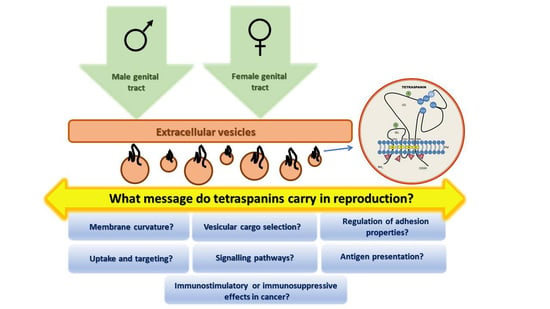
Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction
Abstract
1. Introduction
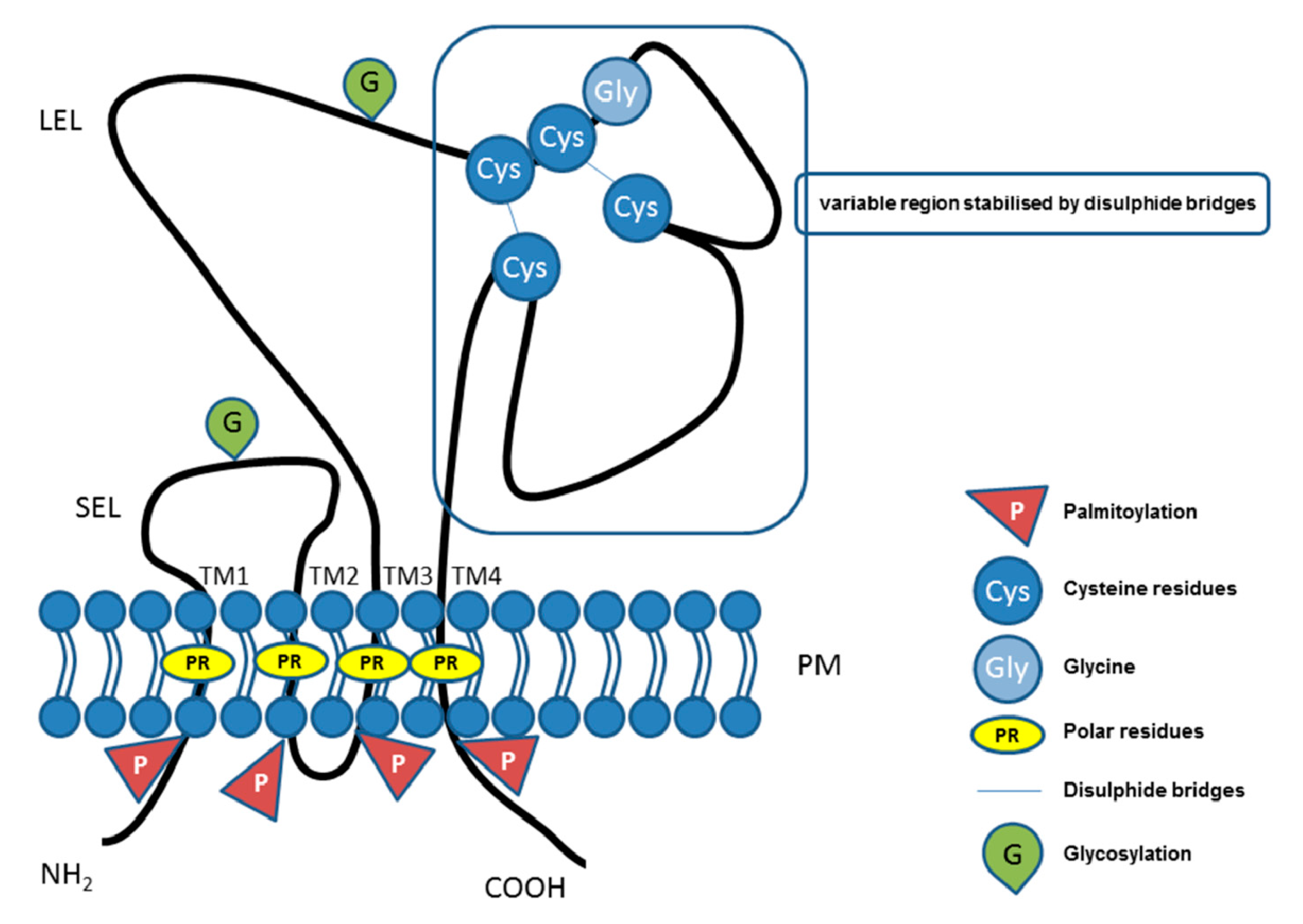
2. Tetraspanin Family Proteins
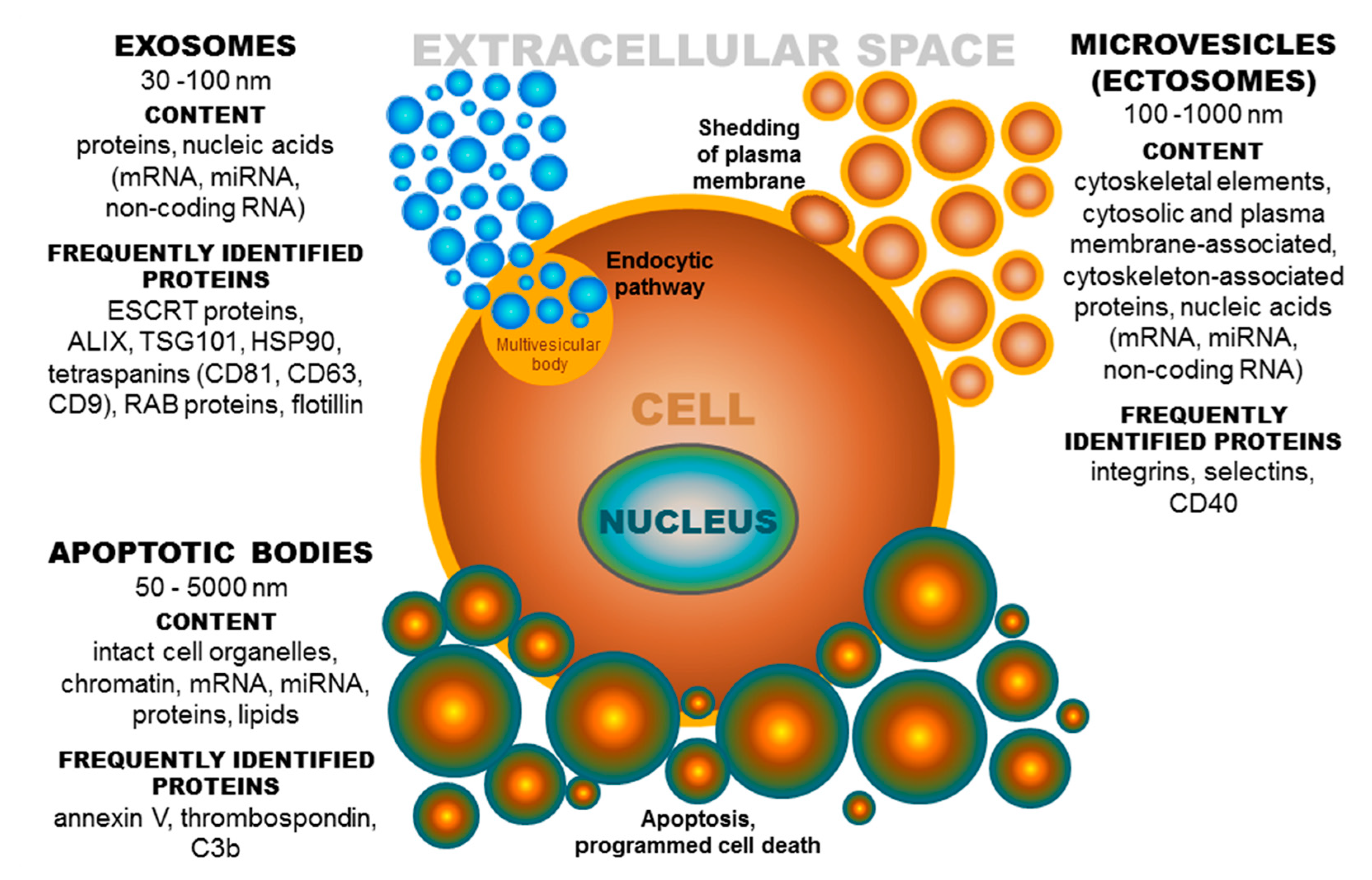
3. Tetraspanins and Other Proteins in Extracellular Vesicles (EV Tetraspanin Network?)
4. Role of EV Tetraspanins in Somatic Cells
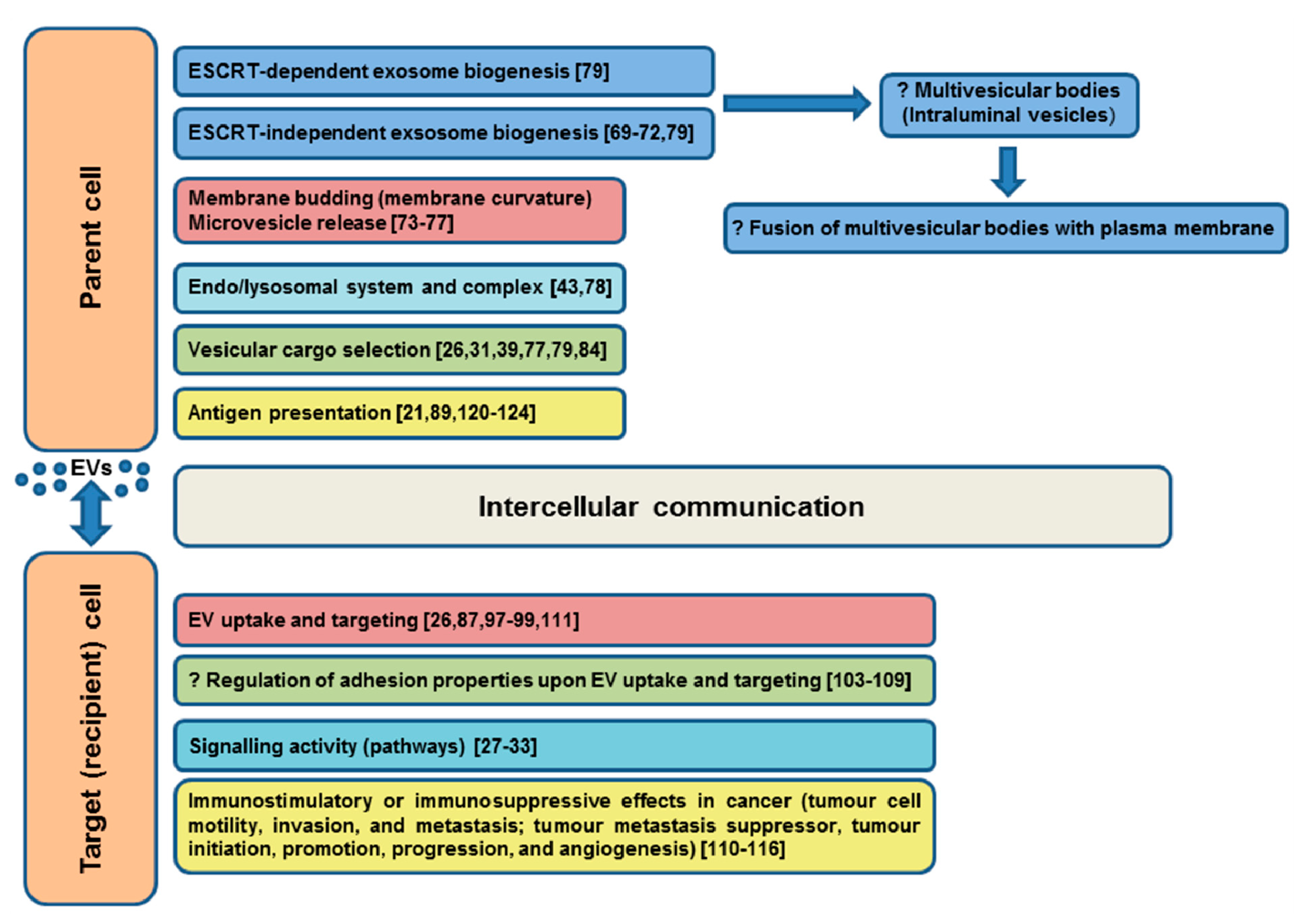
4.1. Role of Tetraspanins in EV Formation
4.2. Role of Tetraspanins in EV Cargo Selection, Targeting, and Uptake
4.3. Role of Tetraspanins in Immunostimulatory and Immunosuppressive Properties of EV Subpopulations in Cancer
4.4. Role of EV Tetraspanins in Antigen Presentation
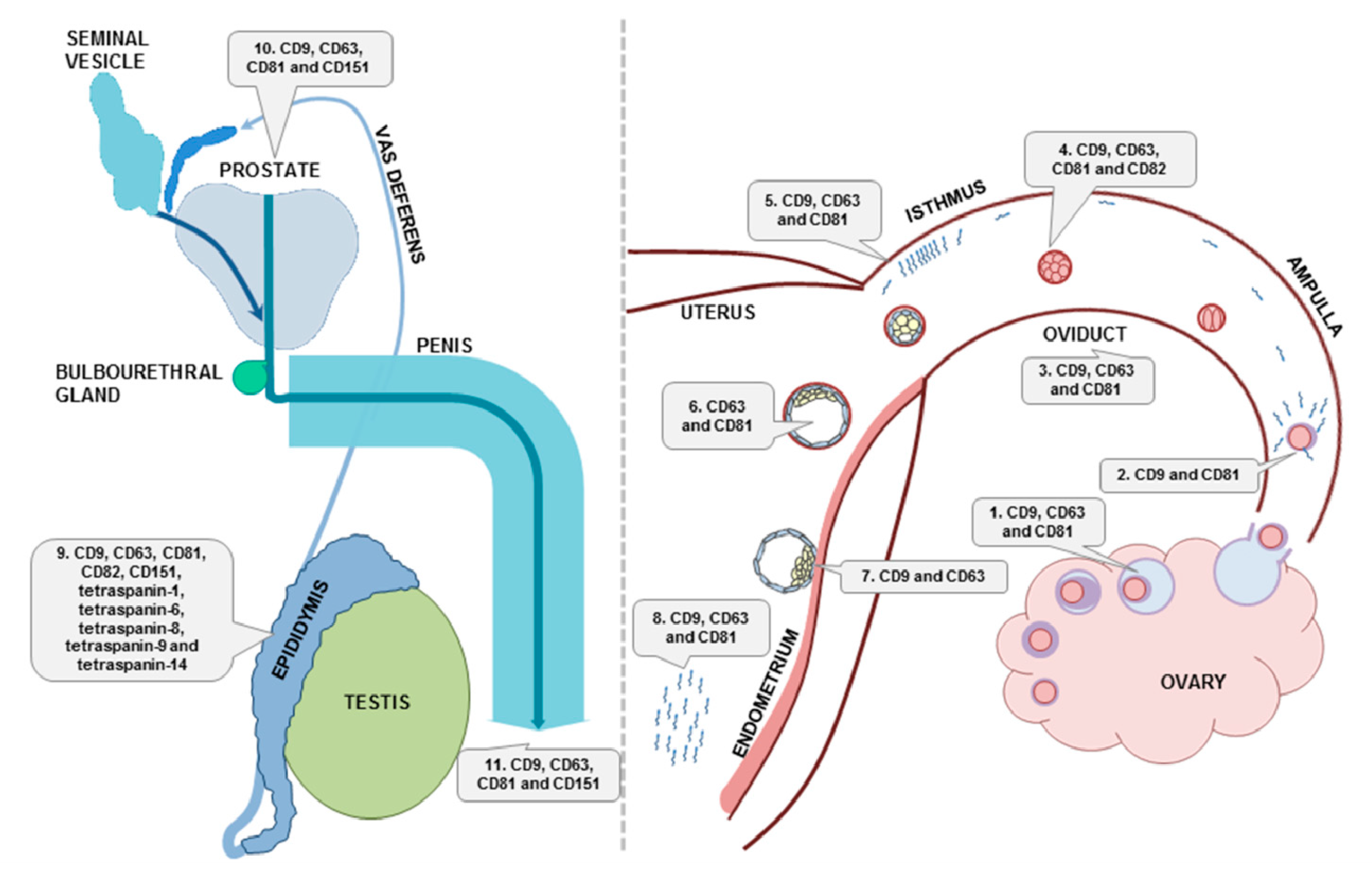
5. Tetraspanins in EVs of the Female and Male Reproductive System
5.1. Female Reproductive System
5.1.1. EVs in Follicular Fluid
5.1.2. Oocyte EVs
5.1.3. Oviductal EVs
5.1.4. EVs of Fertilized Oocytes and Early Embryos
5.1.5. EVs in Later Embryo Development and Implantation
5.2. Male Reproductive Tract
5.2.1. Epididymal EVs-Epididymosomes
5.2.2. EVs in Semen
5.2.3. Prostasomes
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABs | Apoptotic bodies |
| AKR1B1 | Aldo-keto reductase family 1 member B1 |
| ALIX | ALG-2 interacting protein X (programmed cell death 6 interacting protein) |
| APCs | Antigen-presenting cells |
| BLVRA | Biliverdin reductase A |
| CCs | Cumulus cells |
| CD | Cluster of differentiation |
| COC | Cumulus-oocyte complex |
| eEVs | Embryonal extracellular vesicles |
| ELSPBP1 | Epididymal sperm binding protein 1 |
| ESCRT | Endosomal sorting complex required for transport |
| EVs | Extracellular vesicles |
| EWI | Proteins, members of immunoglobulin superfamily |
| EXs | Exosomes |
| FF | Follicular fluid |
| GDM | Gestational diabetes mellitus |
| GliPriL1 | Glioma pathogenesis-related 1-like protein 1 |
| GPI | Glycosylphosphatidylinositol |
| HSP | Heat shock proteins |
| ICAM | Intracellular adhesion molecule |
| LEL | Large extracellular loop |
| MHC | Major histocompatibility complex |
| MIF | Macrophage migration inhibitory factor |
| MII | Metaphase II |
| MVs | Microvesicles |
| oEVs | Oviductal extracellular vesicles |
| PE | Preeclampsia |
| PMCA4 | Plasma membrane calcium ATPase 4 |
| PVS | Perivitelline space |
| RAB | Proteins included in regulation of endocytosis and secretory processes |
| ROS | Reactive oxygen species |
| SEL | Small extracellular loop |
| SPAM | Sperm adhesion molecule |
| SSTs | Sperm storage tubules |
| TEMs | Tetraspanin-enriched microdomains |
| TSG101 | Tumor susceptibility gene 101 protein |
| TZPs | Transzonal projections |
| VDAC1/2 | Voltage-dependent anion-selective channel protein 1/2 |
| ZP | Zona pellucida |
References
- Keller, S.; Ridinger, J.; Rupp, A.-K.; Janssen, J.W.; Altevogt, P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- Silverman, J.M.; Clos, J.; de’Oliveira, C.C.; Shirvani, O.; Fang, Y.; Wang, C.; Foster, L.J.; Reiner, N.E. An exosome-based secretion pathway is responsible for protein export from Leishmania and communication with macrophages. J. Cell. Sci. 2010, 123, 842–852. [Google Scholar] [CrossRef]
- Klibi, J.; Niki, T.; Riedel, A.; Pioche-Durieu, C.; Souquere, S.; Rubinstein, E.; Le Moulec, S.; Moulec, S.L.E.; Guigay, J.; Hirashima, M.; et al. Blood diffusion and Th1-suppressive effects of galectin-9-containing exosomes released by Epstein-Barr virus-infected nasopharyngeal carcinoma cells. Blood 2009, 113, 1957–1966. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Ji, H.; Simpson, R.J. Exosomes: Extracellular organelles important in intercellular communication. J. Proteom. 2010, 73, 1907–1920. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Lässer, C.; Alikhani, V.S.; Ekström, K.; Eldh, M.; Paredes, P.T.; Bossios, A.; Sjöstrand, M.; Gabrielsson, S.; Lötvall, J.; Valadi, H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J. Transl. Med. 2011, 9, 9. [Google Scholar] [CrossRef] [PubMed]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filén, J.-J.; Lahesmaa, R.; Norman, M.; Neve, E.P.A.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef]
- Montoro-García, S.; Shantsila, E.; Marín, F.; Blann, A.; Lip, G.Y.H. Circulating microparticles: New insights into the biochemical basis of microparticle release and activity. Basic Res. Cardiol. 2011, 106, 911–923. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef]
- Mathivanan, S.; Fahner, C.J.; Reid, G.E.; Simpson, R.J. ExoCarta 2012: Database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012, 40, 1241–1244. [Google Scholar] [CrossRef]
- Mathivanan, S.; Simpson, R.J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009, 9, 4997–5000. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [PubMed]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef] [PubMed]
- Barrachina, M.N.; Calderón-Cruz, B.; Fernandez-Rocca, L.; García, Á. Application of Extracellular Vesicles Proteomics to Cardiovascular Disease: Guidelines, Data Analysis, and Future Perspectives. Proteomics 2019, 19, 1800247. [Google Scholar] [CrossRef]
- György, B.; Szabó, T.G.; Pásztói, M.; Pál, Z.; Misják, P.; Aradi, B.; László, V.; Pállinger, É.; Pap, E.; Kittel, Á.; et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell. Mol. Life Sci. 2011, 68, 2667–2688. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.F.R.; Wyllie, A.H.; Currie, A.R. Apoptosis: A Basic Biological Phenomenon with Wideranging Implications in Tissue Kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef]
- Février, B.; Raposo, G. Exosomes: Endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 2004, 16, 415–421. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef] [PubMed]
- van der Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef]
- Hood, J.L.; San, R.S.; Wickline, S.A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011, 71, 3792–3801. [Google Scholar] [CrossRef]
- Peinado, H.; Alečković, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; García-Santos, G.; Nitadori-Hoshino, A.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.Á.; Bernad, A.; Sánchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef] [PubMed]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef]
- van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A common pathway for a specialized function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212. [Google Scholar] [CrossRef]
- Laulagnier, K.; Motta, C.; Hamdi, S.; Roy, S.; Fauvelle, F.; Pageaux, J.-F.; Kobayashi, T.; Salles, J.-P.; Perret, B.; Bonnerot, C.; et al. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 2004, 380, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Cufaro, M.C.; Pieragostino, D.; Lanuti, P.; Rossi, C.; Cicalini, I.; Federici, L.; De Laurenzi, V.; Del Boccio, P. Extracellular Vesicles and Their Potential Use in Monitoring Cancer Progression and Therapy: The Contribution of Proteomics. J. Oncol. 2019, 2019, 1–19. [Google Scholar] [CrossRef]
- Ahmed, K.A.; Xiang, J. Mechanisms of cellular communication through intercellular protein transfer. J. Cell Mol. Med. 2011, 15, 1458–1473. [Google Scholar] [CrossRef] [PubMed]
- Machtinger, R.; Laurent, L.C.; Baccarelli, A.A. Extracellular vesicles: Roles in gamete maturation, fertilization and embryo implantation. Hum. Reprod. Update 2016, 22, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Zöller, M. Tetraspanins: Push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 2009, 9, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Zöller, M. Exosome target cell selection and the importance of exosomal tetraspanins: A hypothesis. Biochem. Soc. Trans. 2011, 39, 559–562. [Google Scholar] [CrossRef]
- Rana, S.; Yue, S.; Stadel, D.; Zöller, M. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012, 44, 1574–1584. [Google Scholar] [CrossRef]
