Abstract
G-quadruplexes are four-stranded helical nucleic acid structures formed by guanine-rich sequences. A considerable number of studies have revealed that these noncanonical structural motifs are widespread throughout the genome and transcriptome of numerous organisms, including humans. In particular, G-quadruplexes occupy strategic locations in genomic DNA and both coding and noncoding RNA molecules, being involved in many essential cellular and organismal functions. In this review, we first outline the fundamental structural features of G-quadruplexes and then focus on the concept that these DNA and RNA structures convey a distinctive layer of epigenetic information that is critical for the complex regulation, either positive or negative, of biological activities in different contexts. In this framework, we summarize and discuss the proposed mechanisms underlying the functions of G-quadruplexes and their interacting factors. Furthermore, we give special emphasis to the interplay between G-quadruplex formation/disruption and other epigenetic marks, including biochemical modifications of DNA bases and histones, nucleosome positioning, and three-dimensional organization of chromatin. Finally, epigenetic roles of RNA G-quadruplexes in post-transcriptional regulation of gene expression are also discussed. Undoubtedly, the issues addressed in this review take on particular importance in the field of comparative epigenetics, as well as in translational research.
1. Introduction
More than one century ago, seminal observations from Phoebus Levene’s laboratory revealed that guanosine forms polycrystalline gels at high concentrations, providing the first indirect evidence that guanine-rich sequences in nucleic acids may form higher-order structures [1]. Curiously, many authors have attributed the achievement of this discovery to the Scandinavian biochemist Ivar Bang, who described a similar gelling process one year after Levene’s paper, using aqueous solutions of guanylic acid isolated from ox pancreas [2].
More than five decades later, crystallographic analysis of guanosine-5′-monophosphate gels has provided the structural explanation of the jellification phenomenon, revealing the association of guanine bases in tetrameric arrays (referred to as G-tetrads) via non-Watson–Crick base pairing [3]. Further X-ray fiber diffraction and biophysical studies on guanine-rich polynucleotides have confirmed and extended these findings, paving the way for the concept of G-quadruplex, a four-stranded continuous helix whose geometric formalism is fully consistent with the G-tetrad structural motif [4,5,6].
Since then, a very large collection of evidence has identified these structures in nucleic acid sequences of a wide spectrum of organisms, highlighting their involvement in the regulation of various key cellular and organismal processes, including embryogenesis, genome maintenance, replication and expression [7,8,9,10,11,12,13,14,15,16]. In this review, we will focus on the epigenetic roles played by both DNA and RNA G-quadruplexes in the modulation of chromatin states and gene expression.
2. Structural Features of G-Quadruplexes and Their Occurrence in Biological Contexts
G-quadruplexes, henceforth referred to as G4s, are four-stranded knot-like structures dynamically folded in guanine-rich regions of nucleic acids. As mentioned, the building block of these noncanonical structures is the G-tetrad or G-quartet, a coplanar cyclic array of four adjacent guanylic nucleotides held together by a network of hydrogen bonds at the edges of the resulting square platform (Figure 1A). In particular, the guanine purine rings simultaneously accomplish hydrogen bond donor and acceptor functionalities, yielding eight Hoogsteen-type interactions per G-quartet [3,17]. In this tetrameric arrangement, distinct combinations of anti and syn conformations, defined by torsion angles around the N-glycosidic bond that connects the guanine base to the sugar, directly determine the parallel/antiparallel directionality of the phosphodiester backbone of each of the four participating polynucleotide strands [18,19].

Figure 1.
Chemical structures of G-quartet and G-quadruplex. (A) Structural arrangement of the G-quartet, highlighting the hydrogen bonding network between the Hoogsteen and Watson–Crick faces of the coplanar guanine bases. The attached deoxyribose sugars are shown together with a centrally placed metal ion. (B) The conventional consensus sequence for a G-quadruplex. (C) Side view of the schematic diagram showing an intramolecular antiparallel G-quadruplex formed by the stacking of three G-quartets. Strand polarity and anticlockwise rotation are indicated.
Since the prototypical G4-forming consensus sequence 5′-G≥3-N1–7-G≥3-N1–7-G≥3-N1–7-G≥3-3′ includes four runs of at least three consecutive guanines (Figure 1B), a given G4 generally, but not always, contains a minimum of three G-quartet layers stacked upon one another by virtue of electron interactions between π orbitals from their aromatic surfaces (Figure 1C) [20,21]. Lone pair electrons at the oxygen atom of each guanine carbonyl group lie in the central core of adjoining G-quartets, creating an electronegative axial tunnel running through the G4 stack (Figure 1C) [22,23]. In this context, the symmetric bipyramidal antiprismatic arrangement for eight oxygen atoms juxtaposed at each level of the stack allows coordination of monovalent alkali metal cations that, in turn, impart further stability to the G4 structure (Figure 1C) [24]. In the cellular environment, potassium ions are preferentially coordinated because they exist in the highest concentration and give the best size match for the accommodation inside the G4 central channel [24,25].
Worth mentioning, although G4s are commonly depicted as having a straight three-dimensional morphology, they are instead helical structures displaying rotational symmetry and four grooves varying in width in the spaces between the four guanine-rich strands (Figure 1) [26]. In the overall G4 structure, G-quartets are stacked perpendicularly to the helix axis, and continuous stretches of guanines establishing the stacked G-quartets are connected by loops of spacer tracts varying in length and nucleotide composition (Figure 1C) [18]. The structural uniformity of the G4 stack is tolerant of bulging out single unpaired bases or embedding them into the core structure, which broadens the definition of G4-forming sequences, as well as the specific nomenclature of G4 themselves [27]. Moreover, G4s can arise from a single polynucleotide chain of DNA or RNA containing an adequate number of guanine-run stretches or can alternatively embrace distinct guanine-rich regions belonging to multiple (either two, three or four) nucleic acid chains [28,29,30]. Adding further complexity, stacking of intra-/intermolecular G-quartets may produce G-wires, high-order thread-like superstructures exhibiting peculiar periodicity and physical properties that are not found in basic G4s [31,32].
In sum, it can be argued that the repertoire of G4 molecular architectures displays extensive geometry and conformational polymorphism, comprehensively according to the variability of the aforementioned intrinsic structural parameters [33,34,35,36,37]. Additional sources for the high topological divergence are given by extrinsic factors, including chemical modification and pH-driven protonation/de-protonation of bases [38,39,40,41], molecular crowding [42], and the presence of chaperone molecules [43,44].
Genomewide computational screenings for sequence motifs that have the propensity to form G4, as well as G4-specific chromatin immunoprecipitation coupled with high-throughput sequencing analysis, have revealed widespread G4 occurrence in a large number of species belonging to all the kingdoms of life, and in diverse ribo- and deoxyribo-viruses [45,46,47,48,49,50,51,52,53,54,55]. These studies have coherently indicated that the global amount of genomic G4s displays considerable variation across species, roughly according to size and guanine-richness of their genome, but not to evolutionary distances [56,57,58,59,60]. Furthermore, they unveiled that G4s are not randomly scattered across the genome, being rather biased toward telomeric sequences, satellite DNA, noncoding transcription units, and gene cis-regulatory regions [61,62,63,64]. Intriguingly, comparative inspections have also highlighted similarities across diverse organisms and species-specific trends of G4 distribution in conserved genomic portions [57,58,59,65]. For example, strong G4 enrichment in gene promoters has been reported in higher vertebrates, including humans, while preferential enrichment in noncoding transcription units has been detected in other metazoans such as the nematode worm Caenorhabditis elegans and zebrafish (Danio rerio) [57]. Unfortunately, it remains poorly understood whether all of the G4s emerging from these stimulating studies actually form in vivo, especially in light of the fact that only a very modest fraction of them have been experimentally validated by immune-fluorescent visualization in different types of living cells [66,67,68,69]. Beyond this direct line of evidence provided by employing G4-specific antibodies generated by independent groups, G4 formation in vivo has been indirectly justified by alternative approaches, such as “in-cell” NMR spectroscopy [70,71], and by the recent identification and functional characterization of G4-specific DNA and RNA helicases from various organisms [72,73,74,75,76,77,78].
In fact, although G4s are generally conceived as energetically favorable and highly stable structures under physiological conditions, their assembly in vivo often requires preliminary local chromatin dismantling and DNA double-helix denaturation. This occurs especially during biological processes, such as replication and transcription, that transiently expose single-strand DNA segments. Once formed, G4s on both DNA and RNA molecules can contribute to the regulation of fundamental cell functions, as described in the following sections.
3. Involvement of G4 Structures in Epigenetic Processes
Based on our actual understanding of epigenetics, an epigenetic trait is broadly intended as a reversible molecular modification associated with changes in gene expression in the absence of variation in genomic DNA sequence, occurring in the course of adaptive responses of a given cell/organism to environmental influence [79,80]. There is growing experimental evidence indicating that G4s entirely fulfill this definition, being, in principle, interconverting DNA structural conformations dynamically adopted by peculiar genome segments. The occurrence of G4 structures in regulatory regions such as promoters and enhancers may influence gene expression either positively or negatively, thereby provoking transcriptome changes [81,82]. Folding of G4 structures is differentially affected, either directly or indirectly, by disparate environmental cues, including dietary molecules and epigenetic drugs [83,84,85,86,87,88]. In addition, G4 structures and other established epigenetic modifications frequently go side by side, and they harmoniously affect each other in many fascinating ways, reshaping genome transcriptional outputs.
3.1. Interplay between G4 Structures and Epigenetic Modifications of DNA Bases
Stability of a conventional G4 structure may be affected by epigenetic biochemical modifications of sequences either forming or flanking the G4 motif in the same DNA strand, such as methylation at carbon 5 on the cytosine pyrimidine ring (5meC, Figure 2). In fact, G4s exhibit much higher thermal stability when sandwiched by stacking forces provided by nearby 5meC-5meC+ base pairs formed by N3-protonation of cytosine under physiological-like conditions [89,90,91]. In certain exceptional cases, however, methylated G4s appear to be less thermally stable than their unmethylated equivalents, suggesting that the influence of methylation on G4 thermodynamic properties is strictly dependent on the DNA sequence being investigated [92]. It can be argued that the connection between G4 stability and cytosine methylation could have relevant consequences in vivo, especially because DNA methylation generally results in a reduction of chromatin accessibility and transcriptional activity. In this respect, recent genomewide approaches have shown that the occurrence of intrinsically stable G4s and 5meC is inversely related at CpG island promoters of different human cell lines and tissues, as well as in peripheral blood samples from distinct individuals, strongly suggesting that G4s are epigenetic marks of active hypomethylated chromatin [93,94,95]. This finding is further supported and extended by separate mechanistic evidence indicating that (i) G4 structures, once formed, avidly interact with DNA methyltransferase (DNMT) enzymes both in vitro and in vivo, and that (ii) G4-dependent recruitment of DNMT1 at lowly methylated CpG islands locally results in significant inhibition of DNMT1 enzymatic activity [95,96]. Taken together, these findings point to the coordinated contribution of two distinct classes of epigenetic players, G4 structures and the enzymes that establish and maintain deoxycytidine methylation, in shaping the methylome.
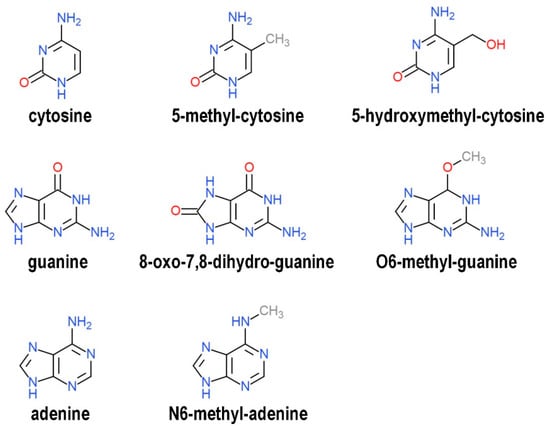
Figure 2.
