Screening and Identification of Novel cGAS Homologues Using a Combination of in Vitro and In Vivo Protein Synthesis
Abstract
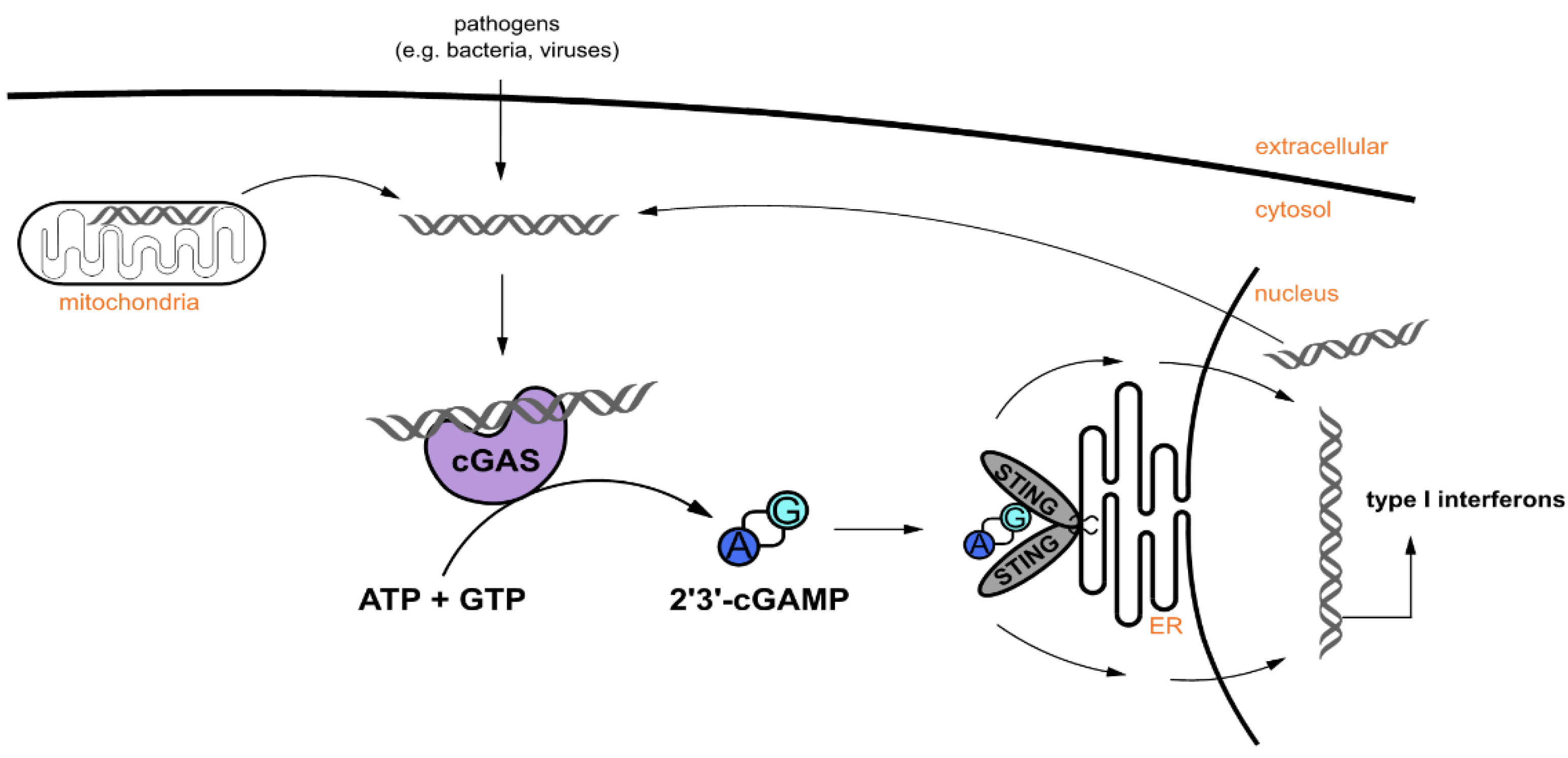
1. Introduction
2. Results and Discussion
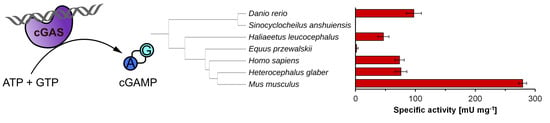
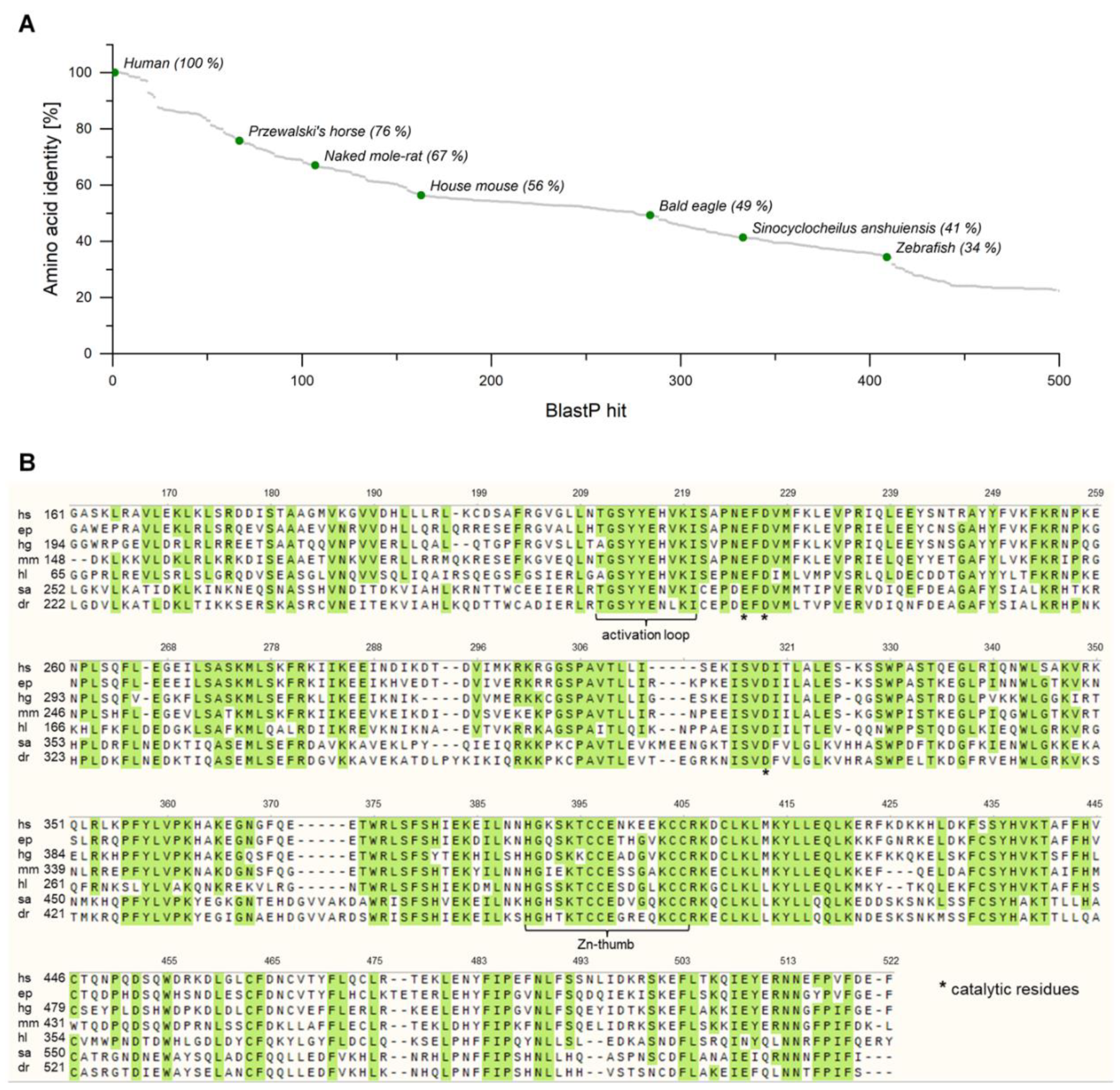
2.1. Identification of Putative cGAS Homologues in Several Metazoans
2.2. Expression of cGAS Genes with an E. coli-Based Cell-Free Protein Synthesis System
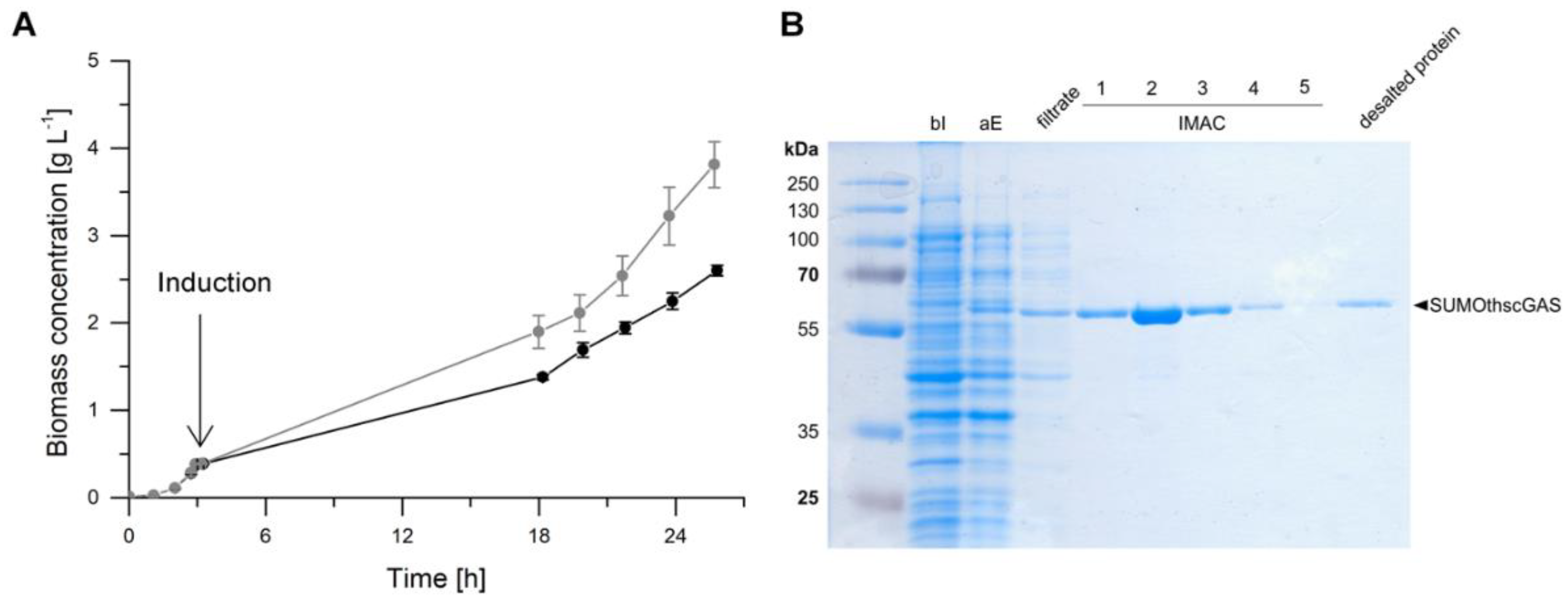
2.3. Heterologous Expression in Escherichia coli
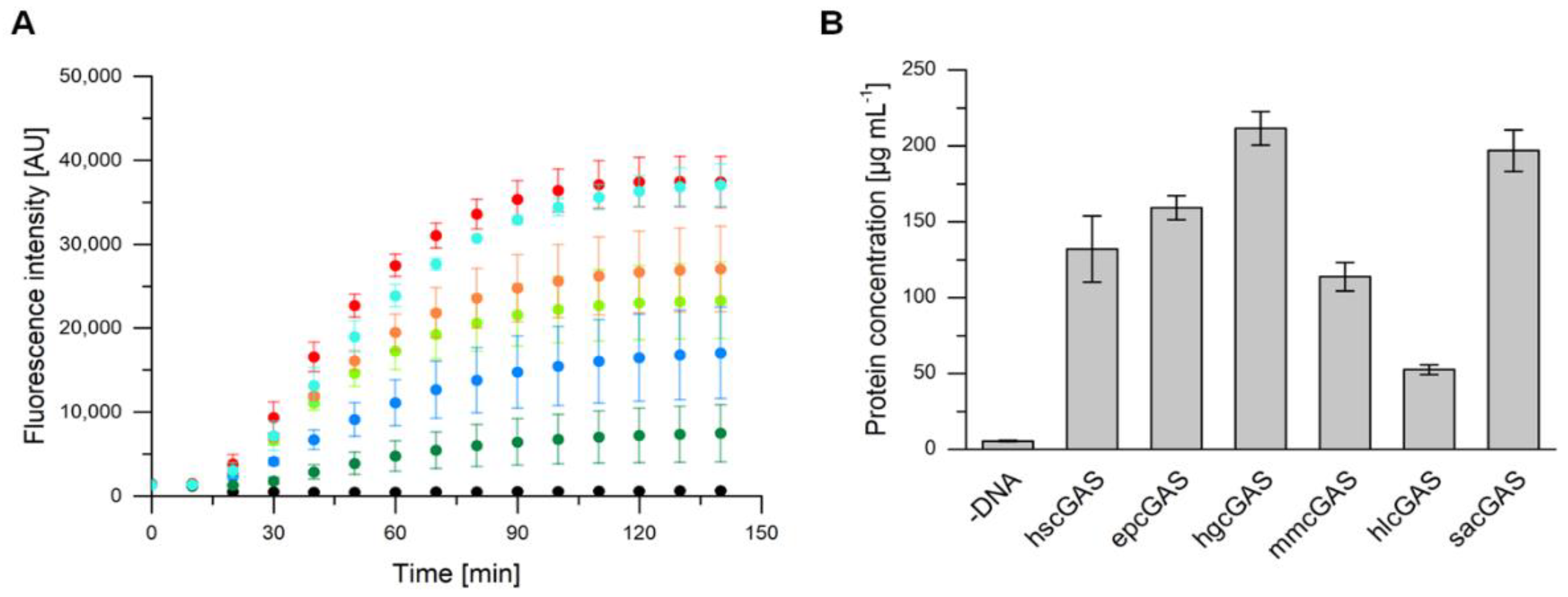
2.4. cGAS Variants Catalyze cGAMP Synthesis
3. Materials and Methods
3.1. Strains and Plasmids
3.2. E. coli Extract Preparation
3.3. Cell-Free Protein Synthesis (CFPS)
3.4. Cell-Based Protein Synthesis
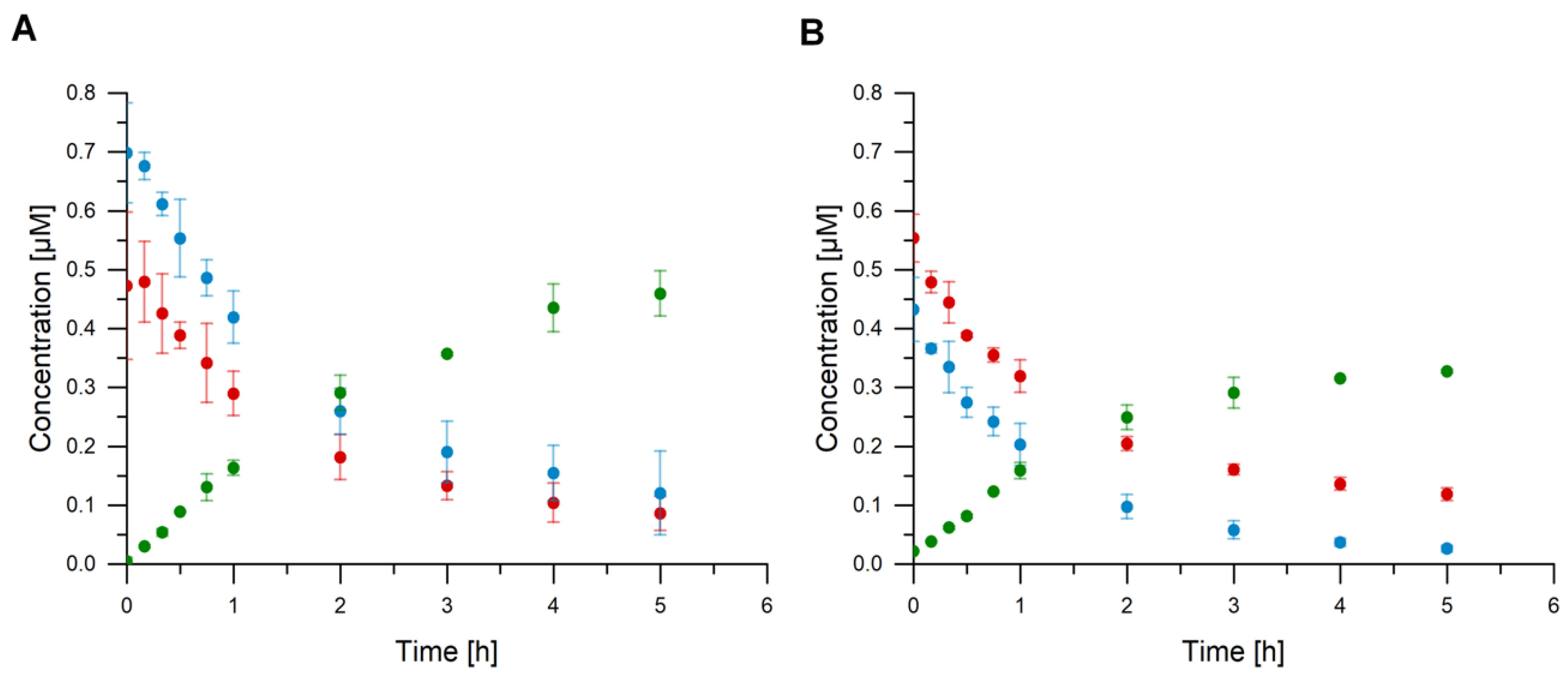
3.5. Activity Measurement
3.6. Quantification of cGAMP, ATP, and GTP
3.7. Verification of cGAMP Production
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| BlastP | Basic local alignment search tool |
| CDN | Cyclic dinucleotide |
| CFPS | Cell-free protein synthesis |
| cGAMP | Cyclic guanosine monophosphate-adenosine monophosphate |
| cGAS | cyclic guanosine monophosphate-adenosine monophosphate synthase |
| dr | Danio rerio |
| ER | Endoplasmic reticulum |
| ep | Equus przewalskii |
| GFP | Green fluorescent protein |
| GST | Glutathione S-transferase |
| hg | Heterocephalus glaber |
| hl | Haliaeetus leucocephalus |
| hs | Homo sapiens |
| IMAC | Immobilized metal affinity chromatography |
| Mab21 | Male abnormal 21 |
| MBP | Maltose binding protein |
| mm | Mus musculus |
| NTase | Nucleotidyltransferase |
| sa | Sinocyclocheilus anshuiensis |
