The Effect of miR-146a on the Gene Expression of Immunoregulatory Cytokines in Human Mesenchymal Stromal Cells
Abstract
1. Introduction
2. Results
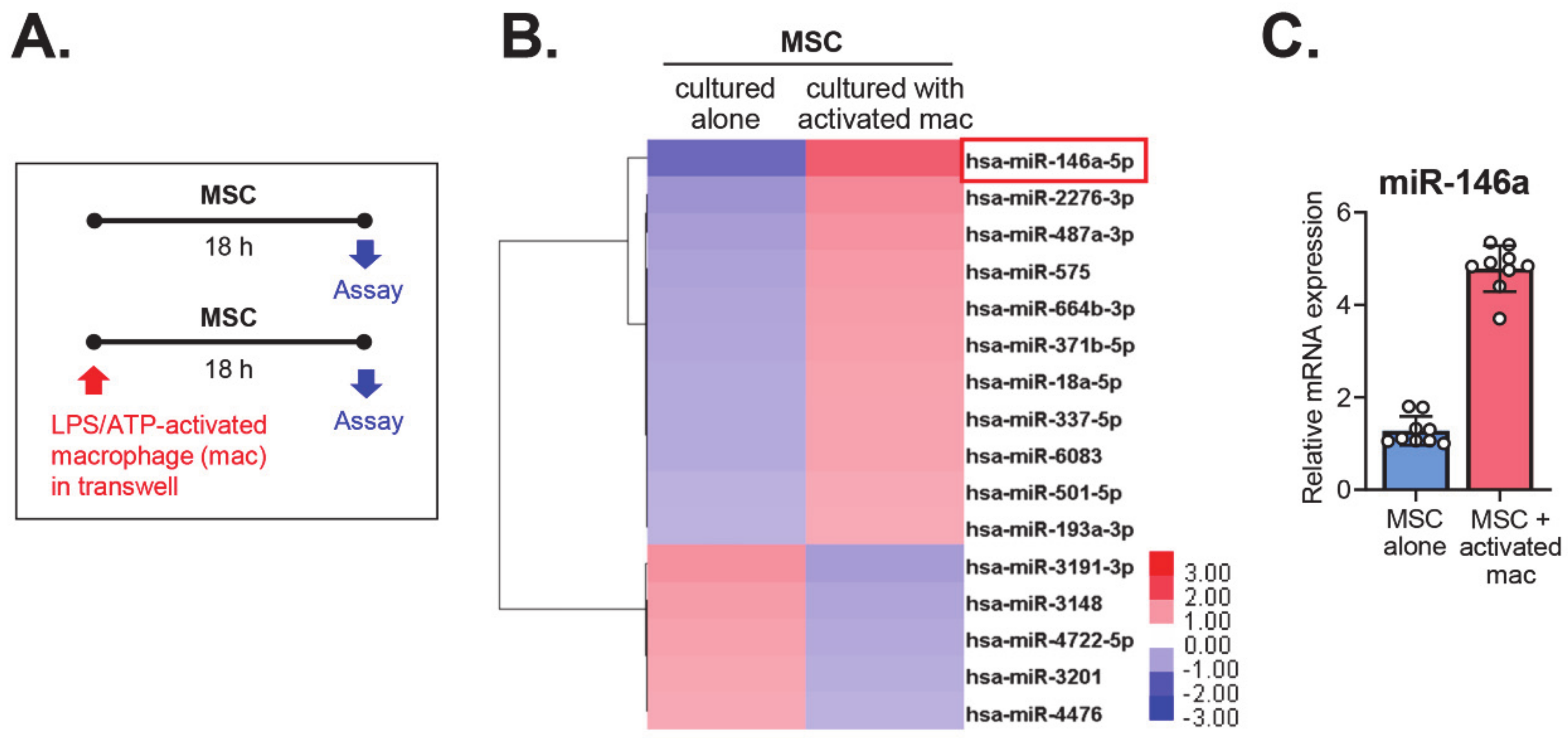
2.1. The MiRNA Expression Profile of MSCs Co-Cultured with Activated Macrophages
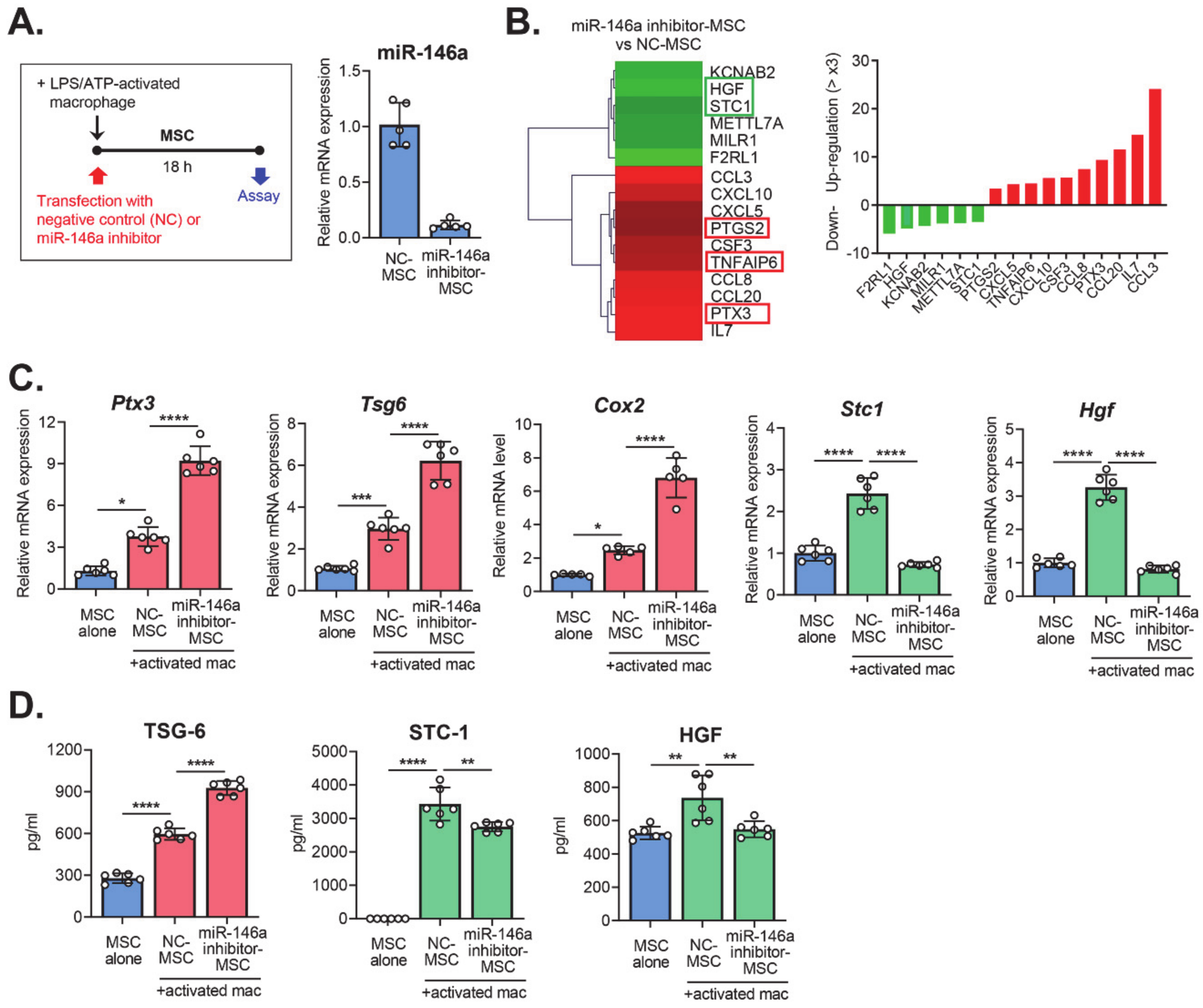
2.2. Transcriptome Profile of MSCs with miR-146a Inhibition
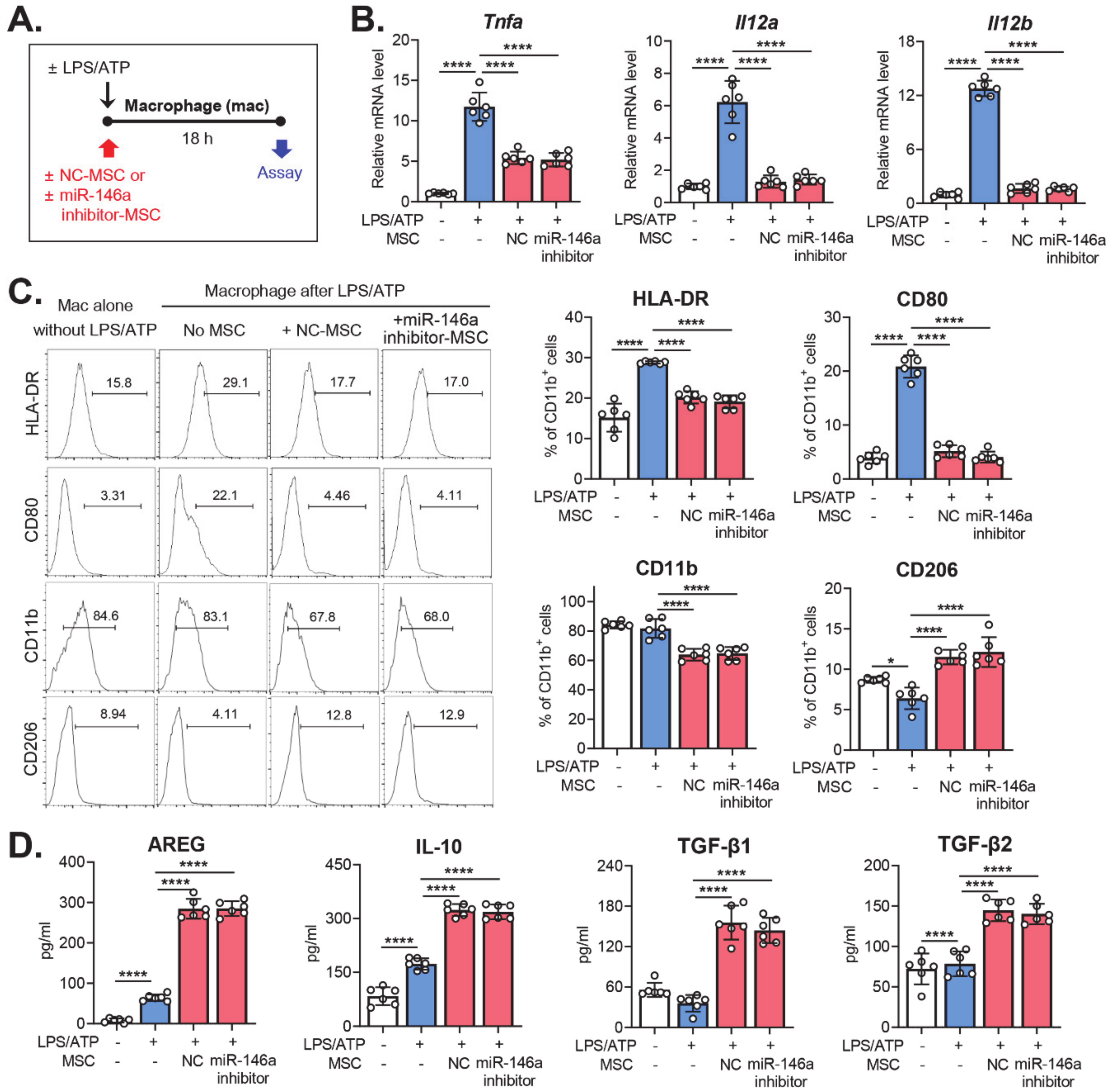
2.3. Effects of miR-146a Inhibition in MSCs on the Modulation of Macrophage Polarization
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Transfection
4.2. RNA Sequencing
4.3. Real-Time RT-PCR
4.4. ELISA
4.5. Flow Cytometry
4.6. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Clark, E.A.; Kalomoiris, S.; Nolta, J.A.; Fierro, F.A. Concise review: MicroRNA function in multipotent mesenchymal stromal cells. Stem Cells 2014, 32, 1074–1082. [Google Scholar] [CrossRef]
- Rostami, Z.; Khorashadizadeh, M.; Naseri, M. Immunoregulatory properties of mesenchymal stem cells: Micro-RNAs. Immunol. Lett. 2020, 219, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Prockop, D.J. Marrow stromal cells as stem cells for nonhematopoietic tissues. Science 1997, 276, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Prockop, D.J.; Oh, J.Y. Mesenchymal stem/stromal cells (MSCs): Role as guardians of inflammation. Mol. Ther. 2012, 20, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.H.; Oh, J.Y.; Choi, H.; Bazhanov, N. Therapeutic factors secreted by mesenchymal stromal cells and tissue repair. J. Cell. Biochem. 2011, 112, 3073–3078. [Google Scholar] [CrossRef]