Chemical structure of canonical and biochemically modified nitrogenous bases discussed in this review.
It is commonly accepted that G4 structures formed on sequences juxtaposed to the gene core promoters may inhibit transcriptional activity by acting as a steric block to RNA polymerase recruitment, an effect that is exacerbated when the G4-forming regions undergo cytosine methylation [97]. Along with this negative impact on transcription, the co-occurrence of G4 formation and cytosine methylation can instead give a stimulatory contribution to gene expression. To cite an example, cytosine hypomethylation in the core promoter region, coupled with methylation in a specific CpG in the first exon, favors hTERT gene expression in tumor cells [98,99]. In fact, exonic 5meC triggers the formation of a stable G4 structure that, in turn, prevents the binding of the CTCF factor, otherwise involved in hTERT transcription repression [99,100].
A major route to CpG demethylation in metazoans involves the sequential enzymatic conversion of 5meC into the progressively higher oxidation states of 5-hydroxymethyl-, 5-formyl-, and 5-carboxyl-cytosine until the base is excised and replaced by an unmodified cytosine [101,102]. Intriguingly, emerging evidence suggests that the oxidation products of 5meC could reflect distinctive epigenetic modifications associated with differential genomic density and specific biological functions. For example, transcriptional activity is negatively correlated with 5meC but positively correlated with 5-hydroxymethyl-cytosine (5hmeC, Figure 2) [103,104]. This is probably due to the poor binding affinity of DNMT1 toward 5hmC, suggesting that 5hmC, as opposite to 5meC, could promote DNA demethylation by excluding DNMT1 from CpG islands [105]. In human stem cells, a very small fraction of G4s harbors 5hmeC in loop regions, and the presence of this modification does not markedly affect G4 formation or stability [106,107]. On the other hand, it has been shown that the oxidized derivatives of cytosine dynamically recruit distinct sets of regulatory proteins in differentiating mouse embryonic stem cells, suggesting that G4-associated cytosine modifications epigenetically influence the propensity of G4 structures to be recognized by DNA-binding effector proteins [108]. Recent studies focused on the G4 from the vegf promoter have revealed that this is indeed the case, where the presence of 5meC significantly decreases the binding ability of the VEGF165 protein, while 5hmeC specifically abrogates nucleolin recruitment [107,109,110].
Epigenetic variations reverberating on G4 structure stability can also be inflicted by environmental stress endured by individual cells or organisms. A pertaining example is provided by the establishment of guanine oxidation, which is induced by reactive oxygen species from both exogenous and endogenous origins [111]. At the DNA level, the redox potential of the guanine heterocycle is particularly low, and it further decreases proportionally when a rising number of adjacent guanine bases stack upon one another [112,113]. It follows that guanine runs embedded in G4-forming sequences are the most prone to oxidation within eukaryotic genomes, where 8-oxo-7,8-dihydro-guanine (8oxoG, Figure 2) is the oxidation product found in the highest yield. To give an idea of scale, 8oxoG is present at a very low frequency in murine embryonic stem cells and adult cortex tissue, being more than three orders of magnitude smaller in concentration than 5meC in the genome [114].
From a structural perspective, the presence of 8oxoG impacts either positively or negatively on G4 integrity, depending on the exact position occupied within the quadruplex [115,116]. For instance, the substitution of a guanine with 8oxoG at the middle tetrad of a conventional G4 disrupts both the Hoogsteen and stacking interactions, thereby hindering G4 folding. Nonetheless, divergent G4-forming sequences, containing excess guanines in their G runs, and/or more than four G runs, more broadly tolerate oxidative modifications, being able to rearrange into alternative topologies in which 8oxoG is looped out and the overall structural integrity is maintained [117,118]. In the case of G4s occurring in promoter sequences, such as an 8oxoG-driven topology switch, this leads to an upregulation of gene expression during oxidative stress. Based on published evidence, this purely epigenetic mechanism is most probably achieved by the recruitment of OGG1, a DNA glycosylase involved in the reversal of guanine oxidation [119,120]. Following specific recognition of 8oxoG, OGG1 would indeed facilitate the assembly of an effector complex that could contain transcription activators, heterogeneous nuclear ribonucleoprotein particles, and RNA polymerase II for the induction of gene transcription [121,122,123,124,125,126]. According to recent models, the abasic site yielded by OGG1-dependent release of 8oxoG would concomitantly favor the structural switch from duplex DNA to G4 and recruitment of a complex, including the apurinic-apyrimidinic endonuclease 1 (APE1) and other partners such as HIF1-α or the RNA polymerase II. The G4 structural context, however, would severely attenuate the endonuclease activity of APE1, thus allowing transcription activation driven by HIF1-α or direct positioning of RNA polymerase II [126]. A differing model suggests that the base excision repair of 8oxoG embedded into G4 would stimulate gene transcription after returning the sequence back to the wild-type state by means of enhanced recruitment of MYC-associated zinc-finger transcription factor and heterogeneous nuclear ribonucleoprotein A1 [126]. Further studies are needed to address the exact choreography of events regarding the reversal of guanine oxidation located in G4-forming sequences on gene promoters and the upregulation of gene expression.
Much less is known about the relationship between the formation or function of G4s and other biochemical modifications of DNA bases. In this regard, the O6-methyl-deoxyguanine (6meG, Figure 2) adduct is formed in response to alkylating environmental pollutants and enzymatically reversed by O6-alkylguanine DNA alkyltransferase [127]. The presence of this atypical modification significantly weakens the overall G4 structure because 6meG is flipped out from the stacked G-quartets and, therefore, prevented from participating in metal cation coordination [128]. Similarly, it has been recently shown that the presence of N6-methyl-deoxyadenosine (6meA, Figure 2) in an unconventional G4-forming sequence from the human c-kit gene is detrimental to G4 folding in vitro, probably due to disruption of Watson–Crick base pairing resulting from the preference for the syn conformation adopted by 6meA [129,130,131,132]. Functional findings in vivo need to be addressed since 6meA is widely distributed, although with low abundance, across the eukaryotic genome and predominantly within genomic deserts [133].
3.2. G4 Structures and the Histone Epigenetic Machinery
A fundamental source of epigenetic information is stored on nucleosomes, in each of four core histones H2A, H2B, H3, and H4 [134,135,136,137,138]. Accurate propagation of the epigenetic information residing into nucleosomes is strictly dependent upon local recycling of modified parental histones to daughter chromatin by histone chaperones during active DNA replication [139,140]. In this respect, structured G4s represent physical obstacles to the replisome progression, and their unfolding is ensured by a number of helicases and/or specialized DNA polymerases involved in translesion synthesis [141,142,143,144]. However, when the unwinding of G4 structures is temporarily delayed, the corresponding genomic regions are bypassed and gaps are completed by postreplicative mechanisms, leading to preferential incorporation of newly synthesized histones that are devoid of parental modifications [145]. Therefore, the G4-dependent decoupling of parental histone recycling from DNA replication ultimately disrupts the local inheritance of epigenetic transmission. This effect has been formerly documented in avian DT40 cells lacking either the Y-family DNA polymerase REV1 or the RecQ-family helicase FANCJ [13,146,147,148]. In both cases, altered transcription of specific loci was well correlated with anomalous histone mark patterns in the vicinity of potential G4-forming sequences (Figure 3A). Strikingly, artificial insertion of a G4-forming sequence into the first intron of the silent lysozyme C (lysC) gene resulted in derepression of lysC in REV1 deficient cells, further highlighting the need for G4-structure processing in the maintenance of histone epigenetic memory and regulation of gene expression [13].
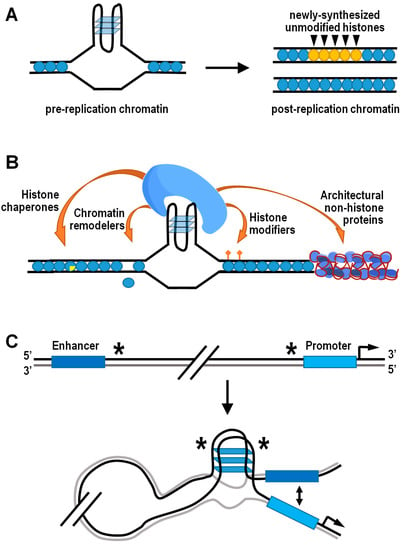
Figure 3.
Interplay between G4 structures and the epigenetic machinery. (A) Schematic drawing indicating loss of histone epigenetic memory following DNA replication at sites with G4 structures in cells lacking either REV1 or FANCJ. (B) Mechanistic model showing that G4 structures can act as docking sites for several classes of epigenetic players. (C) Schematic model of enhancer–promoter interaction mediated by an intermolecular G4 structure formed by two distinct half G4s (indicated by asterisks).
Formation of G4 structures may also provide the docking site for effector protein complexes horboring histone-modifying activities (Figure 3B). For instance, the TLS/FUS/KMT5C ternary complex is able to bind both RNA and DNA G4 structures at telomeric chromatin, where KMT5C specifically trimethylates the lysine 20 residue of nucleosomal histone H4, thus favoring chromatin condensation [149]. Contrarily, the H4K20-specific histone demethylase PHF8 has been associated with G4-containing promoters of genes located in open chromatin and expressed at a significant level [150]. Another well-established example is the REST/coREST repressor complex, which conveys the histone H3K4-specific demethylase LSD1 at peculiar chromatin locations containing G4 structures, including the p21 and hTERT gene promoters [151,152,153,154]. More recently, outcomes from a high-throughput screening assay in vitro added further insights on the interaction between topologically different intramolecular G4 DNA structures and human epigenetic proteins immobilized on microarrays [155]. Top significant hits resulting from this analysis indeed included the peptidylarginine deiminase PADi4, which converts both arginine and monomethyl-arginine histone residues to citrulline [156], and the scaffolding protein ASXL1, which binds to chromatin and recruits polycomb repressive complex 2 (PRC2), thus favoring the locus-specific accumulation of H3K27me3 [157,158]. It is noteworthy that G4 structures in nascent chromatin-associated RNAs are involved in the temporal regulation of PRC2 occupancy at its target genes, as they evict PRC2 from nucleosome particles and inhibit its methyltransferase activity [159,160].
Structured G4s, both in DNA and chromatin-associated RNA, may locally control chromatin compactness and/or composition by directly attracting classic chromatin remodeler and histone chaperones such as SWI/SNF, NuRD, FACT, and BRD3, which are renowned players in nucleosome disassembly/reassembly processes [155]. For example, G4-dependent recruitment of the complex containing the SWI/SNF-like chromatin remodeler ATRX and DAXX histone chaperone permits deposition/exchange of the H3.3 histone variant, which is subsequently targeted for K9 trimethylation to establish a heterochromatic state at telomeric and pericentromeric regions [161,162]. In other chromatin contexts, the FACT-like histone chaperone nucleolin both binds and stabilizes G4 structures, promoting the remodeling of nucleosomes containing macroH2A, and the removal of H2A–H2B dimers from assembled nucleosomes [163,164,165].