| sfGFP | Superfolder green fluorescent protein |
| STING | Stimulator of interferon genes |
| SUMO | Small ubiquitin-like modifier |
Appendix A
| Metazoan | Binomial Name | Abbreviation | Protein Reference Sequence | Nucleotide Reference Sequence |
|---|---|---|---|---|
| Human | Homo Sapiens | hs | NP_612450.2 | NM_138441.3 |
| Przewalski’s horse | Equus przewalskii | ep | XP_008535543.1 | XM_008537321.1 |
| Naked mole-rat | Heterocephalus glaber | hg | XP_010564126.1 | XM_021243735.1 |
| House mouse | Mus musculus | mm | XP_016348574.1 | NM_173386.5 |
| Bald eagle | Haliaeetus leucocephalus | hl | XP_021099394.1 | XM_010565824.1 |
| - | Sinocyclocheilus anshuiensis | sa | NP_775562.2 | XM_016493088.1 |
| Zebrafish | Danio rerio | dr | XP_016348574.1 | XM_680019.5 |
| cGAS Variant | Protein Yield [mg gCDW−1] |
|---|---|
| hs | 6.8 |
| fl hs | 6.7 |
| ep | 3.0 |
| hg | 9.9 |
| mm | 13.6 |
| mm(G379W) | 21.5 |
| hl | 4.1 |
| sa | 9.8 |
| dr | 3.2 |
| cGAS variant | Specific Activity [mU mg−1] | kcat [s−1] | Conversion [%] |
|---|---|---|---|
| hs | 73.5 ± 7.8 | 0.14 ± 0.01 | 93.0 ± 2.8 |
| fl hs | 0.5 ± 0.5 | 9.3 × 10−4 ± 9.3 × 10−4 | n.d. |
| ep | 1.7 ± 2.6 | 3.1 × 10−3 ± 4.8 × 10−3 | 5.5 ± 0.7 |
| hg | 75.8 ± 10.0 | 0.14 ± 0.02 | 68.0 ± 6.2 |
| mm | 279.2 ± 6.7 | 0.52 ± 0.01 | 92.3 ± 0.9 |
| mm(G379W) | n.d. | n.d. | n.d. |
| hl | 46.6 ± 9.2 | 0.09 ± 0.02 | 92.7 ± 10.3 |
| sa | n.d. | n.d. | n.d. |
| dr | 97.4 ± 12.6 | 0.18 ± 0.02 | 76.0 ± 3.5 |
References
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]
- Wu, J.; Sun, L.; Chen, X.; Du, F.; Shi, H.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP Is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science 2013, 339, 826–830. [Google Scholar] [CrossRef]
