Regulatory Mechanisms of Somatostatin Expression
Abstract
1. Introduction
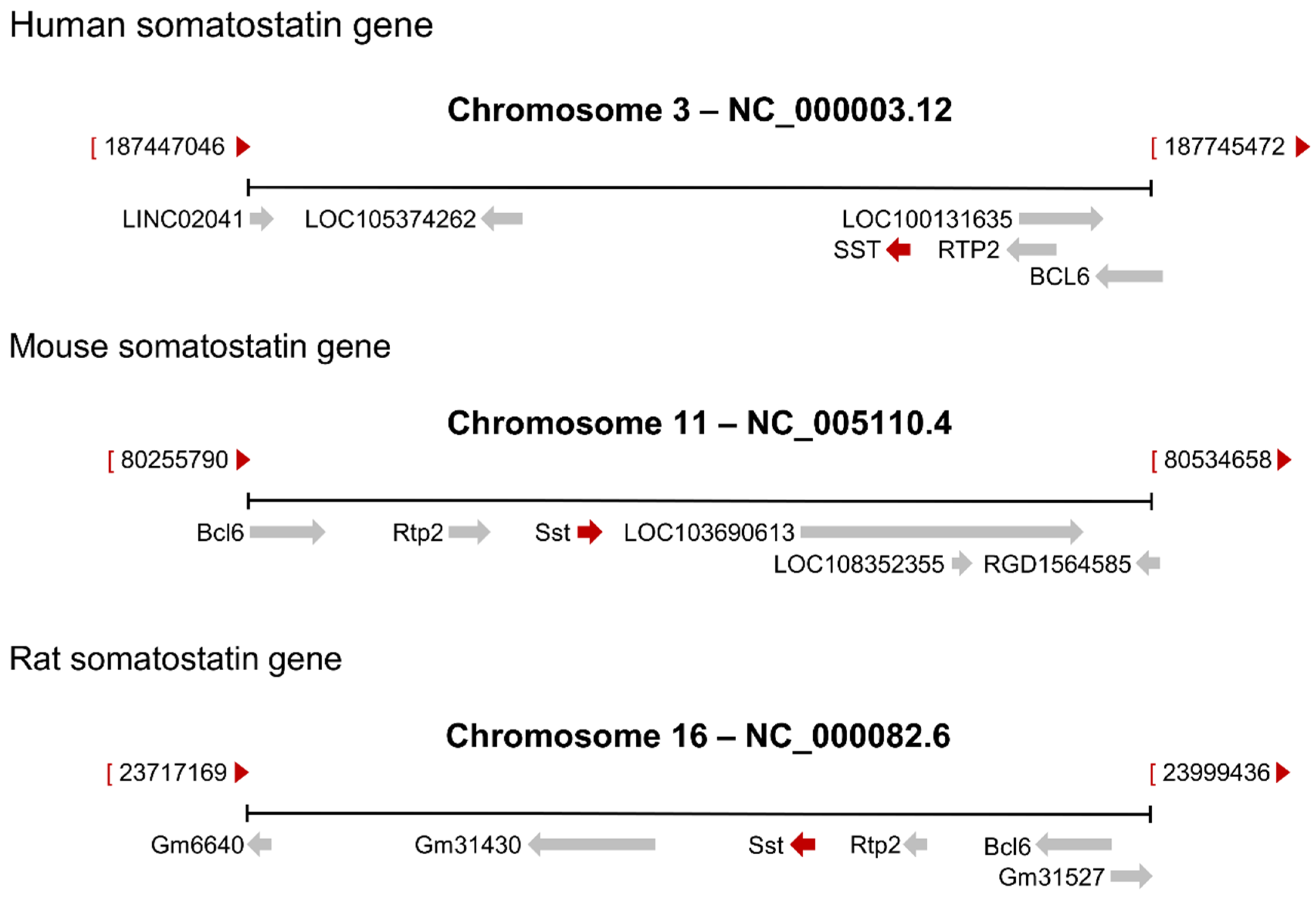
2. The Somatostatin Gene
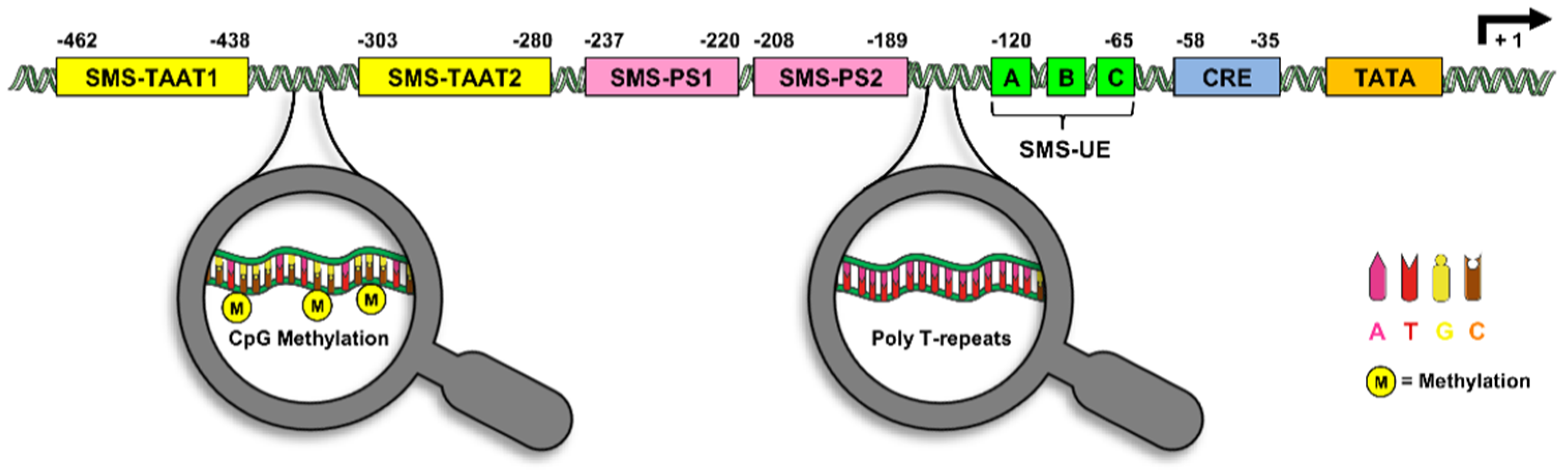
3. The Somatostatin Promoter
4. Transcription Factors Regulating Somatostatin Expression
5. Exogenous Factors Regulating Somatostatin Expression
6. Putative Autocrine Feedback
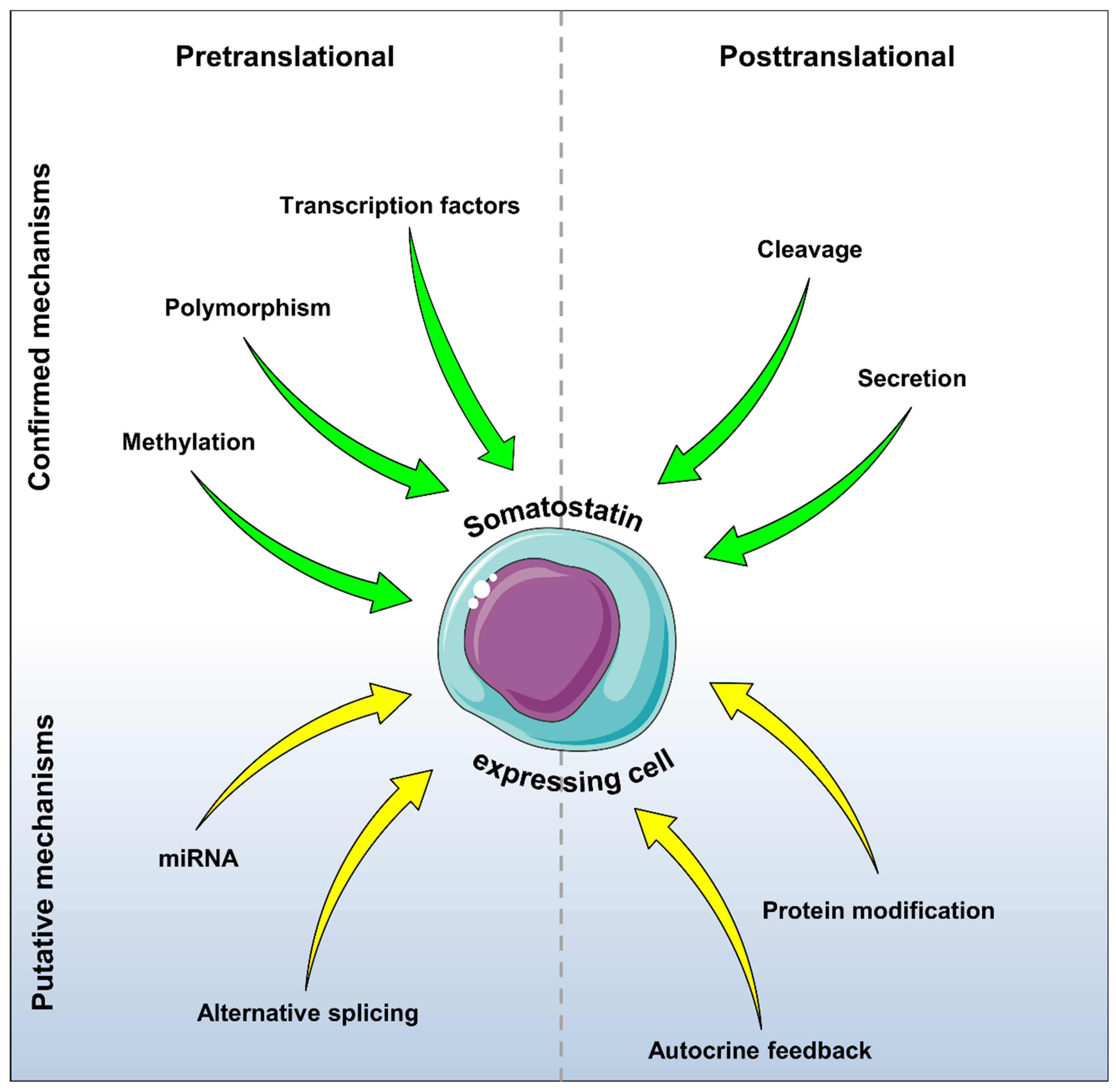
7. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Brazeau, P.; Vale, W.; Burgus, R.; Ling, N.; Butcher, M.; Rivier, J.; Guillemin, R. Hypothalamic polypeptide that inhibits the secretion of immunoreactive pituitary growth hormone. Science 1973, 179, 77–79. [Google Scholar] [CrossRef]
- Shen, L.P.; Rutter, W.J. Sequence of the human somatostatin I gene. Science 1984, 224, 168–171. [Google Scholar] [CrossRef]
- Cote, G.J.; Gagel, R.F. Different mechanisms of calcitonin, calcitonin gene-related peptide, and somatostatin regulation by glucocorticoids in a cell culture of human medullary thyroid carcinoma. Henry Ford Hosp. Med. J. 1987, 35, 149–152. [Google Scholar] [PubMed]
- Warren, T.G.; Shields, D. Expression of preprosomatostatin in heterologous cells: Biosynthesis, posttranslational processing, and secretion of mature somatostatin. Cell 1984, 39, 547–555. [Google Scholar] [CrossRef]
- Pradayrol, L.; Jornvall, H.; Mutt, V.; Ribet, A. N-terminally extended somatostatin: The primary structure of somatostatin-28. FEBS Lett. 1980, 109, 55–58. [Google Scholar] [CrossRef]
- Francis, B.H.; Baskin, D.G.; Saunders, D.R.; Ensinck, J.W. Distribution of somatostatin-14 and somatostatin-28 gastrointestinal-pancreatic cells of rats and humans. Gastroenterology 1990, 99, 1283–1291. [Google Scholar] [CrossRef]
- Samson, W.K.; Zhang, J.V.; Avsian-Kretchmer, O.; Cui, K.; Yosten, G.L.; Klein, C.; Lyu, R.M.; Wang, Y.X.; Chen, X.Q.; Yang, J.; et al. Neuronostatin encoded by the somatostatin gene regulates neuronal, cardiovascular, and metabolic functions. J. Biol. Chem. 2008, 283, 31949–31959. [Google Scholar] [CrossRef]
- Vainio, L.; Perjes, A.; Ryti, N.; Magga, J.; Alakoski, T.; Serpi, R.; Kaikkonen, L.; Piuhola, J.; Szokodi, I.; Ruskoaho, H.; et al. Neuronostatin, a novel peptide encoded by somatostatin gene, regulates cardiac contractile function and cardiomyocyte survival. J. Biol. Chem. 2012, 287, 4572–4580. [Google Scholar] [CrossRef] [PubMed]
- DiGruccio, M.R.; Mawla, A.M.; Donaldson, C.J.; Noguchi, G.M.; Vaughan, J.; Cowing-Zitron, C.; van der Meulen, T.; Huising, M.O. Comprehensive alpha, beta and delta cell transcriptomes reveal that ghrelin selectively activates delta cells and promotes somatostatin release from pancreatic islets. Mol. Metab. 2016, 5, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, C.S.; Salehi, A.; Gopel, S.O.; Holm, C.; Rorsman, P. Palmitate stimulation of glucagon secretion in mouse pancreatic alpha-cells results from activation of L-type calcium channels and elevation of cytoplasmic calcium. Diabetes 2004, 53, 2836–2843. [Google Scholar] [CrossRef]
- Ipp, E.; Dobbs, R.E.; Arimura, A.; Vale, W.; Harris, V.; Unger, R.H. Release of immunoreactive somatostatin from the pancreas in response to glucose, amino acids, pancreozymin-cholecystokinin, and tolbutamide. J. Clin. Investig. 1977, 60, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Ahren, B.; Paquette, T.L.; Taborsky, G.J., Jr. Effect and mechanism of vagal nerve stimulation on somatostatin secretion in dogs. Am. J. Physiol. 1986, 250, E212–E217. [Google Scholar] [CrossRef] [PubMed]
- Hokfelt, T.; Elfvin, L.G.; Elde, R.; Schultzberg, M.; Goldstein, M.; Luft, R. Occurrence of somatostatin-like immunoreactivity in some peripheral sympathetic noradrenergic neurons. Proc. Natl. Acad. Sci. USA 1977, 74, 3587–3591. [Google Scholar] [CrossRef]
- Patel, Y.C.; Wheatley, T. In vivo and in vitro plasma disappearance and metabolism of somatostatin-28 and somatostatin-14 in the rat. Endocrinology 1983, 112, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Rorsman, P.; Huising, M.O. The somatostatin-secreting pancreatic delta-cell in health and disease. Nat. Rev. Endocrinol. 2018, 14, 404–414. [Google Scholar] [CrossRef]
