Myc as a Regulator of Ribosome Biogenesis and Cell Competition: A Link to Cancer
Abstract
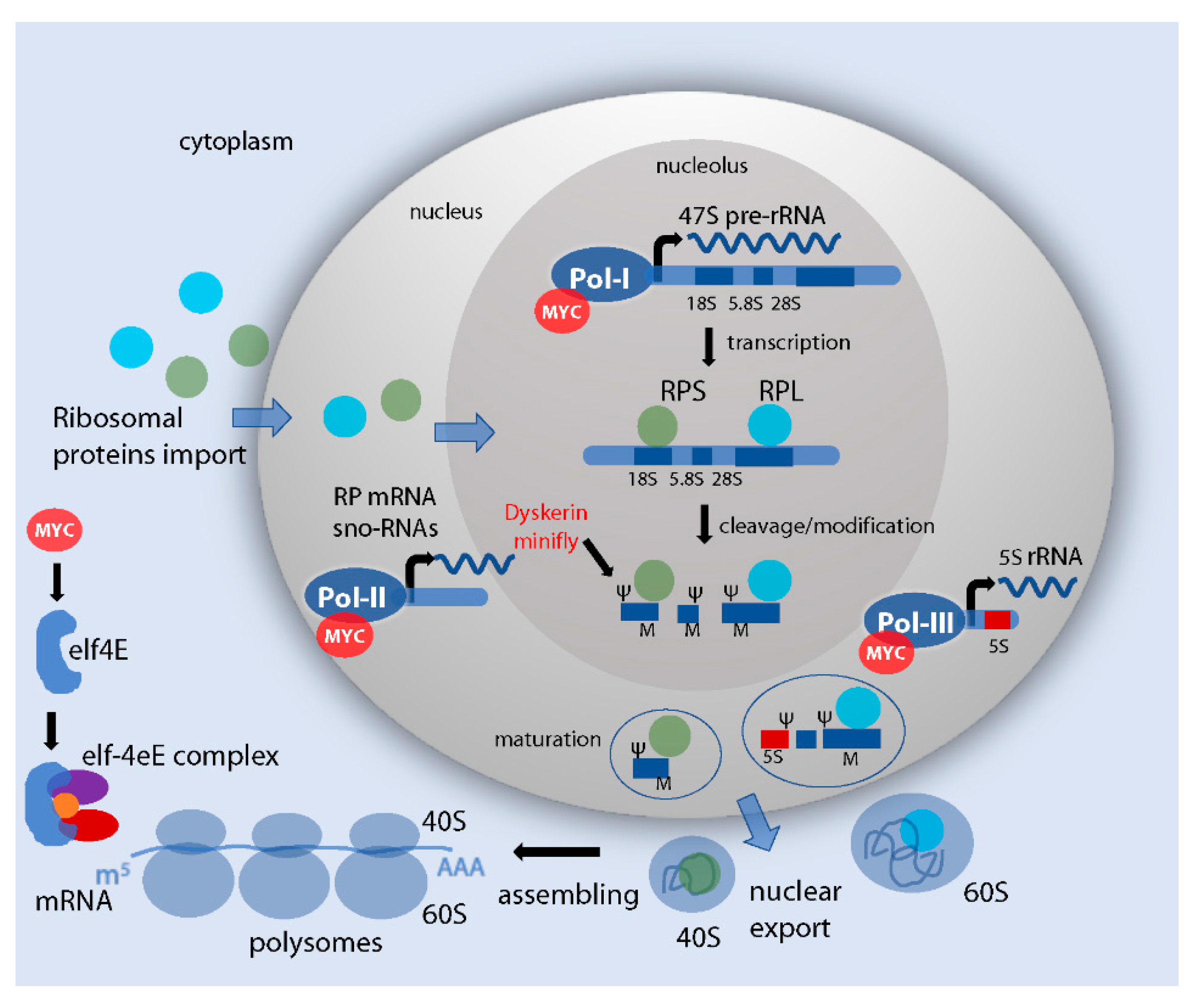
1. Ribosome Biogenesis
2. Role of Myc in Ribosome Biogenesis
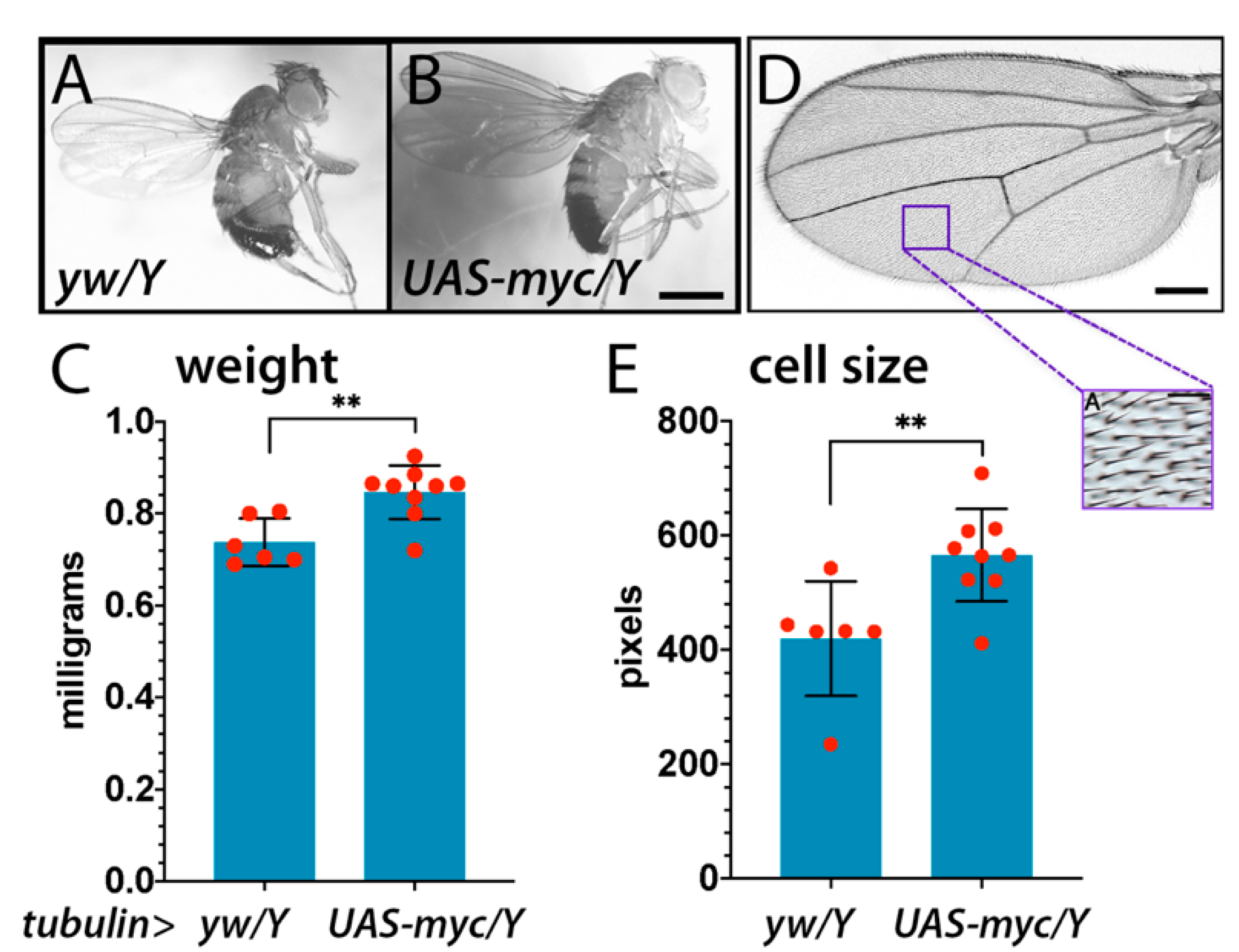
2.1. Drosophila Myc and the Regulation of Size
2.2. Myc Controls the Transcription of rDNA, Ribosomal Proteins and snoRNAs Genes
2.3. Myc Controls rRNA Processing and Assembly of the Ribosomes
2.4. Myc-Dependent Regulation of Ribosome Biogenesis Can Be Influenced by Growth Factors Signaling
2.5. Myc Control of Metabolism
3. Cell Competition
3.1. Early Discoveries in Cell Competition
3.2. Cell Competition Mechanisms
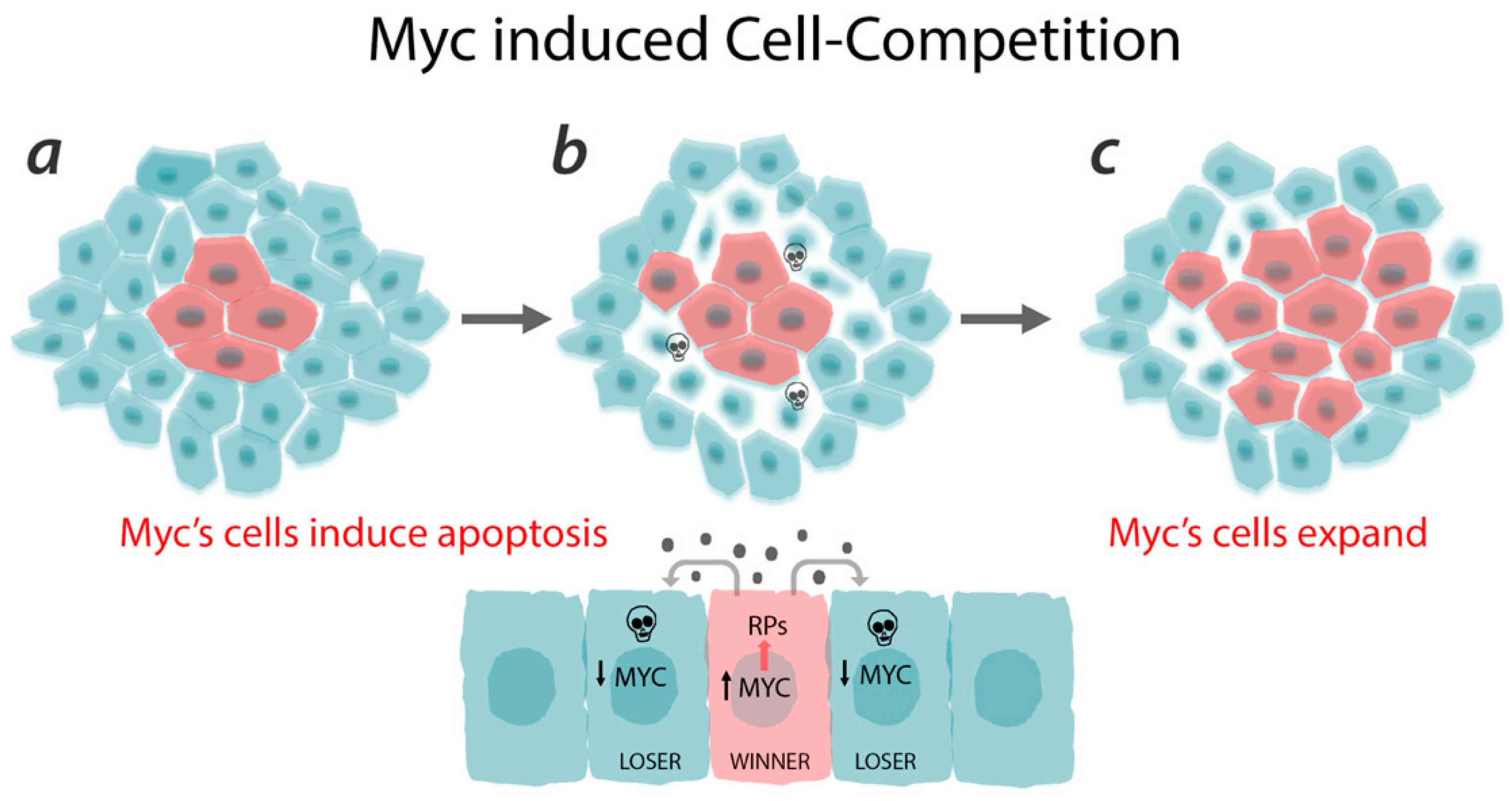
3.3. Myc and Ribosomes as Drivers of Cell Fitness
3.4. Physiological Role of Cell Competition
3.5. Cell Competition and Cancer
4. Myc and Ribosomes in Cancer
4.1. The Contribution of Ribosomes in Cancer
4.2. The Cooperation between Myc and Ribosomes in Cancer
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| snoRNP | small nucleolar ribonucleoproteins |
| DREF | DNA replication-related element binding factor |
| TFIIB | Transcription factor II B |
| TRAPP | Trafficking protein particle complex |
| GCN5 | Histone acetyltransferase GCN5 |
| TAFs | TATA box-binding protein-associated factor-s |
| TIF-IA | Transcription intermediary factor-1A |
| UBF/SL-1 | Nucleolar transcription factor 1/ |
| TIF-IB | Transcription intermediary factor-1B |
| Xrp1 | Transcription factor/bZip DNA binding protein |
References
- Anger, A.M.; Armache, J.P.; Berninghausen, O.; Habeck, M.; Subklewe, M.; Wilson, D.N.; Beckmann, R. Structures of the human and Drosophila 80S ribosome. Nature 2013, 497, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Yoshihama, M.; Uechi, T.; Asakawa, S.; Kawasaki, K.; Kato, S.; Higa, S.; Maeda, N.; Minoshima, S.; Tanaka, T.; Shimizu, N.; et al. The human ribosomal protein genes: Sequencing and comparative analysis of 73 genes. Genome Res. 2002, 12, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, F.M.; Van Koningsbruggen, S.; Navascues, J.; Lamond, A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007, 8, 574–585. [Google Scholar] [CrossRef] [PubMed]
- Fromont-Racine, M.; Senger, B.; Saveanu, C.; Fasiolo, F. Ribosome assembly in eukaryotes. Gene 2003, 313, 17–42. [Google Scholar] [CrossRef]
- Orsolic, I.; Jurada, D.; Pullen, N.; Oren, M.; Eliopoulos, A.G.; Volarevic, S. The relationship between the nucleolus and cancer: Current evidence and emerging paradigms. Semin. Cancer Biol. 2016, 37–38, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Thomson, E.; Ferreira-Cerca, S.; Hurt, E. Eukaryotic ribosome biogenesis at a glance. J. Cell Sci. 2013, 126, 4815–4821. [Google Scholar] [CrossRef]
- Weeks, S.E.; Metge, B.J.; Samant, R.S. The nucleolus: A central response hub for the stressors that drive cancer progression. Cell. Mol. Life Sci. 2019, 76, 4511–4524. [Google Scholar] [CrossRef]
- Russell, J.; Zomerdijk, J.C. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 2005, 30, 87–96. [Google Scholar] [CrossRef]
- Ciganda, M.; Williams, N. Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip. Rev. RNA 2011, 2, 523–533. [Google Scholar] [CrossRef]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef]
- Sharma, S.; Lafontaine, D.L.J. View From A Bridge: A New Perspective on Eukaryotic rRNA Base Modification. Trends Biochem. Sci. 2015, 40, 560–575. [Google Scholar] [CrossRef]
- Penzo, M.; Guerrieri, A.N.; Zacchini, F.; Trere, D.; Montanaro, L. RNA Pseudouridylation in Physiology and Medicine: For Better and for Worse. Genes 2017, 8, 301. [Google Scholar] [CrossRef] [PubMed]
- Monaco, P.L.; Marcel, V.; Diaz, J.J.; Catez, F. 2′-O-Methylation of Ribosomal RNA: Towards an Epitranscriptomic Control of Translation? Biomolecules 2018, 8, 106. [Google Scholar] [CrossRef] [PubMed]
