Emerging Gene-Editing Modalities for Osteoarthritis
Abstract
1. Introduction
2. Current Treatment Options and Limitations
3. CRISPR/Cas
4. Mesenchymal Stem Cells and Tissue Regeneration
5. Extracellular Vesicles
6. Potential CRISPR/Cas9 Molecular Targets for the Treatment of Osteoarthritis
7. Additional Emerging Targets for the Treatment of Osteoarthritis Using CRISPR/Cas9
8. Limitations and Future Considerations
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| OA | Osteoarthritis |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| Cas | Cas nuclease |
| crRNA | CRISPR RNA |
| tracrRNA | trans-activating crRNA |
| PAM | protospacer adjacent motif |
| sgRNA | single guide RNA |
| MSC | Mesenchymal stem cells |
| miRNA | Micro RNA |
| EV | Extracellular vesicles |
| IL-1β | Interleukin 1 beta |
| TNFα | Tumor Necrosis Factor |
| Hac | Human articular cartilage |
| IL1-R1 | IL-1β cytokine receptor 1 |
| BGLAP | Osteocalcin |
| Has2 | Hyaluronan synthase 2 |
| PRG4 | Lubricin |
| Runx2 | Runt Related Transcription Factor 2 |
| Hrdl | E3 ubiquitin-protein ligase hrd-like protein 1 |
| Mmp13 | Matrix metalloprotein 13 |
| Cx43 | Connexin 43 |
| Cbx4 | Chromobox 4 |
| Foxd1 | Forkhead box D1 |
| YAP | Yes-associated protein 1 |
| NHEJ | Non-homologous end joining |
| HDR | Homology Directed Repair |
| TALENs | Transcription activator-like effector nucleases |
| ZFN | Zinc finger nucleases |
References
- Bitton, R. The economic burden of osteoarthritis. Am. J. Manag. Care 2009, 15, S230–S235. [Google Scholar] [PubMed]
- Murphy, L.B.; Cisternas, M.G.; Pasta, D.J.; Helmick, C.G.; Yelin, E.H. Medical Expenditures and Earnings Losses Among US Adults with Arthritis in 2013. Arthritis Care Res. 2018, 70, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Barbour, K.E.; Helmick, C.G.; Boring, M.; Brady, T.J. Vital Signs: Prevalence of Doctor-Diagnosed Arthritis and Arthritis-Attributable Activity Limitation-United States, 2013–2015. MMWR. Morb. Mortal. Wkly. Rep. 2017, 66, 246–253. [Google Scholar] [CrossRef] [PubMed]
- Hootman, J.M.; Helmick, C.G.; Barbour, K.E.; Theis, K.A.; Boring, M.A. Updated Projected Prevalence of Self-Reported Doctor-Diagnosed Arthritis and Arthritis-Attributable Activity Limitation Among US Adults, 2015–2040. Arthritis Rheumatol. 2016, 68, 1582–1587. [Google Scholar] [CrossRef]
- Sophia Fox, A.J.; Bedi, A.; Rodeo, S.A. The basic science of articular cartilage: Structure, composition, and function. Sports Health 2009, 1, 461–468. [Google Scholar] [CrossRef]
- Medvedeva, E.V.; Grebenik, E.A.; Gornostaeva, S.N.; Telpuhov, V.I.; Lychagin, A.V.; Timashev, P.S.; Chagin, A.S. Repair of damaged articular cartilage: Current approaches and future directions. Int. J. Mol. Sci. 2018, 19, 2366. [Google Scholar] [CrossRef] [PubMed]
- Karuppal, R. Current concepts in the articular cartilage repair and regeneration. J. Orthop. 2017, 14, A1–A3. [Google Scholar] [CrossRef]
- Kreuz, P.C.; Steinwachs, M.R.; Erggelet, C.; Krause, S.J.; Konrad, G.; Uhl, M.; Südkamp, N. Results after microfracture of full-thickness chondral defects in different compartments in the knee. Osteoarthr. Cartil. 2006, 14, 1119–1125. [Google Scholar] [CrossRef]
- Armiento, A.R.; Alini, M.; Stoddart, M.J. Articular fibrocartilage—Why does hyaline cartilage fail to repair? Adv. Drug Deliv. Rev. 2019, 146, 289–305. [Google Scholar] [CrossRef]
- Tamer, T.M. Hyaluronan and synovial joint: Function, distribution and healing. Interdiscip. Toxicol. 2013, 6, 111–125. [Google Scholar] [CrossRef]
- Seror, J.; Zhu, L.; Goldberg, R.; Day, A.J.; Klein, J. Supramolecular synergy in the boundary lubrication of synovial joints. Nat. Commun. 2015, 6, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Odgren, P.R.; Witwicka, H.; Reyes-Gutierrez, P. The cast of clasts: Catabolism and vascular invasion during bone growth, repair, and disease by osteoclasts, chondroclasts, and septoclasts. Connect. Tissue Res. 2016, 57, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Buckwalter, J.A.; Mankin, H.J. Articular cartilage: Degeneration and osteoarthritis, repair, regeneration, and transplantation. Instr. Course Lect. 1998, 47, 487–504. [Google Scholar] [PubMed]