- Di Pietro, M.; Filardo, S.; Falasca, F.; Turriziani, O.; Sessa, R. cGAS/STING Pathway in Cancer: Jekyll and Hyde Story of Cancer Immune Response. Int. J. Mol. Sci. 2017, 18, 2459. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Cheng, H.; Yuan, H.; Xu, Q.; Shu, C.; Zhang, Y.; Xu, P.; Tan, J.; Rui, Y.; Li, P.; et al. Antitumor Activity of cGAMP via Stimulation of cGAS-cGAMP-STING-IRF3 Mediated Innate Immune Response. Sci. Rep. 2016, 6, 19049. [Google Scholar] [CrossRef] [PubMed]
- Ablasser, A.; Chen, Z.J. cGAS in action: Expanding roles in immunity and inflammation. Science 2019, 363, eaat8657. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Ascano, M.; Wu, Y.; Barchet, W.; Gaffney, B.L.; Zillinger, T.; Serganov, A.A.; Liu, Y.; Jones, R.A.; Hartmann, G.; et al. Cyclic [G(2′,5′)pA(3′,5′)p] Is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell 2013, 153, 1094–1107. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, H.; Wu, J.; Zhang, X.; Sun, L.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is An Endogenous High-Affinity Ligand for STING. Mol. Cell 2013, 51, 226–235. [Google Scholar] [CrossRef]
- Gaffney, B.L.; Veliath, E.; Zhao, J.; Jones, R.A. One-Flask Syntheses of c-di-GMP and the [ R p, R p ] and [ R p, S p ] Thiophosphate Analogues. Org. Lett. 2010, 12, 3269–3271. [Google Scholar] [CrossRef]
- Gaffney, B.L.; Jones, R.A. One-Flask Synthesis of Cyclic Diguanosine Monophosphate (c-di-GMP). Curr. Protoc. Nucleic Acid Chem. 2012, 48, 14–18. [Google Scholar] [CrossRef]
- Civril, F.; Deimling, T.; de Oliveira Mann, C.C.; Ablasser, A.; Moldt, M.; Witte, G.; Hornung, V.; Hopfner, K.-P. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 2013, 498, 332–337. [Google Scholar] [CrossRef]
