A Comprehensive Analysis of MicroRNAs Expressed in Susceptible and Resistant Rice Cultivars during Rhizoctonia solani AG1-IA Infection Causing Sheath Blight Disease
Abstract
:1. Introduction
2. Results
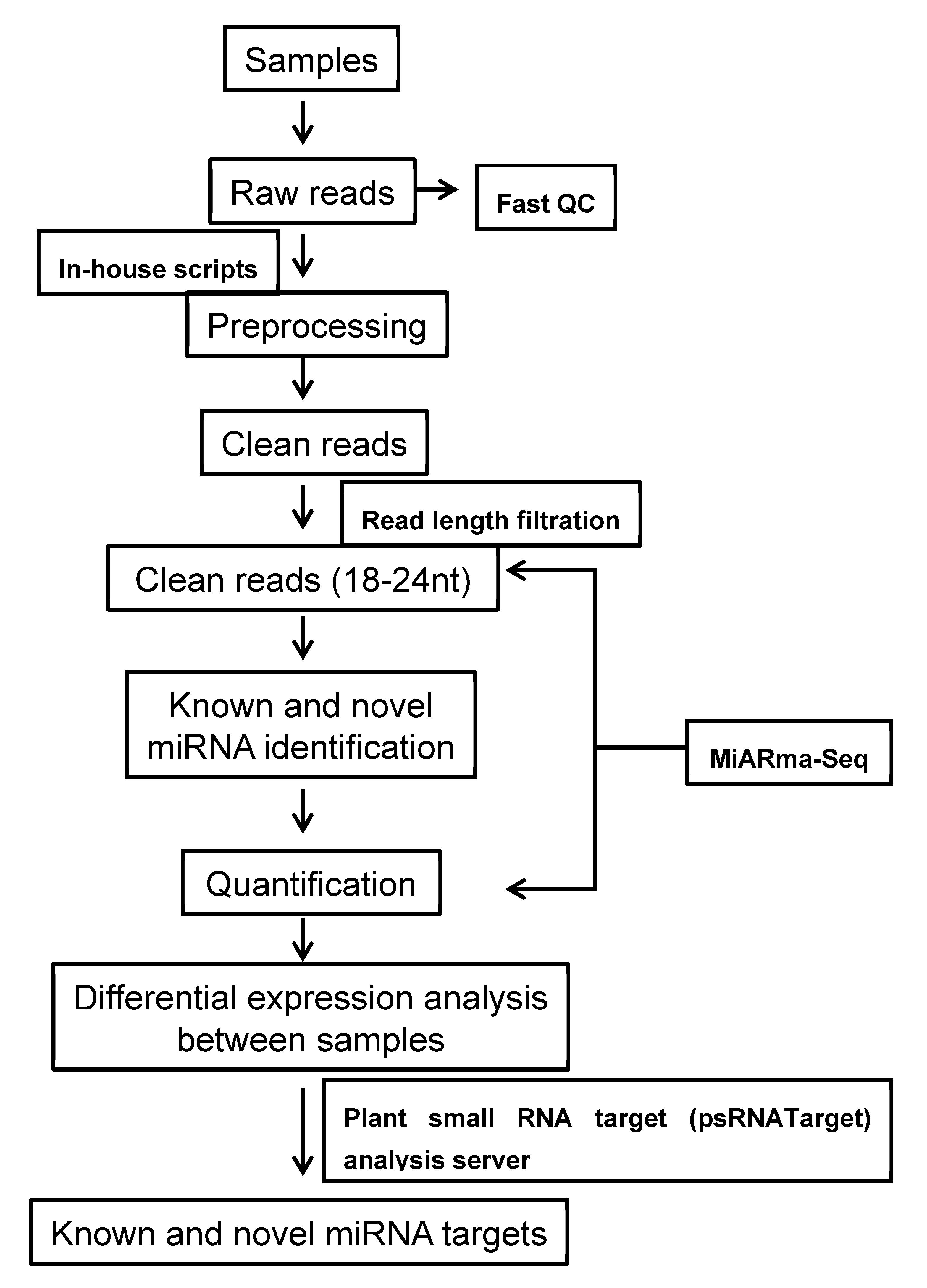
2.1. Sequencing of Small RNAs
2.2. Expression of Known miRNAs
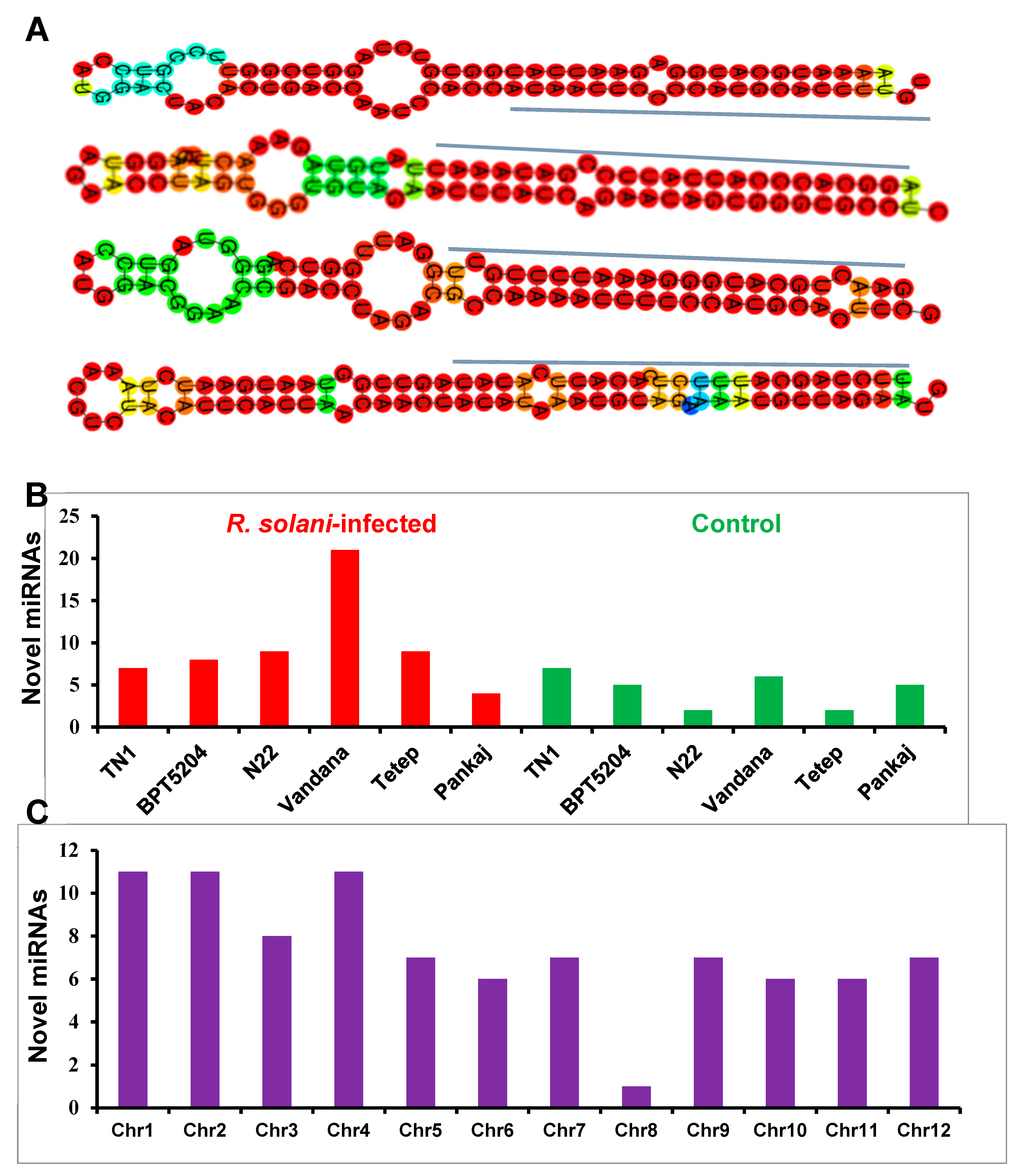
2.3. Identification and Distribution of Novel miRNAs
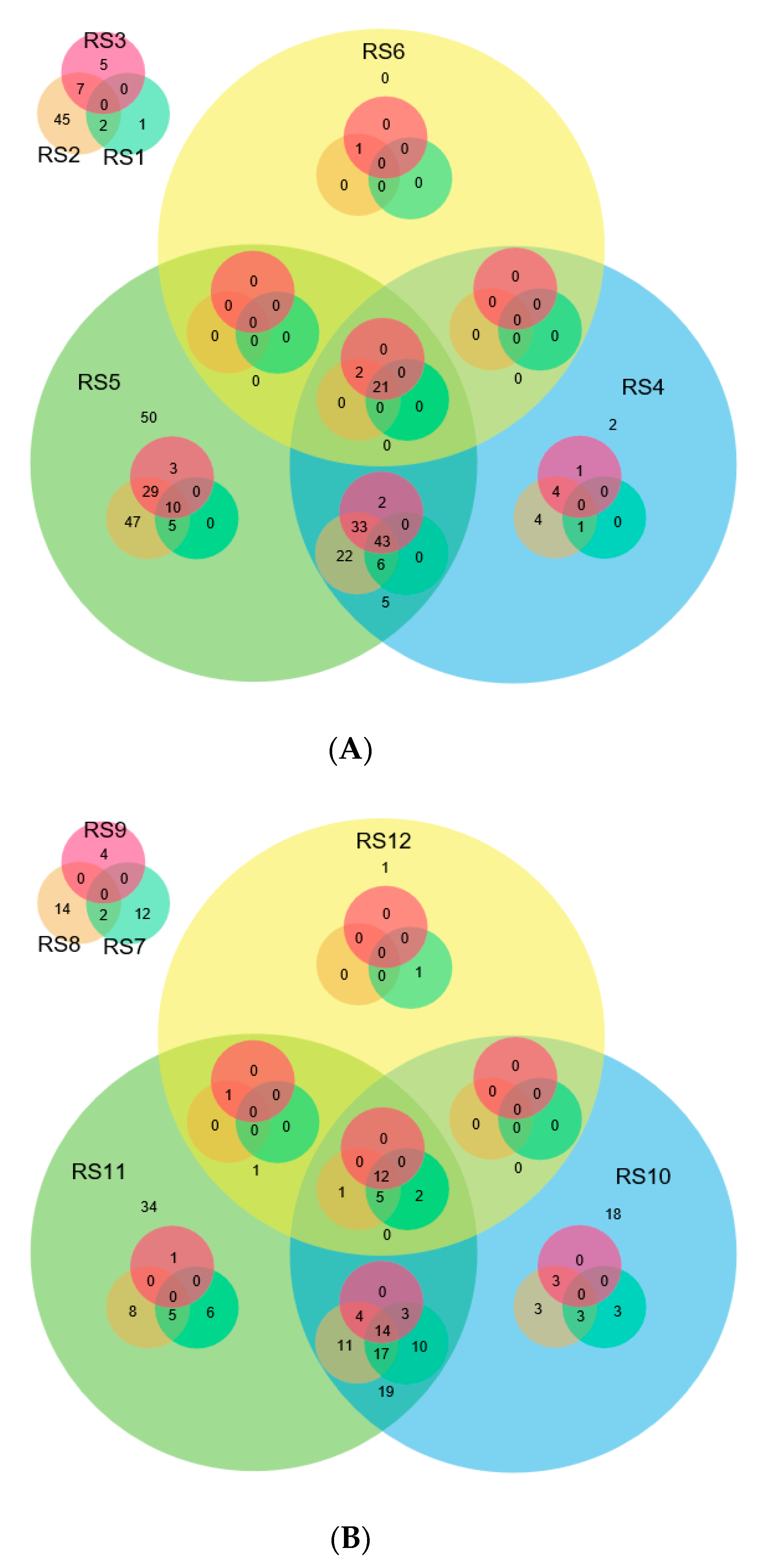
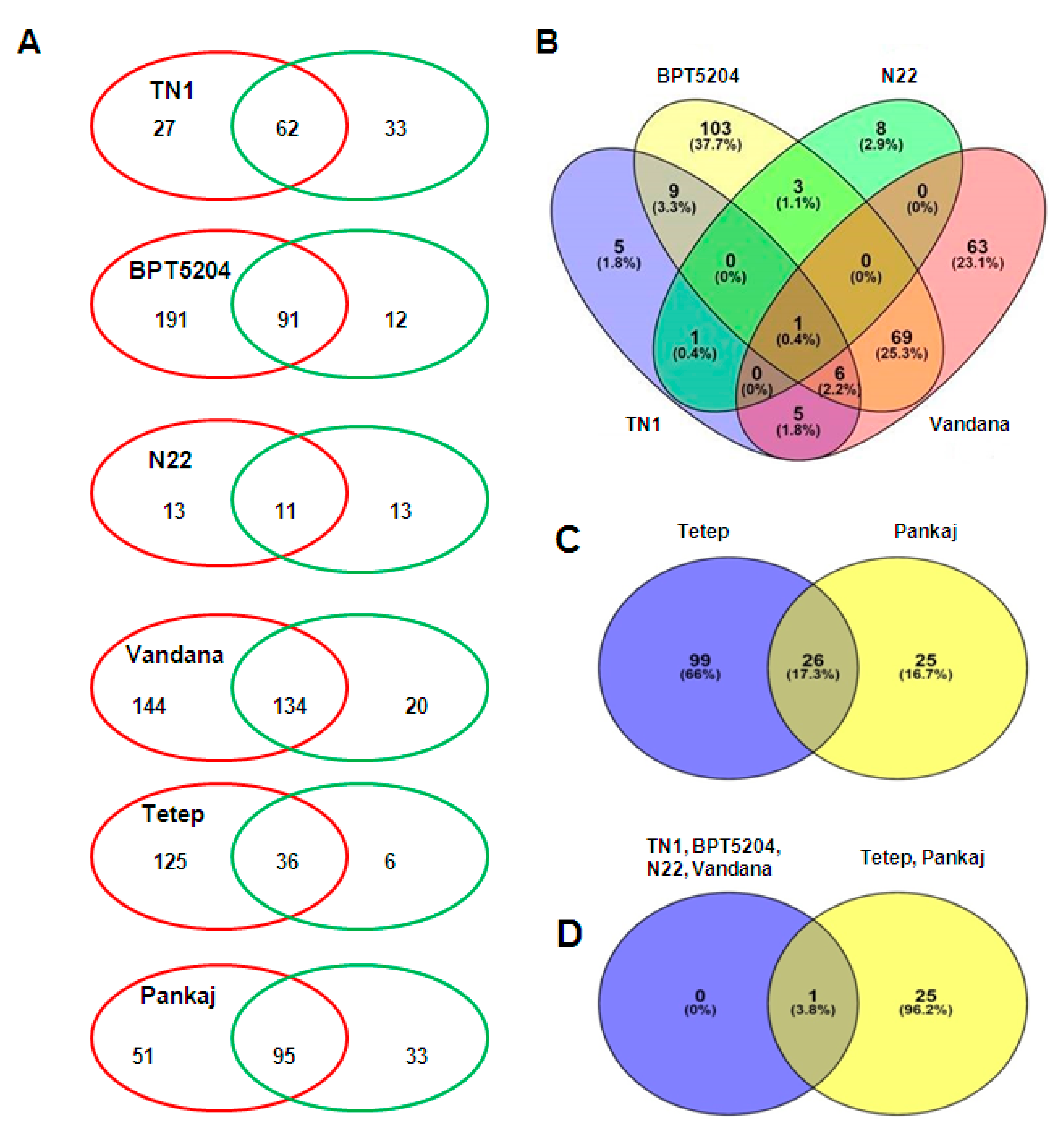
2.4. Comparison of miRNAs Expression in Control and Infected Samples
2.5. Differential Regulation of miRNAs and Target Genes
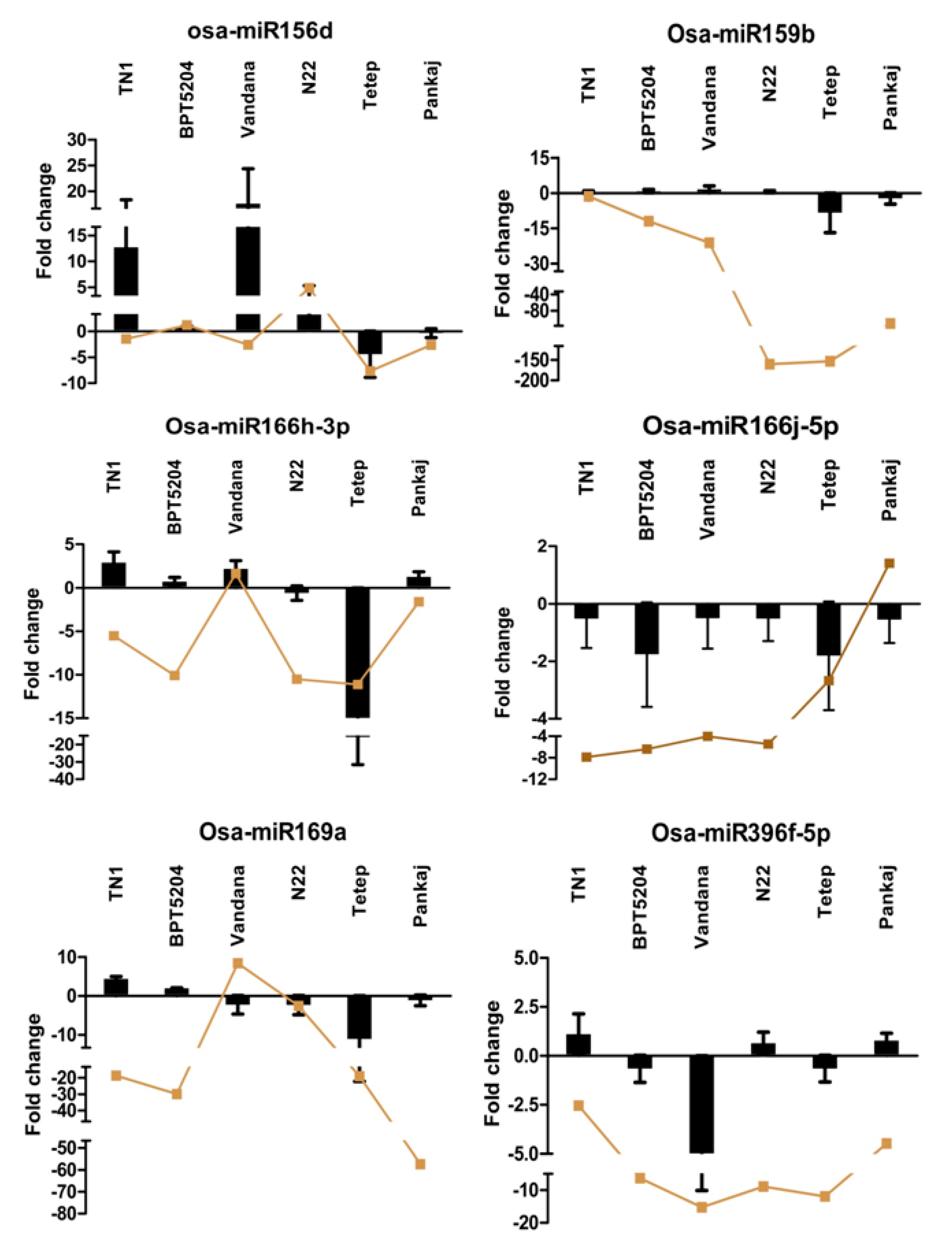
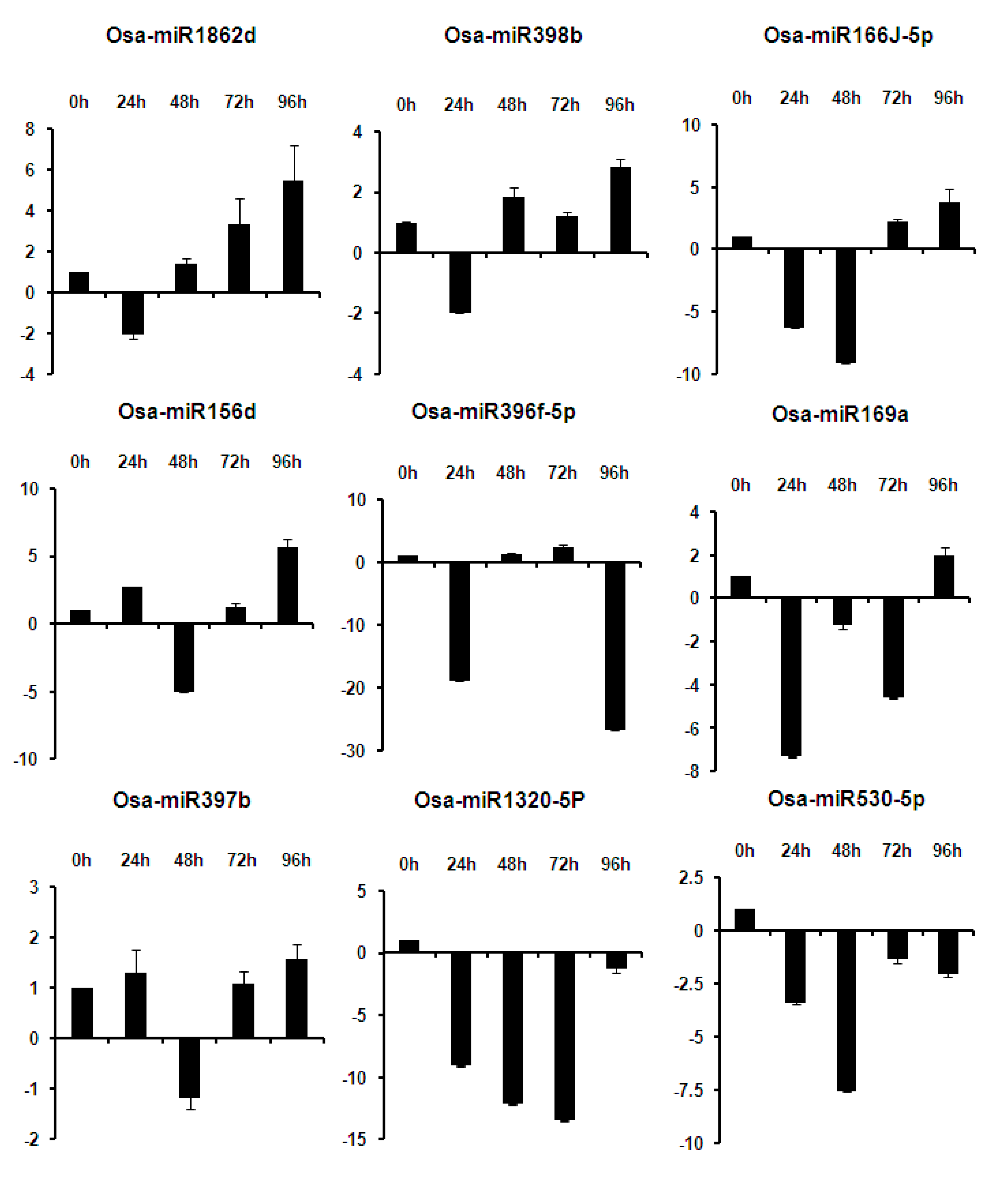
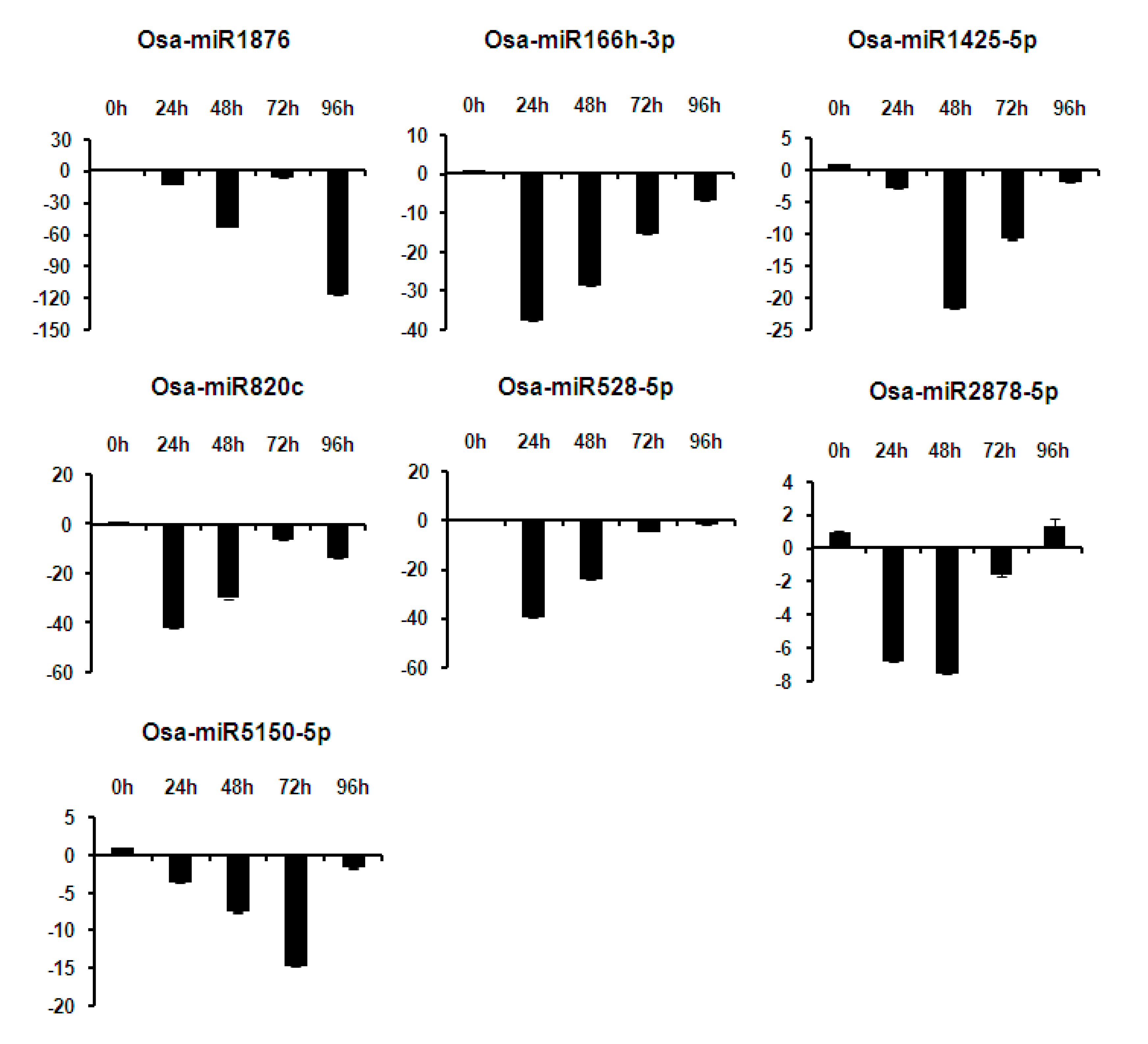
2.6. Expression Analysis of miRNAs at Different Time Points of Infection
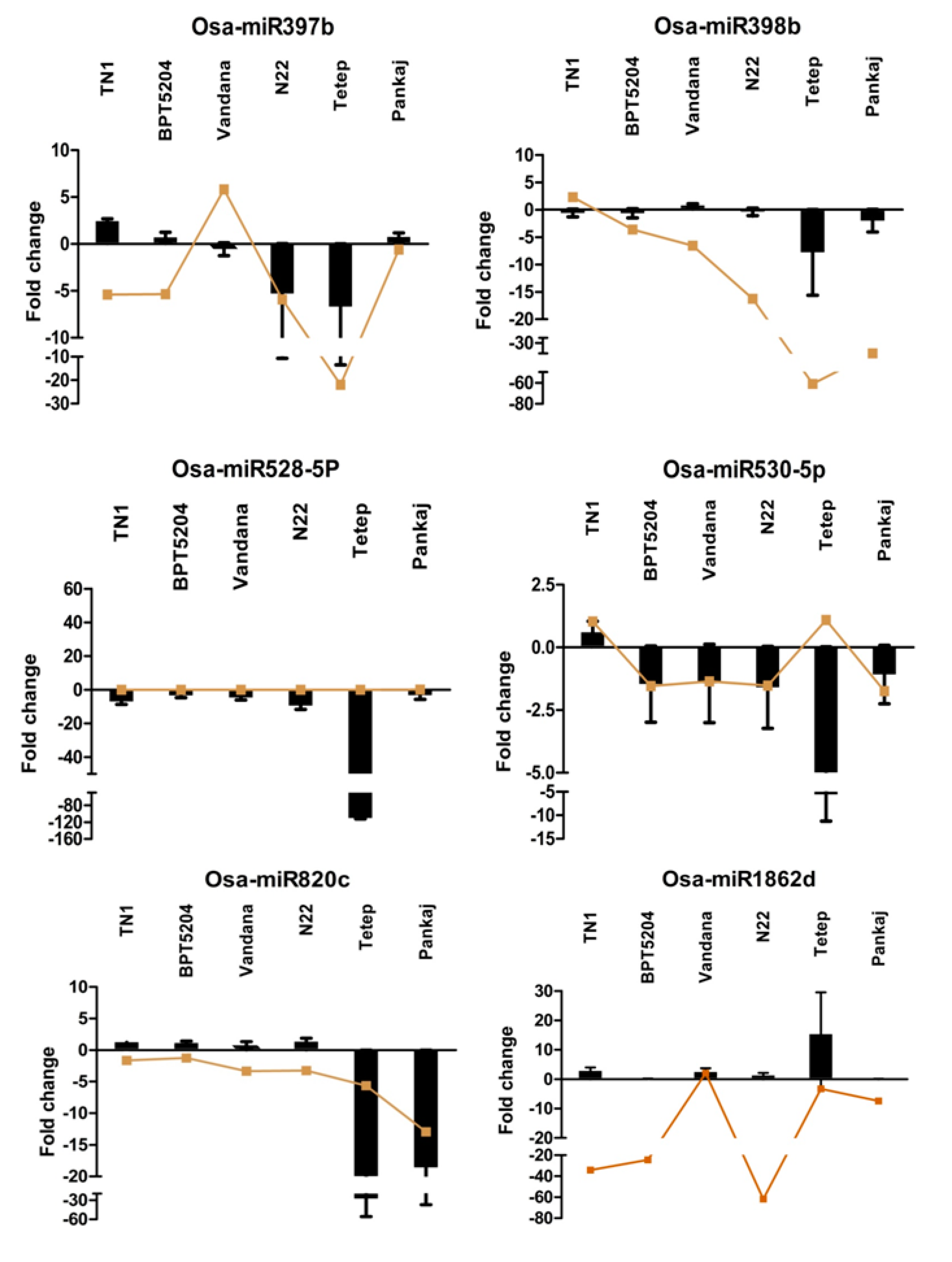
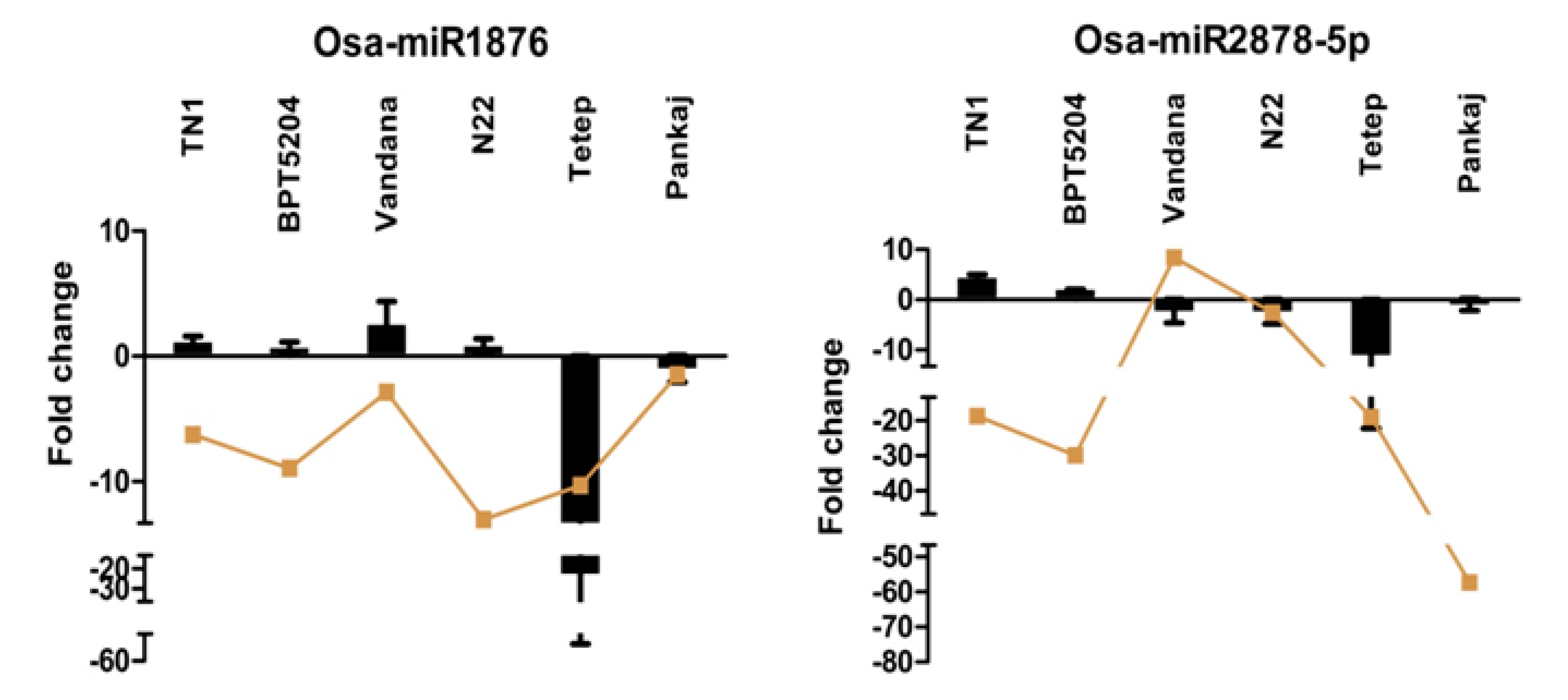
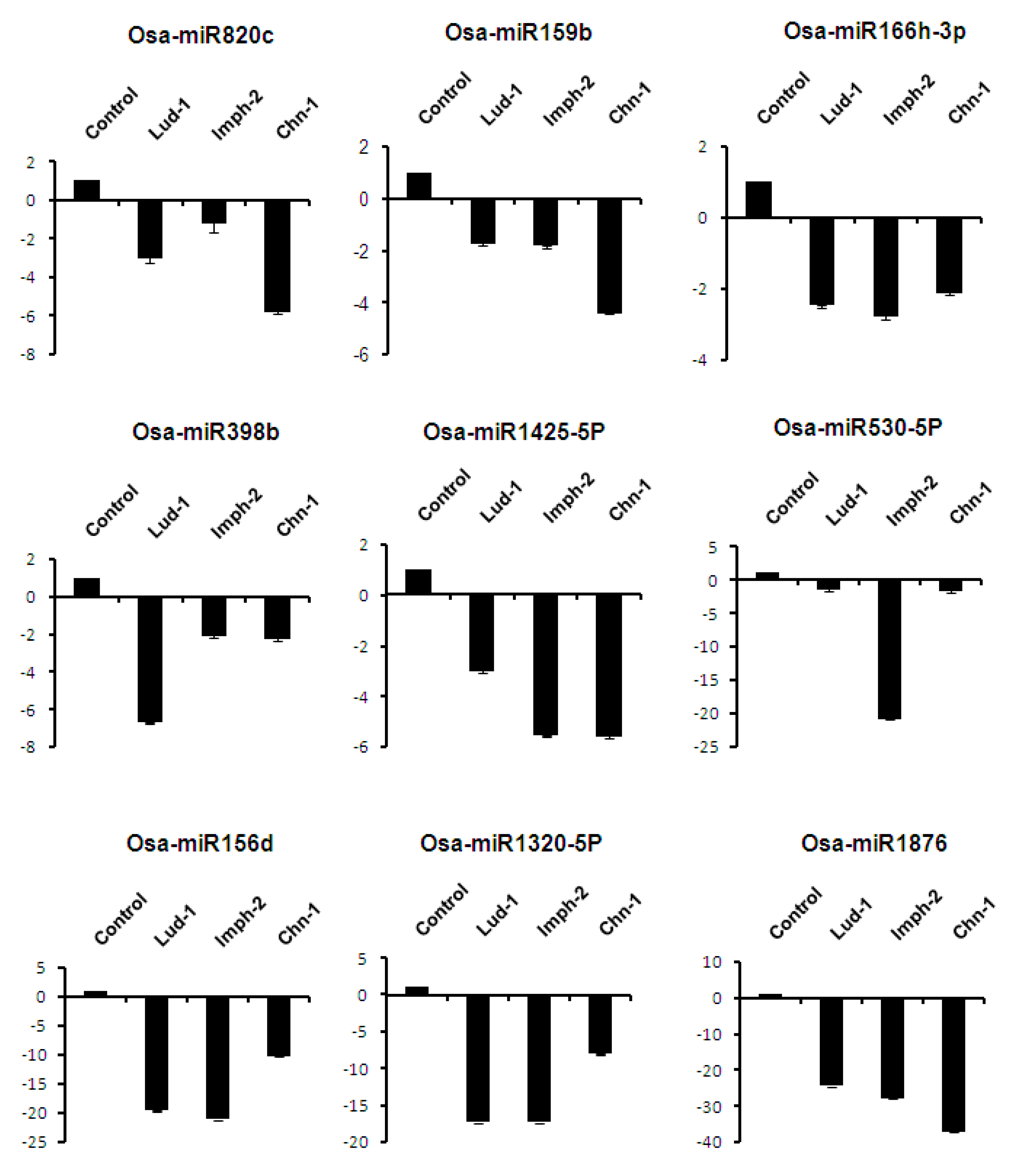
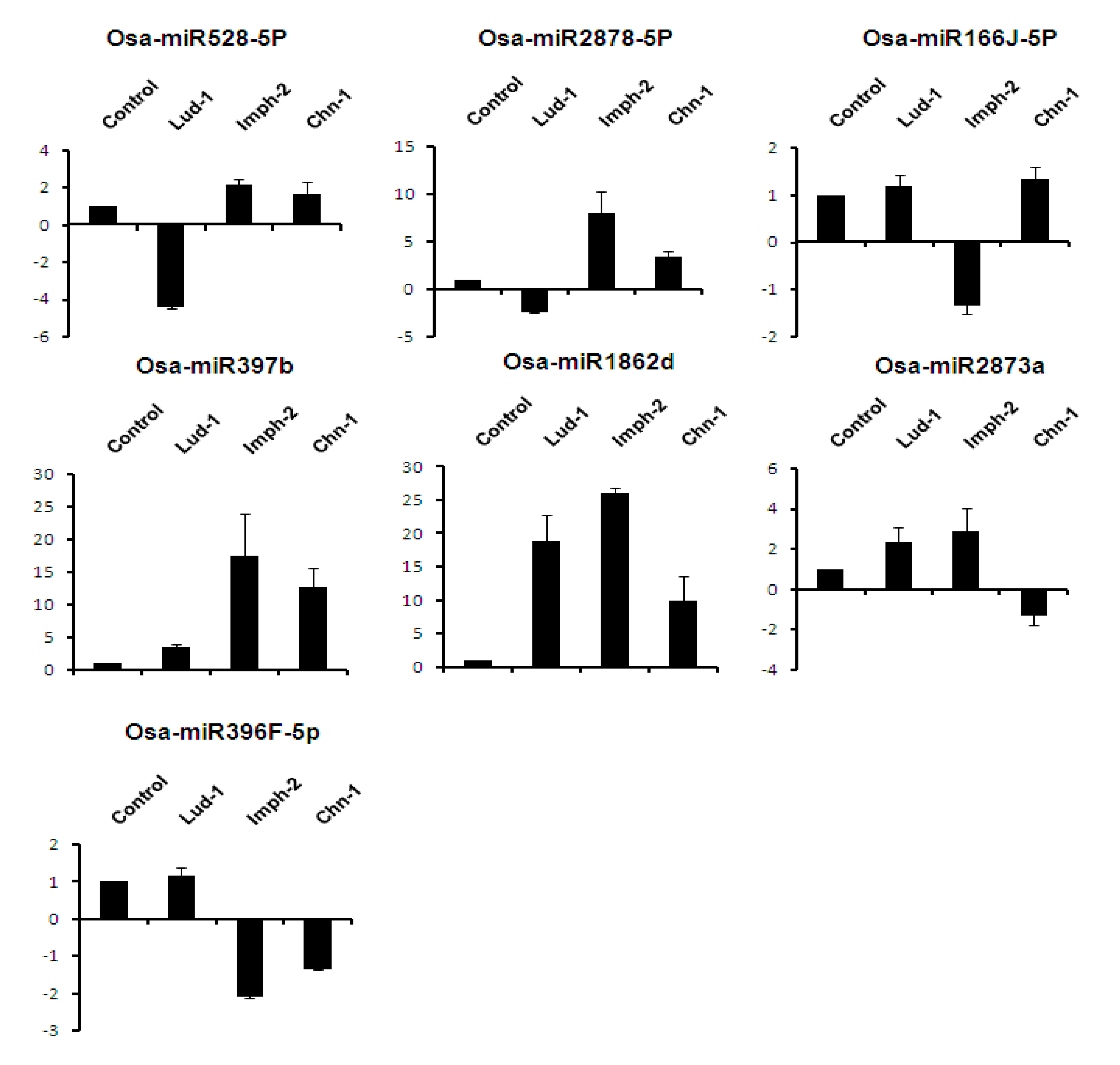
2.7. Expression Analysis of miRNAs Induced by Different Strains of R. solani
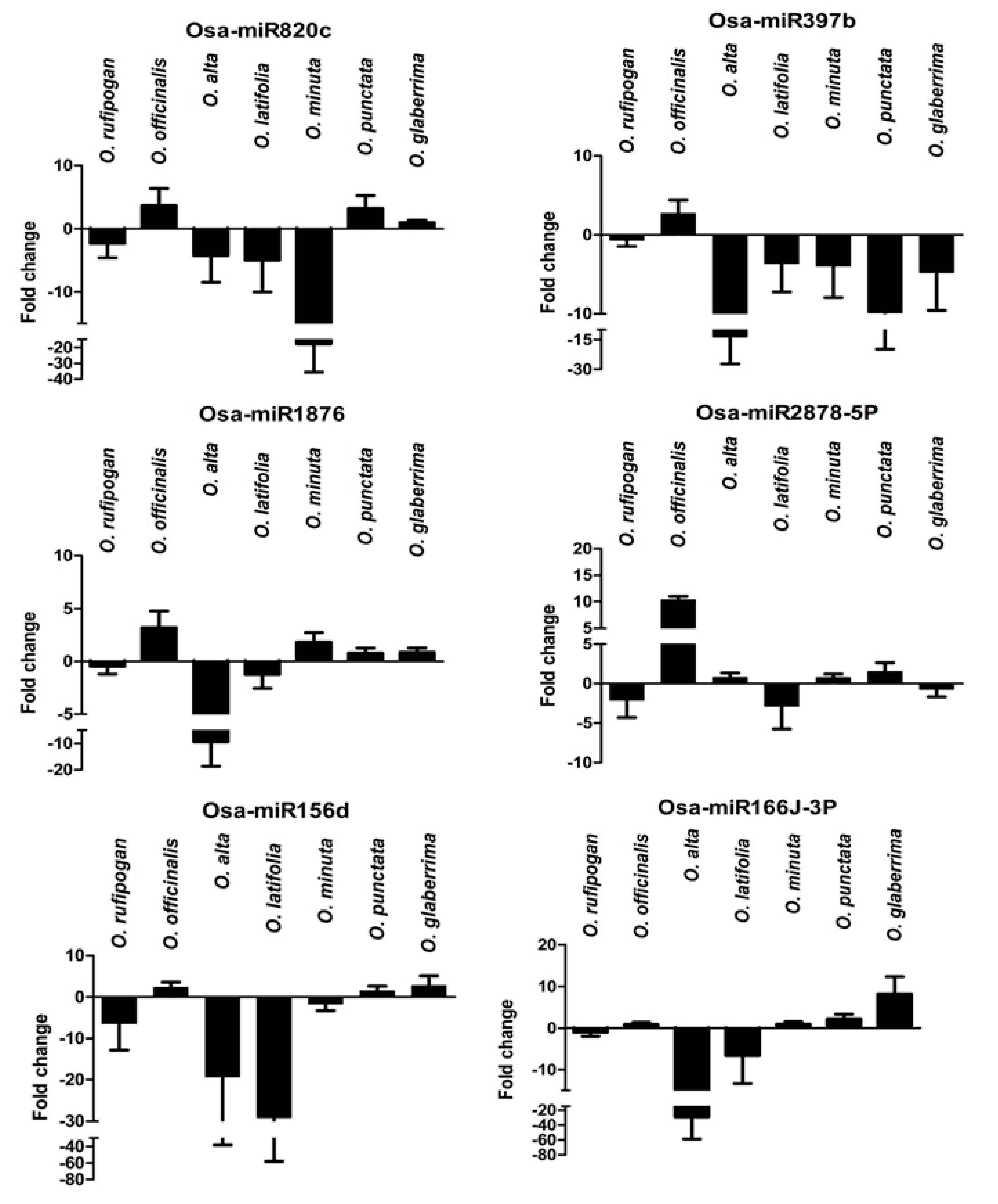
2.8. Expression Analysis of R. solani Responsive miRNAs in Wild Relatives of Rice
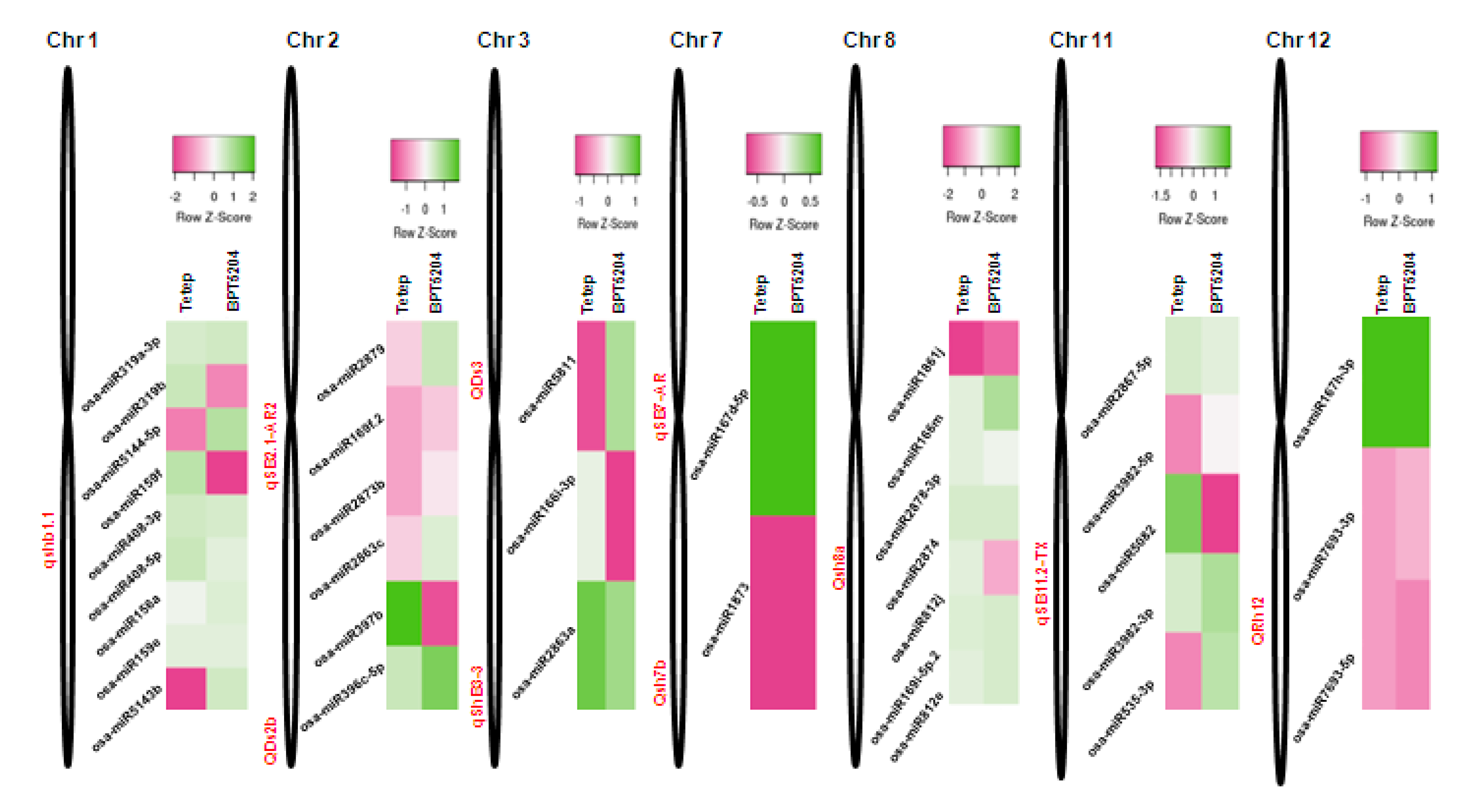
2.9. Mapping of R. solani-Induced miRNAs to Sheath Blight-Resistant QTL Regions
3. Discussion
4. Materials and Methods
4.1. Plant Material, Fungal Strains, and Sample Preparation
4.2. Small RNA Library Construction and Sequencing
4.3. Sequence Analysis
4.4. RT-qPCR Analysis of miRNAs and Target Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data and Materials
References
- Cu, R.M.; Mew, T.W.; Cassman, K.G.; Teng, P.S. Effect of sheath blight on yield in tropical, intensive rice production system. Plant Dis. 1996, 1103–1108. [Google Scholar] [CrossRef]
- Margani, R.; Widadi, S. Utilizing Bacillus to inhibit the growth and infection by sheath blight pathogen Rhizoctonia solani in rice. In IOP Conference Series: Earth and Environmental Science; IOP Publishing Ltd.: Bristol, UK, 2018; Volume 142. [Google Scholar] [CrossRef] [Green Version]
- Ogoshi, A. Introduction—The genus Rhizoctonia. In Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control; Sneh, B., Jabaji-Hare, S., Neate, S., Dijst, G., Eds.; Springer: Dordrecht, The Netherlands, 1996; pp. 1–9. [Google Scholar] [CrossRef]