- Harrell, C.R.; Markovic, B.S.; Fellabaum, C.; Arsenijevic, A.; Volarevic, V. Mesenchymal stem cell-based therapy of osteoarthritis: Current knowledge and future perspectives. Biomed. Pharmacother. 2019, 109, 2318–2326. [Google Scholar] [CrossRef]
- Sandell, L.J.; Aigner, T. Articular cartilage and changes in arthritis. An introduction: Cell biology of osteoarthritis. Arthritis Res. 2001, 3, 107–113. [Google Scholar] [CrossRef]
- Berenbaum, F. Osteoarthritis as an inflammatory disease (osteoarthritis is not osteoarthrosis!). Osteoarthr. Cartil. 2013, 21, 16–21. [Google Scholar] [CrossRef]
- Fukui, N.; Purple, C.R.; Sandell, L.J. Cell biology of osteoarthritis: The chondrocyte’s response to injury. Curr. Rheumatol. Rep. 2001, 3, 496–505. [Google Scholar] [CrossRef]
- Andriacchi, T.P.; Mündermann, A.; Smith, R.L.; Alexander, E.J.; Dyrby, C.O.; Koo, S. A framework for the in vivo pathomechanics of osteoarthritis at the knee. Ann. Biomed. Eng. 2004, 32, 447–457. [Google Scholar] [CrossRef]
- DeFrate, L.E.; Kim-Wang, S.Y.; Englander, Z.A.; McNulty, A.L. Osteoarthritis year in review 2018: Mechanics. Osteoarthr. Cartil. 2019, 27, 392–400. [Google Scholar] [CrossRef]
- Musumeci, G.; Aiello, F.C.; Szychlinska, M.A.; Di Rosa, M.; Castrogiovanni, P.; Mobasheri, A. Osteoarthritis in the XXIst century: Risk factors and behaviours that influence disease onset and progression. Int. J. Mol. Sci. 2015, 16, 6093–6112. [Google Scholar] [CrossRef]
- Qin, Y.X.; Lam, H.Y. Intramedullary pressure and matrix strain induced by oscillatory skeletal muscle stimulation and its potential in adaptation. J. Biomech. 2009, 42, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Anandacoomarasamy, A.; March, L. Current evidence for osteoarthritis treatments. Ther. Adv. Musculoskelet. Dis. 2010, 2, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Songer, T.J.; LaPorte, R.E. Disabilities due to injury in the military. Am. J. Prev. Med. 2000, 18, 33–40. [Google Scholar] [CrossRef]
- Ma, V.Y.; Chan, L.; Carruthers, K.J. Incidence, prevalence, costs, and impact on disability of common conditions requiring rehabilitation in the united states: Stroke, spinal cord injury, traumatic brain injury, multiple sclerosis, osteoarthritis, rheumatoid arthritis, limb loss, and back pa. Arch. Phys. Med. Rehabil. 2014, 95, 986–995. [Google Scholar] [CrossRef]
- Losina, E.; Walensky, R.P.; Reichmann, W.M.; Holt, H.L.; Gerlovin, H.; Solomon, D.H.; Jordan, J.M.; Hunter, D.J.; Suter, L.G.; Weinstein, A.M.; et al. Impact of obesity and knee osteoarthritis on morbidity and mortality in older Americans. Ann. Intern. Med. 2011, 154, 217–226. [Google Scholar] [CrossRef]
- Park, H.M.; Kim, H.S.; Lee, Y.J. Knee osteoarthritis and its association with mental health and health-related quality of life: A nationwide cross-sectional study. Geriatr. Gerontol. Int. 2020, 20, 379–383. [Google Scholar] [CrossRef]
- Sharma, M.; Jamieson, C.; Johnson, M.; Molloy, M.P.; Henderson, B.R. Specific Armadillo Repeat Sequences Facilitate β-Catenin Nuclear Transport in Live Cells via Direct Binding to Nucleoporins Nup62, Nup153, and RanBP2/Nup358. J. Biol. Chem. 2012, 287, 819–831. [Google Scholar] [CrossRef]
- Luong, M.L.N.; Cleveland, R.J.; Nyrop, K.A.; Callahan, L.F. Social determinants and osteoarthritis outcomes. Aging Health 2012, 8, 413–437. [Google Scholar] [CrossRef]
- Grässel, S.; Muschter, D. Recent advances in the treatment of osteoarthritis. F1000Research 2020, 9, 325. [Google Scholar] [CrossRef]
- Brittberg, M.; Lindahl, A.; Nilsson, A.; Ohlsson, C.; Isaksson, O.; Peterson, L. Treatment of deep cartilage defects in the knee with autologous chondrocyte transplantation. N. Engl. J. Med. 1994, 331, 889–895. [Google Scholar] [CrossRef]