- Frohman, L.A.; Downs, T.R.; Kelijman, M.; Clarke, I.J.; Thomas, G. Somatostatin secretion and action in the regulation of growth hormone secretion. Metabolism 1990, 39, 43–45. [Google Scholar] [CrossRef]
- Saiz-Sanchez, D.; Ubeda-Banon, I.; Flores-Cuadrado, A.; Gonzalez-Rodriguez, M.; Villar-Conde, S.; Astillero-Lopez, V.; Martinez-Marcos, A. Somatostatin, Olfaction, and Neurodegeneration. Front. Neurosci. 2020, 14, 96. [Google Scholar] [CrossRef]
- Patel, Y.C.; Greenwood, M.T.; Warszynska, A.; Panetta, R.; Srikant, C.B. All five cloned human somatostatin receptors (hSSTR1−5) are functionally coupled to adenylyl cyclase. Biochem. Biophys. Res. Commun. 1994, 198, 605–612. [Google Scholar] [CrossRef]
- Kossut, M.; Lukomska, A.; Dobrzanski, G.; Liguz-Lecznar, M. Somatostatin receptors in the brain. Postepy Biochem. 2018, 64, 213–221. [Google Scholar] [CrossRef]
- Klisovic, D.D.; O’Dorisio, M.S.; Katz, S.E.; Sall, J.W.; Balster, D.; O’Dorisio, T.M.; Craig, E.; Lubow, M. Somatostatin receptor gene expression in human ocular tissues: RT-PCR and immunohistochemical study. Investig. Ophthalmol. Vis. Sci. 2001, 42, 2193–2201. [Google Scholar]
- Csaba, Z.; Peineau, S.; Dournaud, P. Molecular mechanisms of somatostatin receptor trafficking. J. Mol. Endocrinol. 2012, 48, R1–R12. [Google Scholar] [CrossRef]
- Koerker, D.J.; Ruch, W.; Chideckel, E.; Palmer, J.; Goodner, C.J.; Ensinck, J.; Gale, C.C. Somatostatin: Hypothalamic inhibitor of the endocrine pancreas. Science 1974, 184, 482–484. [Google Scholar] [CrossRef] [PubMed]
- Strowski, M.Z.; Parmar, R.M.; Blake, A.D.; Schaeffer, J.M. Somatostatin inhibits insulin and glucagon secretion via two receptors subtypes: An in vitro study of pancreatic islets from somatostatin receptor 2 knockout mice. Endocrinology 2000, 141, 111–117. [Google Scholar] [CrossRef]
- Van Hagen, P.M.; Krenning, E.P.; Kwekkeboom, D.J.; Reubi, J.C.; Anker-Lugtenburg, P.J.; Lowenberg, B.; Lamberts, S.W. Somatostatin and the immune and haematopoetic system; A review. Eur. J. Clin. Investig. 1994, 24, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Tani, S. Inhibitory action of somatostatin on cAMP dependent pepsinogen secretion from rat gastric chief cells: Involvement of pertussis toxin-sensitive G-protein. Biol. Pharm. Bull. 1994, 17, 415–418. [Google Scholar] [CrossRef]
- Oddsdottir, M.; Ballantyne, G.H.; Adrian, T.E.; Zdon, M.J.; Zucker, K.A.; Modlin, I.M. Somatostatin inhibition of intrinsic factor secretion from isolated guinea pig gastric glands. Scand. J. Gastroenterol. 1987, 22, 233–238. [Google Scholar] [CrossRef]
- Grauslund, J.; Frydkjaer-Olsen, U.; Peto, T.; Fernandez-Carneado, J.; Ponsati, B.; Hernandez, C.; Cunha-Vaz, J.; Simo, R. Topical Treatment With Brimonidine and Somatostatin Causes Retinal Vascular Dilation in Patients With Early Diabetic Retinopathy From the EUROCONDOR. Investig. Ophthalmol. Vis. Sci. 2019, 60, 2257–2262. [Google Scholar] [CrossRef]
- Zhang, C.; Xu, J.M.; Kong, D.R.; Min, X.K.; Chen, R. Immediate effects of different schedules of somatostatin on portal pressure in patients with liver cirrhosis. J. Clin. Pharm. Ther. 2013, 38, 206–211. [Google Scholar] [CrossRef]
- Ruscica, M.; Arvigo, M.; Steffani, L.; Ferone, D.; Magni, P. Somatostatin, somatostatin analogs and somatostatin receptor dynamics in the biology of cancer progression. Curr. Mol. Med. 2013, 13, 555–571. [Google Scholar] [CrossRef]
- Barbieri, F.; Bajetto, A.; Pattarozzi, A.; Gatti, M.; Wurth, R.; Thellung, S.; Corsaro, A.; Villa, V.; Nizzari, M.; Florio, T. Peptide receptor targeting in cancer: The somatostatin paradigm. Int. J. Pept. 2013, 2013, 926295. [Google Scholar] [CrossRef]
- Theodoropoulou, M.; Stalla, G.K. Somatostatin receptors: From signaling to clinical practice. Front. Neuroendocrinol. 2013, 34, 228–252. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Porras, M.; Cardenas-Salas, J.; Alvarez-Escola, C. Somatostatin Analogs in Clinical Practice: A Review. Int. J. Mol. Sci. 2020, 21, 1682. [Google Scholar] [CrossRef]