- Kaji, K.; Oda, S.; Shikano, T.; Ohnuki, T.; Uematsu, Y.; Sakagami, J.; Tada, N.; Miyazaki, S.; Kudo, A. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat. Genet. 2000, 24, 279–282. [Google Scholar] [CrossRef]
- Le Naour, F.; Rubinstein, E.; Jasmin, C.; Prenant, M.; Boucheix, C. Severely reduced female fertility in CD9-deficient mice. Science 2000, 287, 319–321. [Google Scholar] [CrossRef]
- Miyado, K. Requirement of CD9 on the Egg Plasma Membrane for Fertilization. Science 2000, 287, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Stipp, C.S.; Kolesnikova, T.V.; Hemler, M.E. Functional domains in tetraspanin proteins. Trends Biochem. Sci. 2003, 28, 106–112. [Google Scholar] [CrossRef]
- Berditchevski, F.; Odintsova, E.; Sawada, S.; Gilbert, E. Expression of the palmitoylation-deficient CD151 weakens the association of alpha 3 beta 1 integrin with the tetraspanin-enriched microdomains and affects integrin-dependent signaling. J. Biol. Chem. 2002, 277, 36991–37000. [Google Scholar] [CrossRef] [PubMed]
- Charrin, S.; Manié, S.; Oualid, M.; Billard, M.; Boucheix, C.; Rubinstein, E. Differential stability of tetraspanin/tetraspanin interactions: Role of palmitoylation. FEBS Lett. 2002, 516, 139–144. [Google Scholar] [CrossRef]
- Yang, X.; Claas, C.; Kraeft, S.-K.; Chen, L.B.; Wang, Z.; Kreidberg, J.A.; Hemler, M.E. Palmitoylation of Tetraspanin Proteins: Modulation of CD151 Lateral Interactions, Subcellular Distribution, and Integrin-dependent Cell Morphology. Mol. Biol Cell 2002, 13, 767–781. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Yuan, S.; Dong, M.; Su, J.; Yu, C.; Shen, Y.; Xie, X.; Yu, Y.; Yu, X.; Chen, S.; et al. The phylogenetic analysis of tetraspanins projects the evolution of cell–cell interactions from unicellular to multicellular organisms. Genomics 2005, 86, 674–684. [Google Scholar] [CrossRef]
- Boucheix, C.; Rubinstein, E. Tetraspanins. Cell. Mol. Life Sci. 2001, 58, 1189–1205. [Google Scholar] [CrossRef]
- Tang, M.; Yin, G.; Wang, F.; Liu, H.; Zhou, S.; Ni, J.; Chen, C.; Zhou, Y.; Zhao, Y. Downregulation of CD9 promotes pancreatic cancer growth and metastasis through upregulation of epidermal growth factor on the cell surface. Oncol. Rep. 2015, 34, 350–358. [Google Scholar] [CrossRef][Green Version]
- Berditchevski, F.; Odintsova, E. Tetraspanins as regulators of protein trafficking. Traffic 2007, 8, 89–96. [Google Scholar] [CrossRef]
- Hemler, M.E. Specific tetraspanin functions. J. Cell Biol. 2001, 155, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Hemler, M.E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 2005, 6, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Charrin, S.; le Naour, F.; Silvie, O.; Milhiet, P.-E.; Boucheix, C.; Rubinstein, E. Lateral organization of membrane proteins: Tetraspanins spin their web. Biochem. J. 2009, 420, 133–154. [Google Scholar] [CrossRef] [PubMed]
- Hemler, M.E. Integrin associated proteins. Curr. Opin. Cell Biol. 1998, 10, 578–585. [Google Scholar] [CrossRef]
- Winterwood, N.E.; Varzavand, A.; Meland, M.N.; Ashman, L.K.; Stipp, C.S. A Critical Role for Tetraspanin CD151 in α3β1 and α6β4 Integrin–dependent Tumor Cell Functions on Laminin-5. Mol. Biol Cell 2006, 17, 2707–2721. [Google Scholar] [CrossRef]
- Boucheix, C.; Duc, G.H.; Jasmin, C.; Rubinstein, E. Tetraspanins and malignancy. Expert Rev. Mol. Med. 2001, 3. [Google Scholar] [CrossRef]
- Berditchevski, F. Complexes of tetraspanins with integrins: More than meets the eye. J. Cell Sci. 2001, 114, 4143–4151. [Google Scholar]
- Yáñez-Mó, M.; Mittelbrunn, M.; Sánchez-Madrid, F. Tetraspanins and Intercellular Interactions. Microcirculation 2001, 8, 153–168. [Google Scholar] [CrossRef]
- Hemler, M.E. Targeting of tetraspanin proteins—Potential benefits and strategies. Nat. Rev. Drug Discov. 2008, 7, 747–758. [Google Scholar] [CrossRef] [PubMed]
- van Spriel, A.B.; Figdor, C.G. The role of tetraspanins in the pathogenesis of infectious diseases. Microbes Infect. 2010, 12, 106–112. [Google Scholar] [CrossRef]
- Martin, F.; Roth, D.M.; Jans, D.A.; Pouton, C.W.; Partridge, L.J.; Monk, P.N.; Moseley, G.W. Tetraspanins in Viral Infections: A Fundamental Role in Viral Biology? J. Virol. 2005, 79, 10839–10851. [Google Scholar] [CrossRef] [PubMed]
- Florin, L.; Lang, T. Tetraspanin Assemblies in Virus Infection. Front. Immunol. 2018, 9, 1140. [Google Scholar] [CrossRef] [PubMed]
- Hemler, M.E. Tetraspanin proteins promote multiple cancer stages. Nat. Rev. Cancer 2014, 14, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Detchokul, S.; Williams, E.D.; Parker, M.W.; Frauman, A.G. Tetraspanins as regulators of the tumour microenvironment: Implications for metastasis and therapeutic strategies. Br. J. Pharmacol. 2014, 171, 5462–5490. [Google Scholar] [CrossRef]
- Escola, J.M.; Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar] [CrossRef]
- Simpson, R.J.; Kalra, H.; Mathivanan, S. ExoCarta as a resource for exosomal research. J. Extracell. Vesicles 2012, 1, 18374. [Google Scholar] [CrossRef]
- Kim, M.; Park, H.J.; Seol, J.W.; Jang, J.Y.; Cho, Y.; Kim, K.R.; Choi, Y.; Lydon, J.P.; DeMayo, F.J.; Shibuya, M.; et al. VEGF—A regulated by progesterone governs uterine angiogenesis and vascular remodelling during pregnancy. EMBO Mol. Med. 2013, 5, 1415–1430. [Google Scholar] [CrossRef]
- Lozano-Andrés, E.; Libregts, S.F.; Toribio, V.; Royo, F.; Morales, S.; López-Martín, S.; Valés-Gómez, M.; Reyburn, H.T.; Falcón-Pérez, J.M.; Wauben, M.H.; et al. Tetraspanin-decorated extracellular vesicle-mimetics as a novel adaptable reference material. J. Extracell. Vesicles 2019, 8, 1573052. [Google Scholar] [CrossRef]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The Tetraspanin CD63 Regulates ESCRT-Independent and -Dependent Endosomal Sorting during Melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef] [PubMed]
- Chairoungdua, A.; Smith, D.L.; Pochard, P.; Hull, M.; Caplan, M.J. Exosome release of β-catenin: A novel mechanism that antagonizes Wnt signaling. J. Cell. Biol. 2010, 190, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Guo, Q.; Xia, B.; Zhang, Y.H.; Giesert, E.E.; Levy, S.; Zheng, J.J.; Zhang, X.A. Tetraspanins regulate the protrusive activities of cell membrane. Biochem. Biophys. Res. Commun. 2011, 415, 619–626. [Google Scholar] [CrossRef]
- Umeda, R.; Satouh, Y.; Takemoto, M.; Nakada-Nakura, Y.; Liu, K.; Yokoyama, T.; Shirouzu, M.; Iwata, S.; Nomura, N.; Sato, K.; et al. Structural insights into tetraspanin CD9 function. Nat. Commun. 2020, 11, 1606. [Google Scholar] [CrossRef] [PubMed]
- Frolikova, M.; Manaskova-Postlerova, P.; Cerny, J.; Jankovicova, J.; Simonik, O.; Pohlova, A.; Secova, P.; Antalikova, J.; Dvorakova-Hortova, K. CD9 and CD81 Interactions and Their Structural Modelling in Sperm Prior to Fertilization. Int. J. Mol. Sci. 2018, 19, 1236. [Google Scholar] [CrossRef] [PubMed]
- Jankovicova, J.; Frolikova, M.; Palenikova, V.; Valaskova, E.; Cerny, J.; Secova, P.; Bartokova, M.; Horovska, L.; Manaskova-Postlerova, P.; Antalikova, J.; et al. Expression and distribution of CD151 as a partner of alpha6 integrin in male germ cells. Sci. Rep. 2020, 10, 4374. [Google Scholar] [CrossRef] [PubMed]
- Sala-Valdés, M.; Ursa, Á.; Charrin, S.; Rubinstein, E.; Hemler, M.E.; Sánchez-Madrid, F.; Yáñez-Mó, M. EWI-2 and EWI-F Link the Tetraspanin Web to the Actin Cytoskeleton through Their Direct Association with Ezrin-Radixin-Moesin Proteins. J. Biol. Chem. 2006, 281, 19665–19675. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Claas, C.; Kretz, C.C.; Nazarenko, I.; Zoeller, M. Activation-induced internalization differs for the tetraspanins CD9 and Tspan8: Impact on tumor cell motility. Int. J. Biochem. Cell Biol. 2011, 43, 106–119. [Google Scholar] [CrossRef]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in Extracellular Vesicle Formation and Function. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef]