- Sonenberg, N. eIF4E, the mRNA cap-binding protein: From basic discovery to translational research. Biochem. Cell Biol. 2008, 86, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, D.; Montanaro, L.; Ma, L.; Xu, W.; Londei, P.; Cordon-Cardo, C.; Pandolfi, P.P. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat. Med. 2004, 10, 484–486. [Google Scholar] [CrossRef]
- Carroll, P.A.; Freie, B.W.; Mathsyaraja, H.; Eisenman, R.N. The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 2018, 12, 412–425. [Google Scholar] [CrossRef]
- Eilers, M.; Eisenman, R.N. Myc’s broad reach. Genes Dev. 2008, 22, 2755–2766. [Google Scholar] [CrossRef]
- van Riggelen, J.; Yetil, A.; Felsher, D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 2010, 10, 301–309. [Google Scholar] [CrossRef]
- Campbell, K.J.; White, R.J. MYC regulation of cell growth through control of transcription by RNA polymerases I and III. Cold Spring Harb. Perspect. Med. 2014, 4. [Google Scholar] [CrossRef]
- Gallant, P. Myc/Max/Mad in invertebrates: The evolution of the Max network. Curr. Top. Microbiol. Immunol. 2006, 302, 235–253. [Google Scholar] [CrossRef]
- Hulf, T.; Bellosta, P.; Furrer, M.; Steiger, D.; Svensson, D.; Barbour, A.; Gallant, P. Whole-genome analysis reveals a strong positional bias of conserved dMyc-dependent E-boxes. Mol. Cell Biol. 2005, 25, 3401–3410. [Google Scholar] [CrossRef] [PubMed]
- Orian, A.; Grewal, S.S.; Knoepfler, P.S.; Edgar, B.A.; Parkhurst, S.M.; Eisenman, R.N. Genomic binding and transcriptional regulation by the Drosophila Myc and Mnt transcription factors. Cold Spring Harb. Symp. Quant. Biol. 2005, 70, 299–307. [Google Scholar] [CrossRef] [PubMed]
- Bellosta, P.; Hulf, T.; Balla Diop, S.; Usseglio, F.; Pradel, J.; Aragnol, D.; Gallant, P. Myc interacts genetically with Tip48/Reptin and Tip49/Pontin to control growth and proliferation during Drosophila development. Proc. Natl. Acad. Sci. USA 2005, 102, 11799–11804. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.A.; McMahon, S.B.; Cole, M.D. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 2000, 5, 321–330. [Google Scholar] [CrossRef]
- Meyer, N.; Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 2008, 8, 976–990. [Google Scholar] [CrossRef]
- Gallant, P.; Shiio, Y.; Cheng, P.F.; Parkhurst, S.M.; Eisenman, R.N. Myc and Max homologs in Drosophila. Science 1996, 274, 1523–1527. [Google Scholar] [CrossRef]
- Pierce, S.B.; Yost, C.; Britton, J.S.; Loo, L.W.; Flynn, E.M.; Edgar, B.A.; Eisenman, R.N. dMyc is required for larval growth and endoreplication in Drosophila. Development 2004, 131, 2317–2327. [Google Scholar] [CrossRef]
- Peyrefitte, S.; Kahn, D.; Haenlin, M. New members of the Drosophila Myc transcription factor subfamily revealed by a genome-wide examination for basic helix-loop-helix genes. Mech. Dev. 2001, 104, 99–104. [Google Scholar] [CrossRef]
- Gallant, P. Drosophila Myc. Adv. Cancer Res. 2009, 103, 111–144. [Google Scholar]
- Bellosta, P.; Gallant, P. Myc Function in Drosophila. Genes Cancer 2010, 1, 542–546. [Google Scholar] [CrossRef]
- Schreiber-Agus, N.; Stein, D.; Chen, K.; Goltz, J.S.; Stevens, L.; DePinho, R.A. Drosophila Myc is oncogenic in mammalian cells and plays a role in the diminutive phenotype. Proc. Natl. Acad. Sci. USA 1997, 94, 1235–1240. [Google Scholar] [CrossRef] [PubMed]
- Johnston, L.A.; Prober, D.A.; Edgar, B.A.; Eisenman, R.N.; Gallant, P. Drosophila myc regulates cellular growth during development. Cell 1999, 98, 779–790. [Google Scholar] [CrossRef]
- de la Cova, C.; Abril, M.; Bellosta, P.; Gallant, P.; Johnston, L.A. Drosophila myc regulates organ size by inducing cell competition. Cell 2004, 117, 107–116. [Google Scholar] [CrossRef]
- Pierce, S.B.; Yost, C.; Anderson, S.A.; Flynn, E.M.; Delrow, J.; Eisenman, R.N. Drosophila growth and development in the absence of dMyc and dMnt. Dev. Biol. 2008, 315, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Brand, A.H.; Manoukian, A.S.; Perrimon, N. Ectopic expression in Drosophila. Methods Cell Biol. 1994, 44, 635–654. [Google Scholar] [PubMed]