- Godara, A.; Siddiqui, N.S.; Byrne, M.M.; Saif, M.W. The safety of lanreotide for neuroendocrine tumor. Expert Opin. Drug Saf. 2019, 18, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yau, H.; Kinaan, M.; Quinn, S.L.; Moraitis, A.G. Octreotide long-acting repeatable in the treatment of neuroendocrine tumors: Patient selection and perspectives. Biol. Targets Ther. 2017, 11, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Montminy, M.R.; Sevarino, K.A.; Wagner, J.A.; Mandel, G.; Goodman, R.H. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc. Natl. Acad. Sci. USA 1986, 83, 6682–6686. [Google Scholar] [CrossRef]
- Andrisani, O.M.; Hayes, T.E.; Roos, B.; Dixon, J.E. Identification of the promoter sequences involved in the cell specific expression of the rat somatostatin gene. Nucleic Acids Res. 1987, 15, 5715–5728. [Google Scholar] [CrossRef]
- Montminy, M.; Brindle, P.; Arias, J.; Ferreri, K.; Armstrong, R. Regulation of somatostatin gene transcription by cAMP. Adv. Pharmacol. 1996, 36, 1–13. [Google Scholar]
- Shen, L.P.; Pictet, R.L.; Rutter, W.J. Human somatostatin I: Sequence of the cDNA. Proc. Natl. Acad. Sci. USA 1982, 79, 4575–4579. [Google Scholar] [CrossRef]
- Tostivint, H.; Lihrmann, I.; Vaudry, H. New insight into the molecular evolution of the somatostatin family. Mol. Cell. Endocrinol. 2008, 286, 5–17. [Google Scholar] [CrossRef]
- Liu, Y.; Lu, D.; Zhang, Y.; Li, S.; Liu, X.; Lin, H. The evolution of somatostatin in vertebrates. Gene 2010, 463, 21–28. [Google Scholar] [CrossRef]
- De Lecea, L. Cortistatin: A natural somatostatin analog. J. Endocrinol. Investig. 2005, 28, 10–14. [Google Scholar]
- Tostivint, H.; Gaillard, A.L.; Mazan, S.; Pezeron, G. Revisiting the evolution of the somatostatin family: Already five genes in the gnathostome ancestor. Gen. Comp. Endocrinol. 2019, 279, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Sui, C.; Chen, J.; Ma, J.; Zhao, W.; Canário, A.V.M.; Martins, R.S.T. Somatostatin 4 regulates growth and modulates gametogenesis in zebrafish. Aquac. Fish. 2019, 4, 239–246. [Google Scholar] [CrossRef]
- Sobrido-Camean, D.; Tostivint, H.; Mazan, S.; Rodicio, M.C.; Rodriguez-Moldes, I.; Candal, E.; Anadon, R.; Barreiro-Iglesias, A. Differential expression of five prosomatostatin genes in the central nervous system of the catshark Scyliorhinus canicula. J. Comp. Neurol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Magadum, S.; Banerjee, U.; Murugan, P.; Gangapur, D.; Ravikesavan, R. Gene duplication as a major force in evolution. J. Genet. 2013, 92, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Ayoubi, T.A.; Van De Ven, W.J. Regulation of gene expression by alternative promoters. FASEB J. 1996, 10, 453–460. [Google Scholar] [CrossRef]
- Ohneda, K.; Mirmira, R.G.; Wang, J.; Johnson, J.D.; German, M.S. The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol. Cell Biol. 2000, 20, 900–911. [Google Scholar] [CrossRef][Green Version]
- Gosmain, Y.; Cheyssac, C.; Heddad Masson, M.; Dibner, C.; Philippe, J. Glucagon gene expression in the endocrine pancreas: The role of the transcription factor Pax6 in alpha-cell differentiation, glucagon biosynthesis and secretion. Diabetes Obes. Metab. 2011, 13 (Suppl. 1), 31–38. [Google Scholar] [CrossRef]
- Fukuda, H.; Iritani, N. Transcriptional regulation of leptin gene promoter in rat. FEBS Lett. 1999, 455, 165–169. [Google Scholar] [CrossRef]
- Powers, A.C.; Tedeschi, F.; Wright, K.E.; Chan, J.S.; Habener, J.F. Somatostatin gene expression in pancreatic islet cells is directed by cell-specific DNA control elements and DNA-binding proteins. J. Biol. Chem. 1989, 264, 10048–10056. [Google Scholar]
- Vallejo, M.; Miller, C.P.; Habener, J.F. Somatostatin gene transcription regulated by a bipartite pancreatic islet D-cell-specific enhancer coupled synergetically to a cAMP response element. J. Biol. Chem. 1992, 267, 12868–12875. [Google Scholar] [PubMed]
- Vallejo, M.; Penchuk, L.; Habener, J.F. Somatostatin gene upstream enhancer element activated by a protein complex consisting of CREB, Isl-1-like, and alpha-CBF-like transcription factors. J. Biol. Chem. 1992, 267, 12876–12884. [Google Scholar] [PubMed]
- Beimesche, S.; Neubauer, A.; Herzig, S.; Grzeskowiak, R.; Diedrich, T.; Cierny, I.; Scholz, D.; Alejel, T.; Knepel, W. Tissue-specific transcriptional activity of a pancreatic islet cell-specific enhancer sequence/Pax6-binding site determined in normal adult tissues in vivo using transgenic mice. Mol. Endocrinol. 1999, 13, 718–728. [Google Scholar] [CrossRef] [PubMed]