- Perez-Hernandez, D.; Gutiérrez-Vázquez, C.; Jorge, I.; López-Martín, S.; Ursa, A.; Sánchez-Madrid, F.; Vázquez, J.; Yáñez-Mó, M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem. 2013, 288, 11649–11661. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Barreiro, O.; Gordon-Alonso, M.; Sala-Valdés, M.; Sánchez-Madrid, F. Tetraspanin-enriched microdomains: A functional unit in cell plasma membranes. Trends Cell Biol. 2009, 19, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Bassani, S.; Cingolani, L.A. Tetraspanins: Interactions and interplay with integrins. Int. J. Biochem. Cell Biol. 2012, 44, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Fanaei, M.; Monk, P.N.; Partridge, L.J. The role of tetraspanins in fusion. Biochem. Soc. Trans. 2011, 39, 524–528. [Google Scholar] [CrossRef]
- Kolesnikova, T.V.; Kazarov, A.R.; Lemieux, M.E.; Lafleur, M.A.; Kesari, S.; Kung, A.L.; Hemler, M.E. Glioblastoma Inhibition by Cell Surface Immunoglobulin Protein EWI-2, In Vitro and In Vivo. Neoplasia 2009, 11, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular vesicles in cancer: Exosomes, microvesicles and the emerging role of large oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef]
- Tian, T.; Zhu, Y.-L.; Hu, F.-H.; Wang, Y.-Y.; Huang, N.-P.; Xiao, Z.-D. Dynamics of exosome internalization and trafficking. J. Cell. Physiol. 2013, 228, 1487–1495. [Google Scholar] [CrossRef]
- Levy, S.; Shoham, T. The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 2005, 5, 136–148. [Google Scholar] [CrossRef]
- Lakkaraju, A.; Rodriguez-Boulan, E. Itinerant exosomes: Emerging roles in cell and tissue polarity. Trends Cell Biol. 2008, 18, 199–209. [Google Scholar] [CrossRef]
- Johnstone, R.M. Exosomes biological significance: A concise review. Blood Cells Mol. Dis. 2006, 36, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Lebreton, A.; Séraphin, B. Exosome-mediated quality control: Substrate recruitment and molecular activity. Biochim. Biophys. Acta 2008, 1779, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Bhatnagar, S. Exosome function: From tumor immunology to pathogen biology. Traffic 2008, 9, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Nazarenko, I.; Rana, S.; Baumann, A.; McAlear, J.; Hellwig, A.; Trendelenburg, M.; Lochnit, G.; Preissner, K.T.; Zöller, M. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 2010, 70, 1668–1678. [Google Scholar] [CrossRef] [PubMed]
- Morelli, A.E.; Larregina, A.T.; Shufesky, W.J.; Sullivan, M.L.G.; Stolz, D.B.; Papworth, G.D.; Zahorchak, A.F.; Logar, A.J.; Wang, Z.; Watkins, S.C.; et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 2004, 104, 3257–3266. [Google Scholar] [CrossRef] [PubMed]
- Zech, D.; Rana, S.; Büchler, M.W.; Zöller, M. Tumor-exosomes and leukocyte activation: An ambivalent crosstalk. Cell Commun. Signal. 2012, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Stoeck, A.; Keller, S.; Riedle, S.; Sanderson, M.P.; Runz, S.; Le Naour, F.; Gutwein, P.; Ludwig, A.; Rubinstein, E.; Altevogt, P. A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem. J. 2006, 393, 609–618. [Google Scholar] [CrossRef]
- Hawari, F.I.; Rouhani, F.N.; Cui, X.; Yu, Z.-X.; Buckley, C.; Kaler, M.; Levine, S.J. Release of full-length 55-kDa TNF receptor 1 in exosome-like vesicles: A mechanism for generation of soluble cytokine receptors. Proc. Natl. Acad. Sci. USA 2004, 101, 1297–1302. [Google Scholar] [CrossRef]
- Hakulinen, J.; Junnikkala, S.; Sorsa, T.; Meri, S. Complement inhibitor membrane cofactor protein (MCP.; CD46) is constitutively shed from cancer cell membranes in vesicles and converted by a metalloproteinase to a functionally active soluble form. Eur. J. Immunol. 2004, 34, 2620–2629. [Google Scholar] [CrossRef]
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.-P.; Belting, M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385. [Google Scholar] [CrossRef] [PubMed]
- Menck, K.; Klemm, F.; Gross, J.C.; Pukrop, T.; Wenzel, D.; Binder, C. Induction and transport of Wnt 5a during macrophage-induced malignant invasion is mediated by two types of extracellular vesicles. Oncotarget 2013, 4, 2057–2066. [Google Scholar] [CrossRef] [PubMed]
- Atay, S.; Gercel-Taylor, C.; Taylor, D.D. Human trophoblast-derived exosomal fibronectin induces pro-inflammatory IL-1β production by macrophages. Am. J. Reprod. Immunol. 2011, 66, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Otto, G.P.; Nichols, B.J. The roles of flotillin microdomains--endocytosis and beyond. J. Cell. Sci. 2011, 124, 3933–3940. [Google Scholar] [CrossRef] [PubMed]
- Volonte, D.; Galbiati, F.; Li, S.; Nishiyama, K.; Okamoto, T.; Lisanti, M.P. Flotillins/cavatellins are differentially expressed in cells and tissues and form a hetero-oligomeric complex with caveolins in vivo. Characterization and epitope-mapping of a novel flotillin-1 monoclonal antibody probe. J. Biol. Chem. 1999, 274, 12702–12709. [Google Scholar] [CrossRef]
- Bickel, P.E.; Scherer, P.E.; Schnitzer, J.E.; Oh, P.; Lisanti, M.P.; Lodish, H.F. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J. Biol. Chem. 1997, 272, 13793–13802. [Google Scholar] [CrossRef]
- Frick, M.; Bright, N.A.; Riento, K.; Bray, A.; Merrified, C.; Nichols, B.J. Coassembly of flotillins induces formation of membrane microdomains, membrane curvature, and vesicle budding. Curr. Biol. 2007, 17, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Jahn, R.; Lang, T.; Südhof, T.C. Membrane fusion. Cell 2003, 112, 519–533. [Google Scholar] [CrossRef]
- Logozzi, M.; De Milito, A.; Lugini, L.; Borghi, M.; Calabrò, L.; Spada, M.; Perdicchio, M.; Marino, M.L.; Federici, C.; Iessi, E.; et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS ONE 2009, 4, e5219. [Google Scholar] [CrossRef]
- Chernomordik, L.V.; Kozlov, M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008, 15, 675–683. [Google Scholar] [CrossRef]
- Naito, Y.; Yoshioka, Y.; Yamamoto, Y.; Ochiya, T. How cancer cells dictate their microenvironment: Present roles of extracellular vesicles. Cell. Mol. Life Sci. 2017, 74, 697–713. [Google Scholar] [CrossRef]
- Lu, J.; Li, J.; Liu, S.; Wang, T.; Ianni, A.; Bober, E.; Braun, T.; Xiang, R.; Yue, S. Exosomal tetraspanins mediate cancer metastasis by altering host microenvironment. Oncotarget 2017, 8, 62803–62815. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. Exosomes and tumor-mediated immune suppression. J. Clin. Invest. 2016, 126, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Mu, W.; Erb, U.; Zöller, M. The tetraspanins CD151 and Tspan8 are essential exosome components for the crosstalk between cancer initiating cells and their surrounding. Oncotarget 2014, 6, 2366–2384. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Rana, S.; Zöller, M. Host Matrix Modulation by Tumor Exosomes Promotes Motility and Invasiveness. Neoplasia 2013, 15, 875–887. [Google Scholar] [CrossRef]
- Yang, C.; Robbins, P.D. The Roles of Tumor-Derived Exosomes in Cancer Pathogenesis. Clin. Dev. Immunol. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.-L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef]
- Lindenbergh, M.F.S.; Stoorvogel, W. Antigen Presentation by Extracellular Vesicles from Professional Antigen-Presenting Cells. Annu. Rev. Immunol. 2018, 36, 435–459. [Google Scholar] [CrossRef]
- Szajnik, M.; Czystowska, M.; Szczepanski, M.J.; Mandapathil, M.; Whiteside, T.L. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg). PLoS ONE 2010, 5, e11469. [Google Scholar] [CrossRef]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: Dendritic cell-derived exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Yáñez-Mó, M.; Sancho, D.; Ursa, A.; Sánchez-Madrid, F. Cutting edge: Dynamic redistribution of tetraspanin CD81 at the central zone of the immune synapse in both T lymphocytes and APC. J. Immunol. 2002, 169, 6691–6695. [Google Scholar] [CrossRef]