- Ko, J.H.; Kim, H.J.; Jeong, H.J.; Lee, H.J.; Oh, J.Y. Mesenchymal Stem and Stromal Cells Harness Macrophage-Derived Amphiregulin to Maintain Tissue Homeostasis. Cell Rep. 2020, 30, 3806–3820. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kwon, J.H.; Bae, Y.K.; Kim, G.H.; Um, S.; Ha, J.; Choi, S.J.; Oh, W.; Jin, H.J. Soluble PTX3 of Human Umbilical Cord Blood-Derived Mesenchymal Stem Cells Attenuates Hyperoxic Lung Injury by Activating Macrophage Polarization in Neonatal Rat Model. Stem Cells Int. 2020, 2020, 1802976. [Google Scholar] [CrossRef]
- Kim, H.; Lee, M.J.; Bae, E.H.; Ryu, J.S.; Kaur, G.; Kim, H.J.; Kim, J.Y.; Barreda, H.; Jung, S.Y.; Choi, J.M.; et al. Comprehensive Molecular Profiles of Functionally Effective MSC-Derived Extracellular Vesicles in Immunomodulation. Mol. Ther. 2020, 28, 1628–1644. [Google Scholar] [CrossRef]
- Lee, R.H.; Pulin, A.A.; Seo, M.J.; Kota, D.J.; Ylostalo, J.; Larson, B.L.; Semprun-Prieto, L.; Delafontaine, P.; Prockop, D.J. Intravenous hMSCs improve myocardial infarction in mice because cells embolized in lung are activated to secrete the anti-inflammatory protein TSG-6. Cell Stem Cell. 2009, 5, 54–63. [Google Scholar] [CrossRef]
- Choi, H.; Lee, R.H.; Bazhanov, N.; Oh, J.Y.; Prockop, D.J. Anti-inflammatory protein TSG-6 secreted by activated MSCs attenuates zymosan-induced mouse peritonitis by decreasing TLR2/NF-κB signaling in resident macrophages. Blood 2011, 118, 330–338. [Google Scholar] [CrossRef]