- Schinner, S.; Dellas, C.; Schroder, M.; Heinlein, C.A.; Chang, C.; Fischer, J.; Knepel, W. Repression of glucagon gene transcription by peroxisome proliferator-activated receptor gamma through inhibition of Pax6 transcriptional activity. J. Biol. Chem. 2002, 277, 1941–1948. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.P.; McGehee, R.E., Jr.; Habener, J.F. IDX-1: A new homeodomain transcription factor expressed in rat pancreatic islets and duodenum that transactivates the somatostatin gene. EMBO J. 1994, 13, 1145–1156. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, M.; Miller, C.P.; Beckman, W.; Habener, J.F. Repression of somatostatin gene transcription mediated by two promoter silencer elements. Mol. Cell. Endocrinol. 1995, 113, 61–72. [Google Scholar] [CrossRef]
- Schagdarsurengin, U.; Gimm, O.; Hoang-Vu, C.; Dralle, H.; Pfeifer, G.P.; Dammann, R. Frequent epigenetic silencing of the CpG island promoter of RASSF1A in thyroid carcinoma. Cancer Res. 2002, 62, 3698–3701. [Google Scholar] [PubMed]
- Elisei, R.; Shiohara, M.; Koeffler, H.P.; Fagin, J.A. Genetic and epigenetic alterations of the cyclin-dependent kinase inhibitors p15INK4b and p16INK4a in human thyroid carcinoma cell lines and primary thyroid carcinomas. Cancer 1998, 83, 2185–2193. [Google Scholar] [CrossRef]
- Bird, A.P. CpG-rich islands and the function of DNA methylation. Nature 1986, 321, 209–213. [Google Scholar] [CrossRef]
- Jin, Z.; Mori, Y.; Hamilton, J.P.; Olaru, A.; Sato, F.; Yang, J.; Ito, T.; Kan, T.; Agarwal, R.; Meltzer, S.J. Hypermethylation of the somatostatin promoter is a common, early event in human esophageal carcinogenesis. Cancer 2008, 112, 43–49. [Google Scholar] [CrossRef]
- Shi, X.; Li, X.; Chen, L.; Wang, C. Analysis of somatostatin receptors and somatostatin promoter methylation in human gastric cancer. Oncol. Lett. 2013, 6, 1794–1798. [Google Scholar] [CrossRef] [PubMed]
- Leiszter, K.; Sipos, F.; Galamb, O.; Krenacs, T.; Veres, G.; Wichmann, B.; Furi, I.; Kalmar, A.; Patai, A.V.; Toth, K.; et al. Promoter hypermethylation-related reduced somatostatin production promotes uncontrolled cell proliferation in colorectal cancer. PLoS ONE 2015, 10, e0118332. [Google Scholar] [CrossRef]
- Hoogendoorn, B.; Coleman, S.L.; Guy, C.A.; Smith, K.; Bowen, T.; Buckland, P.R.; O’Donovan, M.C. Functional analysis of human promoter polymorphisms. Hum. Mol. Genet. 2003, 12, 2249–2254. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, M.; Brisson, D.; Gaudet, D. Association between a polymorphic poly-T repeat sequence in the promoter of the somatostatin gene and hypertension. Hypertens. Res. Off. J. Jpn. Soc. Hypertens. 2016, 39, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wang, C.; Lehman, M.L.; He, M.; An, J.; Svingen, T.; Spiller, C.M.; Ng, E.T.; Nelson, C.C.; Koopman, P. Transcriptomic analysis of mRNA expression and alternative splicing during mouse sex determination. Mol. Cell. Endocrinol. 2018, 478, 84–96. [Google Scholar] [CrossRef]
- Li, W.; Shi, Y.H.; Yang, R.L.; Cui, J.; Xiao, Y.; Wang, B.; Le, G.W. Effect of somatostatin analog on high-fat diet-induced metabolic syndrome: Involvement of reactive oxygen species. Peptides 2010, 31, 625–629. [Google Scholar] [CrossRef]
- Montminy, M.R.; Bilezikjian, L.M. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature 1987, 328, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Andrisani, O.M.; Pot, D.A.; Zhu, Z.; Dixon, J.E. Three sequence-specific DNA-protein complexes are formed with the same promoter element essential for expression of the rat somatostatin gene. Mol. Cell Biol. 1988, 8, 1947–1956. [Google Scholar] [CrossRef]
- Andrisani, O.M.; Zhu, Z.N.; Pot, D.A.; Dixon, J.E. In vitro transcription directed from the somatostatin promoter is dependent upon a purified 43-kDa DNA-binding protein. Proc. Natl. Acad. Sci. USA 1989, 86, 2181–2185. [Google Scholar] [CrossRef]
- Mantamadiotis, T.; Lemberger, T.; Bleckmann, S.C.; Kern, H.; Kretz, O.; Martin Villalba, A.; Tronche, F.; Kellendonk, C.; Gau, D.; Kapfhammer, J.; et al. Disruption of CREB function in brain leads to neurodegeneration. Nat. Genet. 2002, 31, 47–54. [Google Scholar] [CrossRef]