- Zhou, W.; Whiteley, A.T.; de Oliveira Mann, C.C.; Morehouse, B.R.; Nowak, R.P.; Fischer, E.S.; Gray, N.S.; Mekalanos, J.J.; Kranzusch, P.J. Structure of the Human cGAS–DNA Complex Reveals Enhanced Control of Immune Surveillance. Cell 2018, 174, 300–311. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.; Ralph, E.C.; Shanker, S.; Wang, H.; Byrnes, L.J.; Horst, R.; Wong, J.; Brault, A.; Dumlao, D.; Smith, J.F.; et al. The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition. Protein Sci. 2017, 26, 2367–2380. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chu, B.; Du, L.; Han, Y.; Zhang, X.; Fan, S.; Wang, Y.; Yang, G. Molecular cloning and functional characterization of porcine cyclic GMP–AMP synthase. Mol. Immunol. 2015, 65, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Novotná, B.; Vaneková, L.; Zavřel, M.; Buděšínský, M.; Dejmek, M.; Smola, M.; Gutten, O.; Tehrani, Z.A.; Pimková Polidarová, M.; Brázdová, A.; et al. Enzymatic Preparation of 2’-5’, 3’-5’Cyclic Dinucleotides, Their Binding Properties to STING Adaptor Protein, and Structure/Activity Correlations. J. Med. Chem. 2019, 62, 10676–10690. [Google Scholar] [CrossRef]
- Tao, J.; Zhang, X.-W.; Jin, J.; Du, X.-X.; Lian, T.; Yang, J.; Zhou, X.; Jiang, Z.; Su, X.-D. Nonspecific DNA Binding of cGAS N Terminus Promotes cGAS Activation. J. Immunol. 2017, 198, 3627–3636. [Google Scholar] [CrossRef]
- Kranzusch, P.J.; Lee, A.S.-Y.; Berger, J.M.; Doudna, J.A. Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity. Cell Rep. 2013, 3, 1362–1368. [Google Scholar] [CrossRef]