As a further possibility, G4s can impact on chromatin dynamics by engaging architectural nonhistone proteins, including some members of the HMG-B and -N families, which alter the pattern of histone modifications and modulate the binding of linker histones to chromatin [166,167,168]. For example, the biological outcome of the HMGB1-mediated G4 stabilization in the promoter of the human kras gene is transcriptional silencing, and the importance of HMGB1 in this mechanism is further supported by the increase in kras gene expression induced following HMGB1 knockdown [169]. A complementary role to HMGB1 could be played by the poly ADP-ribose polymerase 1 (PARP1) protein, which has been positively associated with G4 formation and kras gene expression. In fact, in its activated form, PARP1 specifically binds G4s at the murine kras gene promoter, where it locally behaves as a histone chaperone for chromatin relaxation, triggering nucleosome eviction by histone PARylation [170,171].
On a whole-genome scale, the coalescence of G4 structures and architectural proteins has further relevant epigenetic implications in terms of nucleosome positioning, as well as three-dimensional chromatin organization and functions. Captivating findings have indeed confirmed that G4-forming sequences are located within non-nucleosomal DNA regions in organisms as diverse as yeasts, nematodes, mice, and humans [62,172,173,174,175]. The hypothesis that G4 structures may function as nucleosome exclusion signals is further supported by the observation of inverse phasing patterns between G4 motifs and nucleosomes around a subset of human DNA replication origins [176]. More specifically, the nucleosome-depleted regions containing G4-forming sequences quite frequently coincide with the boundaries of topologically associated domains [150]. These consist of nucleoprotein complexes highly enriched in architectural proteins, such as CTCF and cohesin, involved in three-dimensional partitioning of the eukaryotic genome and correlated with gene regulation [177,178,179,180,181]. Otherwise stated, there is consistent evidence suggesting that G4s may mediate the preferential establishment of long-distance contacts between different regions of interphase chromosomes (Figure 3C). While doing so, G4s could also affect gene expression by facilitating interaction between promoters and their cis-regulatory sequences via looping [150,182]. In this regard, it is worth noting that the so-called “half G4s” are significantly enriched exactly at the abovementioned genomic regions [182]. Individual half G4 sequences are unable to fold into autonomous G4 structures, being composed by only two shortly interspaced runs of at least three consecutive guanines. Nonetheless, two distinct half G4 may join into an intermolecular G4 structure, thereby bringing together two distant genomic regions [182].
3.3. Epigenetic Roles of G4 Structures Formed in Coding and Noncoding RNA Transcripts
Although, to date, structural and functional studies have been mainly focused on DNA G4s, in the last decade, there has been growing attention on RNA G4s in light of the wide variety of functions attained by these structures in multiple physiopathological processes. A fundamental concept that has emerged from several studies is that RNA and DNA G4 structures, despite numerous similarities, are not merely counterparts of each other. In fact, due to the presence of a C2′-hydroxyl group in the ribose sugar and uracil in the spacer loops, RNA G4s are often more compact, less hydrated, and more thermodynamically stable structures than their DNA analogs [183,184,185,186]. On the other hand, RNA G4s almost exclusively adopt a monomorphic parallel topology due to the steric constraints imposed by sugar puckering and C2′-hydroxyl groups, which lock the riboguanosines in an anti conformation [187]. Within eukaryotic cells, RNA G4s are widely distributed in both nuclear and cytoplasmic compartments, implying that they have a greater assortment of protein-binding partners compared to DNA G4s and that they can potentially fold into hybrid intermolecular RNA:DNA structures in the nucleoplasm [30,188].
In general, switchable RNA G4s potentially provide an important contribution to epigenetic regulation of gene expression, and their involvement in a given biological function is strictly context-dependent and antagonized by specialized helicase activities (Figure 4). For example, negative cotranscriptional regulation of reporter gene expression by an impediment to RNA polymerase passage has been reported following the formation of hybrid RNA:DNA G4s between the nascent RNA and the template, but not the coding, DNA strand [189]. As already mentioned, this mechanism could be further refined by dynamic G4 unfolding and refolding governed by helicases. However, in silico predictions have revealed that this cotranscriptional mechanism could be underrepresented in warm-blooded animals, whose genes exhibit a strong bias toward G4 motifs on the coding DNA strand, and therefore, on the corresponding mRNA [190].
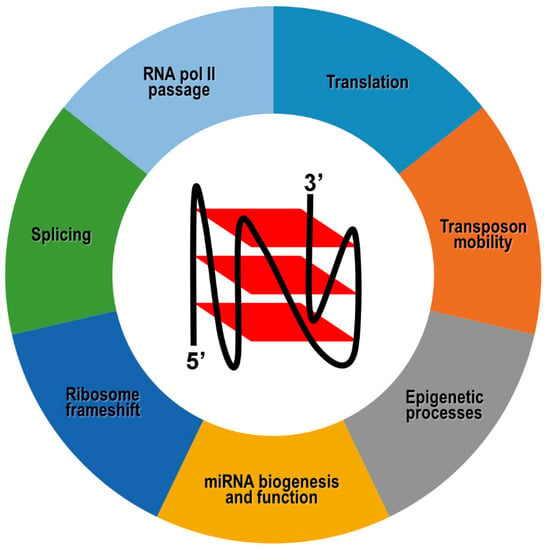
Figure 4.
Molecular processes modulated by G4 structures formed in coding and noncoding RNA transcripts. A schematic drawing indicating the parallel topology adopted by RNA G4s is shown.
In this case, the differential location of G4s in exons, introns, 5′- and 3′-UTRs may exert totally distinct outcomes on gene expression. More specifically, G4s located in 5′-UTR of coding transcripts normally inhibit translation initiation, probably through sterically hindering the recruitment and/or scanning of the 40S ribosome subunit on the mRNA [191,192,193,194,195]. Based on these findings, and considering that G4s have been identified in the 5′-UTR of numerous proto-oncogene transcripts, it has been proposed that such RNA motifs could be suitable targets for small molecules, with the view to interfering with gene expression at the translation level [196]. Indeed, small molecules could, in principle, affect G4 stability and/or compete with G4-binding proteins normally involved in translation stimulation. However, there are no approved anticancer drugs targeting RNA or DNA G-quadruplexes, probably due to nonspecific binding events of small molecules to nucleic acid structures other than G4s.
In a few alternative cases, G4 structures present in 5′-UTR were proposed to be necessary for atypical forms of translation, although the molecular mechanisms underlying these events are not completely understood [197,198,199,200,201,202,203]. Formation of G4 structures at the exon-intron boundaries or near to polyadenylation signals of pre-mRNAs may affect either positively or negatively the binding of regulatory proteins, ultimately leading to the expression of alternative protein isoforms [204,205,206,207]. Similarly, G4 structures within mRNAs may act as translational recoding signals, forcing the ribosome to shift one ribonucleotide backward with respect to the former open reading frame, thereby decoding a distinct protein [208,209].
Another exquisitely epigenetic function accomplished by RNA G4s pertains the transport of mRNAs to subcellular compartments, which allows localized protein synthesis. In particular, G4 structures located at the 3′-UTR represent a very common and effective dendritic localization signal of several neuronal mRNAs that travel in a centrifugal direction along axons. G4 structure stability being differentially influenced by monovalent cations, it has been proposed that sudden variation of intracellular K+ and Na+ concentrations occurring during neuronal activity could govern G4 folding/unfolding, thereby controlling protein-dependent transport of these dendritic mRNAs and translation at their subcellular destination [210,211,212,213,214].
G4 structures have also been mapped in short and long noncoding RNAs, although their biological implication is still unclear in most circumstances [215,216]. Bioinformatics analysis revealed that about 16% of human microRNA precursors present G4s either in the passenger or guide strand [217]. Overall, these G4s could exist in equilibrium with stem-loop secondary structures typically formed by pre-miRNAs, thereby playing a role in microRNA biogenesis by modulation of Dicer cleavage [217,218,219]. In the case of pre-miR-26a-1, G4 unwinding achieved by the RNA helicase DHX36 favors the physiological accumulation of mature miR-26a, and impairing DHX36 activity leads to low miR-26a abundance and obesity [220]. It has also been reported that G4 structures are retained in a number of biologically relevant human microRNAs in their mature form, suggesting that G4 formation could impact microRNA-directed post-transcriptional regulation of gene expression by affecting microRNA-target mRNA interactions [221].
Recent works have also shown that RNA G4s are involved in the regulation of transposable genetic elements mobility. Indeed, human retrotransposon remnants belonging to the LINE-1 family harbor distinct types of G4-forming sequences in their 3′UTR and there is a positive correlation between G4 structure stability and LINE-1 mobilization activity [222]. On the other hand, separate findings have shown that piR-48164, belonging to the PIWI-interacting family of human small noncoding RNAs, similarly bears a stable G4 structure [223]. Once formed, such an RNA G4 is detrimental to PIWI binding and prevents the piR-48164-directed transposon control at the transcriptional and post-transcriptional level [223].
In addition to these mechanisms occurring on RNA, multiple studies have suggested that G4 formation in long noncoding RNAs affect epigenetic processes on DNA, as demonstrated for G4s at telomeric repeat-containing RNAs (TERRAs) [224,225]. Indeed, it was noted that G4 structures formed by TERRAs specifically recruit the histone H3K4-specific demethylase LSD1, which is associated with telomeric heterochromatinization [151].
4. Conclusions and Perspectives
G4 structures have been extensively studied both in silico and in vitro, and in recent years have witnessed a considerable increase in experimental data, substantiating the versatile epigenetic roles of these unconventional structural motifs within cells. In a nutshell, G4s are widespread throughout the genome and transcriptome of various organisms, where they overall influence DNA and histone modifications, nucleosome positioning, and three-dimensional organization of chromatin, as well as post-transcriptional modulation of gene expression. Therefore, it could be argued that context-specific formation of G4s in the genome might direct when and where epigenetic modifications are imposed. On the other hand, the concerted balance between G4 formation/disruption controlled by helicases during DNA replication aids in preserving epigenetic memory. Unfortunately, whether and how G4-mediated epigenetic marks imposed during post-replication are linked to the next round of DNA duplication remains to be confirmed. Similarly, another challenging issue so far unexplored pertains to the potential relationship between G4 and transgenerational inheritance of epigenetic information via the germline of organisms with sexual reproduction.
As discussed, G4 structures are recognized by a plethora of trans-acting factors, which can modulate their stability or serve as a scaffold for the recruitment of further epigenetic effectors. These findings, coupled with the insights linking G4 deregulation to numerous human diseases, suggest promising therapeutic interventions. In this respect, several classes of small molecule ligands have been described to induce G4 conformations and modulate G4–protein interactions. However, further investigations are required to overcome nonspecific binding events of G4 ligands to nucleic acid structures other than G4s. A better understanding of the exact targets of these ligands will potentially improve bona fide patient-specific clinical applications. In parallel, the multitude of G4-interacting proteins could be used to design novel drug molecules. Most probably, the combined use of ligands directed against G4 structures and G4-interacting proteins could be even more effective and specific for therapeutic purposes. However, the epigenetic effects of all these known and potential molecules remain to be confirmed in physiologically relevant settings. From this standpoint, an advancement of in vivo studies implementing suitable animal models is absolutely required for a comprehensive understanding of biological roles played by G4s in both normal and pathological contexts.
Author Contributions
V.C. was the major contributor in reviewing the literature, conceiving and writing the manuscript. C.R. participated in writing during the final stage of manuscript preparation. All authors have read and agreed to the final version of the manuscript.
Funding
The APC was partly funded by PhD grants from the University of Palermo.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study or the writing of the manuscript.