- Singh, P.; Mazumdar, P.; Harikrishna, J.A.; Babu, S. Sheath blight of rice: A review and identification of priorities for future research. Planta 2019, 250, 1387–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molla, K.A.; Karmakar, S.; Molla, J.; Bajaj, P.; Varshney, R.K.; Datta, S.K.; Datta, K. Understanding sheath blight resistance in rice: The road behind and the road ahead. Plant Biotechnol. J. 2020, 18, 895–915. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Yan, J.; Liang, Y.; Shi, Y.; He, Z.; Wu, Y.; Zeng, Q.; Liu, X.; Peng, J. Resistance Genes and their Interactions with Bacterial Blight/Leaf Streak Pathogens (Xanthomonas oryzae) in Rice (Oryza sativa L.)-an Updated Review. Rice 2020, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Chern, M.; Yin, J.; Wang, J.; Chen, X. Recent advances in broad-spectrum resistance to the rice blast disease. Curr. Opin. Plant Biol. 2019, 50, 114–120. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, S.; Yuan, M. An update on molecular mechanism of disease resistance genes and their application for genetic improvement of rice. Mol. Breed. 2019, 39, 154. [Google Scholar] [CrossRef]
- Rao, T.B.; Chopperla, R.; Methre, R.; Punniakotti, E.; Venkatesh, V.; Sailaja, B.; Reddy, M.R.; Yugander, A.; Laha, G.S.; Madhav, M.S.; et al. Pectin induced transcriptome of a Rhizoctonia solani strain causing sheath blight disease in rice reveals insights on key genes and RNAi machinery for development of pathogen derived resistance. Plant Mol. Biol. 2019, 100, 59–71. [Google Scholar] [CrossRef]
- Katiyar-Agarwal, S.; Jin, H. Role of small RNAs in host-microbe interactions. Annu. Rev. phytopathol. 2010, 48, 225–246. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Huang, H. Roles of small RNAs in plant disease resistance. J. Integr. Plant Biol. 2014, 56, 962–970. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Sunkar, R.; Girke, T.; Jain, P.K.; Zhu, J.K. Cloning and characterization of microRNAs from rice. Plant Cell 2005, 17, 1397–1411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Genes Dev. 2002, 16, 1616–1626. [Google Scholar] [CrossRef] [Green Version]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangrauthia, S.K.; Bhogireddy, S.; Agarwal, S.; Prasanth, V.V.; Voleti, S.R.; Neelamraju, S.; Subrahmanyam, D. Genome-wide changes in microRNA expression during short and prolonged heat stress and recovery in contrasting rice cultivars. J. Exp. Bot. 2017, 68, 2399–2412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.; Li, Y.; Cao, X.; Qi, Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.H.; Kumar, V.; Khare, T.; Tripathi, P.; Shah, T.; Ramakrishna, C.; Aglawe, S.; Mangrauthia, S.K. miRNA applications for engineering abiotic stress tolerance in plants. Biologia 2020, 75, 1063–1081. [Google Scholar] [CrossRef]