- Dozin, B.; Malpeli, M.; Cancedda, R.; Bruzzi, P.; Calcagno, S.; Molfetta, L.; Priano, F.; Kon, E.; Marcacci, M. Comparative evaluation of autologous chondrocyte implantation and mosaicplasty: A multicentered randomized clinical trial. Clin. J. Sport Med. 2005, 15, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Dewan, A.K.; Gibson, M.A.; Elisseeff, J.H.; Trice, M.E. Evolution of autologous chondrocyte repair and comparison to other cartilage repair techniques. Biomed Res. Int. 2014, 2014, 272481. [Google Scholar] [CrossRef] [PubMed]
- Stein, S.; Strauss, E.; Bosco, J. Advances in the Surgical Management of Articular Cartilage Defects: Autologous Chondrocyte Implantation Techniques in the Pipeline. Cartilage 2013, 4, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Hubka, K.M.; Dahlin, R.L.; Meretoja, V.V.; Kasper, F.K.; Mikos, A.G. Enhancing Chondrogenic Phenotype for Cartilage Tissue Engineering: Monoculture and Coculture of Articular Chondrocytes and Mesenchymal Stem Cells. Tissue Eng. Part B Rev. 2014, 20, 641–654. [Google Scholar] [CrossRef]
- Goldring, M.B. Articular Cartilage Degradation in Osteoarthritis. HSS J. 2012, 8, 7–9. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, S.; Huang, J.; Guo, W.; Chen, J.; Zhang, L.; Zhao, B.; Peng, J.; Wang, A.; Wang, Y.; et al. The ECM-Cell Interaction of Cartilage Extracellular Matrix on Chondrocytes. Biomed Res. Int. 2014, 2014, 648459. [Google Scholar] [CrossRef]
- Gesslein, M.; Merkl, C.; Bail, H.J.; Krutsch, V.; Biber, R.; Schuster, P. Refixation of Large Osteochondral Fractures After Patella Dislocation Shows Better Mid- to Long-Term Outcome Compared with Debridement. Cartilage 2019, 13. [Google Scholar] [CrossRef]
- Ravaud, P.; Moulinier, L.; Giraudeau, B.; Ayral, X.; Guerin, C.; Noel, E.; Thomas, P.; Fautrel, B.; Mazieres, B.; Dougados, M. Effects of joint lavage and steroid injection in patients with osteoarthritis of the knee: Results of a multicenter, randomized, controlled trial. Arthritis Rheum. 1999, 42, 475–482. [Google Scholar] [CrossRef]
- Ghouri, A.; Conaghan, P.G. Treating osteoarthritis pain: Recent approaches using pharmacological therapies. Clin. Exp. Rheumatol. 2019, 37, 124–129. [Google Scholar]
- Jaswal, A.P.; Bandyopadhyay, A. Re-examining osteoarthritis therapy from a developmental biologist’s perspective. Biochem. Pharmacol. 2019, 165, 17–23. [Google Scholar] [CrossRef]
- Honvo, G.; Leclercq, V.; Geerinck, A.; Thomas, T.; Veronese, N.; Charles, A.; Rabenda, V.; Beaudart, C.; Cooper, C.; Reginster, J.Y.; et al. Safety of Topical Non-steroidal Anti-Inflammatory Drugs in Osteoarthritis: Outcomes of a Systematic Review and Meta-Analysis. Drugs Aging 2019, 36, 45–64. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Robertson, W.B.; Zhao, J.; Chen, W.; Xu, J. Emerging trend in the pharmacotherapy of osteoarthritis. Front. Endocrinol. 2019, 10, 431. [Google Scholar] [CrossRef] [PubMed]
- Palsis, J.A.; Brehmer, T.S.; Pellegrini, V.D.; Drew, J.M.; Sachs, B.L. The cost of joint replacement comparing two approaches to evaluating costs of total hip and knee arthroplasty. J. Bone Jt. Surg.-Am. Vol. 2018, 100, 326–333. [Google Scholar] [CrossRef] [PubMed]
- DeRogatis, M.; Anis, H.K.; Sodhi, N.; Ehiorobo, J.O.; Chughtai, M.; Bhave, A.; Mont, M.A. Non-operative treatment options for knee osteoarthritis. Ann. Transl. Med. 2019, 7 (Suppl 7), S245. [Google Scholar] [CrossRef]
- Deyle, G.D.; Allen, C.S.; Allison, S.C.; Gill, N.W.; Hando, B.R.; Petersen, E.J.; Dusenberry, D.I.; Rhon, D.I. Physical therapy versus glucocorticoid injection for osteoarthritis of the knee. N. Engl. J. Med. 2020, 382, 1420–1429. [Google Scholar] [CrossRef] [PubMed]
- Hermann, W.; Lambova, S.; Müller- Ladner, U. Current Treatment Options for Osteoarthritis. Curr. Rheumatol. Rev. 2018, 14, 108–116. [Google Scholar] [CrossRef]