- Galletti, M.; Riccardo, S.; Parisi, F.; Lora, C.; Saqcena, M.K.; Rivas, L.; Wong, B.; Serra, A.; Serras, F.; Grifoni, D.; et al. Identification of domains responsible for ubiquitin-dependent degradation of dMyc by glycogen synthase kinase 3beta and casein kinase 1 kinases. Mol. Cell Biol. 2009, 29, 3424–3434. [Google Scholar] [CrossRef]
- Grewal, S.S.; Li, L.; Orian, A.; Eisenman, R.N.; Edgar, B.A. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 2005, 7, 295–302. [Google Scholar] [CrossRef]
- Grandori, C.; Gomez-Roman, N.; Felton-Edkins, Z.A.; Ngouenet, C.; Galloway, D.A.; Eisenman, R.N.; White, R.J. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 2005, 7, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Sahoo, D.; Arvanitis, C.; Bradon, N.; Dill, D.L.; Felsher, D.W. Combined analysis of murine and human microarrays and ChIP analysis reveals genes associated with the ability of MYC to maintain tumorigenesis. PLoS Genet. 2008, 4, e1000090. [Google Scholar] [CrossRef] [PubMed]
- Arabi, A.; Wu, S.; Ridderstrale, K.; Bierhoff, H.; Shiue, C.; Fatyol, K.; Fahlen, S.; Hydbring, P.; Soderberg, O.; Grummt, I.; et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005, 7, 303–310. [Google Scholar] [CrossRef]
- Oskarsson, T.; Trumpp, A. The Myc trilogy: Lord of RNA polymerases. Nat. Cell Biol. 2005, 7, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Stine, Z.E.; Walton, Z.E.; Altman, B.J.; Hsieh, A.L.; Dang, C.V. MYC, Metabolism, and Cancer. Cancer Discov. 2015, 5, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Kenneth, N.S.; Ramsbottom, B.A.; Gomez-Roman, N.; Marshall, L.; Cole, P.A.; White, R.J. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 14917–14922. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Roman, N.; Grandori, C.; Eisenman, R.N.; White, R.J. Direct activation of RNA polymerase III transcription by c-Myc. Nature 2003, 421, 290–294. [Google Scholar] [CrossRef]
- Schlosser, I.; Holzel, M.; Murnseer, M.; Burtscher, H.; Weidle, U.H.; Eick, D. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003, 31, 6148–6156. [Google Scholar] [CrossRef] [PubMed]
- Herter, E.K.; Stauch, M.; Gallant, M.; Wolf, E.; Raabe, T.; Gallant, P. snoRNAs are a novel class of biologically relevant Myc targets. BMC Biol. 2015, 13, 25. [Google Scholar] [CrossRef] [PubMed]
- Romano, G.; Veneziano, D.; Acunzo, M.; Croce, C.M. Small non-coding RNA and cancer. Carcinogenesis 2017, 38, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Zirin, J.; Ni, X.; Sack, L.M.; Yang-Zhou, D.; Hu, Y.; Brathwaite, R.; Bulyk, M.L.; Elledge, S.J.; Perrimon, N. Interspecies analysis of MYC targets identifies tRNA synthetases as mediators of growth and survival in MYC-overexpressing cells. Proc. Natl. Acad. Sci. USA 2019, 116, 14614–14619. [Google Scholar] [CrossRef]
- Nemeth, A.; Perez-Fernandez, J.; Merkl, P.; Hamperl, S.; Gerber, J.; Griesenbeck, J.; Tschochner, H. RNA polymerase I termination: Where is the end? Biochim. Biophys. Acta 2013, 1829, 306–317. [Google Scholar] [CrossRef]
- Boon, K.; Caron, H.N.; van Asperen, R.; Valentijn, L.; Hermus, M.C.; Van Sluis, P.; Roobeek, I.; Weis, I.; Voute, P.A.; Schwab, M.; et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001, 20, 1383–1393. [Google Scholar] [CrossRef]
- Kaiser, C.; Dobrikova, E.Y.; Bradrick, S.S.; Shveygert, M.; Herbert, J.T.; Gromeier, M. Activation of cap-independent translation by variant eukaryotic initiation factor 4G in vivo. RNA 2008, 14, 2170–2182. [Google Scholar] [CrossRef]
- Thoma, C.; Fraterman, S.; Gentzel, M.; Wilm, M.; Hentze, M.W. Translation initiation by the c-myc mRNA internal ribosome entry sequence and the poly(A) tail. RNA 2008, 14, 1579–1589. [Google Scholar] [CrossRef] [PubMed]