- Rolf, J.; Rosenthal, K.; Lütz, S. Application of Cell-Free Protein Synthesis for Faster Biocatalyst Development. Catalysts 2019, 9, 190. [Google Scholar] [CrossRef]
- Caschera, F.; Noireaux, V. Synthesis of 2.3 mg/ml of protein with an all Escherichia coli cell-free transcription-translation system. Biochimie 2014, 99, 162–168. [Google Scholar] [CrossRef]
- Fenz, S.F.; Sachse, R.; Schmidt, T.; Kubick, S. Cell-free synthesis of membrane proteins: Tailored cell models out of microsomes. Biochim. Biophys. Acta -Biomembranes 2014, 1838, 1382–1388. [Google Scholar] [CrossRef]
- Adachi, J.; Katsura, K.; Seki, E.; Takemoto, C.; Shirouzu, M.; Terada, T.; Mukai, T.; Sakamoto, K.; Yokoyama, S. Cell-Free Protein Synthesis Using S30 Extracts from Escherichia coli RFzero Strains for Efficient Incorporation of Non-Natural Amino Acids into Proteins. Int. J. Mol. Sci. 2019, 20, 492. [Google Scholar] [CrossRef]
- Orth, J.H.C.; Schorch, B.; Boundy, S.; Ffrench-Constant, R.; Kubick, S.; Aktories, K. Cell-free synthesis and characterization of a novel cytotoxic pierisin-like protein from the cabbage butterfly Pieris rapae. Toxicon 2011, 57, 199–207. [Google Scholar] [CrossRef]
- Failmezger, J.; Rauter, M.; Nitschel, R.; Kraml, M.; Siemann-Herzberg, M. Cell-free protein synthesis from non-growing, stressed Escherichia coli. Sci. Rep. 2017, 7, 16524. [Google Scholar] [CrossRef] [PubMed]
- Nieß, A.; Failmezger, J.; Kuschel, M.; Siemann-Herzberg, M.; Takors, R. Experimentally Validated Model Enables Debottlenecking of in Vitro Protein Synthesis and Identifies a Control Shift under in Vivo Conditions. ACS Synth. Biol. 2017, 6, 1913–1921. [Google Scholar] [CrossRef]