- Bleckmann, S.C.; Blendy, J.A.; Rudolph, D.; Monaghan, A.P.; Schmid, W.; Schutz, G. Activating transcription factor 1 and CREB are important for cell survival during early mouse development. Mol. Cell Biol. 2002, 22, 1919–1925. [Google Scholar] [CrossRef] [PubMed]
- Walton, K.M.; Rehfuss, R.P.; Chrivia, J.C.; Lochner, J.E.; Goodman, R.H. A dominant repressor of cyclic adenosine 3′,5′-monophosphate (cAMP)-regulated enhancer-binding protein activity inhibits the cAMP-mediated induction of the somatostatin promoter in vivo. Mol. Endocrinol. 1992, 6, 647–655. [Google Scholar]
- Gonzalez, G.A.; Montminy, M.R. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 1989, 6, 675–680. [Google Scholar] [CrossRef]
- Wang, H.M.; Dong, J.H.; Li, Q.; Hu, Q.; Ning, S.L.; Zheng, W.; Cui, M.; Chen, T.S.; Xie, X.; Sun, J.P.; et al. A stress response pathway in mice upregulates somatostatin level and transcription in pancreatic delta cells through Gs and beta-arrestin 1. Diabetologia 2014, 57, 1899–1910. [Google Scholar] [CrossRef][Green Version]
- Vallejo, M.; Gosse, M.E.; Beckman, W.; Habener, J.F. Impaired cyclic AMP-dependent phosphorylation renders CREB a repressor of C/EBP-induced transcription of the somatostatin gene in an insulinoma cell line. Mol. Cell Biol. 1995, 15, 415–424. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gachon, F.; Thebault, S.; Peleraux, A.; Devaux, C.; Mesnard, J.M. Molecular interactions involved in the transactivation of the human T-cell leukemia virus type 1 promoter mediated by Tax and CREB-2 (ATF-4). Mol. Cell Biol. 2000, 20, 3470–3481. [Google Scholar] [CrossRef]
- Marshak, S.; Benshushan, E.; Shoshkes, M.; Leibovitz, G.; Kaiser, N.; Gross, D.; Bertuzzi, F.; Cerasi, E.; Melloul, D. beta-cell-specific expression of insulin and PDX-1 genes. Diabetes 2001, 50 (Suppl. 1), S131–S132. [Google Scholar] [CrossRef]
- Melloul, D.; Tsur, A.; Zangen, D. Pancreatic Duodenal Homeobox (PDX-1) in health and disease. J. Pediatr. Endocrinol. Metab. 2002, 15, 1461–1472. [Google Scholar] [CrossRef]
- Piran, R.; Lee, S.H.; Li, C.R.; Charbono, A.; Bradley, L.M.; Levine, F. Pharmacological induction of pancreatic islet cell transdifferentiation: Relevance to type I diabetes. Cell Death Dis. 2014, 5, e1357. [Google Scholar] [CrossRef]
- Leonard, J.; Peers, B.; Johnson, T.; Ferreri, K.; Lee, S.; Montminy, M.R. Characterization of somatostatin transactivating factor-1, a novel homeobox factor that stimulates somatostatin expression in pancreatic islet cells. Mol. Endocrinol. 1993, 7, 1275–1283. [Google Scholar]
- Lu, M.; Miller, C.; Habener, J.F. Functional regions of the homeodomain protein IDX-1 required for transactivation of the rat somatostatin gene. Endocrinology 1996, 137, 2959–2967. [Google Scholar] [CrossRef]
- Peers, B.; Sharma, S.; Johnson, T.; Kamps, M.; Montminy, M. The pancreatic islet factor STF-1 binds cooperatively with Pbx to a regulatory element in the somatostatin promoter: Importance of the FPWMK motif and of the homeodomain. Mol. Cell Biol. 1995, 15, 7091–7097. [Google Scholar] [CrossRef] [PubMed]
- Goudet, G.; Delhalle, S.; Biemar, F.; Martial, J.A.; Peers, B. Functional and cooperative interactions between the homeodomain PDX1, Pbx, and Prep1 factors on the somatostatin promoter. J. Biol. Chem. 1999, 274, 4067–4073. [Google Scholar] [CrossRef]
- Epstein, J.A.; Glaser, T.; Cai, J.; Jepeal, L.; Walton, D.S.; Maas, R.L. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994, 8, 2022–2034. [Google Scholar] [CrossRef]
- Wolf, G.; Hessabi, B.; Karkour, A.; Henrion, U.; Dahlhaus, M.; Ostmann, A.; Giese, B.; Fraunholz, M.; Grabarczyk, P.; Jack, R.; et al. The activation of the rat insulin gene II by BETA2 and PDX-1 in rat insulinoma cells is repressed by Pax6. Mol. Endocrinol. 2010, 24, 2331–2342. [Google Scholar] [CrossRef][Green Version]
- Ritz-Laser, B.; Gauthier, B.R.; Estreicher, A.; Mamin, A.; Brun, T.; Ris, F.; Salmon, P.; Halban, P.A.; Trono, D.; Philippe, J. Ectopic expression of the beta-cell specific transcription factor Pdx1 inhibits glucagon gene transcription. Diabetologia 2003, 46, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Andersen, F.G.; Jensen, J.; Heller, R.S.; Petersen, H.V.; Larsson, L.I.; Madsen, O.D.; Serup, P. Pax6 and Pdx1 form a functional complex on the rat somatostatin gene upstream enhancer. FEBS Lett. 1999, 445, 315–320. [Google Scholar] [CrossRef]