- Giordano, E.; Peluso, I.; Senger, S.; Furia, M. minifly, a Drosophila gene required for ribosome biogenesis. J. Cell Biol. 1999, 144, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Tortoriello, G.; de Celis, J.F.; Furia, M. Linking pseudouridine synthases to growth, development and cell competition. FEBS J. 2010, 277, 3249–3263. [Google Scholar] [CrossRef] [PubMed]
- Dalla Venezia, N.; Vincent, A.; Marcel, V.; Catez, F.; Diaz, J.J. Emerging Role of Eukaryote Ribosomes in Translational Control. Int. J. Mol. Sci. 2019, 20, 1226. [Google Scholar] [CrossRef] [PubMed]
- Milkereit, P.; Gadal, O.; Podtelejnikov, A.; Trumtel, S.; Gas, N.; Petfalski, E.; Tollervey, D.; Mann, M.; Hurt, E.; Tschochner, H. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell 2001, 105, 499–509. [Google Scholar] [CrossRef]
- Kressler, D.; Hurt, E.; Bassler, J. Driving ribosome assembly. Biochim. Biophys. Acta 2010, 1803, 673–683. [Google Scholar] [CrossRef]
- Hierlmeier, T.; Merl, J.; Sauert, M.; Perez-Fernandez, J.; Schultz, P.; Bruckmann, A.; Hamperl, S.; Ohmayer, U.; Rachel, R.; Jacob, A.; et al. Rrp5p, Noc1p and Noc2p form a protein module which is part of early large ribosomal subunit precursors in S. cerevisiae. Nucleic Acids Res. 2013, 41, 1191–1210. [Google Scholar] [CrossRef]
- Chauvin, C.; Koka, V.; Nouschi, A.; Mieulet, V.; Hoareau-Aveilla, C.; Dreazen, A.; Cagnard, N.; Carpentier, W.; Kiss, T.; Meyuhas, O.; et al. Ribosomal protein S6 kinase activity controls the ribosome biogenesis transcriptional program. Oncogene 2014, 33, 474–483. [Google Scholar] [CrossRef]
- Parisi, F.; Riccardo, S.; Daniel, M.; Saqcena, M.; Kundu, N.; Pession, A.; Grifoni, D.; Stocker, H.; Tabak, E.; Bellosta, P. Drosophila insulin and target of rapamycin (TOR) pathways regulate GSK3 beta activity to control Myc stability and determine Myc expression in vivo. BMC Biol. 2011, 9, 65. [Google Scholar] [CrossRef]
- Yeh, E.; Cunningham, M.; Arnold, H.; Chasse, D.; Monteith, T.; Ivaldi, G.; Hahn, W.C.; Stukenberg, P.T.; Shenolikar, S.; Uchida, T.; et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 2004, 6, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.C.; Hannan, K.M.; Riddell, K.; Ng, P.Y.; Peck, A.; Lee, R.S.; Hung, S.; Astle, M.V.; Bywater, M.; Wall, M.; et al. AKT promotes rRNA synthesis and cooperates with c-MYC to stimulate ribosome biogenesis in cancer. Sci. Signal. 2011, 4, ra56. [Google Scholar] [CrossRef]
- Liu, F.; Jin, R.; Liu, X.; Huang, H.; Wilkinson, S.C.; Zhong, D.; Khuri, F.R.; Fu, H.; Marcus, A.; He, Y.; et al. LKB1 promotes cell survival by modulating TIF-IA-mediated pre-ribosomal RNA synthesis under uridine downregulated conditions. Oncotarget 2016, 7, 2519–2531. [Google Scholar] [CrossRef] [PubMed]
- Ilic, N.; Utermark, T.; Widlund, H.R.; Roberts, T.M. PI3K-targeted therapy can be evaded by gene amplification along the MYC-eukaryotic translation initiation factor 4E (eIF4E) axis. Proc. Natl. Acad. Sci. USA 2011, 108, E699–E708. [Google Scholar] [CrossRef]
- Robichaud, N.; Sonenberg, N.; Ruggero, D.; Schneider, R.J. Translational Control in Cancer. Cold Spring Harb. Perspect. Biol. 2019, 11. [Google Scholar] [CrossRef]
- Liu, P.; Cheng, H.; Santiago, S.; Raeder, M.; Zhang, F.; Isabella, A.; Yang, J.; Semaan, D.J.; Chen, C.; Fox, E.A.; et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat. Med. 2011, 17, 1116–1120. [Google Scholar] [CrossRef] [PubMed]
- Hayes, T.K.; Neel, N.F.; Hu, C.; Gautam, P.; Chenard, M.; Long, B.; Aziz, M.; Kassner, M.; Bryant, K.L.; Pierobon, M.; et al. Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer Cell 2016, 29, 75–89. [Google Scholar] [CrossRef]
- Ryan, R.J.H.; Petrovic, J.; Rausch, D.M.; Zhou, Y.; Lareau, C.A.; Kluk, M.J.; Christie, A.L.; Lee, W.Y.; Tarjan, D.R.; Guo, B.; et al. A B Cell Regulome Links Notch to Downstream Oncogenic Pathways in Small B Cell Lymphomas. Cell Rep. 2017, 21, 784–797. [Google Scholar] [CrossRef] [PubMed]