Abbreviations
| 5meC | C5-methyl-deoxycitosine |
| 6meA | N6-methyl-deoxyadenosine |
| 6meG | O6-methyl-deoxyguanine |
| 8oxoG | 8-oxo-7,8-dihydro-guanine |
| APE1 | apurinic-apyrimidinic endonuclease 1 |
| ASXL1 | Additional Sex combs-Like 1 |
| ATRX | ATP- dependent X-linked helicase |
| BRD3 | Bromodomain Containing 3 |
| CTCF | CCCTC-binding Factor |
| DAXX | Death Domain Associated Protein |
| DHX36 | DEAH-Box Helicase 36 |
| DNMT | DNA methyltransferase |
| FACT | Facilitates Chromatin Transcription |
| FANCJ | Fanconi Anemia group J |
| FUS | Fused in Sarcoma protein |
| G4 | G-quadruplex |
| HIF1-α | Hypoxia-inducible factor 1-α |
| HMGB1 | Chromosomal High Mobility Group Box 1 |
| hTERT | human Telomerase Reverse Transcriptase |
| KMT5C | Histone-lysine N-Methyltransferase KMT5C |
| LINE-1 | Long Interspersed Nuclear Element-1 |
| LSD1 | Lysine Specific Demethylase 1 |
| lysC | lysozyme C |
| NuRD | Nucleosome Remodeling and Deacetylase complex |
| OGG1 | 8-oxoguanine DNA glycosylase |
| PADi4 | Peptidylarginine Deiminase 4 |
| PARP1 | Poly ADP-Ribose Polymerase 1 |
| PHF8 | Histone Lysine Demethylase 8 |
| PRC2 | Polycomb Repressive Complex 2 |
| REST | RE1-Silencing Transcription Factor |
| SWI/SNF | SWItch/Sucrose Non-Fermentable |
| TERRA | Telomeric Repeat-containing RNA |
| TLS | Translocated in Liposarcoma protein |
| UTR | Untranslated Region |
| VEGF | Vascular Endothelial Growth Factor |
References
- Levene, P.A.; Jacobs, W.A. Über Guanylsäure. Ber. Dtsch. Chem. Ges. 1909, 42, 2469–2473. [Google Scholar] [CrossRef][Green Version]
- Bang, I. Untersuchungen über die Guanylsäure. Biochem. Z. 1910, 26, 293–311. [Google Scholar]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef]
- Arnott, S.; Chandrasekaran, R.; Marttila, C.M. Structures for polyinosinic acid and polyguanylic acid. Biochem. J. 1974, 141, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.B.; Cohen, G.H.; Davies, D.R. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J. Mol. Biol. 1975, 92, 181–192. [Google Scholar] [CrossRef]
- Howard, F.B.; Frazier, J.; Miles, H.T. Stable and metastable forms of poly (G). Biopolymers 1977, 16, 791–809. [Google Scholar] [CrossRef]
- Besnard, E.; Babled, A.; Lapasset, L.; Milhavet, O.; Parrinello, H.; Dantec, C.; Marin, J.M.; Lemaitre, J.M. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012, 19, 837–844. [Google Scholar] [CrossRef]
- Fernando, H.; Sewitz, S.; Darot, J.; Tavare, S.; Huppert, J.L.; Balasubramanian, S. Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 2009, 37, 6716–6722. [Google Scholar] [CrossRef]
- Paeschke, K.; Simonsson, T.; Postberg, J.; Lipps, H.J. Telomere end-binding proteins control the formation of G-quadruplex DNA structures in vivo. Nat. Struct. Mol. Biol. 2005, 12, 847–854. [Google Scholar] [CrossRef]
- Cavalieri, V.; Di Bernardo, M.; Anello, L.; Spinelli, G. Cis-Regulatory sequences driving the expression of the Hbox12 homeobox-containing gene in the presumptive aboral ectoderm territory of the Paracentrotus lividus sea urchin embryo. Dev. Biol. 2008, 321, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Turturici, G.; La Fiora, V.; Terenzi, A.; Barone, G.; Cavalieri, V. Perturbation of developmental regulatory gene expression by a G-quadruplex DNA inducer in the Sea Urchin embryo. Biochemistry 2018, 57, 4391–4394. [Google Scholar] [CrossRef] [PubMed]
- Ribeyre, C.; Lopes, J.; Boule, J.B.; Piazza, A.; Guedin, A.; Zakian, V.A.; Mergny, J.L.; Nicolas, A. The Yeast Pif1 helicase prevents genomic instability caused by G-Quadruplex-Forming CEB1 sequences in vivo. PLoS Genet. 2009, 5, e1000475. [Google Scholar] [CrossRef] [PubMed]
- Sarkies, P.; Reams, C.; Simpson, L.J.; Sale, J.E. Epigenetic instability due to defective replication of structured DNA. Mol. Cell 2010, 40, 703–713. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef]
- Valton, A.L.; Hassan-Zadeh, V.; Lema, I.; Boggetto, N.; Alberti, P.; Saintome, C.; Riou, J.F.; Prioleau, M.N. G4 motifs affect origin positioning and efficiency in two vertebrate replicators. EMBO J. 2014, 33, 732–746. [Google Scholar] [CrossRef] [PubMed]
- Wieland, M.; Hartig, J.S. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007, 14, 757–763. [Google Scholar] [CrossRef]
- Sen, D.; Gilbert, W. Formtaion of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, 364–366. [Google Scholar] [CrossRef]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef]
- Webba da Silva, M. Geometric formalism for DNA quadruplex folding. Chemistry 2007, 13, 9738–9745. [Google Scholar] [CrossRef]
- Huppert, J.L. Structure, location and interactions of G-quadruplexes. FEBS J. 2010, 277, 3452–3458. [Google Scholar] [CrossRef]
- Mergny, J.L.; Sen, D. DNA Quadruple helices in nanotechnology. Chem. Rev. 2019, 119, 6290–6325. [Google Scholar] [CrossRef] [PubMed]
- Neidle, S.; Balasubramanian, S. Quadruplex Nucleic Acids; Royal Society of Chemistry: London, UK, 2006. [Google Scholar]
- Huppert, J.L. Four-stranded nucleic acids: Structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008, 37, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, D.; Mirihana Arachchilage, G.; Basu, S. metal cations in G-quadruplex folding and stability. Front. Chem. 2016, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Mergny, J.L.; Gabelica, V. Role of Alkali Metal Ions in G-Quadruplex Nucleic Acid Structure and Stability. Met. Ions Life Sci. 2016, 16, 203–258. [Google Scholar] [CrossRef]
- Tran, P.L.; De Cian, A.; Gros, J.; Moriyama, R.; Mergny, J.L. Tetramolecular quadruplex stability and assembly. Top. Curr. Chem. 2013, 330, 243–273. [Google Scholar] [CrossRef]
- Mukundan, V.T.; Phan, A.T. Bulges in G-quadruplexes: Broadening the definition of G-quadruplex-forming sequences. J. Am. Chem. Soc. 2013, 135, 5017–5028. [Google Scholar] [CrossRef]
- Kolesnikova, S.; Curtis, E.A. Structure and Function of Multimeric G-Quadruplexes. Molecules 2019, 24, 3074. [Google Scholar] [CrossRef]
- Zhou, J.; Amrane, S.; Korkut, D.N.; Bourdoncle, A.; He, H.Z.; Ma, D.L.; Mergny, J.L. Combination of i-motif and G-quadruplex structures within the same strand: Formation and application. Angew. Chem. 2013, 52, 7742–7746. [Google Scholar] [CrossRef]
- Zheng, K.W.; Xiao, S.; Liu, J.Q.; Zhang, J.Y.; Hao, Y.H.; Tan, Z. Co-transcriptional formation of DNA:RNA hybrid G-quadruplex and potential function as constitutional cis element for transcription control. Nucleic Acids Res. 2013, 41, 5533–5541. [Google Scholar] [CrossRef]
- Bose, K.; Lech, C.J.; Heddi, B.; Phan, A.T. High-resolution AFM structure of DNA G-wires in aqueous solution. Nat. Commun. 2018, 9, 1959. [Google Scholar] [CrossRef]
- Sondermann, A.; Holste, C.; Moller, R.; Fritzsche, W. Assembly of G-quartet based DNA superstructures (G-wires). AIP Conf. Proc. 2002, 640, 93–98. [Google Scholar]
- Guedin, A.; Gros, J.; Alberti, P.; Mergny, J.L. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010, 38, 7858–7868. [Google Scholar] [CrossRef]
- Lightfoot, H.L.; Hagen, T.; Tatum, N.J.; Hall, J. The diverse structural landscape of quadruplexes. FEBS Lett. 2019, 593, 2083–2102. [Google Scholar] [CrossRef] [PubMed]
- Rachwal, P.A.; Findlow, I.S.; Werner, J.M.; Brown, T.; Fox, K.R. Intramolecular DNA quadruplexes with different arrangements of short and long loops. Nucleic Acids Res. 2007, 35, 4214–4222. [Google Scholar] [CrossRef] [PubMed]
- Simonsson, T. G-quadruplex DNA structures—Variations on a theme. Biol. Chem. 2001, 382, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Wlodarczyk, A.; Grzybowski, P.; Patkowski, A.; Dobek, A. Effect of ions on the polymorphism, effective charge, and stability of human telomeric DNA. Photon correlation spectroscopy and circular dichroism studies. J. Phys. Chem. B 2005, 109, 3594–3605. [Google Scholar] [CrossRef] [PubMed]
- Saccà, B.; Lacroix, L.; Mergny, J.L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005, 33, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.L.; Virgilio, A.; Esposito, V.; Citarella, G.; Mergny, J.L.; Galeone, A. Effects of 8-methylguanine on structure, stability and kinetics of formation of tetramolecular quadruplexes. Biochimie 2011, 93, 399–408. [Google Scholar] [CrossRef]
- Virgilio, A.; Esposito, V.; Citarella, G.; Pepe, A.; Mayol, L.; Galeone, A. The insertion of two 8-methyl-2′-deoxyguanosine residues in tetramolecular quadruplex structures: Trying to orientate the strands. Nucleic Acids Res. 2012, 40, 461–475. [Google Scholar] [CrossRef]
- Yan, Y.Y.; Tan, J.H.; Lu, Y.J.; Yan, S.C.; Wong, K.Y.; Li, D.; Gu, L.Q.; Huang, Z.S. G-Quadruplex conformational change driven by pH variation with potential application as a nanoswitch. Biochim. et Biophys. Acta 2013, 1830, 4935–4942. [Google Scholar] [CrossRef]
- Miyoshi, D.; Nakao, A.; Sugimoto, N. Molecular crowding regulates the structural switch of the DNA G-quadruplex. Biochemistry 2002, 41, 15017–15024. [Google Scholar] [CrossRef] [PubMed]
- Borgognone, M.; Armas, P.; Calcaterra, N.B. Cellular nucleic-acid-binding protein, a transcriptional enhancer of c-Myc, promotes the formation of parallel G-quadruplexes. Biochem. J. 2010, 428, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, A.; Endo, M.; Hidaka, K.; Tran, P.L.; Mergny, J.L.; Gorelick, R.J.; Sugiyama, H. HIV-1 nucleocapsid proteins as molecular chaperones for tetramolecular antiparallel G-quadruplex formation. J. Am. Chem. Soc. 2013, 135, 18575–18585. [Google Scholar] [CrossRef] [PubMed]
- Amrane, S.; Kerkour, A.; Bedrat, A.; Vialet, B.; Andreola, M.L.; Mergny, J.L. Topology of a DNA G-quadruplex structure formed in the HIV-1 promoter: A potential target for anti-HIV drug development. J. Am. Chem. Soc. 2014, 136, 5249–5252. [Google Scholar] [CrossRef]