- Campo, S.; Peris-Peris, C.; Sire, C.; Moreno, A.B.; Donaire, L.; Zytnicki, M.; Notredame, C.; Llave, C.; San Segundo, B. Identification of a novel microRNA (miRNA) from rice that targets an alternatively spliced transcript of the Nramp6 (Natural resistance-associated macrophage protein 6) gene involved in pathogen resistance. New Phytol. 2013, 199, 212–227. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, M.; Tang, M.; Dong, B.; Wu, D.; Zhang, Z.; Zhou, B. Repression of microRNA biogenesis by silencing of OsDCL1 activates the basal resistance to Magnaporthe oryzae in rice. Plant Sci. 2015, 237, 24–32. [Google Scholar] [CrossRef]
- Li, Z.Y.; Xia, J.; Chen, Z.; Yu, Y.; Li, Q.F.; Zhang, Y.C.; Zhang, J.P.; Wang, C.Y.; Zhu, X.Y.; Zhang, W.; et al. Large-scale rewiring of innate immunity circuitry and microRNA regulation during initial rice blast infection. Sci. Rep. 2016, 6, 25493. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, S.L.; Li, J.L.; Hu, X.H.; Wang, H.; Cao, X.L.; Xu, Y.J.; Zhao, Z.X.; Xiao, Z.Y.; Yang, N.; et al. Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front. Plant Sci. 2017, 8, 2. [Google Scholar] [CrossRef] [Green Version]
- Salvador-Guirao, R.; Baldrich, P.; Weigel, D.; Rubio-Somoza, I.; San Segundo, B. The microRNA miR773 is involved in the Arabidopsis immune response to fungal pathogens. Mol. Plant Microbe Interact. 2018, 31, 249–259. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Xia, Y.; Lin, S.; Wang, Y.; Guo, B.; Song, X.; Ding, S.; Zheng, L.; Feng, R.; Chen, S.; et al. Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Plant J. 2018, 95, 584–597. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Jeyakumar, J.M.; Feng, Q.; Zhao, Z.X.; Fan, J.; Khaskheli, M.I.; Wang, W.M. The roles of rice microRNAs in rice-Magnaporthe oryzae interaction. Phytopathol. Res. 2019, 1, 33. [Google Scholar] [CrossRef]
- Chandran, V.; Wang, H.; Gao, F.; Cao, X.L.; Chen, Y.P.; Li, G.B.; Zhu, Y.; Yang, X.M.; Zhang, L.L.; Zhao, Z.X.; et al. miR396-OsGRFs module balances growth and rice blast disease-resistance. Front. Plant Sci. 2019, 9, 1999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wenlei, C.; Xinxin, C.; Jianhua, Z.; Zhaoyang, Z.; Zhiming, F.; Shouqiang, O.; Shimin, Z. Comprehensive characteristics of microRNA expression profile conferring to Rhizoctonia solani in Rice. Rice Sci. 2020, 27, 101–112. [Google Scholar] [CrossRef]