- Rocha-Perugini, V.; Zamai, M.; Gonzalez-Granado, J.M.; Barreiro, O.; Tejera, E.; Yanez-Mo, M.; Caiolfa, V.R.; Sanchez-Madrid, F. CD81 Controls Sustained T Cell Activation Signaling and Defines the Maturation Stages of Cognate Immunological Synapses. Mol. Cell. Biol. 2013, 33, 3644–3658. [Google Scholar] [CrossRef] [PubMed]
- Kropshofer, H.; Spindeldreher, S.; Röhn, T.A.; Platania, N.; Grygar, C.; Daniel, N.; Wölpl, A.; Langen, H.; Horejsi, V.; Vogt, A.B. Tetraspan microdomains distinct from lipid rafts enrich select peptide-MHC class II complexes. Nat. Immunol. 2002, 3, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Imai, T.; Kakizaki, M.; Nishimura, M.; Yoshie, O. Molecular analyses of the association of CD4 with two members of the transmembrane 4 superfamily, CD81 and CD82. J. Immunol. 1995, 155, 1229–1239. [Google Scholar] [PubMed]
- Maecker, H.T.; Levy, S. Normal lymphocyte development but delayed humoral immune response in CD81-null mice. J. Exp. Med. 1997, 185, 1505–1510. [Google Scholar] [CrossRef] [PubMed]
- Rocha-Perugini, V.; González-Granado, J.M.; Tejera, E.; López-Martín, S.; Yañez-Mó, M.; Sanchez-Madrid, F. Tetraspanins CD9 and CD151 at the immune synapse support T-cell integrin signaling. Eur. J. Immunol. 2014, 44, 1967–1975. [Google Scholar] [CrossRef] [PubMed]
- Unternaehrer, J.J.; Chow, A.; Pypaert, M.; Inaba, K.; Mellman, I. The tetraspanin CD9 mediates lateral association of MHC class II molecules on the dendritic cell surface. Proc. Natl. Acad. Sci. USA 2007, 104, 234–239. [Google Scholar] [CrossRef]
- Nj, P.; Lk, D.; Pa, R. CDw78 defines MHC class II-peptide complexes that require Ii chain-dependent lysosomal trafficking, not localization to a specific tetraspanin membrane microdomain. J. Immunol. 2006, 177, 5451–5458. [Google Scholar] [CrossRef]
- Wright, M.D.; Moseley, G.W.; van Spriel, A.B. Tetraspanin microdomains in immune cell signalling and malignant disease. Tissue Antigens 2004, 64, 533–542. [Google Scholar] [CrossRef]
- Sheng, K.-C.; van Spriel, A.B.; Gartlan, K.H.; Sofi, M.; Apostolopoulos, V.; Ashman, L.; Wright, M.D. Tetraspanins CD37 and CD151 differentially regulate Ag presentation and T-cell co-stimulation by DC. Eur. J. Immunol. 2009, 39, 50–55. [Google Scholar] [CrossRef]
- Matzuk, M.M. Intercellular Communication in the Mammalian Ovary: Oocytes Carry the Conversation. Science 2002, 296, 2178–2180. [Google Scholar] [CrossRef]
- Robker, R.L.; Hennebold, J.D.; Russell, D.L. Coordination of Ovulation and Oocyte Maturation: A Good Egg at the Right Time. Endocrinology 2018, 159, 3209–3218. [Google Scholar] [CrossRef] [PubMed]
- Georgiou, A.S.; Sostaric, E.; Wong, C.H.; Snijders, A.P.L.; Wright, P.C.; Moore, H.D.; Fazeli, A. Gametes Alter the Oviductal Secretory Proteome. Mol. Cell Proteom. 2005, 4, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
- Sutton, M.L. Effects of in-vivo and in-vitro environments on the metabolism of the cumulus-oocyte complex and its influence on oocyte developmental capacity. Hum. Reprod. Update 2003, 9, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Eppig, J.J. Intercommunication between mammalian oocytes and companion somatic cells. Bioessays 1991, 13, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Albertini, D.F.; Combelles, C.M.; Benecchi, E.; Carabatsos, M.J. Cellular basis for paracrine regulation of ovarian follicle development. Reproduction 2001, 121, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, A.D.; Gilbert, I.; Scantland, S.; Fournier, E.; Ashkar, F.; Bastien, A.; Saadi, H.A.S.; Gagné, D.; Sirard, M.-A.; Khandjian, É.W.; et al. Cumulus Cell Transcripts Transit to the Bovine Oocyte in Preparation for Maturation1. Biol. Reprod. 2016, 94. [Google Scholar] [CrossRef] [PubMed]
- del Collado, M.; da Silveira, J.C.; Sangalli, J.R.; Andrade, G.M.; da Sousa, L.R.S.; Silva, L.A.; Meirelles, F.V.; Perecin, F. Fatty Acid Binding Protein 3 And Transzonal Projections Are Involved In Lipid Accumulation During In Vitro Maturation Of Bovine Oocytes. Sci. Rep. 2017, 7, 2645. [Google Scholar] [CrossRef]
- da Silveira, J.C.; Veeramachaneni, D.N.R.; Winger, Q.A.; Carnevale, E.M.; Bouma, G.J. Cell-Secreted Vesicles in Equine Ovarian Follicular Fluid Contain miRNAs and Proteins: A Possible New Form of Cell Communication Within the Ovarian Follicle1. Biol. Reprod. 2012, 86. [Google Scholar] [CrossRef]
- da Silveira, J.C.; Carnevale, E.M.; Winger, Q.A.; Bouma, G.J. Regulation of ACVR1 and ID2 by cell-secreted exosomes during follicle maturation in the mare. Reprod. Biol. Endocrinol. 2014, 12, 44. [Google Scholar] [CrossRef]
- Sohel, M.M.H.; Hoelker, M.; Noferesti, S.S.; Salilew-Wondim, D.; Tholen, E.; Looft, C.; Rings, F.; Uddin, M.J.; Spencer, T.E.; Schellander, K.; et al. Exosomal and Non-Exosomal Transport of Extra-Cellular microRNAs in Follicular Fluid: Implications for Bovine Oocyte Developmental Competence. PLoS ONE 2013, 8, e78505. [Google Scholar] [CrossRef]
- Matsuno, Y.; Kanke, T.; Maruyama, N.; Fujii, W.; Naito, K.; Sugiura, K. Characterization of mRNA profiles of the exosome-like vesicles in porcine follicular fluid. PLoS ONE 2019, 14, e0217760. [Google Scholar] [CrossRef] [PubMed]
- da Silveira, J.C.; Andrade, G.M.; del Collado, M.; Sampaio, R.V.; Sangalli, J.R.; Silva, L.A.; Pinaffi, F.V.L.; Jardim, I.B.; Cesar, M.C.; Nogueira, M.F.G.; et al. Supplementation with small-extracellular vesicles from ovarian follicular fluid during in vitro production modulates bovine embryo development. PLoS ONE 2017, 12, e0179451. [Google Scholar] [CrossRef] [PubMed]
- Santonocito, M.; Vento, M.; Guglielmino, M.R.; Battaglia, R.; Wahlgren, J.; Ragusa, M.; Barbagallo, D.; Borzì, P.; Rizzari, S.; Maugeri, M.; et al. Molecular characterization of exosomes and their microRNA cargo in human follicular fluid: Bioinformatic analysis reveals that exosomal microRNAs control pathways involved in follicular maturation. Fertil. Steril. 2014, 102, 1751.e1–1761.e1. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Tang, T.; Zeng, Z.; Wu, J.; Tan, X.; Yan, J. The expression of small RNAs in exosomes of follicular fluid altered in human polycystic ovarian syndrome. PeerJ 2020, 8, e8640. [Google Scholar] [CrossRef] [PubMed]
- Hung, W.-T.; Hong, X.; Christenson, L.K.; McGinnis, L.K. Extracellular Vesicles from Bovine Follicular Fluid Support Cumulus Expansion1. Biol. Reprod. 2015, 93. [Google Scholar] [CrossRef] [PubMed]
- Navakanitworakul, R.; Hung, W.-T.; Gunewardena, S.; Davis, J.S.; Chotigeat, W.; Christenson, L.K. Characterization and Small RNA Content of Extracellular Vesicles in Follicular Fluid of Developing Bovine Antral Follicles. Sci. Rep. 2016, 6, 25486. [Google Scholar] [CrossRef] [PubMed]
- Miyado, K.; Yoshida, K.; Yamagata, K.; Sakakibara, K.; Okabe, M.; Wang, X.; Miyamoto, K.; Akutsu, H.; Kondo, T.; Takahashi, Y.; et al. The fusing ability of sperm is bestowed by CD9-containing vesicles released from eggs in mice. Proc. Natl. Acad. Sci. USA 2008, 105, 12921–12926. [Google Scholar] [CrossRef] [PubMed]
- Ohnami, N.; Nakamura, A.; Miyado, M.; Sato, M.; Kawano, N.; Yoshida, K.; Harada, Y.; Takezawa, Y.; Kanai, S.; Ono, C.; et al. CD81 and CD9 work independently as extracellular components upon fusion of sperm and oocyte. Biol. Open 2012, 1, 640–647. [Google Scholar] [CrossRef]
- Jankovicova, J.; Secova, P.; Manaskova-Postlerova, P.; Simonik, O.; Frolikova, M.; Chmelikova, E.; Horovska, L.; Michalkova, K.; Dvorakova-Hortova, K.; Antalikova, J. Detection of CD9 and CD81 tetraspanins in bovine and porcine oocytes and embryos. Int. J. Biol. Macromol. 2019, 123, 931–938. [Google Scholar] [CrossRef]