- Bhartiya, D.; Chawla, V.; Ghosh, S.; Shankar, R.; Kumar, N. Genome-wide regulatory dynamics of G-quadruplexes in human malaria parasite Plasmodium falciparum. Genomics 2016, 108, 224–231. [Google Scholar] [CrossRef]
- Biswas, B.; Kandpal, M.; Jauhari, U.K.; Vivekanandan, P. Genome-wide analysis of G-quadruplexes in herpesvirus genomes. BMC Genom. 2016, 17, 949. [Google Scholar] [CrossRef]
- Chambers, V.S.; Marsico, G.; Boutell, J.M.; Di Antonio, M.; Smith, G.P.; Balasubramanian, S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015, 33, 877–881. [Google Scholar] [CrossRef]
- Fleming, A.M.; Ding, Y.; Alenko, A.; Burrows, C.J. Zika virus genomic RNA possesses conserved G-quadruplexes characteristic of the flaviviridae family. ACS Infect. Dis. 2016, 2, 674–681. [Google Scholar] [CrossRef]
- Garg, R.; Aggarwal, J.; Thakkar, B. Genome-wide discovery of G-quadruplex forming sequences and their functional relevance in plants. Sci. Rep. 2016, 6, 28211. [Google Scholar] [CrossRef]
- Hershman, S.G.; Chen, Q.; Lee, J.Y.; Kozak, M.L.; Yue, P.; Wang, L.S.; Johnson, F.B. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008, 36, 144–156. [Google Scholar] [CrossRef]
- Huppert, J.L.; Balasubramanian, S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005, 33, 2908–2916. [Google Scholar] [CrossRef]
- Perrone, R.; Nadai, M.; Frasson, I.; Poe, J.A.; Butovskaja, E.; Smithgall, T.E.; Palumbo, M.; Palù, G.; Richter, S.N. A Dynamic G-quadruplex region regulates the HIV-1 long terminal repeat promoter. J. Med. Chem. 2013, 56, 6521–6530. [Google Scholar] [CrossRef] [PubMed]
- Rawal, P.; Kummarasetti, V.B.; Ravindran, J.; Kumar, N.; Halder, K.; Sharma, R.; Mukerji, M.; Das, S.K.; Chowdhury, S. Genome-wide prediction of G4 DNA as regulatory motifs: Role in Escherichia coli global regulation. Genome Res. 2006, 16, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.; Guédin, A.; Amor, S.; Bedrat, A.; Tourasse, N.J.; Fayyad-Kazan, H.; Pratviel, G.; Lacroix, L.; Mergny, J.L. Mapping and characterization of G-quadruplexes in the genome of the social amoeba Dictyostelium discoideum. Nucleic Acids Res. 2019, 47, 4363–4374. [Google Scholar] [CrossRef] [PubMed]
- Capra, J.A.; Paeschke, K.; Singh, M.; Zakian, V.A. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 2010, 6, e1000861. [Google Scholar] [CrossRef]
- Marsico, G.; Chambers, V.S.; Sahakyan, A.B.; McCauley, P.; Boutell, J.M.; Antonio, M.D.; Balasubramanian, S. Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 2019, 47, 3862–3874. [Google Scholar] [CrossRef] [PubMed]
- Puig Lombardi, E.; Holmes, A.; Verga, D.; Teulade-Fichou, M.P.; Nicolas, A.; Londoño-Vallejo, A. Thermodynamically stable and genetically unstable G-quadruplexes are depleted in genomes across species. Nucleic Acids Res. 2019, 47, 6098–6113. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Halder, K.; Halder, R.; Yadav, V.K.; Rawal, P.; Thakur, R.K.; Mohd, F.; Sharma, A.; Chowdhury, S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008, 51, 5641–5649. [Google Scholar] [CrossRef]
- Ravichandran, S.; Kim, Y.E.; Bansal, V.; Ghosh, A.; Hur, J.; Subramani, V.K.; Pradhan, S.; Lee, M.K.; Kim, K.K.; Ahn, J.H. Genome-wide analysis of regulatory G-quadruplexes affecting gene expression in human cytomegalovirus. PLoS Pathog. 2018, 14, e1007334. [Google Scholar] [CrossRef]
- Fleming, A.M.; Zhu, J.; Ding, Y.; Visser, J.A.; Zhu, J.; Burrows, C.J. Human DNA repair genes possess potential G-quadruplex sequences in their promoters and 5′-untranslated regions. Biochemistry 2018, 57, 991–1002. [Google Scholar] [CrossRef]
- Hansel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Maizels, N.; Gray, L.T. The G4 genome. PLoS Genet. 2013, 9, e1003468. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Adrian, M.; Samazan, F.; Heddi, B.; Hamon, F.; Serero, A.; Lopes, J.; Teulade-Fichou, M.P.; Phan, A.T.; Nicolas, A. Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. EMBO J. 2015, 34, 1718–1734. [Google Scholar] [CrossRef]
- Yadav, V.K.; Abraham, J.K.; Mani, P.; Kulshrestha, R.; Chowdhury, S. QuadBase: Genome-wide database of G4 DNAeoccurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008, 36, D381–D385. [Google Scholar] [CrossRef]
- Artusi, S.; Perrone, R.; Lago, S.; Raffa, P.; Di Iorio, E.; Palù, G.; Richter, S.N. Visualization of DNA G-quadruplexes in herpes simplex virus 1-infected cells. Nucleic Acids Res. 2016, 44, 10343–10353. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Henderson, A.; Wu, Y.; Huang, Y.C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M., Jr.; Sen, D.; Lansdorp, P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014, 42, 860–869. [Google Scholar] [CrossRef]
- Schaffitzel, C.; Berger, I.; Postberg, J.; Hanes, J.; Lipps, H.J.; Plückthun, A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. USA 2001, 98, 8572–8577. [Google Scholar] [CrossRef]
- Bao, H.L.; Ishizuka, T.; Sakamoto, T.; Fujimoto, K.; Uechi, T.; Kenmochi, N.; Xu, Y. Characterization of human telomere RNA G-quadruplex structures in vitro and in living cells using 19F NMR spectroscopy. Nucleic Acids Res. 2017, 45, 5501–5511. [Google Scholar] [CrossRef]
- Hänsel, R.; Foldynová-Trantírková, S.; Dötsch, V.; Trantírek, L. Investigation of quadruplex structure under physiological conditions using in-cell NMR. Top. Curr. Chem. 2013, 330, 47–65. [Google Scholar] [CrossRef]
- Chen, M.C.; Murat, P.; Abecassis, K.; Ferré-D’Amaré, A.R.; Balasubramanian, S. Insights into the mechanism of a Gquadruplex-unwinding DEAH-box helicase. Nucleic Acids Res. 2015, 43, 2223–2231. [Google Scholar] [CrossRef] [PubMed]
- Gray, L.T.; Vallur, A.C.; Eddy, J.; Maizels, N. G-quadruplexes are genomewide targets of transcriptional helicases XPB and XPD. Nat. Chem. Biol. 2014, 10, 313–318. [Google Scholar] [CrossRef] [PubMed]
- Saha, T.; Shukla, K.; Thakur, R.S.; Desingu, A.; Nagaraju, G. Mycobacterium tuberculosis UvrD1 and UvrD2 helicases unwind G-quadruplex DNA. FEBS J. 2019, 286, 2062–2086. [Google Scholar] [CrossRef]
- Tippana, R.; Chen, M.C.; Demeshkina, N.A.; Ferré-D’Amaré, A.R.; Myong, S. RNA G-quadruplex is resolved by repetitive and ATP-dependent mechanism of DHX36. Nat. Commun. 2019, 10, 1855. [Google Scholar] [CrossRef] [PubMed]
- Voter, A.F.; Qiu, Y.; Tippana, R.; Myong, S.; Keck, J.L. A guanine-flipping and sequestration mechanism for G-quadruplex unwinding by RecQ helicases. Nat. Commun. 2018, 9, 4201. [Google Scholar] [CrossRef]
- Wu, G.; Xing, Z.; Tran, E.J.; Yang, D. DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl. Acad. Sci. USA 2019, 116, 20453–20461. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zhang, J.; Bochman, M.L.; Zakian, V.A.; Ha, T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife 2014, 3, e02190. [Google Scholar] [CrossRef]
- Cavalieri, V.; Spinelli, G. Environmental epigenetics in zebrafish. Epigenet. Chromatin 2017, 10, 46. [Google Scholar] [CrossRef]
- Cavalieri, V.; Spinelli, G. Histone-mediated transgenerational epigenetics. In Translational Epigenetics, 2nd ed.; Academic Press: Cambridge, MA, USA, 2007; pp. 157–183. [Google Scholar] [CrossRef]
- Halder, R.; Riou, J.F.; Teulade-Fichou, M.P.; Frickey, T.; Hartig, J.S. Bisquinolinium compounds induce quadruplex-specific transcriptome changes in HeLa S3 cell lines. BMC Res. Notes 2012, 5, 138. [Google Scholar] [CrossRef]
- Mikami-Terao, Y.; Akiyama, M.; Yuza, Y.; Yanagisawa, T.; Yamada, O.; Yamada, H. Antitumor activity of G-quadruplex-interactive agent TMPyP4 in K562 leukemic cells. Cancer Lett. 2008, 261, 226–234. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Chakraborty, S.; Sengupta, P.K.; Bhowmik, S. Exploring the interactions of the dietary plant flavonoids fisetin and naringenin with G-quadruplex and duplex DNA, showing contrasting binding behavior: Spectroscopic and molecular modeling approaches. J. Phys. Chem. B 2016, 120, 8942–8952. [Google Scholar] [CrossRef] [PubMed]
- François, M.; Leifert, W.; Tellam, R.; Fenech, M. G-quadruplexes: A possible epigenetic target for nutrition. Mutat. Res. Rev. Mutat. Res. 2015, 764, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, M.; Zhang, J.L.; Li, H.Q.; Zhang, X.C.; Sun, Q.; Qiu, C.M. Interactions of daidzin with intramolecular G-quadruplex. FEBS Lett. 2006, 580, 4905–4910. [Google Scholar] [CrossRef] [PubMed]
- Mikutis, G.; Karaköse, H.; Jaiswal, R.; LeGresley, A.; Islam, T.; Fernandez-Lahore, M.; Kuhnert, N. Phenolic promiscuity in the cell nucleus—Epigallocatechingallate (EGCG) and theaflavin-3,30-digallate from green and black tea bind to model cell nuclear structures including histone proteins, double stranded DNA and telomeric quadruplex DNA. Food Funct. 2013, 4, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Pattanayak, R.; Basak, P.; Sen, S.; Bhattacharyya, M. Interaction of KRAS G-quadruplex with natural polyphenols: A spectroscopic analysis with molecular modeling. Int. J. Biol. Macromol. 2016, 89, 228–237. [Google Scholar] [CrossRef]
- Platella, C.; Guida, S.; Bonmassar, L.; Aquino, A.; Bonmassar, E.; Ravagnan, G.; Montesarchio, D.; Roviello, G.N.; Musumeci, D.; Fuggetta, M.P. Antitumour activity of resveratrol on human melanoma cells: A possible mechanism related to its interaction with malignant cell telomerase. Biochim. et Biophys. Acta Gen. Subj. 2017, 1861, 2843–2851. [Google Scholar] [CrossRef]
- Benabou, S.; Mazzini, S.; Avino, A.; Eritja, R.; Gargallo, R.A. pH-dependent bolt involving cytosine bases located in the lateral loops of antiparallel G-quadruplex structures within the SMARCA4 gene promotor. Sci. Rep. 2019, 9, 15807. [Google Scholar] [CrossRef]
- Hardin, C.C.; Corregan, M.; Brown, B.A., II.; Frederick, L.N. Cytosine-cytosine+ base pairing stabilizes DNA quadruplexes and cytosine methylation greatly enhances the effect. Biochemistry 1993, 32, 5870–5880. [Google Scholar] [CrossRef]