- Zhou, Y.; Sun, H.; Danila, D.C.; Johnson, S.R.; Sigai, D.P.; Zhang, X.; Klibanski, A. Truncated activin type I receptor Alk4 isoforms are dominant negative receptors inhibiting activin signaling. Mol. Endocrinol. 2000, 14, 2066–2075. [Google Scholar] [CrossRef]
- Gongrich, C.; Krapacher, F.A.; Munguba, H.; Fernandez-Suarez, D.; Andersson, A.; Hjerling-Leffler, J.; Ibanez, C.F. ALK4 coordinates extracellular and intrinsic signals to regulate development of cortical somatostatin interneurons. J. Cell Biol. 2020, 219, e201905002. [Google Scholar] [CrossRef]
- Munoz, W.; Tremblay, R.; Levenstein, D.; Rudy, B. Layer-specific modulation of neocortical dendritic inhibition during active wakefulness. Science 2017, 355, 954–959. [Google Scholar] [CrossRef]
- Denaxa, M.; Kalaitzidou, M.; Garefalaki, A.; Achimastou, A.; Lasrado, R.; Maes, T.; Pachnis, V. Maturation-promoting activity of SATB1 in MGE-derived cortical interneurons. Cell Rep. 2012, 2, 1351–1362. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.I.; Young, R.A. Transcriptional regulation and its misregulation in disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef]
- Rage, F.; Alonso, G.; Tapia-Arancibia, L. Stimulatory effect of N-methyl-D-aspartate on somatostatin gene expression in cultured hypothalamic neurons. Brain Res. Mol. Brain Res. 1993, 17, 287–294. [Google Scholar] [CrossRef]
- Rage, F.; Rougeot, C.; Tapia-Arancibia, L. GABAA and NMDA receptor activation controls somatostatin messenger RNA expression in primary cultures of hypothalamic neurons. Neuroendocrinology 1994, 60, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Rage, F.; Jalaguier, S.; Rougeot, C.; Tapia-Arancibia, L. GABA inhibition of somatostatin gene expression in cultured hypothalamic neurones. Neuroreport 1993, 4, 320–322. [Google Scholar] [CrossRef]
- Cammalleri, M.; Bagnoli, P.; Bigiani, A. Molecular and Cellular Mechanisms Underlying Somatostatin-Based Signaling in Two Model Neural Networks, the Retina and the Hippocampus. Int. J. Mol. Sci 2019, 20, 2506. [Google Scholar] [CrossRef]
- Juretic, N.; Urzua, U.; Munroe, D.J.; Jaimovich, E.; Riveros, N. Differential gene expression in skeletal muscle cells after membrane depolarization. J. Cell Physiol. 2007, 210, 819–830. [Google Scholar] [CrossRef]
- Tyssowski, K.M.; DeStefino, N.R.; Cho, J.H.; Dunn, C.J.; Poston, R.G.; Carty, C.E.; Jones, R.D.; Chang, S.M.; Romeo, P.; Wurzelmann, M.K.; et al. Different Neuronal Activity Patterns Induce Different Gene Expression Programs. Neuron 2018, 98, 530–546. [Google Scholar] [CrossRef]
- Tolon, R.M.; Sanchez-Franco, F.; Lopez Fernandez, J.; Lorenzo, M.J.; Vazquez, G.F.; Cacicedo, L. Regulation of somatostatin gene expression by veratridine-induced depolarization in cultured fetal cerebrocortical cells. Brain Res. Mol. Brain Res. 1996, 35, 103–110. [Google Scholar] [CrossRef]
- Tolon, R.M.; Sanchez Franco, F.; de los Frailes, M.T.; Lorenzo, M.J.; Cacicedo, L. Effect of potassium-induced depolarization on somatostatin gene expression in cultured fetal rat cerebrocortical cells. J. Neurosci. 1994, 14, 1053–1059. [Google Scholar] [CrossRef]
- Cacicedo, L.; Tolon, R.M.; Lorenzo, M.J.; Lopez, J.; Sanchez Franco, F. Potassium-induced depolarization stimulates somatostatin gene expression in cultured fetal rat cerebrocortical cells. J. Pediatric Endocrinol. 1993, 6, 219–223. [Google Scholar] [CrossRef]
- Sanchez-Munoz, I.; Sanchez-Franco, F.; Vallejo, M.; Fernandez, A.; Palacios, N.; Fernandez, M.; Cacicedo, L. Activity-dependent somatostatin gene expression is regulated by cAMP-dependent protein kinase and Ca2+-calmodulin kinase pathways. J. Neurosci. Res. 2010, 88, 825–836. [Google Scholar] [CrossRef]
- Ehrman, M.M.; Melroe, G.T.; Kittilson, J.D.; Sheridan, M.A. Glucose-stimulated somatostatin gene expression in the Brockmann bodies of rainbow trout (Oncorhynchus mykiss) results from increased mRNA transcription and not from altered mRNA stability. Zool. Sci. 2004, 21, 87–91. [Google Scholar] [CrossRef][Green Version]
