google Scholar] [CrossRef]
- Wen, H.; Ma, H.; Li, P.; Zheng, J.; Yu, Y.; Lv, G. Expression of far upstream element-binding protein 1 correlates with c-Myc expression in sacral chordomas and is associated with tumor progression and poor prognosis. Biochem. Biophys. Res. Commun. 2017, 491, 1047–1054. [Google Scholar] [CrossRef]
- Wang, B.; Fan, P.; Zhao, J.; Wu, H.; Jin, X.; Wu, H. FBP1 loss contributes to BET inhibitors resistance by undermining c-Myc expression in pancreatic ductal adenocarcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 224. [Google Scholar] [CrossRef]
- Rabenhorst, U.; Beinoraviciute-Kellner, R.; Brezniceanu, M.-L.; Joos, S.; Devens, F.; Lichter, P.; Rieker, R.J.; Trojan, J.; Chung, H.-J.; Levens, D.L.; et al. Overexpression of the far upstream element binding protein 1 in hepatocellular carcinoma is required for tumor growth. Hepatology 2009, 50, 1121–1129. [Google Scholar] [CrossRef]
- Ruijtenberg, S.; van den Heuvel, S. Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle 2016, 15, 196–212. [Google Scholar] [CrossRef]
- Balic, A.; Thesleff, I. Tissue Interactions Regulating Tooth Development and Renewal. In Current Topics in Developmental Biology; Academic Press: Cambridge, MA, USA, 2015; pp. 157–186. [Google Scholar]
- Jani, P.; Liu, C.; Zhang, H.; Younes, K.; Benson, M.D.; Qin, C. The role of bone morphogenetic proteins 2 and 4 in mouse dentinogenesis. Arch. Oral Biol. 2018, 90, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Gluhak-Heinrich, J.; Guo, D.; Yang, W.; Harris, M.A.; Lichtler, A.; Kream, B.; Zhang, J.; Feng, J.Q.; Smith, L.C.; Dechow, P.; et al. New roles and mechanism of action of BMP4 in postnatal tooth cytodifferentiation. Bone 2010, 46, 1533–1545. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Yang, G.; Yuan, G.; Gluhak-Heinrich, J.; Yang, W.; Wang, L.; Chen, Z.; Schulze McDaniel, J.; Donly, K.J.; Harris, S.E.; et al. Abnormalities in the Enamel in Bmp2-Deficient Mice. Cells Tissues Organs 2011, 194, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Tang, Q.; Kwon, H.-J.E.; Wu, Z.; Kim, E.-J.; Jung, H.-S. An Explanation for How FGFs Predict Species-Specific Tooth Cusp Patterns. J. Dent. Res. 2018, 97, 828–834. [Google Scholar] [CrossRef]
- Hamidi, K.; Darvish, J.; Matin, M.M.; Javanmard, A.S.; Kilpatrick, C.W. Tooth Morphogenesis and FGF4 Expression During Development of Molar Tooth in Three Muroid Rodents: Calomyscus elburzensis (Calomyscidae), Mesocricetus auratus (Cricetidae) and Mus musculus (Muridae). Anat. Rec. 2017, 300, 2138–2149. [Google Scholar] [CrossRef]
- Cho, S.-W.; Lee, H.-A.; Cai, J.; Lee, M.-J.; Kim, J.-Y.; Ohshima, H.; Jung, H.-S. The primary enamel knot determines the position of the first buccal cusp in developing mice molars. Differentiation 2007, 75, 441–451. [Google Scholar] [CrossRef]
- Shi, C.; Yuan, Y.; Guo, Y.; Jing, J.; Ho, T.V.; Han, X.; Li, J.; Feng, J.; Chai, Y. BMP Signalling in Regulating Mesenchymal Stem Cells in Incisor Homeostasis. J. Dent. Res. 2019, 98, 904–911. [Google Scholar] [CrossRef]