- Choi, Y.R.; Collins, K.H.; Lee, J.W.; Kang, H.J.; Guilak, F. Genome Engineering for Osteoarthritis: From Designer Cells to Disease-Modifying Drugs. Tissue Eng. Regen. Med. 2019, 1, 1–9. [Google Scholar] [CrossRef]
- Shen, J.; Abu-Amer, Y.; O’Keefe, R.J.; McAlinden, A. Inflammation and epigenetic regulation in osteoarthritis. Connect. Tissue Res. 2017, 58, 49–63. [Google Scholar] [CrossRef]
- Im, G.-I.; Choi, Y.-J. Epigenetics in osteoarthritis and its implication for future therapeutics. Expert Opin. Biol. Ther. 2013, 13, 713–721. [Google Scholar] [CrossRef]
- García-Ibarbia, C.; Delgado-Calle, J.; Casafont, I.; Velasco, J.; Arozamena, J.; Pérez-Núñez, M.I.; Alonso, M.A.; Berciano, M.T.; Ortiz, F.; Pérez-Castrillón, J.L.; et al. Contribution of genetic and epigenetic mechanisms to Wnt pathway activity in prevalent skeletal disorders. Gene 2013, 532, 165–172. [Google Scholar] [CrossRef]
- Simon, T.C.; Jeffries, M.A. The Epigenomic Landscape in Osteoarthritis. Curr. Rheumatol. Rep. 2017, 19, 30. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, D.; Veronesi, F.; Carina, V.; Costa, V.; Raimondi, L.; De Luca, A.; Alessandro, R.; Fini, M.; Giavaresi, G. Gene therapy for chondral and osteochondral regeneration: Is the future now? Cell. Mol. Life Sci. 2018, 75, 649–667. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Huang, J.; Fan, Y.; Li, J.; You, T.; He, S.; Xiao, G.; Chen, D. Exploration of CRISPR/Cas9-based gene editing as therapy for osteoarthritis. Ann. Rheum. Dis. 2019, 78, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Adkar, S.S.; Brunger, J.M.; Willard, V.P.; Wu, C.-L.; Gersbach, C.A.; Guilak, F. Genome Engineering for Personalized Arthritis Therapeutics. Trends Mol. Med. 2017, 23, 917–931. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Esvelt, K.M.; Church, G.M. Cas9 for engineering biology. Nat. Methods 2013, 10, 957–963. [Google Scholar] [CrossRef]
- Karimian, A.; Azizian, K.; Parsian, H.; Rafieian, S.; Shafiei-Irannejad, V.; Kheyrollah, M.; Yousefi, M.; Majidinia, M.; Yousefi, B. CRISPR/Cas9 technology as a potent molecular tool for gene therapy. J. Cell. Physiol. 2019, 234, 12267–12277. [Google Scholar] [CrossRef]
- Magee, C.L.; Kleyn, P.W.; Monks, B.M.; Betz, U.; Basnet, S. Pre-existing technological core and roots for the CRISPR breakthrough. PLoS ONE 2018, 13, e0198541. [Google Scholar] [CrossRef]
- Broeders, M.; Herrero-Hernandez, P.; Ernst, M.P.T.; van der Ploeg, A.T.; Pijnappel, W.W.M.P. Sharpening the Molecular Scissors: Advances in Gene-Editing Technology. Iscience 2020, 23, 100789. [Google Scholar] [CrossRef]
- Ratan, Z.A.; Son, Y.J.; Haidere, M.F.; Uddin, B.M.M.; Yusuf, M.A.; Zaman, S.B.; Kim, J.H.; Banu, L.A.; Cho, J.Y. CRISPR-Cas9: A promising genetic engineering approach in cancer research. Ther. Adv. Med. Oncol. 2018, 10, 1758834018755089. [Google Scholar] [CrossRef]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1–3. [Google Scholar] [CrossRef]
- Khan, S.; Mahmood, M.S.; Rahman, S.U.; Zafar, H.; Habibullah, S.; Khan, Z.; Ahmad, A. CRISPR/Cas9: The Jedi against the dark empire of diseases. J. Biomed. Sci. 2018, 25, 29. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Wang, Y.; Chen, H.; Sun, Z.S.; Ju, X.-D. Recent Progress in CRISPR/Cas9 Technology. J. Genet. Genom. 2016, 43, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Sahel, D.K.; Mittal, A.; Chitkara, D. CRISPR/Cas System for Genome Editing: Progress and Prospects as a Therapeutic Tool. J. Pharmacol. Exp. Ther. 2019, 370, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef]
- Iwamoto, M.; Ohta, Y.; Larmour, C.; Enomoto-Iwamoto, M. Toward regeneration of articular cartilage. Birth Defects Res. Part C-Embryo Today Rev. 2013, 99, 192–202. [Google Scholar] [CrossRef]
- Grimaud, E.; Blanchard, F.; Charrier, C.; Gouin, F.; Redini, F.; Heymann, D. Leukaemia inhibitory factor (lif) is expressed in hypertrophic chondrocytes and vascular sprouts during osteogenesis. Cytokine 2002, 20, 224–230. [Google Scholar] [CrossRef]