- Basu, A.; Chowdhury, S.; Ray Chaudhuri, T.; Kundu, S. Differential behaviour of sheath blight pathogen Rhizoctonia solani in tolerant and susceptible rice varieties before and during infection. Plant Pathol. 2016, 65, 1333–1346. [Google Scholar] [CrossRef] [Green Version]
- Lin, R.; He, L.; He, J.; Qin, P.; Wang, Y.; Deng, Q.; Yang, X.; Li, S.; Wang, S.; Wang, W.; et al. Comprehensive analysis of microRNA-Seq and target mRNAs of rice sheath blight pathogen provides new insights into pathogenic regulatory mechanisms. DNA Res. 2016, 23, 415–425. [Google Scholar] [CrossRef]
- Garris, A.J.; Tai, T.H.; Coburn, J.; Kresovich, S.; McCouch, S. Genetic structure and diversity in Oryza sativa L. Genetics 2005, 169, 1631–1638. [Google Scholar] [CrossRef] [Green Version]
- Prathi, N.B.; Palit, P.; Madhu, P.; Ramesh, M.; Laha, G.S.; Balachandran, S.M.; Madhav, M.S.; Sundaram, R.M.; Mangrauthia, S.K. Proteomic and transcriptomic approaches to identify resistance and susceptibility related proteins in contrasting rice genotypes infected with fungal pathogen Rhizoctonia solani. Plant Physiol. Biochem. 2018, 130, 258–266. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, X.; Sun, B.; Hao, L.; Liu, C.; Zhang, D.; Tang, H.; Li, C.; Li, Y.; Shi, Y.; et al. Genome-wide identification and comparative analysis of drought-related microRNAs in two maize inbred lines with contrasting drought tolerance by deep sequencing. PLoS ONE 2019, 14, e0219176. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Lilly, J.J.; Subramanian, B. Gene network mediated by WRKY13 to regulate resistance against sheath infecting fungi in rice (Oryza sativa L.). Plant Sci. 2019, 280, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Goad, D.M.; Jia, Y.; Gibbons, A.; Liu, Y.; Gealy, D.; Caicedo, A.L.; Olsen, K.M. Identification of Novel QTL Conferring Sheath Blight Resistance in Two Weedy Rice Mapping Populations. Rice 2020, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- Baldrich, P.; Campo, S.; Wu, M.T.; Liu, T.T.; Hsing, Y.I.; San Segundo, B. MicroRNA-mediated regulation of gene expression in the response of rice plants to fungal elicitors. RNA Biol. 2015, 12, 847–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouyang, S.; Park, G.; Atamian, H.S.; Han, C.S.; Stajich, J.E.; Kaloshian, I.; Borkovich, K.A. MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog. 2014, 10, e1004464. [Google Scholar] [CrossRef] [PubMed]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [Green Version]