- Dumonteil, E.; Magnan, C.; Ritz-Laser, B.; Ktorza, A.; Meda, P.; Philippe, J. Glucose regulates proinsulin and prosomatostatin but not proglucagon messenger ribonucleic acid levels in rat pancreatic islets. Endocrinology 2000, 141, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Ehrman, M.M.; Melroe, G.T.; Kittilson, J.D.; Sheridan, M.A. Regulation of pancreatic somatostatin gene expression by insulin and glucagon. Mol. Cell. Endocrinol. 2005, 235, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Papachristou, D.N.; Pham, K.; Zingg, H.H.; Patel, Y.C. Tissue-specific alterations in somatostatin mRNA accumulation in streptozocin-induced diabetes. Diabetes 1989, 38, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Patel, Y.C.; Cameron, D.P.; Bankier, A.; Malaisse-Lagae, F.; Ravazzola, M.; Studer, P.; Orci, L. Changes in somatostatin concentration in pancreas and other tissues of streptozotocin diabetic rats. Endocrinology 1978, 103, 917–923. [Google Scholar] [CrossRef]
- Julien, S.; Laine, J.; Morisset, J. Regulation of rat pancreatic CCKB receptor and somatostatin expression by insulin. Diabetes 2004, 53, 1526–1534. [Google Scholar] [CrossRef]
- Tannenbaum, G.S.; Martin, J.B.; Colle, E. Ultradian growth hormone rhythm in the rat: Effects of feeding, hyperglycemia, and insulin-induced hypoglycemia. Endocrinology 1976, 99, 720–727. [Google Scholar] [CrossRef]
- Santiago, J.A.; Kadowitz, P.J. Analysis of responses to pituitary adenylate cyclase activating polypeptide-38 in the feline hindquarters vascular bed. Eur J. Pharm. 1993, 243, 291–294. [Google Scholar] [CrossRef]
- Hunter, W.M.; Willoughby, J.M.; Strong, J.A. Plasma insulin and growth hormone during 22-hour fasts and after graded glucose loads in six healthy adults. J. Endocrinol. 1968, 40, 297–311. [Google Scholar] [CrossRef]
- Roth, J.; Glick, S.M.; Yalow, R.S.; Bersonsa, S.A. Hypoglycemia: A potent stimulus to secretion of growth hormone. Science 1963, 140, 987–988. [Google Scholar] [CrossRef] [PubMed]
- Katoh-Semba, R.; Takeuchi, I.K.; Semba, R.; Kato, K. Distribution of brain-derived neurotrophic factor in rats and its changes with development in the brain. J. Neurochem. 1997, 69, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Jin, W. Regulation of BDNF-TrkB Signaling and Potential Therapeutic Strategies for Parkinson’s Disease. J. Clin. Med. 2020, 9, 257. [Google Scholar] [CrossRef]
- Andero, R.; Choi, D.C.; Ressler, K.J. BDNF-TrkB receptor regulation of distributed adult neural plasticity, memory formation, and psychiatric disorders. Prog. Mol. Biol. Transl. Sci. 2014, 122, 169–192. [Google Scholar] [PubMed]
- Rage, F.; Riteau, B.; Alonso, G.; Tapia-Arancibia, L. Brain-derived neurotrophic factor and neurotrophin-3 enhance somatostatin gene expression through a likely direct effect on hypothalamic somatostatin neurons. Endocrinology 1999, 140, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Munoz, I.; Sanchez-Franco, F.; Vallejo, M.; Fernandez, A.; Palacios, N.; Fernandez, M.; Sanchez-Grande, M.; Cacicedo, L. Regulation of somatostatin gene expression by brain derived neurotrophic factor in fetal rat cerebrocortical cells. Brain Res. 2011, 1375, 28–40. [Google Scholar] [CrossRef]
- Spencer, T.K.; Mellado, W.; Filbin, M.T. BDNF activates CaMKIV and PKA in parallel to block MAG-mediated inhibition of neurite outgrowth. Mol. Cell. Neurosci. 2008, 38, 110–116. [Google Scholar] [CrossRef]
- Impey, S.; Fong, A.L.; Wang, Y.; Cardinaux, J.R.; Fass, D.M.; Obrietan, K.; Wayman, G.A.; Storm, D.R.; Soderling, T.R.; Goodman, R.H. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron 2002, 34, 235–244. [Google Scholar] [CrossRef]
- Villuendas, G.; Sanchez-Franco, F.; Palacios, N.; Fernandez, M.; Cacicedo, L. Involvement of VIP on BDNF-induced somatostatin gene expression in cultured fetal rat cerebral cortical cells. Brain Res. Mol. Brain Res. 2001, 94, 59–66. [Google Scholar] [CrossRef]
- Kovac, S.; Xiao, L.; Shulkes, A.; Patel, O.; Baldwin, G.S. Gastrin increases its own synthesis in gastrointestinal cancer cells via the CCK2 receptor. FEBS Lett. 2010, 584, 4413–4418. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Kim, C.H.; Yim, Y.S.; Ahn, Y.S. Autocrine function of erythropoietin in IGF-1-induced erythropoietin biosynthesis. Neuroreport 2008, 19, 1699–1703. [Google Scholar] [CrossRef] [PubMed]