- Loeser, R.F. Aging and osteoarthritis: The role of chondrocyte senescence and aging changes in the cartilage matrix. Osteoarthr. Cartil. 2009, 17, 971–979. [Google Scholar] [CrossRef]
- Shi, Y.; Hu, X.; Cheng, J.; Zhang, X.; Zhao, F.; Shi, W.; Ren, B.; Yu, H.; Yang, P.; Li, Z.; et al. A small molecule promotes cartilage extracellular matrix generation and inhibits osteoarthritis development. Nat. Commun. 2019, 10, 1–4. [Google Scholar] [CrossRef]
- Akkiraju, H.; Nohe, A. Role of chondrocytes in cartilage formation, progression of osteoarthritis and cartilage regeneration. J. Dev. Biol. 2015, 3, 177–192. [Google Scholar] [CrossRef]
- Chittiboyina, S.; Bai, Y.; Lelièvre, S.A. Microenvironment-cell nucleus relationship in the context of oxidative stress. Front. Cell Dev. Biol. 2018, 6, 23. [Google Scholar] [CrossRef]
- Jiang, Y.; Tuan, R.S. Origin and function of cartilage stem/progenitor cells in osteoarthritis. Nat. Rev. Rheumatol. 2015, 11, 206. [Google Scholar] [CrossRef] [PubMed]
- de Crombrugghe, B.; Lefebvre, V.; Nakashima, K. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr. Opin. Cell Biol. 2001, 13, 721–727. [Google Scholar] [CrossRef]
- Guilak, F.; Pferdehirt, L.; Ross, A.K.; Choi, Y.; Collins, K.; Nims, R.J.; Katz, D.B.; Klimak, M.; Tabbaa, S.; Pham, C.T.N. Designer Stem Cells: Genome Engineering and the Next Generation of Cell-Based Therapies. J. Orthop. Res. 2019, 37, 1287–1293. [Google Scholar] [CrossRef] [PubMed]
- García-Álvarez, F.; Alegre-Aguarón, E.; Desportes, P.; Royo-Cañas, M.; Castiella, T.; Larrad, L.; Martínez-Lorenzo, M.J. Chondrogenic differentiation in femoral bone marrow-derived mesenchymal cells (MSC) from elderly patients suffering osteoarthritis or femoral fracture. Arch. Gerontol. Geriatr. 2011, 52, 239–242. [Google Scholar] [CrossRef]
- Tuan, R.S.; Boland, G.; Tuli, R. Adult mesenchymal stem cells and cell-based tissue engineering. Arthritis Res. Ther. 2003, 5, 1–4. [Google Scholar] [CrossRef]
- Fellows, C.R.; Matta, C.; Zakany, R.; Khan, I.M.; Mobasheri, A. Adipose, bone marrow and synovial joint-derived mesenchymal stem cells for cartilage repair. Front. Genet. 2016, 7, 213. [Google Scholar] [CrossRef]
- Wang, H.; Leng, Y.; Gong, Y. Bone Marrow Fat and Hematopoiesis. Front. Endocrinol. 2018, 9, 694. [Google Scholar] [CrossRef]
- O’Donoghue, K.; Fisk, N.M. Fetal stem cells. Best Pract. Res. Clin. Obstet. Gynaecol. 2004, 18, 853–875. [Google Scholar] [CrossRef]
- Choi, W.H.; Kim, H.R.; Lee, S.J.; Jeong, N.; Park, S.R.; Choi, B.H.; Min, B.H. Fetal cartilage-derived cells have stem cell properties and are a highly potent cell source for cartilage regeneration. Cell Transpl. 2016, 25, 449–461. [Google Scholar] [CrossRef]
- Park, D.Y.; Min, B.H.; Park, S.R.; Oh, H.J.; Truong, M.D.; Kim, M.; Choi, J.Y.; Park, I.S.; Choi, B.H. Engineered cartilage utilizing fetal cartilage-derived progenitor cells for cartilage repair. Sci. Rep. 2020, 10, 5722. [Google Scholar] [CrossRef]
- Zhang, R.; Ma, J.; Han, J.; Zhang, W.; Ma, J. Mesenchymal stem cell related therapies for cartilage lesions and osteoarthritis. Am. J. Transl. Res. 2019, 11, 6275. [Google Scholar] [PubMed]
- McGonagle, D.; Baboolal, T.G.; Jones, E. Native joint-resident mesenchymal stem cells for cartilage repair in osteoarthritis. Nat. Rev. Rheumatol. 2017, 13, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Jo, C.H.; Lee, Y.G.; Shin, W.H.; Kim, H.; Chai, J.W.; Jeong, E.C.; Kim, J.E.; Shim, H.; Shin, J.S.; Shin, I.S.; et al. Intra-articular injection of mesenchymal stem cells for the treatment of osteoarthritis of the knee: A proof-of-concept clinical trial. Stem Cells 2014, 32, 1254–1266. [Google Scholar] [CrossRef] [PubMed]