- Yochum, G.S.; Sherrick, C.M.; Macpartlin, M.; Goodman, R.H. A beta-catenin/TCF-coordinated chromatin loop at MYC integrates 5’ and 3’ Wnt responsive enhancers. Proc. Natl. Acad. Sci. USA 2010, 107, 145–150. [Google Scholar] [CrossRef]
- Wolpaw, A.J.; Dang, C.V. MYC-induced metabolic stress and tumorigenesis. Biochim. Biophys. Acta Rev. Cancer 2018, 1870, 43–50. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014, 4. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, A.L.; Dang, C.V. MYC, Metabolic Synthetic Lethality, and Cancer. Recent Results Cancer Res. 2016, 207, 73–91. [Google Scholar] [CrossRef]
- Parisi, F.; Riccardo, S.; Zola, S.; Lora, C.; Grifoni, D.; Brown, L.M.; Bellosta, P. dMyc expression in the fat body affects DILP2 release and increases the expression of the fat desaturase Desat1 resulting in organismal growth. Dev. Biol. 2013, 379, 64–75. [Google Scholar] [CrossRef]
- Paiardi, C.; Mirzoyan, Z.; Zola, S.; Parisi, F.; Vingiani, A.; Pasini, M.E.; Bellosta, P. The Stearoyl-CoA Desaturase-1 (Desat1) in Drosophila cooperated with Myc to Induce Autophagy and Growth, a Potential New Link to Tumor Survival. Genes 2017, 8, 131. [Google Scholar] [CrossRef]
- De la Cova, C.; Senoo-Matsuda, N.; Ziosi, M.; Wu, D.C.; Bellosta, P.; Quinzii, C.M.; Johnston, L.A. Supercompetitor status of Drosophila Myc cells requires p53 as a fitness sensor to reprogram metabolism and promote viability. Cell Metab. 2014, 19, 470–483. [Google Scholar] [CrossRef]
- Hsieh, A.L.; Walton, Z.E.; Altman, B.J.; Stine, Z.E.; Dang, C.V. MYC and metabolism on the path to cancer. Semin. Cell Dev. Biol. 2015, 43, 11–21. [Google Scholar] [CrossRef]
- Saeboe-Larssen, S.; Lyamouri, M.; Merriam, J.; Oksvold, M.P.; Lambertsson, A. Ribosomal protein insufficiency and the minute syndrome in Drosophila: A dose-response relationship. Genetics 1998, 148, 1215–1224. [Google Scholar] [PubMed]
- Marygold, S.J.; Roote, J.; Reuter, G.; Lambertsson, A.; Ashburner, M.; Millburn, G.H.; Harrison, P.M.; Yu, Z.; Kenmochi, N.; Kaufman, T.C.; et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007, 8, R216. [Google Scholar] [CrossRef] [PubMed]
- Moreno, E.; Basler, K. dMyc transforms cells into super-competitors. Cell 2004, 117, 117–129. [Google Scholar] [CrossRef]
- Barna, M.; Pusic, A.; Zollo, O.; Costa, M.; Kondrashov, N.; Rego, E.; Rao, P.H.; Ruggero, D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 2008, 456, 971–975. [Google Scholar] [CrossRef] [PubMed]
- Claveria, C.; Torres, M. Cell Competition: Mechanisms and Physiological Roles. Annu Rev. Cell Dev. Biol. 2016, 32, 411–439. [Google Scholar] [CrossRef]
- Amoyel, M.; Bach, E.A. Cell competition: How to eliminate your neighbours. Development 2014, 141, 988–1000. [Google Scholar] [CrossRef]
- Di Gregorio, A.; Bowling, S.; Rodriguez, T.A. Cell Competition and Its Role in the Regulation of Cell Fitness from Development to Cancer. Dev. Cell 2016, 38, 621–634. [Google Scholar] [CrossRef]
- Nagata, R.; Igaki, T. Cell competition: Emerging mechanisms to eliminate neighbors. Dev. Growth Differ. 2018, 60, 522–530. [Google Scholar] [CrossRef]
- Morata, G.; Ripoll, P. Minutes: Mutants of drosophila autonomously affecting cell division rate. Dev. Biol. 1975, 42, 211–221. [Google Scholar] [CrossRef]
- Simpson, P.; Morata, G. Differential mitotic rates and patterns of growth in compartments in the Drosophila wing. Dev. Biol. 1981, 85, 299–308. [Google Scholar] [CrossRef]
- Senoo-Matsuda, N.; Johnston, L.A. Soluble factors mediate competitive and cooperative interactions between cells expressing different levels of Drosophila Myc. Proc. Natl. Acad. Sci. USA 2007, 104, 18543–18548. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Diaz, C.; Fernandez de Manuel, L.; Jimenez-Carretero, D.; Montoya, M.C.; Claveria, C.; Torres, M. Pluripotency Surveillance by Myc-Driven Competitive Elimination of Differentiating Cells. Dev. Cell 2017, 42, 585–599 e584. [Google Scholar] [CrossRef]