- Goerke, A.R.; Swartz, J.R. Development of cell-free protein synthesis platforms for disulfide bonded proteins. Biotechnol. Bioeng. 2008, 99, 351–367. [Google Scholar] [CrossRef] [PubMed]
- Oza, J.P.; Aerni, H.R.; Pirman, N.L.; Barber, K.W.; ter Haar, C.M.; Rogulina, S.; Amrofell, M.B.; Isaacs, F.J.; Rinehart, J.; Jewett, M.C. Robust production of recombinant phosphoproteins using cell-free protein synthesis. Nat. Commun. 2015, 6, 8168. [Google Scholar] [CrossRef] [PubMed]
- Quertinmont, L.T.; Orru, R.; Lutz, S. RApid Parallel Protein EvaluatoR (RAPPER), from gene to enzyme function in one day. Chem. Commun. 2015, 51, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Pédelacq, J.-D.; Cabantous, S.; Tran, T.; Terwilliger, T.C.; Waldo, G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006, 24, 79–88. [Google Scholar] [CrossRef]
- Gregorio, N.E.; Levine, M.Z.; Oza, J.P. A User’s Guide to Cell-Free Protein Synthesis. Methods Protoc. 2019, 2, 24. [Google Scholar] [CrossRef] [PubMed]
- Cole, S.D.; Beabout, K.; Turner, K.B.; Smith, Z.K.; Funk, V.L.; Harbaugh, S.V.; Liem, A.T.; Roth, P.A.; Geier, B.A.; Emanuel, P.A.; et al. Quantification of Interlaboratory Cell-Free Protein Synthesis Variability. ACS Synth. Biol. 2019, 8, 2080–2091. [Google Scholar] [CrossRef]
- Levine, M.Z.; Gregorio, N.E.; Jewett, M.C.; Watts, K.R.; Oza, J.P. Escherichia coli-Based Cell-Free Protein Synthesis: Protocols for a robust, flexible, and accessible platform technology. J. Vis. Exp. 2019. [Google Scholar] [CrossRef]
- Wu, G.; Yan, Q.; Jones, J.A.; Tang, Y.J.; Fong, S.S.; Koffas, M.A.G. Metabolic Burden: Cornerstones in Synthetic Biology and Metabolic Engineering Applications. Trends Biotechnol. 2016, 34, 652–664. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wu, F.-H.; Wang, X.; Wang, L.; Siedow, J.N.; Zhang, W.; Pei, Z.-M. Molecular evolutionary and structural analysis of the cytosolic DNA sensor cGAS and STING. Nucleic Acids Res. 2014, 42, 8243–8257. [Google Scholar] [CrossRef] [PubMed]