- Damia, E.; Chicharro, D.; Lopez, S.; Cuervo, B.; Rubio, M.; Sopena, J.J.; Vilar, J.M.; Carrillo, J.M. Adipose-derived mesenchymal stem cells: Are they a good therapeutic strategy for osteoarthritis? Int. J. Mol. Sci. 2018, 19, 1926. [Google Scholar] [CrossRef]
- Lee, W.Y.-w.; Wang, B. Cartilage repair by mesenchymal stem cells: Clinical trial update and perspectives. J. Orthop. Transl. 2017, 9, 76–88. [Google Scholar] [CrossRef]
- Gee, P.; Lung, M.S.Y.; Okuzaki, Y.; Sasakawa, N.; Iguchi, T.; Makita, Y.; Hozumi, H.; Miura, Y.; Yang, L.F.; Iwasaki, M.; et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat. Commun. 2020, 11, 1–18. [Google Scholar] [CrossRef]
- de Jong, O.G.; Murphy, D.E.; Mäger, I.; Willms, E.; Garcia-Guerra, A.; Gitz-Francois, J.J.; Lefferts, J.; Gupta, D.; Steenbeek, S.C.; van Rheenen, J.; et al. A CRISPR-Cas9-based reporter system for single-cell detection of extracellular vesicle-mediated functional transfer of RNA. Nat. Commun. 2020, 11, 1–3. [Google Scholar]
- Campbell, L.A.; Coke, L.M.; Richie, C.T.; Fortuno, L.V.; Park, A.Y.; Harvey, B.K. Gesicle-Mediated Delivery of CRISPR/Cas9 Ribonucleoprotein Complex for Inactivating the HIV Provirus. Mol. Ther. 2019, 27, 151–163. [Google Scholar] [CrossRef]
- Murphy, D.E.; de Jong, O.G.; Brouwer, M.; Wood, M.J.; Lavieu, G.; Schiffelers, R.M.; Vader, P. Extracellular vesicle-based therapeutics: Natural versus engineered targeting and trafficking. Exp. Mol. Med. 2019, 51, 1–2. [Google Scholar] [CrossRef]
- Ramirez, S.H.; Andrews, A.M.; Paul, D.; Pachter, J.S. Extracellular vesicles: Mediators and biomarkers of pathology along CNS barriers. Fluids Barriers CNS 2018, 15, 19. [Google Scholar] [CrossRef]
- Levy, O.; Zhao, W.; Mortensen, L.J.; LeBlanc, S.; Tsang, K.; Fu, M.; Phillips, J.A.; Sagar, V.; Anandakumaran, P.; Ngai, J.; et al. MRNA-engineered mesenchymal stem cells for targeted delivery of interleukin-10 to sites of inflammation. Blood 2013, 122, e23–e32. [Google Scholar] [CrossRef] [PubMed]
- Ragni, E.; Banfi, F.; Barilani, M.; Cherubini, A.; Parazzi, V.; Larghi, P.; Dolo, V.; Bollati, V.; Lazzari, L. Extracellular Vesicle-Shuttled mRNA in Mesenchymal Stem Cell Communication. Stem Cells 2017, 35, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Martin-Rufino, J.D.; Espinosa-Lara, N.; Osugui, L.; Sanchez-Guijo, F. Targeting the Immune System with Mesenchymal Stromal Cell-Derived Extracellular Vesicles: What Is the Cargo’s Mechanism of Action? Front. Bioeng. Biotechnol. 2019, 7, 308. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, D.; Liu, Z.; Zhou, F.; Dai, J.; Wu, B.; Zhou, J.; Heng, B.C.; Zou, X.H.; Ouyang, H.; et al. Exosomes from embryonic mesenchymal stem cells alleviate osteoarthritis through balancing synthesis and degradation of cartilage extracellular matrix. Stem Cell Res. Ther. 2017, 8, 189. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.; Withrow, J.; Hunter, M.; Liu, Y.; Tang, Y.L.; Fulzele, S.; Hamrick, M.W. Emerging role of extracellular vesicles in musculoskeletal diseases. Mol. Aspects Med. 2018, 60, 123–128. [Google Scholar] [CrossRef]
- Bellavia, D.; Raimondi, L.; Costa, V.; De Luca, A.; Carina, V.; Maglio, M.; Fini, M.; Alessandro, R.; Giavaresi, G. Engineered exosomes: A new promise for the management of musculoskeletal diseases. Biochim. Biophys. Acta-Gen. Subj. 2018, 1862, 1893–1901. [Google Scholar] [CrossRef]
- Hiemer, B.; Krogull, M.; Bender, T.; Ziebart, J.; Krueger, S.; Bader, R.; Jonitz-Heincke, A. Effect of electric stimulation on human chondrocytes and mesenchymal stem cells under normoxia and hypoxia. Mol. Med. Rep. 2018, 18, 2133–2141. [Google Scholar] [CrossRef]
- Lo Monaco, M.; Merckx, G.; Ratajczak, J.; Gervois, P.; Hilkens, P.; Clegg, P.; Bronckaers, A.; Vandeweerd, J.M.; Lambrichts, I. Stem Cells for Cartilage Repair: Preclinical Studies and Insights in Translational Animal Models and Outcome Measures. Stem Cells Int. 2018, 2018. [Google Scholar] [CrossRef]
- De Bari, C.; Roelofs, A.J. Stem cell-based therapeutic strategies for cartilage defects and osteoarthritis. Curr. Opin. Pharmacol. 2018, 40, 74–80. [Google Scholar] [CrossRef]