- Moreno, E.; Basler, K.; Morata, G. Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature 2002, 416, 755–759. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Baker, N.E. Engulfment is required for cell competition. Cell 2007, 129, 1215–1225. [Google Scholar] [CrossRef]
- Rhiner, C.; Lopez-Gay, J.M.; Soldini, D.; Casas-Tinto, S.; Martin, F.A.; Lombardia, L.; Moreno, E. Flower forms an extracellular code that reveals the fitness of a cell to its neighbors in Drosophila. Dev. Cell 2010, 18, 985–998. [Google Scholar] [CrossRef] [PubMed]
- Merino, M.M.; Rhiner, C.; Lopez-Gay, J.M.; Buechel, D.; Hauert, B.; Moreno, E. Elimination of unfit cells maintains tissue health and prolongs lifespan. Cell 2015, 160, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Portela, M.; Casas-Tinto, S.; Rhiner, C.; Lopez-Gay, J.M.; Dominguez, O.; Soldini, D.; Moreno, E. Drosophila SPARC is a self-protective signal expressed by loser cells during cell competition. Dev. Cell 2010, 19, 562–573. [Google Scholar] [CrossRef]
- Meyer, S.N.; Amoyel, M.; Bergantinos, C.; de la Cova, C.; Schertel, C.; Basler, K.; Johnston, L.A. An ancient defense system eliminates unfit cells from developing tissues during cell competition. Science 2014, 346, 1258236. [Google Scholar] [CrossRef]
- Alpar, L.; Bergantinos, C.; Johnston, L.A. Spatially Restricted Regulation of Spatzle/Toll Signaling during Cell Competition. Dev. Cell 2018, 46, 706–719 e705. [Google Scholar] [CrossRef]
- Lee, C.H.; Kiparaki, M.; Blanco, J.; Folgado, V.; Ji, Z.; Kumar, A.; Rimesso, G.; Baker, N.E. A Regulatory Response to Ribosomal Protein Mutations Controls Translation, Growth, and Cell Competition. Dev. Cell 2018, 46, 456–469.e4. [Google Scholar] [CrossRef]
- Orian, A.; van Steensel, B.; Delrow, J.; Bussemaker, H.J.; Li, L.; Sawado, T.; Williams, E.; Loo, L.W.; Cowley, S.M.; Yost, C.; et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003, 17, 1101–1114. [Google Scholar] [CrossRef] [PubMed]
- Baillon, L.; Germani, F.; Rockel, C.; Hilchenbach, J.; Basler, K. Xrp1 is a transcription factor required for cell competition-driven elimination of loser cells. Sci. Rep. 2018, 8, 17712. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Kiparaki, M.; Folgado, V.; Kumar, A.; Blanco, J.; Rimesso, G.; Chuen, J.; Liu, Y.; Zheng, D.; Baker, N.E. Drosophila RpS12 controls translation, growth, and cell competition through Xrp1. PLoS Genet. 2019, 15, e1008513. [Google Scholar] [CrossRef] [PubMed]
- Leevers, S.J.; McNeill, H. Controlling the size of organs and organisms. Curr. Opin. Cell Biol. 2005, 17, 604–609. [Google Scholar] [CrossRef]
- Claveria, C.; Giovinazzo, G.; Sierra, R.; Torres, M. Myc-driven endogenous cell competition in the early mammalian embryo. Nature 2013, 500, 39–44. [Google Scholar] [CrossRef]
- Zimmerman, K.A.; Yancopoulos, G.D.; Collum, R.G.; Smith, R.K.; Kohl, N.E.; Denis, K.A.; Nau, M.M.; Witte, O.N.; Toran-Allerand, D.; Gee, C.E.; et al. Differential expression of myc family genes during murine development. Nature 1986, 319, 780–783. [Google Scholar] [CrossRef]
- Munoz-Martin, N.; Sierra, R.; Schimmang, T.; Villa Del Campo, C.; Torres, M. Myc is dispensable for cardiomyocyte development but rescues Mycn-deficient hearts through functional replacement and cell competition. Development 2019, 146. [Google Scholar] [CrossRef]
- Bras-Pereira, C.; Moreno, E. Mechanical cell competition. Curr. Opin. Cell Biol. 2018, 51, 15–21. [Google Scholar] [CrossRef]
- Liu, N.; Matsumura, H.; Kato, T.; Ichinose, S.; Takada, A.; Namiki, T.; Asakawa, K.; Morinaga, H.; Mohri, Y.; De Arcangelis, A.; et al. Stem cell competition orchestrates skin homeostasis and ageing. Nature 2019, 568, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Ellis, S.J.; Gomez, N.C.; Levorse, J.; Mertz, A.F.; Ge, Y.; Fuchs, E. Distinct modes of cell competition shape mammalian tissue morphogenesis. Nature 2019, 569, 497–502. [Google Scholar] [CrossRef]