- Kuo, M.H.; Wang, Z.F.; Tseng, T.Y.; Li, M.H.; Hsu, S.T.; Lin, J.J.; Chang, T.C. Conformational transition of a hairpin structure to G-quadruplex within the WNT1 gene promoter. J. Am. Chem. Soc. 2015, 137, 210–218. [Google Scholar] [CrossRef]
- Stevens, A.J.; Stuffrein-Roberts, S.; Cree, S.L.; Gibb, A.; Miller, A.L.; Doudney, K.; Aitchison, A.; Eccles, M.R.; Joyce, P.R.; Filichev, V.V.; et al. G-quadruplex structures and CpG methylation cause drop-out of the maternal allele in polymerase chain reaction amplification of the imprinted MEST gene promoter. PLoS ONE 2014, 9, e113955. [Google Scholar] [CrossRef]
- Halder, R.; Halder, K.; Sharma, P.; Garg, G.; Sengupta, S.; Chowdhury, S. Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol. Biosyst. 2010, 6, 2439–2447. [Google Scholar] [CrossRef] [PubMed]
- Jara-Espejo, M.; Line, S.R. DNA G-quadruplex stability, position and chromatin accessibility are associated with CpG island methylation. FEBS J. 2020, 287, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.Q.; Ghanbarian, A.T.; Spiegel, J.; Martínez Cuesta, S.; Beraldi, D.; Di Antonio, M.; Marsico, G.; Hänsel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. DNA G-quadruplex structures mold the DNA methylome. Nat. Struct. Mol. Biol. 2018, 25, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Cree, S.L.; Fredericks, R.; Miller, A.; Pearce, F.G.; Filichev, V.; Fee, C.; Kennedy, M.A. DNA G-quadruplexes show strong interaction with DNA methyltransferases in vitro. FEBS Lett. 2016, 590, 2870–2883. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Hurley, L.H.; Neidle, S. Targeting G-quadruplexes in gene promoters: A novel anticancer strategy? Nat. Rev. Drug Discov. 2011, 10, 261–275. [Google Scholar] [CrossRef]
- Renaud, S.; Loukinov, D.; Abdullaev, Z.; Guilleret, I.; Bosman, F.T.; Lobanenkov, V.; Benhattar, J. Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 2007, 35, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Li, P.T.; Wang, Z.F.; Chu, I.T.; Kuan, Y.M.; Li, M.H.; Huang, M.C.; Chiang, P.C.; Chang, T.C.; Chen, C.T. Expression of the human telomerase reverse transcriptase gene is modulated by quadruplex formation in its first exon due to DNA methylation. J. Biol. Chem. 2017, 292, 20859–20870. [Google Scholar] [CrossRef] [PubMed]
- Renaud, S.; Loukinov, D.; Bosman, F.T.; Lobanenkov, V.; Benhattar, J. CTCF binds the proximal exonic region of hTERT and inhibits its transcription. Nucleic Acids Res. 2005, 33, 6850–6860. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Y. TET-mediated active DNA demethylation: Mechanism, function and beyond. Nat. Rev. Genet. 2017, 18, 517–534. [Google Scholar] [CrossRef]
- Pastor, W.A.; Pape, U.J.; Huang, Y.; Henderson, H.R.; Lister, R.; Ko, M.; McLoughlin, E.M.; Brudno, Y.; Mahapatra, S.; Kapranov, P.; et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature 2011, 473, 394–397. [Google Scholar] [CrossRef] [PubMed]
- Song, C.X.; Szulwach, K.E.; Fu, Y.; Dai, Q.; Yi, C.; Li, X.; Li, Y.; Chen, C.H.; Zhang, W.; Jian, X.; et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011, 29, 68–72. [Google Scholar] [CrossRef] [PubMed]
- Valinluck, V.; Sowers, L.C. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007, 67, 946–950. [Google Scholar] [CrossRef] [PubMed]
- Bhavsar-Jog, Y.P.; Van Dornshuld, E.; Brooks, T.A.; Tschumper, G.S.; Wadkins, R.M. Epigenetic modification, dehydration, and molecular crowding effects on the thermodynamics of i-motif structure formation from C-rich DNA. Biochemistry 2014, 53, 1586–1594. [Google Scholar] [CrossRef]
- Morgan, R.K.; Molnar, M.M.; Batra, H.; Summerford, B.; Wadkins, R.M.; Brooks, T.A. Effects of 5-Hydroxymethylcytosine epigenetic modification on the stability and molecular recognition of VEGF i-motif and G-quadruplex structures. J. Nucleic Acids 2018, 2018, 9281286. [Google Scholar] [CrossRef]
- Spruijt, C.G.; Gnerlich, F.; Smits, A.H.; Pfaffeneder, T.; Jansen, P.W.; Bauer, C.; Münzel, M.; Wagner, M.; Müller, M.; Khan, F.; et al. Dynamic readers for 5-(hydroxy) methylcytosine and its oxidized derivatives. Cell 2013, 152, 1146–1159. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Guo, K.; Shin, Y.J. Evidence of the formation of G-quadruplex structures in the promoter region of the human Vascular Endothelial Growth Factor Gene. Nucleic Acids Res. 2011, 39, 1256–1265. [Google Scholar] [CrossRef]
- Tsukakoshi, K.; Saito, S.; Yoshida, W.; Goto, S.; Ikebukuro, K. CpG methylation changes G-quadruplex structures derived from gene promoters and interaction with VEGF and SP1. Molecules 2018, 23, 944. [Google Scholar] [CrossRef]
- Yu, Y.; Cui, Y.; Niedernhofer, L.J.; Wang, Y. Occurrence, biological consequences, and human health relevance of oxidative stress-induced DNA damage. Chem. Res. Toxicol. 2016, 29, 2008–2039. [Google Scholar] [CrossRef]
- Saito, I.; Takayama, M.; Sugiyama, H.; Nakatani, K.; Tsuchida, A.; Yamamoto, M. Photoinduced DNA cleavage via electron transfer: Demonstration that guanine residues located 5′ to guanine are the most electron-donating sites. J. Am. Chem. Soc. 1995, 117, 6406–6407. [Google Scholar] [CrossRef]
- Steenken, S.; Jovanovic, S.V. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997, 119, 617–618. [Google Scholar] [CrossRef]
- Pfaffeneder, T.; Spada, F.; Wagner, M.; Brandmayr, C.; Laube, S.K.; Eisen, D.; Truss, M.; Steinbacher, J.; Hackner, B.; Kotljarova, O.; et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat. Chem. Biol. 2014, 10, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Bielskutė, S.; Plavec, J.; Podbevšek, P. Impact of Oxidative Lesions on the Human Telomeric G-Quadruplex. J. Am. Chem. Soc. 2019, 141, 2594–2603. [Google Scholar] [CrossRef]
- Lech, C.J.; Cheow Lim, J.K.; Wen Lim, J.M.; Amrane, S.; Heddi, B.; Phan, A.T. Effects of site-specific guanine C8-modifications on an intramolecular DNA G-quadruplex. Biophys. J. 2011, 101, 1987–1998. [Google Scholar] [CrossRef]
- An, N.; Fleming, A.M.; Burrows, C.J. Human Telomere G-Quadruplexes with five repeats accommodate 8-Oxo-7,8-dihydroguanine by looping out the DNA damage. ACS Chem. Biol. 2016, 11, 500–507. [Google Scholar] [CrossRef]
- Fleming, A.M.; Zhou, J.; Wallace, S.S.; Burrows, C.J. A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these “spare tires” have an evolved function? ACS Cent. Sci. 2015, 1, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.; Qi, T.; Pan, L.; Wang, R.; Zhu, B.; Aguilera-Aguirre, L.; Radak, Z.; Hazra, T.K.; Vlahopoulos, S.A.; Bacsi, A.; et al. Effects of the stimuli-dependent enrichment of 8-oxoguanine DNA glycosylase1 on chromatinized DNA. Redox Biol. 2018, 18, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Zhu, B.; Hao, W.; Zeng, X.; Vlahopoulos, S.A.; Hazra, T.K.; Hegde, M.L.; Radak, Z.; Bacsi, A.; Brasier, A.R.; et al. Oxidized guanine base lesions function in 8-Oxoguanine DNA Glycosylase-1-mediated epigenetic regulation of nuclear factor κb-driven gene expression. J. Biol. Chem. 2016, 291, 25553–25566. [Google Scholar] [CrossRef]
- Antoniali, G.; Lirussi, L.; D’Ambrosio, C.; Dal Piaz, F.; Vascotto, C.; Casarano, E.; Marasco, D.; Scaloni, A.; Fogolari, F.; Tell, G. SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol. Biol. Cell 2014, 25, 532–547. [Google Scholar] [CrossRef]
- Cogoi, S.; Xodo, L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006, 34, 2536–2549. [Google Scholar] [CrossRef]
- Cogoi, S.; Ferino, A.; Miglietta, G.; Pedersen, E.B.; Xodo, L.E. The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: Implications on transcription. Nucleic Acids Res. 2018, 46, 661–676. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Ding, Y.; Burrows, C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. USA 2017, 114, 2604–2609. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Zhu, J.; Howpay Manage, S.A.; Burrows, C.J. Human NEIL3 Gene expression regulated by epigenetic-like oxidative DNA modification. J. Am. Chem. Soc. 2019, 141, 11036–11049. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Burrows, C.J. Interplay of guanine oxidation and G-quadruplex folding in gene promoters. J. Am. Chem. Soc. 2020, 142, 1115–1136. [Google Scholar] [CrossRef]
- Pegg, A.E.; Dolan, M.E.; Moschel, R.C. Structure, function, and inhibition of O6-alkylguanine-DNA alkyltransferase. Prog. Nucleic Acid Res. Mol. Biol. 1995, 51, 167–223. [Google Scholar] [CrossRef]
- Mekmaysy, C.S.; Petraccone, L.; Garbett, N.C.; Ragazzon, P.A.; Gray, R.; Trent, J.O.; Chaires, J.B. Effect of O6-methylguanine on the stability of G-quadruplex DNA. J. Am. Chem. Soc. 2008, 130, 6710–6711. [Google Scholar] [CrossRef]
- Engel, J.D.; von Hippel, P.H. Effects of methylation on the stability of nucleic acid conformations: Studies at the monomer level. Biochemistry 1974, 13, 4143–4158. [Google Scholar] [CrossRef]
- Engel, J.D.; von Hippel, P.H. Effects of methylation on the stability of nucleic acid conformations. Studies at the polymer level. J. Biol. Chem. 1978, 253, 927–934. [Google Scholar]
- Laddachote, S.; Nagata, M.; Yoshida, W. Destabilisation of the c-kit1 G-quadruplex structure by N6-methyladenosine modification. Biochem. Biophys. Res. Commun. 2020, 524, 472–476. [Google Scholar] [CrossRef]
- Phan, A.T.; Kuryavyi, V.; Burge, S.; Neidle, S.; Patel, D.J. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007, 129, 4386–4392. [Google Scholar] [CrossRef]
- Koziol, M.J.; Bradshaw, C.R.; Allen, G.E.; Costa, A.S.H.; Frezza, C.; Gurdon, J.B. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat. Struct. Mol. Biol. 2016, 23, 24–30. [Google Scholar] [CrossRef]
- Cavalieri, V.; Melfi, R.; Spinelli, G. Promoter activity of the sea urchin (Paracentrotus lividus) nucleosomal H3 and H2A and linker H1 α-histone genes is modulated by enhancer and chromatin insulator. Nucleic Acids Res. 2009, 37, 7407–7415. [Google Scholar] [CrossRef]