- Zierhut, C.; Yamaguchi, N.; Paredes, M.; Luo, J.-D.; Carroll, T.; Funabiki, H. The Cytoplasmic DNA Sensor cGAS Promotes Mitotic Cell Death. Cell 2019, 178, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Chen, Z.J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 2018, 361, 704–709. [Google Scholar] [CrossRef]
- Xie, W.; Lama, L.; Adura, C.; Tomita, D.; Glickman, J.F.; Tuschl, T.; Patel, D.J. Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation. Proc. Natl. Acad. Sci. USA 2019, 116, 11946–11955. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Li, M.Z.; Elledge, S.J. SLIC: A Method for Sequence- and Ligation-Independent Cloning. In Gene Synthesis; Peccoud, J., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 51–59. [Google Scholar]
- Davanloo, P.; Rosenberg, A.H.; Dunn, J.J.; Studier, F.W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 1984, 81, 2035–2039. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Liu, H.; Jiang, Y.; Luo, Y.; Jiang, W. A Simple and Rapid Determination of ATP, ADP and AMP Concentrations in Pericarp Tissue of Litchi Fruit by High Performance Liquid Chromatography. Food Technol. Biotechnol. 2006, 44, 531–534. [Google Scholar]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef]
- Xia, P.; Ye, B.; Wang, S.; Zhu, X.; Du, Y.; Xiong, Z.; Tian, Y.; Fan, Z. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat. Immunol. 2016, 17, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Ablasser, A. ReGLUation of cGAS. Nat. Immunol. 2016, 17, 347–349. [Google Scholar] [CrossRef] [PubMed]