- Foraker, J.E.; Oh, J.Y.; Ylostalo, J.H.; Lee, R.H.; Watanabe, J.; Prockop, D.J. Cross-talk between human mesenchymal stem/progenitor cells (MSCs) and rat hippocampal slices in LPS-stimulated cocultures: The MSCs are activated to secrete prostaglandin E2. J. Neurochem. 2011, 119, 1052–1063. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Ko, J.H.; Kim, H.J.; Jeong, H.J.; Oh, J.Y. Mesenchymal stromal cells induce distinct myeloid-derived suppressor cells in inflammation. JCI Insight. 2020, 5, 136059. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.K.; Foulsham, W.; Shukla, S.; Elbasiony, E.; Omoto, M.; Chauhan, S.K. Mesenchymal Stromal Cells Modulate Corneal Alloimmunity via Secretion of Hepatocyte Growth Factor. Stem Cells Transl. Med. 2019, 8, 1030–1040. [Google Scholar] [CrossRef]
- Oh, J.Y.; Ko, J.H.; Lee, H.J.; Yu, J.M.; Choi, H.; Kim, M.K.; Wee, W.R.; Prockop, D.J. Mesenchymal stem/stromal cells inhibit the NLRP3 inflammasome by decreasing mitochondrial reactive oxygen species. Stem Cells 2014, 32, 1553–1563. [Google Scholar] [CrossRef] [PubMed]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef]
- Boldin, M.P.; Taganov, K.D.; Rao, D.S.; Yang, L.; Zhao, J.L.; Kalwani, M.; Garcia-Flores, Y.; Luong, M.; Devrekanli, A.; Xu, J.; et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011, 208, 1189–1201. [Google Scholar] [CrossRef]