- Cavalieri, V.; Spinelli, G. Ectopic hbox12 expression evoked by histone deacetylase inhibition disrupts axial specification of the sea urchin embryo. PLoS ONE 2015, 10, e0143860. [Google Scholar] [CrossRef]
- Cavalieri, V. Histones, their variants and post-translational modifications in zebrafish development. Front. Cell Dev. Biol. 2020, 8, 456. [Google Scholar] [CrossRef]
- Di Caro, D.; Melfi, R.; Alessandro, C.; Serio, G.; Di Caro, V.; Cavalieri, V.; Palla, F.; Spinelli, G. Down-regulation of early sea urchin histone H2A gene relies on cis regulative sequences located in the 5′ and 3′ regions and including the enhancer blocker sns. J. Mol. Biol. 2004, 342, 1367–1377. [Google Scholar] [CrossRef]
- Di Caro, V.; Cavalieri, V.; Melfi, R.; Spinelli, G. Regulated sea urchin alpha-H2A histone gene. J. Mol. Biol. 2007, 365, 1285–1297. [Google Scholar] [CrossRef]
- Alabert, C.; Jasencakova, Z.; Groth, A. Chromatin replication and histone dynamics. Adv. Exp. Med. Biol. 2017, 1042, 311–333. [Google Scholar] [CrossRef]
- Hammond, C.M.; Strømme, C.B.; Huang, H.; Patel, D.J.; Groth, A. Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 2017, 18, 141–158. [Google Scholar] [CrossRef]
- Brosh, R.M., Jr.; Majumdar, A.; Desai, S.; Hickson, I.D.; Bohr, V.A.; Seidman, M.M. Unwinding of a DNA triple helix by the Werner and Bloom syndrome helicases. J. Biol. Chem. 2001, 276, 3024–3030. [Google Scholar] [CrossRef]
- Sale, J.E.; Lehmann, A.R.; Woodgate, R. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 2012, 13, 141–152. [Google Scholar] [CrossRef]
- Sanders, C.M. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem. J. 2010, 430, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shin-ya, K.; Brosh, R.M., Jr. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell Biol. 2008, 28, 4116–4128. [Google Scholar] [CrossRef] [PubMed]
- Jasencakova, Z.; Scharf, A.N.; Ask, K.; Corpet, A.; Imhof, A.; Almouzni, G.; Groth, A. Replication stress interferes with histone recycling and predeposition marking of new histones. Mol. Cell 2010, 37, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Sarkies, P.; Murat, P.; Phillips, L.G.; Patel, K.J.; Balasubramanian, S.; Sale, J.E. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012, 40, 1485–1498. [Google Scholar] [CrossRef] [PubMed]
- Schiavone, D.; Guilbaud, G.; Murat, P.; Papadopoulou, C.; Sarkies, P.; Prioleau, M.N.; Balasubramanian, S.; Sale, J.E. Determinants of G quadruplex-induced epigenetic instability in REV1-deficient cells. EMBO J. 2014, 33, 2507–2520. [Google Scholar] [CrossRef]
- Schwab, R.A.; Nieminuszczy, J.; Shin-ya, K.; Niedzwiedz, W. FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J. Cell Biol. 2013, 201, 33–48. [Google Scholar] [CrossRef]
- Takahama, K.; Takada, A.; Tada, S.; Shimizu, M.; Sayama, K.; Kurokawa, R.; Oyoshi, T. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem. Biol. 2013, 20, 341–350. [Google Scholar] [CrossRef]
- Hou, Y.; Li, F.; Zhang, R.; Li, S.; Liu, H.; Qin, Z.S.; Sun, X. Integrative characterization of G-Quadruplexes in the three-dimensional chromatin structure. Epigenetics 2019, 14, 894–911. [Google Scholar] [CrossRef]
- Hirschi, A.; Martin, W.J.; Luka, Z.; Loukachevitch, L.V.; Reiter, N.J. G-quadruplex RNA binding and recognition by the lysine-specific Histone Demethylase-1 Enzyme. RNA 2016, 22, 1250–1260. [Google Scholar] [CrossRef]
- Hussain, T.; Saha, D.; Purohit, G.; Kar, A.; Kishore Mukherjee, A.; Sharma, S.; Sengupta, S.; Dhapola, P.; Maji, B.; Vedagopuram, S.; et al. Transcription regulation of CDKN1A (p21/CIP1/WAF1) by TRF2 is epigenetically controlled through the REST repressor complex. Sci. Rep. 2017, 7, 11541. [Google Scholar] [CrossRef]
- Saha, D.; Singh, A.; Hussain, T.; Srivastava, V.; Sengupta, S.; Kar, A.; Dhapola, P.; Dhople, V.; Ummanni, R.; Chowdhury, S. Epigenetic suppression of human telomerase (hTERT) is mediated by the metastasis suppressor NME2 in a G-quadruplex-dependent fashion. J. Biol. Chem. 2017, 292, 15205–15215. [Google Scholar] [CrossRef] [PubMed]
- Thakur, R.K.; Kumar, P.; Halder, K.; Verma, A.; Kar, A.; Parent, J.L.; Basundra, R.; Kumar, A.; Chowdhury, S. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009, 37, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Vlasenok, M.; Levchenko, O.; Basmanov, D.; Klinov, D.; Varizhuk, A.; Pozmogova, G. Data set on G4 DNA interactions with human proteins. Data Brief 2018, 18, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Cuthbert, G.L.; Daujat, S.; Snowden, A.W.; Erdjument-Bromage, H.; Hagiwara, T.; Yamada, M.; Schneider, R.; Gregory, P.D.; Tempst, P.; Bannister, A.J.; et al. Histone deimination antagonizes arginine methylation. Cell 2004, 118, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Wahab, O.; Adli, M.; LaFave, L.M.; Gao, J.; Hricik, T.; Shih, A.H.; Pandey, S.; Patel, J.P.; Chung, Y.R.; Koche, R.; et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell 2012, 22, 180–193. [Google Scholar] [CrossRef]
- Fisher, C.L.; Berger, J.; Randazzo, F.; Brock, H.W. A human homolog of Additional sex combs, ADDITIONAL SEX COMBS-LIKE 1, maps to chromosome 20q11. Gene 2003, 306, 115–126. [Google Scholar] [CrossRef]
- Beltran, M.; Tavares, M.; Justin, N.; Khandelwal, G.; Ambrose, J.; Foster, B.M.; Worlock, K.B.; Tvardovskiy, A.; Kunzelmann, S.; Herrero, J.; et al. G-tract RNA removes Polycomb repressive complex 2 from genes. Nat. Struct. Mol. Biol. 2019, 26, 899–909. [Google Scholar] [CrossRef]
- Wang, X.; Paucek, R.D.; Gooding, A.R.; Brown, Z.Z.; Ge, E.J.; Muir, T.W.; Cech, T.R. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat. Struct. Mol. Biol. 2017, 24, 1028–1038. [Google Scholar] [CrossRef]
- Nguyen, D.T.; Voon, H.P.J.; Xella, B.; Scott, C.; Clynes, D.; Babbs, C.; Ayyub, H.; Kerry, J.; Sharpe, J.A.; Sloane-Stanley, J.A.; et al. The chromatin remodelling factor ATRX suppresses R-loops in transcribed telomeric repeats. EMBO Rep. 2017, 18, 914–928. [Google Scholar] [CrossRef]
- Udugama, M.; Chang, F.T.; Chan, F.L.; Tang, M.C.; Pickett, H.A.; McGhie, J.D.; Mayne, L.; Collas, P.; Mann, J.R.; Wong, L.H. Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res. 2015, 43, 10227–10237. [Google Scholar] [CrossRef]
- Angelov, D.; Bondarenko, V.A.; Almagro, S.; Menoni, H.; Mongélard, F.; Hans, F.; Mietton, F.; Studitsky, V.M.; Hamiche, A.; Dimitrov, S.; et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006, 25, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, L.A.; Sun, H.; Hanakahi, L.A.; Maizels, N. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, a role for G-G pairing in immunoglobulin switch recombination. J. Biol. Chem. 1999, 274, 1066–1071. [Google Scholar] [CrossRef] [PubMed]
- González, V.; Guo, K.; Hurley, L.; Sun, D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J. Biol. Chem. 2009, 284, 23622–23635. [Google Scholar] [CrossRef] [PubMed]
- Bustin, M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem. Sci. 2001, 26, 152–153. [Google Scholar] [CrossRef]
- Malarkey, C.S.; Churchill, M.E. The high mobility group box: The ultimate utility player of a cell. Trends Biochem. Sci. 2012, 37, 553–562. [Google Scholar] [CrossRef] [PubMed]
- Postnikov, Y.; Bustin, M. Regulation of chromatin structure and function by HMGN proteins. Biochim. et Biophys. Acta 2010, 1799, 62–68. [Google Scholar] [CrossRef]
- Amato, J.; Madanayake, T.W.; Iaccarino, N.; Novellino, E.; Randazzo, A.; Hurley, L.H.; Pagano, B. HMGB1 binds to the KRAS promoter G-quadruplex: A new player in oncogene transcriptional regulation? Chem. Commun. 2018, 54, 9442–9445. [Google Scholar] [CrossRef] [PubMed]
- Cogoi, S.; Paramasivam, M.; Membrino, A.; Yokoyama, K.K.; Xodo, L.E. The KRAS promoter responds to Myc-associated zinc finger and poly (ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010, 285, 22003–22016. [Google Scholar] [CrossRef]
- Muthurajan, U.M.; Hepler, M.R.; Hieb, A.R.; Clark, N.J.; Kramer, M.; Yao, T.; Luger, K. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc. Natl. Acad. Sci. USA 2014, 111, 12752–12757. [Google Scholar] [CrossRef]
- Gushchanskaya, E.S.; Artemov, A.V.; Ulyanov, S.V.; Logacheva, M.D.; Penin, A.A.; Kotova, E.S.; Akopov, S.B.; Nikolaev, L.G.; Iarovaia, O.V.; Sverdlov, E.D.; et al. The clustering of CpG islands may constitute an important determinant of the 3D organization of interphase chromosomes. Epigenetics 2014, 9, 951–963. [Google Scholar] [CrossRef][Green Version]
- Halder, K.; Halder, R.; Chowdhury, S. Genome-wide analysis predicts DNA structural motifs as nucleosome exclusion signals. Mol. Biosyst. 2009, 5, 1703–1712. [Google Scholar] [CrossRef] [PubMed]
- Kouzine, F.; Wojtowicz, D.; Baranello, L.; Yamane, A.; Nelson, S.; Resch, W.; Kieffer-Kwon, K.R.; Benham, C.J.; Casellas, R.; Przytycka, T.M.; et al. Permanganate/S1 Nuclease footprinting reveals non-b dna structures with regulatory potential across a mammalian genome. Cell Syst. 2017, 4, 344–356. [Google Scholar] [CrossRef]
- Wong, H.M.; Huppert, J.L. Stable G-quadruplexes are found outside nucleosome-bound regions. Mol. Biosyst. 2009, 5, 1713–1719. [Google Scholar] [CrossRef]
- Foulk, M.S.; Urban, J.M.; Casella, C.; Gerbi, S.A. Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res. 2015, 25, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Baiamonte, E.; Spinelli, G.; Maggio, A.; Acuto, S.; Cavalieri, V. the sea urchin sns5 chromatin insulator shapes the chromatin architecture of a lentivirus vector integrated in the mammalian genome. Nucleic Acid Ther. 2016, 26, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Cavalieri, V.; Melfi, R.; Spinelli, G. The Compass-like locus, exclusive to the Ambulacrarians, encodes a chromatin insulator binding protein in the Sea Urchin embryo. PLoS Genet. 2013, 9, e1003847. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef]