- Fernandes, T.L.; Kimura, H.A.; Pinheiro, C.C.G.; Shimomura, K.; Nakamura, N.; Ferreira, J.R.; Gomoll, A.H.; Hernandez, A.J.; Bueno, D.F. Human synovial mesenchymal stem cells good manufacturing practices for articular cartilage regeneration. Tissue Eng.-Part C Methods 2018, 24, 709–716. [Google Scholar] [CrossRef]
- Cosenza, S.; Ruiz, M.; Toupet, K.; Jorgensen, C.; Noël, D. Mesenchymal stem cells derived exosomes and microparticles protect cartilage and bone from degradation in osteoarthritis. Sci. Rep. 2017, 7, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Cosenza, S.; Ruiz, M.; Toupet, K.; Bony, C.; Jorgensen, C.; Noel, D. Mesenchymal stem cells produced exosomes and microparticles that exert a similar chondroprotective effect in osteoarthritis. Osteoarthr. Cartil. 2018, 26, S297. [Google Scholar] [CrossRef]
- Farhang, N.; Brunger, J.M.; Stover, J.D.; Thakore, P.I.; Lawrence, B.; Guilak, F.; Gersbach, C.A.; Setton, L.A.; Bowles, R.D. CRISPR-Based Epigenome Editing of Cytokine Receptors for the Promotion of Cell Survival and Tissue Deposition in Inflammatory Environments. Tissue Eng. Part A 2017, 23, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, A.; Yoshinaga, M. Sequential generation of cytokines during the initiative phase of inflammation, with reference to neutrophils. Inflammation Reearchs 1998, 47 (Suppl. 3), S137–S144. [Google Scholar] [CrossRef]
- Cigan, A.D.; Roach, B.L.; Nims, R.J.; Tan, A.R.; Albro, M.B.; Stoker, A.M.; Cook, J.L.; Vunjak-Novakovic, G.; Hung, C.T.; Ateshian, G.A. High seeding density of human chondrocytes in agarose produces tissue-engineered cartilage approaching native mechanical and biochemical properties. J. Biomech. 2016, 49, 1909–1917. [Google Scholar] [CrossRef]
- Karlsen, T.A.; Pernas, P.F.; Staerk, J.; Caglayan, S.; Brinchmann, J.E. Generation of IL1β-resistant chondrocytes using CRISPR-CAS genome editing. Osteoarthr. Cartil. 2016, 24, S325. [Google Scholar] [CrossRef]
- Lambert, L.J.; Challa, A.K.; Niu, A.; Zhou, L.; Tucholski, J.; Johnson, M.S.; Nagy, T.R.; Eberhardt, A.W.; Estep, P.N.; Kesterson, R.A.; et al. Increased trabecular bone and improved biomechanics in an osteocalcin-null rat model created by CRISPR/Cas9 technology. DMM Dis. Model. Mech. 2016, 9, 1169–1179. [Google Scholar] [CrossRef]
- Asahara, H. Current Status and Strategy of microRNA Research for Cartilage Development and Osteoarthritis Pathogenesis. J. Bone Metab. 2016, 23, 121–127. [Google Scholar] [CrossRef]
- Huang, Y.; Askew, E.B.; Knudson, C.B.; Knudson, W. CRISPR/Cas9 knockout of HAS2 in rat chondrosarcoma chondrocytes demonstrates the requirement of hyaluronan for aggrecan retention. Matrix Biol. 2016, 56, 74–94. [Google Scholar] [CrossRef]
- Brunger, J.M.; Zutshi, A.; Willard, V.P.; Gersbach, C.A.; Guilak, F. Genome Engineering of Stem Cells for Autonomously Regulated, Closed-Loop Delivery of Biologic Drugs. Stem Cell Rep. 2017, 8, 1202–1213. [Google Scholar] [CrossRef]
- Khakshooy, A.; Balenton, N.; Chiappelli, F. Lubricin: A Principal Modulator of the Psychoneuroendocrine-Osteoimmune Interactome-Implications for Novel Treatments of Osteoarthritic Pathologies. Bioinformation 2017, 13, 343–346. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rice, S.J.; Aubourg, G.; Sorial, A.K.; Almarza, D.; Tselepi, M.; Deehan, D.J.; Reynard, L.N.; Loughlin, J. Identification of a novel, methylation-dependent, RUNX2 regulatory region associated with osteoarthritis risk. Hum. Mol. Genet. 2018, 27, 3464–3474. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Baek, S.-H.; Ye, Y.; Zhang, T. Proteomic characterization of endogenous substrates of mammalian ubiquitin ligase Hrd1. Cell Biosci. 2018, 8, 46. [Google Scholar] [CrossRef] [PubMed]