- Zoja, C.; Bettoni, S.; Morigi, M.; Remuzzi, G.; Rambaldi, A. Interleukin-1 regulates cytokine gene expression in human mesangial cells through the interleukin-1 receptor type 1. J. Am. Soc. Nephrol. 1992, 2, 1709–1715. [Google Scholar] [PubMed]
- Braun, M.; Ramracheya, R.; Rorsman, P. Autocrine regulation of insulin secretion. Diabetes Obes. Metab. 2012, 14 (Suppl. 3), 143–151. [Google Scholar] [CrossRef] [PubMed]
- Leibiger, B.; Moede, T.; Muhandiramlage, T.P.; Kaiser, D.; Vaca Sanchez, P.; Leibiger, I.B.; Berggren, P.O. Glucagon regulates its own synthesis by autocrine signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 20925–20930. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.G.; Zhou, G.; Liu, S.H.; Li, M.; Jeong, J.W.; DeMayo, F.J.; Gingras, M.C.; Gibbs, R.A.; Fisher, W.E.; Brunicardi, F.C. Microarray analysis of somatostatin receptor 5-regulated gene expression profiles in murine pancreas. World J. Surg. 2009, 33, 630–637. [Google Scholar] [CrossRef]
- Zatelli, M.C.; Tagliati, F.; Taylor, J.E.; Rossi, R.; Culler, M.D.; degli Uberti, E.C. Somatostatin receptor subtypes 2 and 5 differentially affect proliferation in vitro of the human medullary thyroid carcinoma cell line tt. J. Clin. Endocrinol. Metab. 2001, 86, 2161–2169. [Google Scholar] [CrossRef]
- Zhou, G.; Liu, S.H.; Shahi, K.M.; Wang, H.; Duan, X.; Lin, X.; Feng, X.H.; Li, M.; Fisher, W.E.; Demayo, F.J.; et al. Negative regulation of pancreatic and duodenal homeobox-1 by somatostatin receptor subtype 5. Mol. Endocrinol. 2012, 26, 1225–1234. [Google Scholar] [CrossRef]
- Jepsen, S.L.; Grunddal, K.V.; Wewer Albrechtsen, N.J.; Engelstoft, M.S.; Gabe, M.B.N.; Jensen, E.P.; Orskov, C.; Poulsen, S.S.; Rosenkilde, M.M.; Pedersen, J.; et al. Paracrine crosstalk between intestinal L- and D-cells controls secretion of glucagon-like peptide-1 in mice. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E1081–E1093. [Google Scholar] [CrossRef]
- Bos, J.L. Epac: A new cAMP target and new avenues in cAMP research. Nat. Rev. Mol. Cell Biol. 2003, 4, 733–738. [Google Scholar] [CrossRef]
- Patel, Y.C. Somatostatin and its receptor family. Front. Neuroendocrinol. 1999, 20, 157–198. [Google Scholar] [CrossRef] [PubMed]
- Eigler, T.; Ben-Shlomo, A.; Zhou, C.; Khalafi, R.; Ren, S.G.; Melmed, S. Constitutive somatostatin receptor subtype-3 signaling suppresses growth hormone synthesis. Mol. Endocrinol. 2014, 28, 554–564. [Google Scholar] [CrossRef]
- Jacobs, S.; Calebiro, D.; Nikolaev, V.O.; Lohse, M.J.; Schulz, S. Real-time monitoring of somatostatin receptor-cAMP signaling in live pituitary. Endocrinology 2010, 151, 4560–4565. [Google Scholar] [CrossRef] [PubMed]
- Feelders, R.A.; Hofland, L.J.; van Aken, M.O.; Neggers, S.J.; Lamberts, S.W.; de Herder, W.W.; van der Lely, A.J. Medical therapy of acromegaly: Efficacy and safety of somatostatin analogues. Drugs 2009, 69, 2207–2226. [Google Scholar] [CrossRef] [PubMed]
- Patil, V.S.; Zhou, R.; Rana, T.M. Gene regulation by non-coding RNAs. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 16–32. [Google Scholar] [CrossRef]
- Tellier, M.; Maudlin, I.; Murphy, S. Transcription and splicing: A two-way street. Wiley Interdiscip. Rev. RNA 2020, e1593. [Google Scholar] [CrossRef]
- Han, Z.J.; Feng, Y.H.; Gu, B.H.; Li, Y.M.; Chen, H. The post-translational modification, SUMOylation, and cancer (Review). Int. J. Oncol. 2018, 52, 1081–1094. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ampofo, E.; Nalbach, L.; Menger, M.D.; Laschke, M.W. Regulatory Mechanisms of Somatostatin Expression. Int. J. Mol. Sci. 2020, 21, 4170. https://doi.org/10.3390/ijms21114170
Ampofo E, Nalbach L, Menger MD, Laschke MW. Regulatory Mechanisms of Somatostatin Expression. International Journal of Molecular Sciences. 2020; 21(11):4170. https://doi.org/10.3390/ijms21114170
Chicago/Turabian StyleAmpofo, Emmanuel, Lisa Nalbach, Michael D. Menger, and Matthias W. Laschke. 2020. "Regulatory Mechanisms of Somatostatin Expression" International Journal of Molecular Sciences 21, no. 11: 4170. https://doi.org/10.3390/ijms21114170
APA StyleAmpofo, E., Nalbach, L., Menger, M. D., & Laschke, M. W. (2020). Regulatory Mechanisms of Somatostatin Expression. International Journal of Molecular Sciences, 21(11), 4170. https://doi.org/10.3390/ijms21114170