- Zhang, L.; Hua, F.; Yuan, G.-H.; Zhang, Y.-D.; Chen, Z. Sonic hedgehog signalling is critical for cytodifferentiation and cusp formation in developing mouse molars. J. Mol. Histol. 2008, 39, 87–94. [Google Scholar] [CrossRef]
- Otsu, K.; Harada, H. Rho GTPases in ameloblast differentiation. Jpn. Dent. Sci. Rev. 2016, 52, 32–40. [Google Scholar] [CrossRef]
- Huang, X.; Xu, X.; Bringas, P.; Hung, Y.P.; Chai, Y. Smad4-Shh-Nfic signalling cascade-mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J. Bone Miner. Res. 2010, 25, 1167–1178. [Google Scholar] [CrossRef]
- van Dam, S.; Võsa, U.; van der Graaf, A.; Franke, L.; de Magalhães, J.P. Gene co-expression analysis for functional classification and gene-disease predictions. Brief. Bioinform. 2018, 19, 575–592. [Google Scholar] [CrossRef]
- Hatakeyama, J.; Fukumoto, S.; Nakamura, T.; Haruyama, N.; Suzuki, S.; Hatakeyama, Y.; Shum, L.; Gibson, C.W.; Yamada, Y.; Kulkarni, A.B. Synergistic roles of amelogenin and ameloblastin. J. Dent. Res. 2009, 88, 318–322. [Google Scholar] [CrossRef]
- Verdelis, K.; Szabo-Rogers, H.L.; Xu, Y.; Chong, R.; Kang, R.; Cusack, B.J.; Jani, P.; Boskey, A.L.; Qin, C.; Beniash, E. Accelerated enamel mineralization in Dspp mutant mice. Matrix Biol. 2016, 52–54, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xie, X.; Liu, P.; Liang, T.; Lu, Y.; Qin, C. Transgenic expression of dentin phosphoprotein (DPP) partially rescued the dentin defects of DSPP-null mice. PLoS ONE 2018, 13, e0195854. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.L.; Menko, A.S.; Khalil, S.; Rebustini, I.; Hoffman, M.P.; Kreidberg, J.A.; Kukuruzinska, M.A. Diverse roles of E-cadherin in the morphogenesis of the submandibular gland: Insights into the formation of acinar and ductal structures. Dev. Dyn. 2008, 237, 3128–3141. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Shin, J.-H.; Kee, S.-H. E-cadherin expression increases cell proliferation by regulating energy metabolism through nuclear factor-κB in AGS cells. Cancer Sci. 2017, 108, 1769–1777. [Google Scholar] [CrossRef] [PubMed]
- Kilkenny, C.; Browne, W.J.; Cuthill, I.C.; Emerson, M.; Altman, D.G. Improving Bioscience Research Reporting: The ARRIVE Guidelines for Reporting Animal Research. PLoS Biol. 2010, 8, e1000412. [Google Scholar] [CrossRef]
- Cai, J.; Cho, S.-W.; Kim, J.-Y.; Lee, M.-J.; Cha, Y.-G.; Jung, H.-S. Patterning the size and number of tooth and its cusps. Dev. Biol. 2007, 304, 499–507. [Google Scholar] [CrossRef]
- Jung, J.K.; Gwon, G.J.; Neupane, S.; Sohn, W.J.; Kim, K.R.; Kim, J.Y.; An, S.Y.; Kwon, T.Y.; An, C.H.; Lee, Y.; et al. Bortezomib Facilitates Reparative Dentin Formation after Pulp Access Cavity Preparation in Mouse Molar. J. Endod. 2017, 43, 2041–2047. [Google Scholar] [CrossRef]


| Gene | Accession | Primer Sequence | Product Size (bp) | Remarks | |
|---|---|---|---|---|---|
| Fubp1 | NM_001355372.1 | Forward | GCATCAGCAGCAAAGCAGAT | 419 | Probe synthesis |