- Seidl, C.I.; Fulga, T.A.; Murphy, C.L. CRISPR-Cas9 targeting of MMP13 in human chondrocytes leads to significantly reduced levels of the metalloproteinase and enhanced type II collagen accumulation. Osteoarthr. Cartil. 2019, 27, 140–147. [Google Scholar] [CrossRef]
- Varela-Eirín, M.; Varela-Vázquez, A.; Guitián-Caamaño, A.; Paíno, C.L.; Mato, V.; Largo, R.; Aasen, T.; Tabernero, A.; Fonseca, E.; Kandouz, M.; et al. Targeting of chondrocyte plasticity via connexin43 modulation attenuates cellular senescence and fosters a pro-regenerative environment in osteoarthritis. Cell Death Dis. 2018, 9, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Hu, B.; Song, M.; Ding, Z.; Dang, Y.; Liu, Z.; Zhang, W.; Ji, Q.; Ren, R.; Ding, J.; et al. Maintenance of Nucleolar Homeostasis by CBX4 Alleviates Senescence and Osteoarthritis. Cell Rep. 2019, 26, 3643–3656. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Hu, Y.; Song, M.; Liu, Z.; Zhang, W.; Yu, F.X.; Wu, J.; Wang, S.; Belmonte, J.C.I.; Chan, P.; et al. Up-regulation of FOXD1 by yap alleviates senescence and osteoarthritis. PLoS Biol. 2019, 17, e3000201. [Google Scholar] [CrossRef]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Ramos, Y.F.M.; Meulenbelt, I. The role of epigenetics in osteoarthritis: Current perspective. Curr. Opin. Rheumatol. 2017, 29, 119–129. [Google Scholar] [CrossRef]
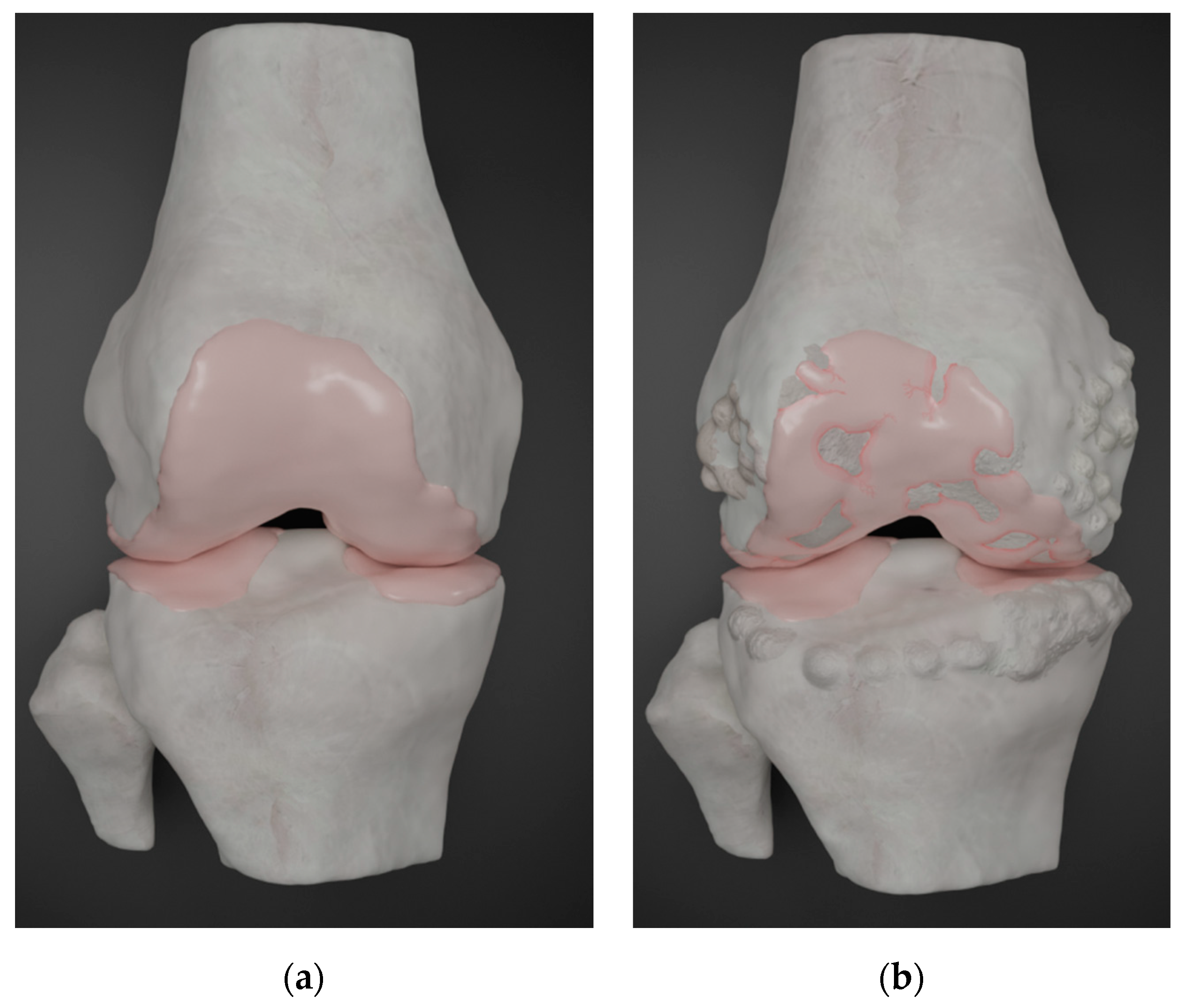
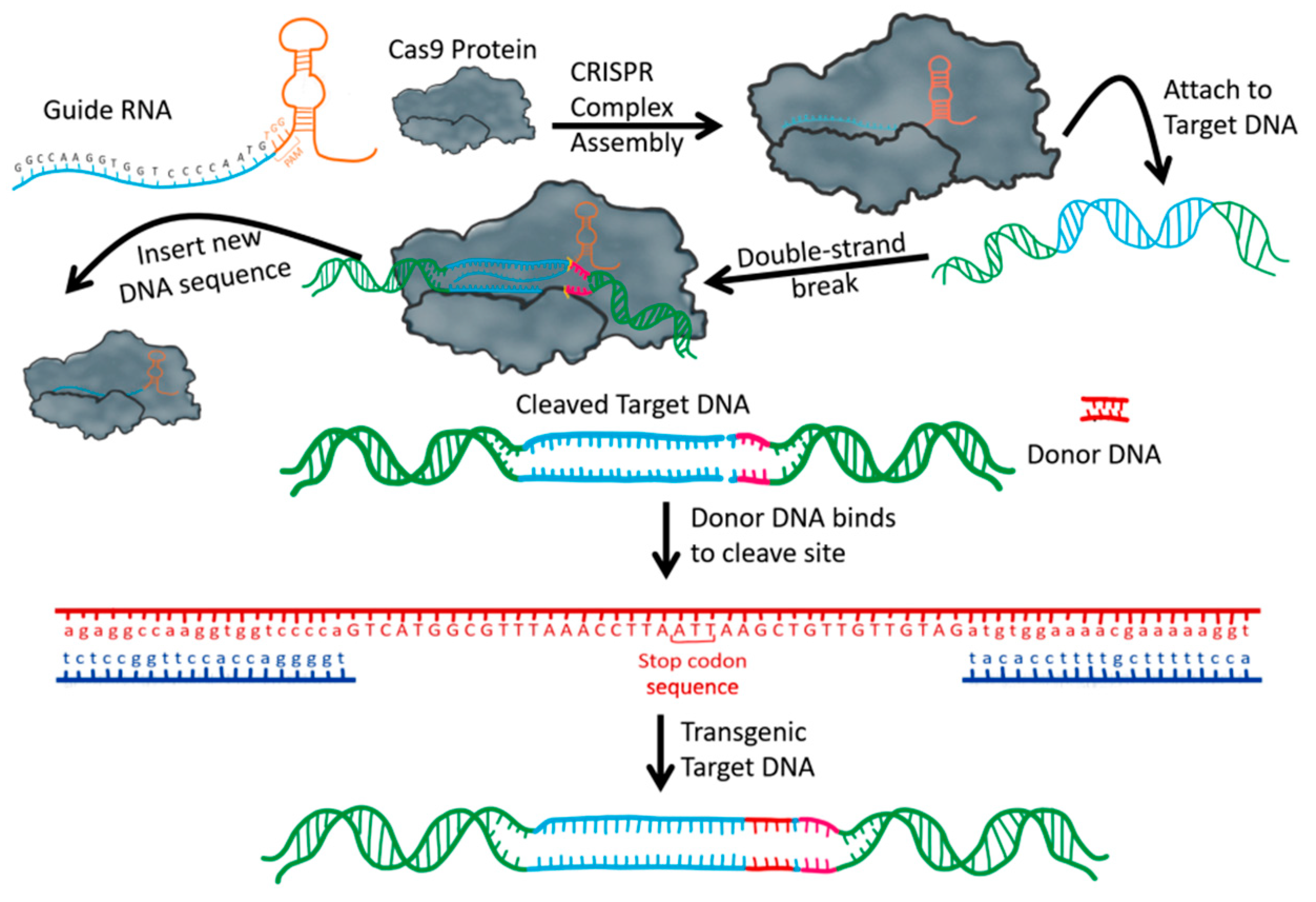
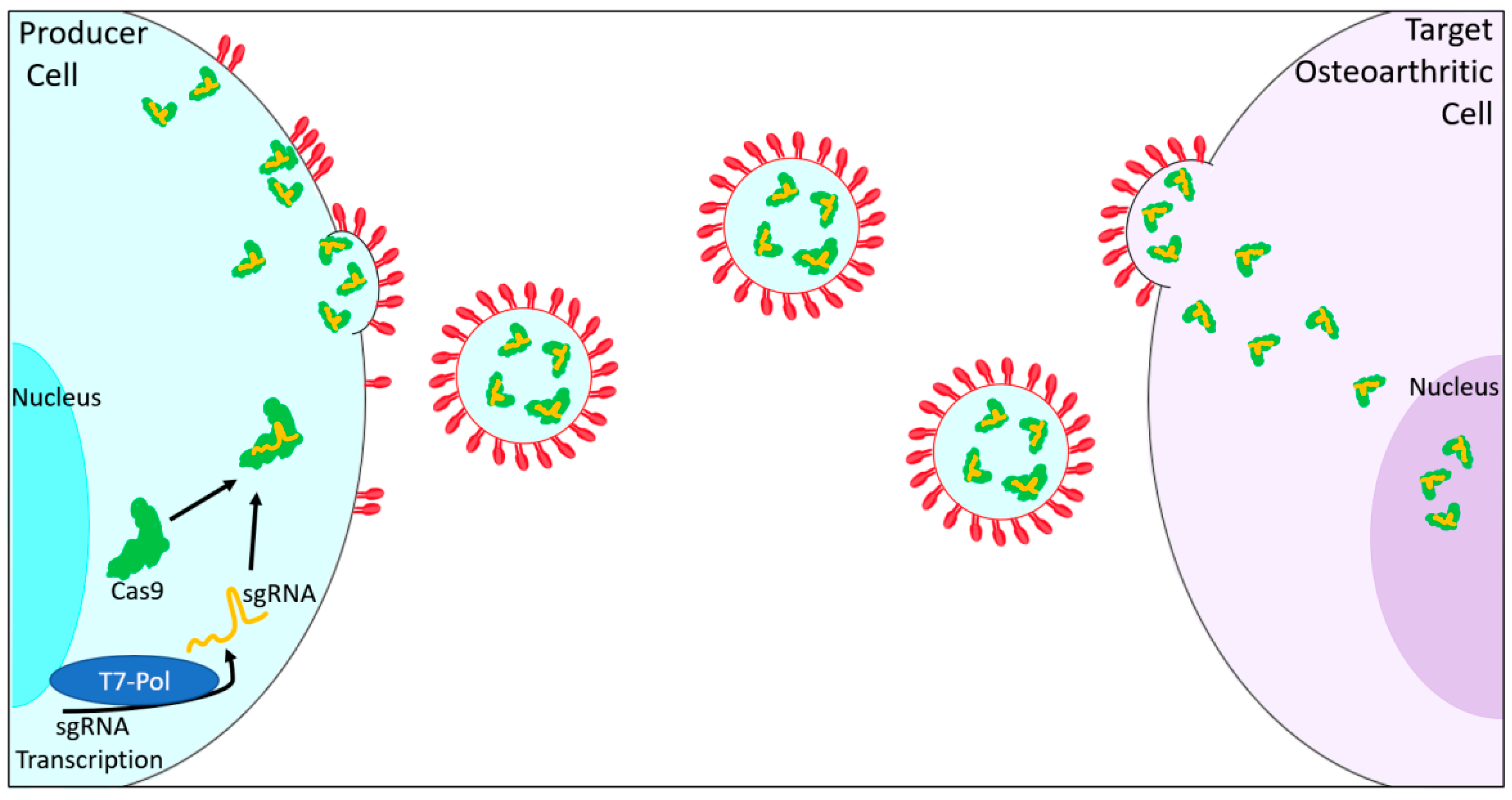
| Gene Symbol | Gene Name | Function | Reference |
|---|---|---|---|
| IL1-β | Interleukin 1 beta | Inflammation | Karlsen, 2016 [106] Zhao, 2019 [53] Guilak [73] |
| IL1-R1 | Interleukin 1 beta receptor | Inflammation | Karlsen, 2016 [106] |
| BGLAP | Osteocalcin | Trabecular bone formation | Lambert, 2016 [107] |
| miR-140 | Micro RNA 140 | Chondrocyte homeostasis | Asahara, 2016 [108] |
| Has2 | Hyaluronan synthase 2 | Chondrocyte accumulation of aggrecan | Huang, 2016 [109] |
| sTNFR1α | Soluble Tumor necrosis factor receptor 1 | TNF antagonist | Brunger, 2017 [110] |
| IL1RA | Interleukin 1 bets receptor antagonist | IL-1 beta antagonist | Brunger, 2017 [110] |
| PRG4 | Lubricin | Joint lubrication | Khakshooy, 2017 [111] |
| Runx2 | Runt Related Transcription Factor 2 | Osteoblast differentiation | Rice, 2018 [112] |
| Hrdl | E3 ubiquitin-protein ligase hrd-like protein 1 | Protein turnover and proteasomal degradation | Ye, 2018 [113] |
| Mmp13 | Matrix metalloprotein 13 | Tissue degradation | Seidl, 2019 [114] Zhao, 2019 [53] |
| Cx43 | Connexin 43 | Gap junction protein | Varela-Eirín M, 2018 [115] |
| NGF | Nerve growth factor | Pain sensitivity | Zhao, 2019 [53] |
| Cbx4 | Chromobox 4 | Nucleolar homeostasis | Ren, 2019 [116] |
| Foxd1 | Forkhead box D1 | Transcription factor | Fu, 2019 [117] |
| YAP | Yes-associated protein 1 | Mechanosensing transcription factor | Fu, 2019 [117] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanikella, A.S.; Hardy, M.J.; Frahs, S.M.; Cormier, A.G.; Gibbons, K.D.; Fitzpatrick, C.K.; Oxford, J.T. Emerging Gene-Editing Modalities for Osteoarthritis. Int. J. Mol. Sci. 2020, 21, 6046. https://doi.org/10.3390/ijms21176046
Tanikella AS, Hardy MJ, Frahs SM, Cormier AG, Gibbons KD, Fitzpatrick CK, Oxford JT. Emerging Gene-Editing Modalities for Osteoarthritis. International Journal of Molecular Sciences. 2020; 21(17):6046. https://doi.org/10.3390/ijms21176046
Chicago/Turabian StyleTanikella, Alekya S., Makenna J. Hardy, Stephanie M. Frahs, Aidan G. Cormier, Kalin D. Gibbons, Clare K. Fitzpatrick, and Julia Thom Oxford. 2020. "Emerging Gene-Editing Modalities for Osteoarthritis" International Journal of Molecular Sciences 21, no. 17: 6046. https://doi.org/10.3390/ijms21176046
APA StyleTanikella, A. S., Hardy, M. J., Frahs, S. M., Cormier, A. G., Gibbons, K. D., Fitzpatrick, C. K., & Oxford, J. T. (2020). Emerging Gene-Editing Modalities for Osteoarthritis. International Journal of Molecular Sciences, 21(17), 6046. https://doi.org/10.3390/ijms21176046