- Lee, B.K.; Iyer, V.R. Genome-wide studies of CCCTC-binding factor (CTCF) and cohesin provide insight into chromatin structure and regulation. J. Biol. Chem. 2012, 287, 30906–30913. [Google Scholar] [CrossRef]
- Ong, C.T.; Corces, V.G. CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014, 15, 234–246. [Google Scholar] [CrossRef]
- Hegyi, H. Enhancer-promoter interaction facilitated by transiently forming G-quadruplexes. Sci. Rep. 2015, 5, 9165. [Google Scholar] [CrossRef]
- Joachimi, A.; Benz, A.; Hartig, J.S. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg. Med. Chem. 2009, 17, 6811–6815. [Google Scholar] [CrossRef]
- Liu, H.; Kugimiya, A.; Sakurai, T.; Katahira, M.; Uesugi, S. A comparison of the properties and the solution structure for RNA and DNA quadruplexes which contain two GGAGG sequences joined with a tetranucleotide linker. Nucleosides Nucleotides Nucleic Acids 2002, 21, 785–801. [Google Scholar] [CrossRef] [PubMed]
- Olsen, C.M.; Marky, L.A. Energetic and hydration contributions of the removal of methyl groups from thymine to form uracil in G-quadruplexes. J. Phys. Chem. B 2009, 113, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Zaccaria, F.; Fonseca Guerra, C. RNA versus DNA G-Quadruplex: The origin of increased stability. Chemistry 2018, 24, 16315–16322. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.F.; Shafer, R.H. Engineering the quadruplex fold: Nucleoside conformation determines both folding topology and molecularity in Guanine quadruplexes. J. Am. Chem. Soc. 2006, 128, 5966–5973. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Xia, Y.; Hao, Y.H.; Tan, Z. DNA:RNA hybrid G-quadruplex formation upstream of transcription start site. Sci. Rep. 2020, 10, 7429. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, T.; Roy, S.; Kumar, S.; Chakraborty, T.K.; Maiti, S. In the sense of transcription regulation by G-quadruplexes: Asymmetric effects in sense and antisense strands. Biochemistry 2014, 53, 3711–3718. [Google Scholar] [CrossRef]
- Xiao, S.; Zhang, J.Y.; Zheng, K.W.; Hao, Y.H.; Tan, Z. Bioinformatic analysis reveals an evolutional selection for DNA: RNA hybrid G-quadruplex structures as putative transcription regulatory elements in warm-blooded animals. Nucleic Acids Res. 2013, 41, 10379–10390. [Google Scholar] [CrossRef]
- Arora, A.; Dutkiewicz, M.; Scaria, V.; Hariharan, M.; Maiti, S.; Kurreck, J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA 2008, 14, 1290–1296. [Google Scholar] [CrossRef]
- Gomez, D.; Guedin, A.; Mergny, J.L.; Salles, B.; Riou, J.F.; Teulade-Fichou, M.P.; Calsou, P.A. G-quadruplex structure within the 50-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res. 2010, 38, 7187–7198. [Google Scholar] [CrossRef]
- Kumari, S.; Bugaut, A.; Huppert, J.L.; Balasubramanian, S. An RNA G-quadruplex in the 5′-UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007, 3, 218–221. [Google Scholar] [CrossRef]
- Morris, M.J.; Basu, S. An unusually stable G-quadruplex within the 5′-UTR of the MT3 matrix metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry 2009, 48, 5313–5319. [Google Scholar] [CrossRef] [PubMed]
- Shahid, R.; Bugaut, A.; Balasubramanian, S. The BCL-2 5′ untranslated region contains an RNA G-quadruplex-forming motif that modulates protein expression. Biochemistry 2010, 49, 8300–8306. [Google Scholar] [CrossRef] [PubMed]
- Bugaut, A. Balasubramanian, S. 5′-UTR RNA G-quadruplexes: Translation regulation and targeting. Nucleic Acids Res. 2012, 40, 4727–4741. [Google Scholar] [CrossRef] [PubMed]
- Agarwala, P.; Pandey, S.; Mapa, K.; Maiti, S. The G-quadruplex augments translation in the 5’ untranslated region of transforming growth factor β2. Biochemistry 2013, 52, 1528–1538. [Google Scholar] [CrossRef] [PubMed]
- Agarwala, P.; Pandey, S.; Maiti, S. Role of G-quadruplex located at 5’ end of mRNAs. Biochim. et Biophys. Acta 2014, 1840, 3503–3510. [Google Scholar] [CrossRef]
- Agarwala, P.; Pandey, S.; Ekka, M.K.; Chakraborty, D.; Maiti, S. Combinatorial role of two G-quadruplexes in 5’ UTR of transforming growth factor β2 (TGFβ2). Biochim. et Biophys. Acta Gen. Subj. 2019, 1863, 129416. [Google Scholar] [CrossRef]
- Bonnal, S.; Schaeffer, C.; Créancier, L.; Clamens, S.; Moine, H.; Prats, A.C.; Vagner, S. A single internal ribosome entry site containing a G-quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003, 278, 39330–39336. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Diamond, P.; Basu, S. An independently folding RNA G-quadruplex domain directly recruits the 40S ribosomal subunit. Biochemistry 2015, 54, 1879–1885. [Google Scholar] [CrossRef]
- Morris, M.J.; Negishi, Y.; Pazsint, C.; Schonhoft, J.D.; Basu, S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc. 2010, 132, 17831–17839. [Google Scholar] [CrossRef]
- Reddy, K.; Zamiri, B.; Stanley, S.Y.; Macgregor, R.B., Jr.; Pearson, C.E. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 2013, 288, 9860–9866. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin, J.D.; Perreault, J.P. Exploring mRNA 3’-UTR G-quadruplexes: Evidence of roles in both alternative polyadenylation and mRNA shortening. Nucleic Acids Res. 2013, 41, 5898–5911. [Google Scholar] [CrossRef] [PubMed]
- Didiot, M.C.; Tian, Z.; Schaeffer, C.; Subramanian, M.; Mandel, J.L.; Moine, H. The G-quartet containing FMRP binding site in FMR1mRNAis a potent exonic splicing enhancer. Nucleic Acids Res. 2008, 36, 4902–4912. [Google Scholar] [CrossRef] [PubMed]
- Gomez, D.; Lemarteleur, T.; Lacroix, L.; Mailliet, P.; Mergny, J.L.; Riou, J.F. Telomerase downregulation induced by the G-quadruplex ligand 12459 in A549 cells is mediated by hTERT RNA alternative splicing. Nucleic Acids Res. 2004, 32, 371–379. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, J.; Harvey, S.E.; Hu, X.; Cheng, C. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev. 2017, 31, 2296–2309. [Google Scholar] [CrossRef] [PubMed]
- Endoh, T.; Sugimoto, N. Unusual -1 ribosomal frameshift caused by stable RNA G-quadruplex in open reading frame. Anal. Chem. 2013, 85, 11435–11439. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.H.; Teulade-Fichou, M.P.; Olsthoorn, R.C. Stimulation of ribosomal frameshifting by RNA G-quadruplex structures. Nucleic Acids Res. 2014, 42, 1887–1892. [Google Scholar] [CrossRef]
- Imperatore, J.A.; McAninch, D.S.; Valdez-Sinon, A.N.; Bassell, G.J.; Mihailescu, M.R. FUS recognizes G-quadruplex structures within neuronal mRNAs. Front. Mol. Biosci. 2020, 7, 6. [Google Scholar] [CrossRef]
- Ishiguro, A.; Kimura, N.; Watanabe, Y.; Watanabe, S.; Ishihama, A. TDP-43 binds and transports G-quadruplex-containing mRNAs into neurites for local translation. Genes Cells 2016, 21, 466–481. [Google Scholar] [CrossRef]
- Rihan, K.; Antoine, E.; Maurin, T.; Bardoni, B.; Bordonné, R.; Soret, J.; Rage, F. A new cis-acting motif is required for the axonal SMN-dependent Anxa2 mRNA localization. RNA 2017, 23, 899–909. [Google Scholar] [CrossRef][Green Version]
- Subramanian, M.; Rage, F.; Tabet, R.; Flatter, E.; Mandel, J.L.; Moine, H. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011, 12, 697–704. [Google Scholar] [CrossRef]
- Zhang, Y.; Gaetano, C.M.; Williams, K.R.; Bassell, G.J.; Mihailescu, M.R. FMRP interacts with G-quadruplex structures in the 3′-UTR of its dendritic target Shank1 mRNA. RNA Biol. 2014, 11, 1364–1374. [Google Scholar] [CrossRef]
- Jayaraj, G.G.; Pandey, S.; Scaria, V.; Maiti, S. Potential G-quadruplexes in the human long non-coding transcriptome. RNA Biol. 2012, 9, 81–86. [Google Scholar] [CrossRef]
- Yang, S.Y.; Lejault, P.; Chevrier, S.; Boidot, R.; Robertson, A.G.; Wong, J.M.Y.; Monchaud, D. Transcriptome-wide identification of transient RNA G-quadruplexes in human cells. Nat. Commun. 2018, 9, 4730. [Google Scholar] [CrossRef] [PubMed]
- Mirihana Arachchilage, G.; Dassanayake, A.C.; Basu, S. A potassium ion-dependent RNA structural switch regulates human pre-miRNA 92b maturation. Chem. Biol. 2015, 22, 262–272. [Google Scholar] [CrossRef]
- Pandey, S.; Agarwala, P.; Jayaraj, G.G.; Gargallo, R.; Maiti, S. The RNA stem-Loop to G-quadruplex equilibrium controls mature microRNA production inside the cell. Biochemistry 2015, 54, 7067–7078. [Google Scholar] [CrossRef]
- Rouleau, S.G.; Garant, J.M.; Bolduc, F.; Bisaillon, M.; Perreault, J.P. G-Quadruplexes influence pri-microRNA processing. RNA Biol. 2018, 15, 198–206. [Google Scholar] [CrossRef]
- Liu, G.; Du, W.; Xu, H.; Sun, Q.; Tang, D.; Zou, S.; Zhang, Y.; Ma, M.; Zhang, G.; Du, X.; et al. RNA G-quadruplex regulates microRNA-26a biogenesis and function. J. Hepatol. 2020. [Google Scholar] [CrossRef]
- Chan, K.L.; Peng, B.; Umar, M.I.; Chan, C.Y.; Sahakyan, A.B.; Le, M.T.N.; Kwok, C.K. Structural analysis reveals the formation and role of RNA G-quadruplex structures in human mature microRNAs. Chem. Commun. 2018, 54, 10878–10881. [Google Scholar] [CrossRef]
- Sahakyan, A.B.; Murat, P.; Mayer, C.; Balasubramanian, S. G-quadruplex structures within the 3′-UTR of LINE-1 elements stimulate retrotransposition. Nat. Struct. Mol. Biol. 2017, 24, 243–247. [Google Scholar] [CrossRef]
- Balaratnam, S.; Hettiarachchilage, M.; West, N.; Piontkivska, H.; Basu, S. A secondary structure within a human piRNA modulates its functionality. Biochimie 2019, 157, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Hirashima, K.; Seimiya, H. Telomeric repeat-containing RNA/G-quadruplex-forming sequences cause genome-wide alteration of gene expression in human cancer cells in vivo. Nucleic Acids Res. 2015, 43, 2022–2032. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Komiyama, M. Structure, function and targeting of human telomere RNA. Methods 2012, 57, 100–105. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).