- Suarez, S.S. Chapter 5 - Gamete and Zygote Transport. In Knobil and Neill’s Physiology of Reproduction, 4th ed.; Plant, T.M., Zeleznik, A.J., Eds.; Academic Press: San Diego, CA, USA, 2015; pp. 197–232. ISBN 978-0-12-397175-3. [Google Scholar]
- Al-Dossary, A.A.; Strehler, E.E.; Martin-DeLeon, P.A. Expression and Secretion of Plasma Membrane Ca2+-ATPase 4a (PMCA4a) during Murine Estrus: Association with Oviductal Exosomes and Uptake in Sperm. PLoS ONE 2013, 8, e80181. [Google Scholar] [CrossRef]
- Fereshteh, Z.; Schmidt, S.A.; Al-Dossary, A.A.; Accerbi, M.; Arighi, C.; Cowart, J.; Song, J.L.; Green, P.J.; Choi, K.; Yoo, S.; et al. Murine Oviductosomes (OVS) microRNA profiling during the estrous cycle: Delivery of OVS-borne microRNAs to sperm where miR-34c-5p localizes at the centrosome. Sci. Rep. 2018, 8, 16094. [Google Scholar] [CrossRef] [PubMed]
- Lopera-Vásquez, R.; Hamdi, M.; Fernandez-Fuertes, B.; Maillo, V.; Beltrán-Breña, P.; Calle, A.; Redruello, A.; López-Martín, S.; Gutierrez-Adán, A.; Yañez-Mó, M.; et al. Extracellular Vesicles from BOEC in In Vitro Embryo Development and Quality. PLoS ONE 2016, 11, e0148083. [Google Scholar] [CrossRef]
- Almiñana, C.; Corbin, E.; Tsikis, G.; Alcântara-Neto, A.S.; Labas, V.; Reynaud, K.; Galio, L.; Uzbekov, R.; Garanina, A.S.; Druart, X.; et al. Oviduct extracellular vesicles protein content and their role during oviduct–embryo cross-talk. Reproduction 2017, 154, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Bathala, P.; Fereshteh, Z.; Li, K.; Al-Dossary, A.A.; Galileo, D.S.; Martin-DeLeon, P.A. Oviductal extracellular vesicles (oviductosomes, OVS) are conserved in humans: Murine OVS play a pivotal role in sperm capacitation and fertility. Mol. Hum. Reprod. 2018, 24, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Isobe, N.; Yoshimura, Y. Changes in localization and density of CD63-positive exosome-like substances in the hen oviduct with artificial insemination and their effect on sperm viability. Theriogenology 2017, 101, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Lange-Consiglio, A.; Perrini, C.; Albini, G.; Modina, S.; Lodde, V.; Orsini, E.; Esposti, P.; Cremonesi, F. Oviductal microvesicles and their effect on in vitro maturation of canine oocytes. Reproduction 2017, 154, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Waqas, M.Y.; Zhang, Q.; Ahmed, N.; Yang, P.; Xing, G.; Akhtar, M.; Basit, A.; Liu, T.; Hong, C.; Arshad, M.; et al. Cellular Evidence of Exosomes in the Reproductive Tract of Chinese Soft-Shelled Turtle Pelodiscus sinensis: Identification of Exosomes in Female Turtle. J. Exp. Zool. 2017, 327, 18–27. [Google Scholar] [CrossRef]
- Nakano, S.; Yamamoto, S.; Okada, A.; Nakajima, T.; Sato, M.; Takagi, T.; Tomooka, Y. Role of extracellular vesicles in the interaction between epithelial and mesenchymal cells during oviductal ciliogenesis. Biochem. Biophys. Res. Commun. 2017, 483, 245–251. [Google Scholar] [CrossRef]
- Almiñana, C.; Tsikis, G.; Labas, V.; Uzbekov, R.; da Silveira, J.C.; Bauersachs, S.; Mermillod, P. Deciphering the oviductal extracellular vesicles content across the estrous cycle: Implications for the gametes-oviduct interactions and the environment of the potential embryo. BMC Genom. 2018, 19, 622. [Google Scholar] [CrossRef]
- Lopera-Vasquez, R.; Hamdi, M.; Maillo, V.; Gutierrez-Adan, A.; Bermejo-Alvarez, P.; Ramírez, M.Á.; Yáñez-Mó, M.; Rizos, D. Effect of bovine oviductal extracellular vesicles on embryo development and quality in vitro. Reproduction 2017, 153, 461–470. [Google Scholar] [CrossRef]
- Florman, H.M.; Fissore, R.A. Chapter 4—Fertilization in Mammals. In Knobil and Neill’s Physiology of Reproduction, 4th ed.; Plant, T.M., Zeleznik, A.J., Eds.; Academic Press: San Diego, CA, USA, 2015; pp. 149–196. ISBN 978-0-12-397175-3. [Google Scholar]
- Al-Dossary, A.A.; Bathala, P.; Caplan, J.L.; Martin-DeLeon, P.A. Oviductosome-Sperm Membrane Interaction in Cargo Delivery: Detection of Fusion and Underlying Molecular Players Using Three-Dimensional Super-Resolution Structured Illumination Microscopy (SR-SIM). J. Biol. Chem. 2015, 290, 17710–17723. [Google Scholar] [CrossRef] [PubMed]
- Griffiths, G.S.; Galileo, D.S.; Reese, K.; Martin-DeLeon, P.A. Investigating the role of murine epididymosomes and uterosomes in GPI-linked protein transfer to sperm using SPAM1 as a model. Mol. Reprod. Dev. 2008, 75, 1627–1636. [Google Scholar] [CrossRef]
- Timothy Smith, T.; Nothnick, W.B. Role of Direct Contact between Spermatozoa and Oviductal Epithelial Cells in Maintaining Rabbit Sperm Viability1. Biol. Reprod. 1997, 56, 83–89. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dobrinski, I.; Timothy Smith, T.; Suarez, S.S.; Ball, B.A. Membrane Contact with Oviductal Epithelium Modulates the Intracellular Calcium Concentration of Equine Spermatozoa in Vitro1. Biol. Reprod. 1997, 56, 861–869. [Google Scholar] [CrossRef]
- Murray, S.C.; Smith, T.T. Sperm interaction with fallopian tube apical membrane enhances sperm motility and delays capacitation. Fertil. Steril. 1997, 68, 351–357. [Google Scholar] [CrossRef]
- Bakst, M.R.; Wishart, G.; Brillard, J.P. Oviducal sperm selection, transport, and storage in poultry. Poult. Sci. Rev. 1994, 5, 117–143. [Google Scholar]
- Barraud-Lange, V.; Naud-Barriant, N.; Bomsel, M.; Wolf, J.; Ziyyat, A. Transfer of oocyte membrane fragments to fertilizing spermatozoa. FASEB J. 2007, 21, 3446–3449. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Primakoff, P.; Myles, D.G. Can the presence of wild-type oocytes during insemination rescue the fusion defect of CD9 null oocytes? Mol. Reprod. Dev. 2009, 76, 602. [Google Scholar] [CrossRef]
- Barraud-Lange, V.; Chalas Boissonnas, C.; Serres, C.; Auer, J.; Schmitt, A.; Lefèvre, B.; Wolf, J.-P.; Ziyyat, A. Membrane transfer from oocyte to sperm occurs in two CD9-independent ways that do not supply the fertilising ability of Cd9-deleted oocytes. Reproduction 2012, 144, 53–66. [Google Scholar] [CrossRef]
- Ravaux, B.; Favier, S.; Perez, E.; Gourier, C. Egg CD9 protein tides correlated with sperm oscillations tune the gamete fusion ability in mammal. J. Mol. Cell Biol. 2018. [Google Scholar] [CrossRef]
- Vyas, P.; Balakier, H.; Librach, C.L. Ultrastructural identification of CD9 positive extracellular vesicles released from human embryos and transported through the zona pellucida. Syst. Biol. Reprod. Med. 2019, 65, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Jankovicova, J.; Frolikova, M.; Sebkova, N.; Simon, M.; Cupperova, P.; Lipcseyova, D.; Michalkova, K.; Horovska, L.; Sedlacek, R.; Stopka, P.; et al. Characterization of tetraspanin protein CD81 in mouse spermatozoa and bovine gametes. Reproduction 2016, 152, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Almiñana, C.; Bauersachs, S. Extracellular vesicles: Multi-signal messengers in the gametes/embryo-oviduct cross-talk. Theriogenology 2020, 150, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Almiñana; Bauersachs Extracellular Vesicles in the Oviduct: Progress, Challenges and Implications for the Reproductive Success. Bioengineering 2019, 6, 32. [CrossRef] [PubMed]
- Ng, Y.H.; Rome, S.; Jalabert, A.; Forterre, A.; Singh, H.; Hincks, C.L.; Salamonsen, L.A. Endometrial Exosomes/Microvesicles in the Uterine Microenvironment: A New Paradigm for Embryo-Endometrial Cross Talk at Implantation. PLoS ONE 2013, 8, e58502. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Nguyen, H.P.T.; Elgass, K.; Simpson, R.J.; Salamonsen, L.A. Human Endometrial Exosomes Contain Hormone-Specific Cargo Modulating Trophoblast Adhesive Capacity: Insights into Endometrial-Embryo Interactions1. Biol. Reprod. 2016, 94, 38. [Google Scholar] [CrossRef]