- Agrawal, N.; Joshi, S.; Kango, M.; Saha, D.; Mishra, A.; Sinha, P. Epithelial hyperplasia of imaginal discs induced by mutations in Drosophila tumor suppressor genes: Growth and pattern formation in genetic mosaics. Dev. Biol. 1995, 169, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Fahey-Lozano, N.; La Marca, J.E.; Portela, M.; Richardson, H.E. Drosophila Models of Cell Polarity and Cell Competition in Tumourigenesis. Adv. Exp. Med. Biol. 2019, 1167, 37–64. [Google Scholar] [CrossRef] [PubMed]
- Brumby, A.M.; Richardson, H.E. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J. 2003, 22, 5769–5779. [Google Scholar] [CrossRef] [PubMed]
- Woods, D.F.; Bryant, P.J. The discs-large tumor suppressor gene of Drosophila encodes a guanylate kinase homolog localized at septate junctions. Cell 1991, 66, 451–464. [Google Scholar] [CrossRef]
- Igaki, T.; Pastor-Pareja, J.C.; Aonuma, H.; Miura, M.; Xu, T. Intrinsic tumor suppression and epithelial maintenance by endocytic activation of Eiger/TNF signaling in Drosophila. Dev. Cell 2009, 16, 458–465. [Google Scholar] [CrossRef]
- Froldi, F.; Ziosi, M.; Garoia, F.; Pession, A.; Grzeschik, N.A.; Bellosta, P.; Strand, D.; Richardson, H.E.; Grifoni, D. The lethal giant larvae tumour suppressor mutation requires dMyc oncoprotein to promote clonal malignancy. BMC Biol. 2010, 8, 33. [Google Scholar] [CrossRef]
- Pan, D. The hippo signaling pathway in development and cancer. Dev. Cell 2010, 19, 491–505. [Google Scholar] [CrossRef]
- Ziosi, M.; Baena-Lopez, L.A.; Grifoni, D.; Froldi, F.; Pession, A.; Garoia, F.; Trotta, V.; Bellosta, P.; Cavicchi, S. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet. 2010, 6. [Google Scholar] [CrossRef]
- Neto-Silva, R.M.; De Beco, S.; Johnston, L.A. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev. Cell 2010, 19, 507–520. [Google Scholar] [CrossRef]
- Johnston, L.A. Socializing with MYC: Cell Competition in Development and as a Model for Premalignant Cancer. Cold Spring Harb. Perspect. Med. 2014, 4. [Google Scholar] [CrossRef]
- Moreno, E. Is cell competition relevant to cancer? Nat. Rev. Cancer 2008, 8, 141–147. [Google Scholar] [CrossRef]
- Rhiner, C.; Moreno, E. Super competition as a possible mechanism to pioneer precancerous fields. Carcinogenesis 2009, 30, 723–728. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.S.; Shah, H.S.; Shrivastava, N. c-Myc-Dependent Cell Competition in Human Cancer Cells. J. Cell Biochem. 2017, 118, 1782–1791. [Google Scholar] [CrossRef] [PubMed]
- Di Giacomo, S.; Sollazzo, M.; de Biase, D.; Ragazzi, M.; Bellosta, P.; Pession, A.; Grifoni, D. Human Cancer Cells Signal Their Competitive Fitness Through MYC Activity. Sci. Rep. 2017, 7, 12568. [Google Scholar] [CrossRef] [PubMed]
- Ulirsch, J.C.; Verboon, J.M.; Kazerounian, S.; Guo, M.H.; Yuan, D.; Ludwig, L.S.; Handsaker, R.E.; Abdulhay, N.J.; Fiorini, C.; Genovese, G.; et al. The Genetic Landscape of Diamond-Blackfan Anemia. Am. J. Hum. Genet. 2018, 103, 930–947. [Google Scholar] [CrossRef] [PubMed]
- Uechi, T.; Tanaka, T.; Kenmochi, N. A complete map of the human ribosomal protein genes: Assignment of 80 genes to the cytogenetic map and implications for human disorders. Genomics 2001, 72, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Kenmochi, N.; Yoshihama, M.; Higa, S.; Tanaka, T. The human ribosomal protein L6 gene in a critical region for Noonan syndrome. J. Hum. Genet. 2000, 45, 290–293. [Google Scholar] [CrossRef]
- Narla, A.; Ebert, B.L. Ribosomopathies: Human disorders of ribosome dysfunction. Blood 2010, 115, 3196–3205. [Google Scholar] [CrossRef]
- Draptchinskaia, N.; Gustavsson, P.; Andersson, B.; Pettersson, M.; Willig, T.N.; Dianzani, I.; Ball, S.; Tchernia, G.; Klar, J.; Matsson, H.; et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat. Genet. 1999, 21, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.W.; Green, R. Ribosomopathies: There’s strength in numbers. Science 2017, 358. [Google Scholar] [CrossRef]