- Hsieh, J.Y.; Huang, T.S.; Cheng, S.M.; Lin, W.S.; Tsai, T.N.; Lee, O.K.; Wang, H.W. miR-146a-5p circuitry uncouples cell proliferation and migration, but not differentiation, in human mesenchymal stem cells. Nucleic Acids Res. 2013, 41, 9753–9763. [Google Scholar] [CrossRef]
- Matysiak, M.; Fortak-Michalska, M.; Szymanska, B.; Orlowski, W.; Jurewicz, A.; Selmaj, K. MicroRNA-146a negatively regulates the immunoregulatory activity of bone marrow stem cells by targeting prostaglandin E2 synthase-2. J. Immunol. 2013, 190, 5102–5109. [Google Scholar] [CrossRef]
- Su, Y.L.; Wang, X.; Mann, M.; Adamus, T.P.; Wang, D.; Moreira, D.F.; Zhang, Z.; Ouyang, C.; He, X.; Zhang, B.; et al. Myeloid cell-targeted miR-146a mimic inhibits NF-κB-driven inflammation and leukemia progression in vivo. Blood 2020, 135, 167–180. [Google Scholar] [CrossRef]
- Li, D.; Duan, M.; Feng, Y.; Geng, L.; Li, X.; Zhang, W. MiR-146a modulates macrophage polarization in systemic juvenile idiopathic arthritis by targeting INHBA. Mol. Immunol. 2016, 77, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Nahid, M.A.; Pauley, K.M.; Satoh, M.; Chan, E.K. miR-146a is critical for endotoxin-induced tolerance: IMPLICATION IN INNATE IMMUNITY. J. Biol. Chem. 2009, 284, 34590–34599. [Google Scholar] [CrossRef] [PubMed]
- Doxaki, C.; Kampranis, S.C.; Eliopoulos, A.G.; Spilianakis, C.; Tsatsanis, C. Coordinated Regulation of miR-155 and miR-146a Genes during Induction of Endotoxin Tolerance in Macrophages. J. Immunol. 2015, 195, 5750–5761. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Dou, H.; Li, X.; Zhao, X.; Li, Y.; Liu, D.; Ji, J.; Liu, F.; Ding, L.; Ni, Y.; et al. Exosomal miR-146a Contributes to the Enhanced Therapeutic Efficacy of Interleukin-1β-Primed Mesenchymal Stem Cells Against Sepsis. Stem Cells 2017, 35, 1208–1221. [Google Scholar] [CrossRef]