- Burns, G.W.; Brooks, K.E.; Spencer, T.E. Extracellular Vesicles Originate from the Conceptus and Uterus During Early Pregnancy in Sheep1. Biol. Reprod. 2016, 94, 1–11. [Google Scholar] [CrossRef]
- Kropp, J.; Salih, S.M.; Khatib, H. Expression of microRNAs in bovine and human pre-implantation embryo culture media. Front. Genet. 2014, 5, 91. [Google Scholar] [CrossRef]
- Mellisho, E.A.; Velásquez, A.E.; Nuñez, M.J.; Cabezas, J.G.; Cueto, J.A.; Fader, C.; Castro, F.O.; Rodríguez-Álvarez, L. Identification and characteristics of extracellular vesicles from bovine blastocysts produced in vitro. PLoS ONE 2017, 12, e0178306. [Google Scholar] [CrossRef]
- Qu, P.; Zhao, Y.; Wang, R.; Zhang, Y.; Li, L.; Fan, J.; Liu, E. Extracellular vesicles derived from donor oviduct fluid improved birth rates after embryo transfer in mice. Reprod. Fertil. Dev. 2019, 31, 324. [Google Scholar] [CrossRef]
- Maillo, V.; Gaora, P.Ó.; Forde, N.; Besenfelder, U.; Havlicek, V.; Burns, G.W.; Spencer, T.E.; Gutierrez-Adan, A.; Lonergan, P.; Rizos, D. Oviduct-Embryo Interactions in Cattle: Two-Way Traffic or a One-Way Street?1. Biol. Reprod. 2015, 92, 144. [Google Scholar] [CrossRef] [PubMed]
- Pavani, K.C.; Alminana, C.; Wydooghe, E.; Catteeuw, M.; Ramírez, M.A.; Mermillod, P.; Rizos, D.; Van Soom, A. Emerging role of extracellular vesicles in communication of preimplantation embryos in vitro. Reprod. Fertil. Dev. 2017, 29, 66. [Google Scholar] [CrossRef] [PubMed]
- Giacomini, E.; Vago, R.; Sanchez, A.M.; Podini, P.; Zarovni, N.; Murdica, V.; Rizzo, R.; Bortolotti, D.; Candiani, M.; Viganò, P. Secretome of in vitro cultured human embryos contains extracellular vesicles that are uptaken by the maternal side. Sci. Rep. 2017, 7, 5210. [Google Scholar] [CrossRef]
- Sheller-Miller, S.; Trivedi, J.; Yellon, S.M.; Menon, R. Exosomes Cause Preterm Birth in Mice: Evidence for Paracrine Signaling in Pregnancy. Sci. Rep. 2019, 9, 608. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, J.; Lee, T.B.; Jun, J.H. Embryotrophic effects of extracellular vesicles derived from outgrowth embryos in pre- and peri-implantation embryonic development in mice. Mol. Reprod Dev. 2019, 86, 187–196. [Google Scholar] [CrossRef]
- Marin, D.; Scott, R.T. Extracellular vesicles: A promising tool for assessment of embryonic competence. Curr. Opin. Obstet. Gynecol. 2018, 30, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Saadeldin, I.M.; Kim, S.J.; Choi, Y.B.; Lee, B.C. Improvement of Cloned Embryos Development by Co-Culturing with Parthenotes: A Possible Role of Exosomes/Microvesicles for Embryos Paracrine Communication. Cell. Reprogram. 2014, 16, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Pavani, K.; Hendrix, A.; Van Den Broeck, W.; Couck, L.; Szymanska, K.; Lin, X.; De Koster, J.; Van Soom, A.; Leemans, B. Isolation and Characterization of Functionally Active Extracellular Vesicles from Culture Medium Conditioned by Bovine Embryos In Vitro. Int. J. Mol. Sci. 2018, 20, 38. [Google Scholar] [CrossRef]
- Qu, P.; Qing, S.; Liu, R.; Qin, H.; Wang, W.; Qiao, F.; Ge, H.; Liu, J.; Zhang, Y.; Cui, W.; et al. Effects of embryo-derived exosomes on the development of bovine cloned embryos. PLoS ONE 2017, 12, e0174535. [Google Scholar] [CrossRef]
- Battaglia, R.; Palini, S.; Vento, M.E.; La Ferlita, A.; Lo Faro, M.J.; Caroppo, E.; Borzì, P.; Falzone, L.; Barbagallo, D.; Ragusa, M.; et al. Identification of extracellular vesicles and characterization of miRNA expression profiles in human blastocoel fluid. Sci. Rep. 2019, 9, 84. [Google Scholar] [CrossRef]
- Dissanayake, K.; Nõmm, M.; Lättekivi, F.; Ressaissi, Y.; Godakumara, K.; Lavrits, A.; Midekessa, G.; Viil, J.; Bæk, R.; Jørgensen, M.M.; et al. Individually cultured bovine embryos produce extracellular vesicles that have the potential to be used as non-invasive embryo quality markers. Theriogenology 2020, 149, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Vilella, F.; Moreno-Moya, J.M.; Balaguer, N.; Grasso, A.; Herrero, M.; Martinez, S.; Marcilla, A.; Simon, C. Hsa-miR-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development 2015, 142, 3210–3221. [Google Scholar] [CrossRef] [PubMed]
- Park, K.R.; Inoue, T.; Ueda, M.; Hirano, T.; Higuchi, T.; Maeda, M.; Konishi, I.; Fujiwara, H.; Fujii, S. CD9 is expressed on human endometrial epithelial cells in association with integrins α6, α3 and β1. Mol. Hum. Reprod. 2000, 6, 252–257. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Domínguez, F.; Simón, C.; Quiñonero, A.; Ramírez, M.Á.; González-Muñoz, E.; Burghardt, H.; Cervero, A.; Martínez, S.; Pellicer, A.; Palacín, M.; et al. Human Endometrial CD98 Is Essential for Blastocyst Adhesion. PLoS ONE 2010, 5, e13380. [Google Scholar] [CrossRef]
- Wynne, F.; Ball, M.; McLellan, A.S.; Dockery, P.; Zimmermann, W.; Moore, T. Mouse pregnancy-specific glycoproteins: Tissue-specific expression and evidence of association with maternal vasculature. Reproduction 2006, 131, 721–732. [Google Scholar] [CrossRef][Green Version]
- Liu, W.M. Tetraspanin CD9 regulates invasion during mouse embryo implantation. J. Mol. Endocrinol. 2006, 36, 121–130. [Google Scholar] [CrossRef][Green Version]
- Lv, C.; Yu, W.-X.; Wang, Y.; Yi, D.-J.; Zeng, M.-H.; Xiao, H.-M. MiR-21 in extracellular vesicles contributes to the growth of fertilized eggs and embryo development in mice. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Nakamura, K.; Kusama, K.; Bai, R.; Sakurai, T.; Isuzugawa, K.; Godkin, J.D.; Suda, Y.; Imakawa, K. Induction of IFNT-Stimulated Genes by Conceptus-Derived Exosomes during the Attachment Period. PLoS ONE 2016, 11, e0158278. [Google Scholar] [CrossRef]
- Kusama, K.; Nakamura, K.; Bai, R.; Nagaoka, K.; Sakurai, T.; Imakawa, K. Intrauterine exosomes are required for bovine conceptus implantation. Biochem. Biophys. Res. Commun. 2018, 495, 1370–1375. [Google Scholar] [CrossRef]
- Burns, G.; Brooks, K.; Wildung, M.; Navakanitworakul, R.; Christenson, L.K.; Spencer, T.E. Extracellular Vesicles in Luminal Fluid of the Ovine Uterus. PLoS ONE 2014, 9, e90913. [Google Scholar] [CrossRef]
- Krawczynski, K.; Najmula, J.; Bauersachs, S.; Kaczmarek, M.M. MicroRNAome of Porcine Conceptuses and Trophoblasts: Expression Profile of microRNAs and Their Potential to Regulate Genes Crucial for Establishment of Pregnancy1. Biol. Reprod. 2015, 92, 21. [Google Scholar] [CrossRef] [PubMed]
- Bidarimath, M.; Khalaj, K.; Kridli, R.T.; Kan, F.W.K.; Koti, M.; Tayade, C. Extracellular vesicle mediated intercellular communication at the porcine maternal-fetal interface: A new paradigm for conceptus-endometrial cross-talk. Sci. Rep. 2017, 7, 40476. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.-S.; Ishibashi, O.; Ishikawa, G.; Ishikawa, T.; Katayama, A.; Mishima, T.; Takizawa, T.; Shigihara, T.; Goto, T.; Izumi, A.; et al. Human Villous Trophoblasts Express and Secrete Placenta-Specific MicroRNAs into Maternal Circulation via Exosomes1. Biol. Reprod. 2009, 81, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Salomon, C.; Torres, M.J.; Kobayashi, M.; Scholz-Romero, K.; Sobrevia, L.; Dobierzewska, A.; Illanes, S.E.; Mitchell, M.D.; Rice, G.E. A Gestational Profile of Placental Exosomes in Maternal Plasma and Their Effects on Endothelial Cell Migration. PLoS ONE 2014, 9, e98667. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.; Scholz-Romero, K.; Perez, A.; Illanes, S.E.; Mitchell, M.D.; Rice, G.E.; Salomon, C. Placenta-derived exosomes continuously increase in maternal circulation over the first trimester of pregnancy. J. Transl. Med. 2014, 12, 204. [Google Scholar] [CrossRef] [PubMed]
- Redman, C.W.G.; Sargent, I.L. Microparticles and immunomodulation in pregnancy and pre-eclampsia. J. Reprod. Immunol. 2007, 76, 61–67. [Google Scholar] [CrossRef]