- Montanaro, L.; Trere, D.; Derenzini, M. Nucleolus, ribosomes, and cancer. Am. J. Pathol. 2008, 173, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Zink, D.; Fischer, A.H.; Nickerson, J.A. Nuclear structure in cancer cells. Nat. Rev. Cancer 2004, 4, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Gentilella, A.; Kozma, S.C.; Thomas, G. A liaison between mTOR signaling, ribosome biogenesis and cancer. Biochim. Biophys. Acta 2015, 1849, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Ruggero, D. Translational control in cancer etiology. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef]
- Bellodi, C.; Kopmar, N.; Ruggero, D. Deregulation of oncogene-induced senescence and p53 translational control in X-linked dyskeratosis congenita. EMBO J. 2010, 29, 1865–1876. [Google Scholar] [CrossRef]
- Yoon, A.; Peng, G.; Brandenburger, Y.; Zollo, O.; Xu, W.; Rego, E.; Ruggero, D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science 2006, 312, 902–906. [Google Scholar] [CrossRef]
- Devlin, J.R.; Hannan, K.M.; Hein, N.; Cullinane, C.; Kusnadi, E.; Ng, P.Y.; George, A.J.; Shortt, J.; Bywater, M.J.; Poortinga, G.; et al. Combination Therapy Targeting Ribosome Biogenesis and mRNA Translation Synergistically Extends Survival in MYC-Driven Lymphoma. Cancer Discov. 2016, 6, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Hald, O.H.; Olsen, L.; Gallo-Oller, G.; Elfman, L.H.M.; Lokke, C.; Kogner, P.; Sveinbjornsson, B.; Flaegstad, T.; Johnsen, J.I.; Einvik, C. Inhibitors of ribosome biogenesis repress the growth of MYCN-amplified neuroblastoma. Oncogene 2019, 38, 2800–2813. [Google Scholar] [CrossRef]
- Farley-Barnes, K.I.; Ogawa, L.M.; Baserga, S.J. Ribosomopathies: Old Concepts, New Controversies. Trends Genet. 2019, 35, 754–767. [Google Scholar] [CrossRef]
- Liu, Y.; Deisenroth, C.; Zhang, Y. RP-MDM2-p53 Pathway: Linking Ribosomal Biogenesis and Tumor Surveillance. Trends Cancer 2016, 2, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Morcelle, C.; Menoyo, S.; Moron-Duran, F.D.; Tauler, A.; Kozma, S.C.; Thomas, G.; Gentilella, A. Oncogenic MYC Induces the Impaired Ribosome Biogenesis Checkpoint and Stabilizes p53 Independent of Increased Ribosome Content. Cancer Res. 2019, 79, 4348–4359. [Google Scholar] [CrossRef] [PubMed]
- Ebright, R.Y.; Lee, S.; Wittner, B.S.; Niederhoffer, K.L.; Nicholson, B.T.; Bardia, A.; Truesdell, S.; Wiley, D.F.; Wesley, B.; Li, S.; et al. Deregulation of ribosomal protein expression and translation promotes breast cancer metastasis. Science 2020, 367, 1468–1473. [Google Scholar] [CrossRef] [PubMed]
- Mirzoyan, Z.; Sollazzo, M.; Allocca, M.; Valenza, A.M.; Grifoni, D.; Bellosta, P. Drosophila melanogaster: A Model Organism to Study Cancer. Front. Genet. 2019, 10, 51. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Destefanis, F.; Manara, V.; Bellosta, P. Myc as a Regulator of Ribosome Biogenesis and Cell Competition: A Link to Cancer. Int. J. Mol. Sci. 2020, 21, 4037. https://doi.org/10.3390/ijms21114037
Destefanis F, Manara V, Bellosta P. Myc as a Regulator of Ribosome Biogenesis and Cell Competition: A Link to Cancer. International Journal of Molecular Sciences. 2020; 21(11):4037. https://doi.org/10.3390/ijms21114037
Chicago/Turabian StyleDestefanis, Francesca, Valeria Manara, and Paola Bellosta. 2020. "Myc as a Regulator of Ribosome Biogenesis and Cell Competition: A Link to Cancer" International Journal of Molecular Sciences 21, no. 11: 4037. https://doi.org/10.3390/ijms21114037
APA StyleDestefanis, F., Manara, V., & Bellosta, P. (2020). Myc as a Regulator of Ribosome Biogenesis and Cell Competition: A Link to Cancer. International Journal of Molecular Sciences, 21(11), 4037. https://doi.org/10.3390/ijms21114037