- Fang, S.B.; Zhang, H.Y.; Wang, C.; He, B.X.; Liu, X.Q.; Meng, X.C.; Peng, Y.Q.; Xu, Z.B.; Fan, X.L.; Wu, Z.J.; et al. Small extracellular vesicles derived from human mesenchymal stromal cells prevent group 2 innate lymphoid cell-dominant allergic airway inflammation through delivery of miR-146a-5p. J. Extracell. Vesicles. 2020, 9, 1723260. [Google Scholar] [CrossRef]
- Miyaji, T.; Takami, T.; Fujisawa, K.; Matsumoto, T.; Yamamoto, N.; Sakaida, I. Bone marrow-derived humoral factors suppress oxidative phosphorylation, upregulate TSG-6, and improve therapeutic effects on liver injury of mesenchymal stem cells. J. Clin. Biochem. Nutr. 2020, 66, 213–223. [Google Scholar] [CrossRef]
- Lei, Q.; Liu, T.; Gao, F.; Xie, H.; Sun, L.; Zhao, A.; Ren, W.; Guo, H.; Zhang, L.; Wang, H.; et al. Microvesicles as Potential Biomarkers for the Identification of Senescence in Human Mesenchymal Stem Cells. Theranostics 2017, 7, 2673–2689. [Google Scholar] [CrossRef]
- Xie, Q.; Wei, W.; Ruan, J.; Ding, Y.; Zhuang, A.; Bi, X.; Sun, H.; Gu, P.; Wang, Z.; Fan, X. Effects of miR-146a on the osteogenesis of adipose-derived mesenchymal stem cells and bone regeneration. Sci. Rep. 2017, 7, 42840. [Google Scholar] [CrossRef]