- Baig, S.; Kothandaraman, N.; Manikandan, J.; Rong, L.; Ee, K.; Hill, J.; Lai, C.; Tan, W.; Yeoh, F.; Kale, A.; et al. Proteomic analysis of human placental syncytiotrophoblast microvesicles in preeclampsia. Clin. Proteom. 2014, 11, 40. [Google Scholar] [CrossRef]
- Pillay, P.; Maharaj, N.; Moodley, J.; Mackraj, I. Placental exosomes and pre-eclampsia: Maternal circulating levels in normal pregnancies and, early and late onset pre-eclamptic pregnancies. Placenta 2016, 46, 18–25. [Google Scholar] [CrossRef]
- Salomon, C.; Scholz-Romero, K.; Sarker, S.; Sweeney, E.; Kobayashi, M.; Correa, P.; Longo, S.; Duncombe, G.; Mitchell, M.D.; Rice, G.E.; et al. Gestational Diabetes Mellitus Is Associated With Changes in the Concentration and Bioactivity of Placenta-Derived Exosomes in Maternal Circulation Across Gestation. Diabetes 2016, 65, 598–609. [Google Scholar] [CrossRef]
- Kshirsagar, S.K.; Alam, S.M.; Jasti, S.; Hodes, H.; Nauser, T.; Gilliam, M.; Billstrand, C.; Hunt, J.S.; Petroff, M.G. Immunomodulatory molecules are released from the first trimester and term placenta via exosomes. Placenta 2012, 33, 982–990. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, S.; Pu, D.; Zhang, H.; Yang, L.; Zeng, P.; Su, F.; Chen, Z.; Guo, M.; Gu, Y.; et al. Detection of fetal trisomy and single gene disease by massively parallel sequencing of extracellular vesicle DNA in maternal plasma: A proof-of-concept validation. BMC Med. Genom. 2019, 12, 151. [Google Scholar] [CrossRef] [PubMed]
- Burkova, E.E.; Dmitrenok, P.S.; Bulgakov, D.V.; Vlassov, V.V.; Ryabchikova, E.I.; Nevinsky, G.A. Exosomes from human placenta purified by affinity chromatography on sepharose bearing immobilized antibodies against CD81 tetraspanin contain many peptides and small proteins: EXOSOMES FROM HUMAN PLACENTA. IUMBM Life 2018, 70, 1144–1155. [Google Scholar] [CrossRef] [PubMed]
- Yeung, C.H.; Cooper, T.G.; Bergmann, M.; Schulze, H. Organization of tubules in the human caput epididymidis and the ultrastructure of their epithelia. Am. J. Anat. 1991, 191, 261–279. [Google Scholar] [CrossRef]
- Akbarsha, M.; Faisal, K.; Radha, A. The Epididymis: Structure and Function. In Mammalian Endocrinology and Male Reproductive Biology; Singh, S., Ed.; CRC Press: Boca Raton, FL, USA, 2015; pp. 115–166. ISBN 978-1-4987-2735-8. [Google Scholar]
- Cornwall, G.A. New insights into epididymal biology and function. Hum. Reprod. Update 2009, 15, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Gervasi, M.G.; Visconti, P.E. Molecular changes and signaling events occurring in spermatozoa during epididymal maturation. Andrology 2017, 5, 204–218. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, R.; Mieusset, R. The human epididymis: Its function in sperm maturation. Hum. Reprod. Update 2016, 22, 574–587. [Google Scholar] [CrossRef]
- Yanagimachi, R.; Kamiguchi, Y.; Mikamo, K.; Suzuki, F.; Yanagimachi, H. Maturation of spermatozoa in the epididymis of the Chinese hamster. Am. J. Anat. 1985, 172, 317–330. [Google Scholar] [CrossRef]
- Fornés, M.W.; De Rosas, J.C. Interactions between rat epididymal epithelium and spermatozoa. Anat. Rec. 1991, 231, 193–200. [Google Scholar] [CrossRef]
- Gatti, J.-L.; Métayer, S.; Moudjou, M.; Andréoletti, O.; Lantier, F.; Dacheux, J.-L.; Sarradin, P. Prion Protein Is Secreted in Soluble Forms in the Epididymal Fluid and Proteolytically Processed and Transported in Seminal Plasma. Biol. Reprod 2002, 67, 393–400. [Google Scholar] [CrossRef]
- Rejraji, H.; Vernet, P.; Drevet, J.R. GPX5 is present in the mouse caput and cauda epididymidis lumen at three different locations. Mol. Reprod. Dev. 2002, 63, 96–103. [Google Scholar] [CrossRef]
- Yeung, C.-H.; Schröter, S.; Wagenfeld, A.; Kirchhoff, C.; Kliesch, S.; Poser, D.; Weinbauer, G.F.; Nieschlag, E.; Cooper, T.G. Interaction of the human epididymal protein CD52 (HE5) with epididymal spermatozoa from men and cynomolgus monkeys. Mol. Reprod. Dev. 1997, 48, 267–275. [Google Scholar] [CrossRef]
- Frenette, G.; Sullivan, R. Prostasome-like particles are involved in the transfer of P25b from the bovine epididymal fluid to the sperm surface. Mol. Reprod. Dev. 2001, 59, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Girouard, J.; Frenette, G.; Sullivan, R. Compartmentalization of Proteins in Epididymosomes Coordinates the Association of Epididymal Proteins with the Different Functional Structures of Bovine Spermatozoa1. Biol. Reprod. 2009, 80, 965–972. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sullivan, R.; Saez, F. Epididymosomes, prostasomes, and liposomes: Their roles in mammalian male reproductive physiology. Reproduction 2013, 146, R21–R35. [Google Scholar] [CrossRef]
- Thimon, V.; Frenette, G.; Saez, F.; Thabet, M.; Sullivan, R. Protein composition of human epididymosomes collected during surgical vasectomy reversal: A proteomic and genomic approach. Hum. Reprod. 2008, 23, 1698–1707. [Google Scholar] [CrossRef]
- Nixon, B.; De Iuliis, G.N.; Hart, H.M.; Zhou, W.; Mathe, A.; Bernstein, I.R.; Anderson, A.L.; Stanger, S.J.; Skerrett-Byrne, D.A.; Jamaluddin, M.F.B.; et al. Proteomic Profiling of Mouse Epididymosomes Reveals their Contributions to Post-testicular Sperm Maturation. Mol. Cell Proteom. 2019, 18, S91–S108. [Google Scholar] [CrossRef]
- Caballero, J.N.; Frenette, G.; Belleannée, C.; Sullivan, R. CD9-Positive Microvesicles Mediate the Transfer of Molecules to Bovine Spermatozoa during Epididymal Maturation. PLoS ONE 2013, 8, e65364. [Google Scholar] [CrossRef]
- Sullivan, R. Epididymosomes: A heterogeneous population of microvesicles with multiple functions in sperm maturation and storage. Asian J. Androl. 2015, 17, 726–729. [Google Scholar] [CrossRef]
- Kirchhoff, C.; Hale, G. Cell-to-cell transfer of glycosylphosphatidylinositol-anchored membrane proteins during sperm maturation. Mol. Hum. Reprod. 1996, 2, 177–184. [Google Scholar] [CrossRef]
- Orlean, P.; Menon, A.K. Thematic review series: Lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: How we learned to stop worrying and love glycophospholipids. J. Lipid Res. 2007, 48, 993–1011. [Google Scholar] [CrossRef]
- Légaré, C.; Bérubé, B.; Boué, F.; Lefièvre, L.; Morales, C.R.; El-Alfy, M.; Sullivan, R. Hamster sperm antigen P26h is a phosphatidylinositol-anchored protein. Mol. Reprod. Dev. 1999, 52, 225–233. [Google Scholar] [CrossRef]
- Michalková, K.; Simon, M.; Antalíková, J.; Klíma, J.; Horovská, L.; Jankovicová, J.; Hluchý, S. Identification of bovine CD52-like molecule by monoclonal antibody IVA-543: Distribution of CD52-like molecule in the bull genital tract. Theriogenology 2010, 74, 1066–1074. [Google Scholar] [CrossRef] [PubMed]
- Lefèvre, B.; Wolf, J.-P.; Ziyyat, A. Sperm-egg interaction: Is there a link between tetraspanin(s) and GPI-anchored protein(s)? Bioessays 2010, 32, 143–152. [Google Scholar] [CrossRef] [PubMed]
- de Gassart, A.; Geminard, C.; Fevrier, B.; Raposo, G.; Vidal, M. Lipid raft-associated protein sorting in exosomes. Blood 2003, 102, 4336–4344. [Google Scholar] [CrossRef] [PubMed]
- Juyena, N.S.; Stelletta, C. Seminal Plasma: An Essential Attribute to Spermatozoa. J. Androl. 2012, 33, 536–551. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Guo, W.-B.; Zhang, W.-S.; Bian, J.; Yang, J.-K.; Zhou, Q.-Z.; Chen, M.-K.; Peng, W.; Qi, T.; Wang, C.-Y.; et al. Comprehensive proteomics analysis of exosomes derived from human seminal plasma. Andrology 2017, 5, 1007–1015. [Google Scholar] [CrossRef] [PubMed]
- Rolland, A.D.; Lavigne, R.; Dauly, C.; Calvel, P.; Kervarrec, C.; Freour, T.; Evrard, B.; Rioux-Leclercq, N.; Auger, J.; Pineau, C. Identification of genital tract markers in the human seminal plasma using an integrative genomics approach. Hum. Reprod. 2013, 28, 199–209. [Google Scholar] [CrossRef]