- Bundo, M.; Campo, S.; San Segundo, B. Plant microRNAs. Concepts and Strategies in Plant Sciences; Miguel, C., Dalmay, T., Chaves, I., Eds.; Springer Nature Switzerland AG: Cham, Switzerland, 2020; pp. 199–220. [Google Scholar] [CrossRef]
- Li, Y.; Lu, Y.G.; Shi, Y.; Wu, L.; Xu, Y.J.; Huang, F.; Guo, X.Y.; Zhang, Y.; Fan, J.; Zhao, J.Q.; et al. Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol. 2014, 164, 1077–1092. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, F.; Li, J.; Chen, J.-P.; Zhang, H.-M. Integrative analysis of the microRNAome and transcriptome illuminates the response of susceptible rice plants to rice stripe virus. PLoS ONE 2016, 11, e0146946. [Google Scholar] [CrossRef] [Green Version]
- Jia, Y.; Li, C.; Li, Q.; Liu, P.; Liu, D.; Liu, Z.; Wang, Y.; Jiang, G.; Zhai, W. Characteristic dissection of Xanthomonas oryzae pv. oryzae responsive microRNAs in Rice. Int. J. Mol. Sci. 2020, 21, 785. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Jia, Y.; Liu, F.; Wang, F.; Fan, F.; Wang, J.; Zhu, J.; Xu, Y.; Zhong, W.; Yang, J. Integration analysis of small RNA and degradome sequencing reveals MicroRNAs responsive to Dickeya zeae in resistant rice. Int. J. Mol. Sci. 2019, 20, 222. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Bao, Y.; Shan, D.; Wang, Z.; Song, X.; Wang, Z.; Wang, J.; He, L.; Wu, L.; Zhang, Z.; et al. Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 2018, 177, 352–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Zhu, S.; Liu, F.; Wang, W.; Wang, X.; Han, G.; Cheng, B. Identification of arbuscular mycorrhiza fungi responsive microRNAs and their regulatory network in maize. Int. J. Mol. Sci. 2018, 19, 3201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, H.; Hong, G.; Li, L.; Zhang, X.; Kong, Y.; Sun, Z.; Li, J.; Chen, J.; He, Y. miR156/SPL9 regulates reactive oxygen species accumulation and immune response in Arabidopsis thaliana. Phytopathology 2019, 109, 632–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Z.; Wang, N.; Jalajakumari, M.; Blackman, L.; Shen, E.; Verma, S.; Wang, M.-B.; Millar, A.A. miR159 represses a constitutive pathogen defense response in tobacco. Plant Physiol. 2020, 182, 2182–2198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhang, Q.; Hao, L.; Wang, S.; Wang, S.; Zhang, W.; Xu, C.; Yu, Y.; Li, T. A novel miRNA negatively regulates resistance to Glomerella leaf spot by suppressing expression of an NBS gene in apple. Hortic. Res. 2019, 6, 93. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.-Y.; Zhang, S.; Yu, Y.; Luo, Y.-C.; Liu, Q.; Ju, C.; Zhang, Y.-C.; Qu, L.-H.; Lucas, W.J.; Wang, X.; et al. MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnol. J. 2014, 12, 1132–1142. [Google Scholar] [CrossRef] [Green Version]
- Lian, S.; Cho, W.K.; Kim, S.-M.; Choi, H.; Kim, K.-H. Time-course small RNA profiling reveals rice miRNAs and their target genes in response to rice stripe virus infection. PLoS ONE 2016, 11, e0162319. [Google Scholar] [CrossRef]
- Li, Y.; Cao, X.-L.; Zhu, Y.; Yang, X.-M.; Zhang, K.-N.; Xiao, Z.-Y.; Wang, H.; Zhao, J.-H.; Zhang, L.-L.; Li, G.-B.; et al. Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol. 2019, 222, 1507–1522. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Qian, Y. Cloning and heterologous expression of SS10, a subtilisin-like protease displaying antifungal activity from Trichoderma harzianum. FEMS Microbiol. Lett. 2009, 290, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Liu, F.; Lechner, E.; Genschik, P.; Crosby, W.L.; Ma, H.; Peng, W.; Huang, D.; Xie, D. The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 2002, 14, 1919–1935. [Google Scholar] [CrossRef] [Green Version]
- Jing, Y.; Liu, J.; Liu, P.; Ming, D.; Sun, J. Overexpression of TaJAZ1 increases powdery mildew resistance through promoting reactive oxygen species accumulation in bread wheat. Sci. Rep. 2019, 9, 5691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, T.B.; Chopperla, R.; Prathi, N.B.; Balakrishnan, M.; Prakasam, V.; Laha, G.S.; Balachandran, S.M.; Mangrauthia, S.K. A Comprehensive Gene Expression Profile of Pectin Degradation Enzymes Reveals the Molecular Events during Cell Wall Degradation and Pathogenesis of Rice Sheath Blight Pathogen Rhizoctonia solani AG1-IA. J. Fungi 2020, 6, 71. [Google Scholar] [CrossRef]
- Wang, Y.; Bai, X.; Yan, C.; Gui, Y.; Wei, X.; Zhu, Q.-H.; Guo, L.; Fan, L. Genomic dissection of small RNAs in wild rice (Oryza rufipogon): Lessons for rice domestication. New Phytol. 2012, 196, 914–925. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-W.; Long, Y.; Xue, M.; Xiao, X.; Pei, X. Identification of microRNAs in response to drought in common wild rice (Oryza rufipogon Griff.) shoots and roots. PLoS ONE 2017, 12, e0170330. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.K.; Panda, A.K.; Rawal, H.C.; Sharma, T.R. Discovery of microRNA-target modules of African rice (Oryza glaberrima) under salinity stress. Sci. Rep. 2018, 8, 570. [Google Scholar] [CrossRef]