- Zhou, X.; Moussa, F.M.; Mankoci, S.; Ustriyana, P.; Zhang, N.; Abdelmagid, S.; Molenda, J.; Murphy, W.L.; Safadi, F.F.; Sahai, N. Orthosilicic acid, Si(OH)4, stimulates osteoblast differentiation in vitro by upregulating miR-146a to antagonize NF-κB activation. Acta Biomater. 2016, 39, 192–202. [Google Scholar] [CrossRef]
- Cho, H.H.; Shin, K.K.; Kim, Y.J.; Song, J.S.; Kim, J.M.; Bae, Y.C.; Kim, C.D.; Jung, J.S. NF-kappaB activation stimulates osteogenic differentiation of mesenchymal stem cells derived from human adipose tissue by increasing TAZ expression. J. Cell. Physiol. 2010, 223, 168–177. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ko, J.H.; Oh, J.Y. The Effect of miR-146a on the Gene Expression of Immunoregulatory Cytokines in Human Mesenchymal Stromal Cells. Int. J. Mol. Sci. 2020, 21, 6809. https://doi.org/10.3390/ijms21186809
Ko JH, Oh JY. The Effect of miR-146a on the Gene Expression of Immunoregulatory Cytokines in Human Mesenchymal Stromal Cells. International Journal of Molecular Sciences. 2020; 21(18):6809. https://doi.org/10.3390/ijms21186809
Chicago/Turabian StyleKo, Jung Hwa, and Joo Youn Oh. 2020. "The Effect of miR-146a on the Gene Expression of Immunoregulatory Cytokines in Human Mesenchymal Stromal Cells" International Journal of Molecular Sciences 21, no. 18: 6809. https://doi.org/10.3390/ijms21186809
APA StyleKo, J. H., & Oh, J. Y. (2020). The Effect of miR-146a on the Gene Expression of Immunoregulatory Cytokines in Human Mesenchymal Stromal Cells. International Journal of Molecular Sciences, 21(18), 6809. https://doi.org/10.3390/ijms21186809