- Höög, J.L.; Lötvall, J. Diversity of extracellular vesicles in human ejaculates revealed by cryo-electron microscopy. J. Extracell. Vesicles 2015, 4, 28680. [Google Scholar] [CrossRef]
- Alvarez-Rodriguez, M.; Ljunggren, S.A.; Karlsson, H.; Rodriguez-Martinez, H. Exosomes in specific fractions of the boar ejaculate contain CD44: A marker for epididymosomes? Theriogenology 2019, 140, 143–152. [Google Scholar] [CrossRef]
- Murdica, V.; Giacomini, E.; Alteri, A.; Bartolacci, A.; Cermisoni, G.C.; Zarovni, N.; Papaleo, E.; Montorsi, F.; Salonia, A.; Viganò, P.; et al. Seminal plasma of men with severe asthenozoospermia contain exosomes that affect spermatozoa motility and capacitation. Fertil. Steril. 2019, 111, 897–908. [Google Scholar] [CrossRef]
- Ronquist, G.; Brody, I.; Gottfries, A.; Stegmayr, B. An Mg2+ and Ca2+-Stimulated Adenosine Triphosphatase in Human Prostatic Fluid: Part I. Andrologia 1978, 10, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, G.; Brody, I.; Gottfries, A.; Stegmayr, B. An Mg2+ and Ca2+-Stimulated Adenosine Triphosphatase in Human Prostatic Fluid—Part II. Andrologia 1978, 10, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Brody, I.; Ronquist, G.; Gottfries, A. Ultrastructural localization of the prostasome—An organelle in human seminal plasma. Ups. J. Med. Sci. 1983, 88, 63–80. [Google Scholar] [CrossRef]
- Ronquist, G.; Brody, I. The prostasome: Its secretion and function in man. Biochim. Biophys. Acta 1985, 822, 203–218. [Google Scholar] [CrossRef]
- Ronquist, G. Prostasomes are mediators of intercellular communication: From basic research to clinical implications: Review: Prostasomes are mediators of intercellular communication. J. Intern. Med. 2012, 271, 400–413. [Google Scholar] [CrossRef] [PubMed]
- Aalberts, M.; van Dissel-Emiliani, F.M.F.; van Adrichem, N.P.H.; van Wijnen, M.; Wauben, M.H.M.; Stout, T.A.E.; Stoorvogel, W. Identification of distinct populations of prostasomes that differentially express prostate stem cell antigen, annexin A1, and GLIPR2 in humans. Biol. Reprod. 2012, 86, 82. [Google Scholar] [CrossRef] [PubMed]
- Aalberts, M.; Sostaric, E.; Wubbolts, R.; Wauben, M.W.M.; Nolte-’t Hoen, E.N.M.; Gadella, B.M.; Stout, T.A.E.; Stoorvogel, W. Spermatozoa recruit prostasomes in response to capacitation induction. Biochim. Biophys. Acta 2013, 1834, 2326–2335. [Google Scholar] [CrossRef]
- Du, J.; Shen, J.; Wang, Y.; Pan, C.; Pang, W.; Diao, H.; Dong, W. Boar seminal plasma exosomes maintain sperm function by infiltrating into the sperm membrane. Oncotarget 2016, 7, 58832–58847. [Google Scholar] [CrossRef] [PubMed]
- Brzozowski, J.S.; Bond, D.R.; Jankowski, H.; Goldie, B.J.; Burchell, R.; Naudin, C.; Smith, N.D.; Scarlett, C.J.; Larsen, M.R.; Dun, M.D.; et al. Extracellular vesicles with altered tetraspanin CD9 and CD151 levels confer increased prostate cell motility and invasion. Sci. Rep. 2018, 8, 8822. [Google Scholar] [CrossRef]
- Barranco, I.; Padilla, L.; Parrilla, I.; Álvarez-Barrientos, A.; Pérez-Patiño, C.; Peña, F.J.; Martínez, E.A.; Rodriguez-Martínez, H.; Roca, J. Extracellular vesicles isolated from porcine seminal plasma exhibit different tetraspanin expression profiles. Sci. Rep. 2019, 9, 11584. [Google Scholar] [CrossRef]
- Alvarez-Rodriguez, M.; Ntzouni, M.; Wright, D.; Khan, K.I.; López-Béjar, M.; Martinez, C.A.; Rodriguez-Martinez, H. Chicken seminal fluid lacks CD9- and CD44-bearing extracellular vesicles. Reprod. Domest. Anim. 2020, 55, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Lötvall, J.; Hill, A.F.; Hochberg, F.; Buzás, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [PubMed]
Publisher's Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
| Protein Name | Alternative Name (s) | Gene Name |
|---|---|---|
| Tetraspanin-1 | Tetraspan NET-1 (neuroepithelial cell-transforming gene 1 protein), Tetraspanin TM4-C | TSPAN1 |
| Tetraspanin-2 | Tetraspan NET-3 | TSPAN2 |
| Tetraspanin-3 | Tetraspanin TM4-A, Transmembrane 4 superfamily member 8 | TSPAN3 |
| Tetraspanin-4 | Novel antigen 2 (NAG-2), Transmembrane 4 superfamily member 7 | TSPAN4 |
| Tetraspanin-5 | Tetraspan NET-4, Transmembrane 4 superfamily member 9 | TSPAN5 |
| Tetraspanin-6 | A15 homologue, Putative NF-kappa-B-activating protein 321, T245 protein, Tetraspanin TM4-D, Transmembrane 4 superfamily member 6 | TSPAN6 |
| Tetraspanin-7 | Cell surface glycoprotein A15, Membrane component chromosome X surface marker 1, T-cell acute lymphoblastic leukemia-associated antigen 1 (TALLA-1), Transmembrane 4 superfamily member 2, CD231 | TSPAN7 |
| Tetraspanin-8 | Transmembrane 4 superfamily member 3, Tumor-associated antigen CO-029 | TSPAN8 |
| Tetraspanin-9 | Tetraspan NET-5 | TSPAN9 |
| Tetraspanin-10 | OCULOSPANIN | TSPAN10 |
| Tetraspanin-11 | - | TSPAN11 |
| Tetraspanin-12 | Tetraspan NET-2, Transmembrane 4 superfamily member 12 | TSPAN12 |
| Tetraspanin-13 | Tetraspan NET-6, Transmembrane 4 superfamily member 13 | TSPAN13 |
| Tetraspanin-14 | DC-TM4F2, Transmembrane 4 superfamily member 14 | TSPAN14 |
| Tetraspanin-15 | Tetraspan NET-7, Transmembrane 4 superfamily member 15 | TSPAN15 |
| Tetraspanin-16 | Tetraspanin TM4-B, Transmembrane 4 superfamily member 16 | TSPAN16 |
| Tetraspanin-17 | F-box only protein 23, Tetraspan protein SB134, Transmembrane 4 superfamily member 17 | TSPAN17 |
| Tetraspanin-18 | - | TSPAN18 |
| Putative tetraspanin-19 | - | TSPAN19 |
| Uroplakin-1b (UP1b) | Tetraspanin-20 (Tspan-20) | UPK1B |
| Uroplakin-1a (UP1a, UPKa) | Tetraspanin-21 (Tspan-21) | UPK1A |
| Peripherin-2 | Retinal degeneration slow protein, Tetraspanin-22 (Tspan-22) | PRPH2 |
| Rod outer segment membrane protein 1 (ROSP1) | Tetraspanin-23 (Tspan-23) | ROM1 |
| CD151 antigen | CD151, GP27, Membrane glycoprotein SFA-1, Platelet-endothelial tetraspan antigen 3 (PETA-3), Tetraspanin-24 (Tspan-24) | CD151 |
| Leucocyte surface antigen CD53 | CD53, Cell surface glycoprotein CD53, Tetraspanin-25 (Tspan-25) | CD53 |
| Leucocyte antigen CD37 | CD37, Tetraspanin-26 (Tspan-26) | CD37 |
| CD82 antigen | CD82, C33 antigen, IA4, Inducible membrane protein R2, Metastasis suppressor Kangai-1, Suppressor of tumorigenicity 6 protein, Tetraspanin-27 (Tspan-27) | CD82 |
| CD81 antigen | CD81, 26 kDa cell surface protein TAPA1, Tetraspanin-28 (Tspan-28), Target of the antiproliferative antibody 1 | CD81 |
| CD9 antigen | CD9, 5H9 antigen, Cell growth-inhibiting gene 2 protein, Leukocyte antigen MIC3, Motility-related protein (MRP-1), p24, Tetraspanin-29 (Tspan-29) | CD9 |
| CD63 antigen | CD63, Granulophysin, Lysosomal-associated membrane protein 3 (LAMP-3), Melanoma-associated antigen ME491, OMA81H, Ocular melanoma-associated antigen, Tetraspanin-30 (Tspan-30) | CD63 |
| Tetraspanin-31 (Tspan-31) | Sarcoma-amplified sequence | TSPAN31 |
| Tetraspanin-32 (Tspan-32) | Protein Phemx | TSPAN32 |
| Tetraspanin-33 (Tspan-33) | Penumbra (hPen), Proerythroblast new membrane | TSPAN33 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction. Int. J. Mol. Sci. 2020, 21, 7568. https://doi.org/10.3390/ijms21207568
Jankovičová J, Sečová P, Michalková K, Antalíková J. Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction. International Journal of Molecular Sciences. 2020; 21(20):7568. https://doi.org/10.3390/ijms21207568
Chicago/Turabian StyleJankovičová, Jana, Petra Sečová, Katarína Michalková, and Jana Antalíková. 2020. "Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction" International Journal of Molecular Sciences 21, no. 20: 7568. https://doi.org/10.3390/ijms21207568
APA StyleJankovičová, J., Sečová, P., Michalková, K., & Antalíková, J. (2020). Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction. International Journal of Molecular Sciences, 21(20), 7568. https://doi.org/10.3390/ijms21207568