- Swetha, C.; Basu, D.; Pachamuthu, K.; Tirumalai, V.; Nair, A.; Prasad, M.; Shivaprasad, P.V. Major domestication-related phenotypes in Indica rice are due to loss of miRNA-mediated laccase silencing. Plant Cell 2018, 30, 2649–2662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yugander, A.; Ladhalakshmi, D.; Prakasham, V.; Mangrauthia, S.K.; Prasad, M.S.; Krishnaveni, D.; Madhav, M.S.; Sundaram, R.M.; Laha, G.S. Pathogenic and genetic variation among the isolates of Rhizoctonia solani (AG 1-IA), the rice sheath blight pathogen. J. Phytopathol. 2015, 163, 465–474. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Andres-Leon, E.; Nunez-Torres, R.; Rojas, A. miARma-Seq: A comprehensive tool for miRNA, mRNA and circRNA analysis. Sci. Rep. 2016, 11, 25749. [Google Scholar] [CrossRef] [Green Version]
- Ossowski, S.; Schwab, R.; Weigel, D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008, 53, 674–690. [Google Scholar] [CrossRef]
- Gao, J.; Luo, M.; Zhang, C.; Peng, H.; Lin, H.; Shen, Y.; Zhao, M.; Pan, G.; Zhang, Z. A putative pathogen-resistant regulatory pathway between MicroRNAs and candidate target genes in maize. J. Plant Biol. 2015, 58, 211–219. [Google Scholar] [CrossRef]
- Ding, Y.; Chen, Z.; Zhu, C. Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J. Exp. Bot. 2011, 62, 3563–3573. [Google Scholar] [CrossRef] [PubMed]
- Sailaja, B.; Voleti, S.R.; Subrahmanyam, D.; Sarla, N.; Prasanth, V.V.; Bhadana, V.P.; Mangrauthia, S.K. Prediction and expression analysis of miRNAs associated with heat stress in Oryza sativa. Rice Sci. 2014, 21, 3–12. [Google Scholar] [CrossRef]
- Lee, S.; Persson, D.P.; Hansen, T.H.; Husted, S.; Schjoerring, J.K.; Kim, Y.-S.; Jeon, U.S.; Kim, Y.-K.; Kakei, Y.; Masuda, H.; et al. Bio-available zinc in rice seeds is increased by activation tagging of nicotianamine synthase. Plant Biotechnol. J. 2011, 9, 865–873. [Google Scholar] [CrossRef] [PubMed]
| Sample Name | Total Number of Reads | Clean Reads before Data Filtering | Q30 (%) | Clean Reads after Data Filtering | Clean Reads (18–24 nt) | Identified Known miRNAs | Identified Novel miRNAs |
|---|---|---|---|---|---|---|---|
| R. solani-infected samples | |||||||
| TN1 | 17,292,908 | 17,277,169 | 96.65 | 16,998,190 | 1,209,465 | 89 | 7 |
| BPT5204 | 19,838,709 | 19,817,284 | 96.7 | 19,603,829 | 3,442,243 | 282 | 8 |