| Reverse | CAGCACCAGTGTTTTGAGGC | ||||
| Forward | TGCAGCAAAAATTGGGGGTG | 173 | qPCR | ||
| Reverse | ATCTGCTTTGCTGCTGATGC | ||||
| Ambn | NM_001303431.1 | Forward | TTCTTGCTTTCCCCAATGAC | 234 | Amelogenesis |
| Reverse | GGTGCACTTTGTTTCCAGGT | ||||
| Amelx | XM_017348358.1 | Forward | GCAGCCGTATCCTTCCTATGGTT | 120 | Amelogenesis |
| Reverse | GGAAGGTGGTGATGAGGCTGAA | ||||
| Axin2 | BC057338.1 | Forward | TGAAGAAGAGGAGTGGACGT | 115 | Wnt signalling |
| Reverse | AGCTGTTTCCGTGGATCTCA | ||||
| β-catenin | NM_007614.3 | Forward | TGACCTGATGGAGTTGGACA | 104 | Wnt signalling |
| Reverse | TGGCACCAGAATGGATTCCA | ||||
| Bmp2 | NM_007553.3 | Forward | AAGTGGCCCATTTAGAGGAG | 104 | Bmp signalling |
| Reverse | CCATGGCCTTATCTGTGACC | ||||
| Bmp4 | NM_007554.3 | Forward | ACCTCAAGGGAGTGGAGATT | 113 | Bmp signalling |
| Reverse | GATGCTTGGGACTACGTTTG | ||||
| Dspp | NM_010080.3 | Forward | GTGGGGTTGCTACACATGAAAC | 169 | Dentinogenesis |
| Reverse | CCATCACCAGAGCCTGTATCTTC | ||||
| Fgf4 | NM_010202.5 | Forward | TCGCCTACCATGAAGGTAAC | 114 | Fgf signalling |
| Reverse | TCTCCATCGAGAGAAAGTGC | ||||
| Lef1 | NM_010703.4 | Forward | ACAGCGACGAGCACTTTTCT | 82 | Shh Signalling |
| Reverse | TGTCTGGACATGCCTTGCTT | ||||
| Rock1 | NM_009071.2 | Forward | GGTATCGTCACAAGTAGCAGCATC | 140 | Wnt signalling |
| Reverse | TAAACCAGGGCATCCAATCCA | ||||
| Shh | NM_009170.3 | Forward | CCAAAGCTCACATCCACTGT | 131 | Shh Signalling |
| Reverse | GGGACGTAAGTCCTTCACCA | ||||
| Hprt | NM_013556.1 | Forward | CCTAAGATGATCGCAAGTTG | 86 | Internal standard |
| Reverse | CCACAGGGACTAGAACACCTGCTAA | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aryal, Y.P.; Neupane, S.; Kim, T.-Y.; Lee, E.-S.; Pokhrel, N.K.; Yeon, C.-Y.; Kim, J.-Y.; An, C.-H.; An, S.-Y.; Park, E.-K.; et al. Developmental Roles of FUSE Binding Protein 1 (Fubp1) in Tooth Morphogenesis. Int. J. Mol. Sci. 2020, 21, 8079. https://doi.org/10.3390/ijms21218079
Aryal YP, Neupane S, Kim T-Y, Lee E-S, Pokhrel NK, Yeon C-Y, Kim J-Y, An C-H, An S-Y, Park E-K, et al. Developmental Roles of FUSE Binding Protein 1 (Fubp1) in Tooth Morphogenesis. International Journal of Molecular Sciences. 2020; 21(21):8079. https://doi.org/10.3390/ijms21218079
Chicago/Turabian StyleAryal, Yam Prasad, Sanjiv Neupane, Tae-Young Kim, Eui-Seon Lee, Nitin Kumar Pokhrel, Chang-Yeol Yeon, Ji-Youn Kim, Chang-Hyeon An, Seo-Young An, Eui-Kyun Park, and et al. 2020. "Developmental Roles of FUSE Binding Protein 1 (Fubp1) in Tooth Morphogenesis" International Journal of Molecular Sciences 21, no. 21: 8079. https://doi.org/10.3390/ijms21218079
APA StyleAryal, Y. P., Neupane, S., Kim, T.-Y., Lee, E.-S., Pokhrel, N. K., Yeon, C.-Y., Kim, J.-Y., An, C.-H., An, S.-Y., Park, E.-K., Ha, J.-H., Jung, J.-K., Yamamoto, H., Cho, S.-W., Lee, S., Kim, D.-Y., Kwon, T.-Y., Lee, Y., Sohn, W.-J., & Kim, J.-Y. (2020). Developmental Roles of FUSE Binding Protein 1 (Fubp1) in Tooth Morphogenesis. International Journal of Molecular Sciences, 21(21), 8079. https://doi.org/10.3390/ijms21218079