| N22 | 12,331,370 | 12,318,143 | 96.45 | 12,028,969 | 1,563,885 | 24 | 9 |
| Vandana | 15,584,456 | 15,566,007 | 96.42 | 15,179,461 | 1,082,338 | 278 | 21 |
| Tetep | 16,772,072 | 16,754,325 | 96.64 | 16,346,243 | 2,601,317 | 161 | 9 |
| Pankaj | 14,666,490 | 14,652,163 | 96.66 | 14,207,919 | 1,023,439 | 147 | 4 |
| Control samples | |||||||
| TN1 | 18,776,171 | 18,757,059 | 96.58 | 18,058,533 | 822,299 | 95 | 7 |
| BPT5204 | 17,754,335 | 17,733,540 | 95.66 | 17,107,679 | 1,202,770 | 104 | 5 |
| N22 | 16,568,740 | 16,549,626 | 96.38 | 15,910,033 | 1,397,952 | 24 | 2 |
| Vandana | 15,698,991 | 15,682,752 | 96.48 | 14,906,413 | 1,062,798 | 154 | 6 |
| Tetep | 15,304,171 | 15,283,850 | 96.11 | 14,364,145 | 986,089 | 43 | 2 |
| Pankaj | 16,132,109 | 16,114,741 | 96.57 | 15,524,879 | 851,888 | 128 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chopperla, R.; Mangrauthia, S.K.; Bhaskar Rao, T.; Balakrishnan, M.; Balachandran, S.M.; Prakasam, V.; Channappa, G. A Comprehensive Analysis of MicroRNAs Expressed in Susceptible and Resistant Rice Cultivars during Rhizoctonia solani AG1-IA Infection Causing Sheath Blight Disease. Int. J. Mol. Sci. 2020, 21, 7974. https://doi.org/10.3390/ijms21217974
Chopperla R, Mangrauthia SK, Bhaskar Rao T, Balakrishnan M, Balachandran SM, Prakasam V, Channappa G. A Comprehensive Analysis of MicroRNAs Expressed in Susceptible and Resistant Rice Cultivars during Rhizoctonia solani AG1-IA Infection Causing Sheath Blight Disease. International Journal of Molecular Sciences. 2020; 21(21):7974. https://doi.org/10.3390/ijms21217974
Chicago/Turabian StyleChopperla, Ramakrishna, Satendra K. Mangrauthia, Talluri Bhaskar Rao, Marudamuthu Balakrishnan, Sena Munuswamy Balachandran, Vellaisamy Prakasam, and Gireesh Channappa. 2020. "A Comprehensive Analysis of MicroRNAs Expressed in Susceptible and Resistant Rice Cultivars during Rhizoctonia solani AG1-IA Infection Causing Sheath Blight Disease" International Journal of Molecular Sciences 21, no. 21: 7974. https://doi.org/10.3390/ijms21217974
APA StyleChopperla, R., Mangrauthia, S. K., Bhaskar Rao, T., Balakrishnan, M., Balachandran, S. M., Prakasam, V., & Channappa, G. (2020). A Comprehensive Analysis of MicroRNAs Expressed in Susceptible and Resistant Rice Cultivars during Rhizoctonia solani AG1-IA Infection Causing Sheath Blight Disease. International Journal of Molecular Sciences, 21(21), 7974. https://doi.org/10.3390/ijms21217974