| Plasmid | Encoded Protein | Source |
|---|---|---|
| pET-28 a (+) SUMOthscGAS | truncated hs cGAS (N-terminal His6SUMO) | this study |
| pET-28 a (+) SUMOflhscGAS | full-length hs cGAS (N-terminal His6SUMO) | this study |
| pET-28 a (+) SUMOtepcGAS | truncated ep cGAS (N-terminal His6SUMO) | this study |
| pET-28 a (+) SUMOthgcGAS | truncated hg cGAS (N-terminal His6SUMO) | this study |
| pET-28 a (+) SUMOtmmcGAS | truncated mm cGAS (N-terminal His6SUMO) | this study |
| pET-28 a (+) SUMOtmmG379WcGAS | truncated mmG379W cGAS (N-terminal His6SUMO) | this study |
| pET-28 a (+) SUMOthlcGAS | truncated hl cGAS (N-terminal His6SUMO) | this study |
| pET-28 a (+) SUMOtsacGAS | truncated sa cGAS (N-terminal His6SUMO) | this study |
| pET-28 a (+) SUMOtdrcGAS | truncated dr cGAS (N-terminal His6SUMO) | this study |
| pET SUMOthscGASsfGFP | truncated hs cGAS (N-terminal His6SUMO, C-terminal sfGFP) | this study |
| pET SUMOtepcGASsfGFP | truncated ep cGAS (N-terminal His6SUMO, C-terminal sfGFP) | this study |
| pET SUMOthgcGASsfGFP | truncated hg cGAS (N-terminal His6SUMO, C-terminal sfGFP) | this study |
| pET SUMOtmmcGASsfGFP | truncated mm cGAS (N-terminal His6SUMO, C-terminal sfGFP) | this study |
| pET SUMOthlcGASsfGFP | truncated hl cGAS (N-terminal His6SUMO, C-terminal sfGFP) | this study |
| pET SUMOtsacGASsfGFP | truncated sa cGAS (N-terminal His6SUMO, C-terminal sfGFP) | this study |
| pET GFP LIC (u-msfGFP) | sfGFP | Addgene_29772 |
| pAR1219 | T7 RNA polymerase | [38] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rolf, J.; Siedentop, R.; Lütz, S.; Rosenthal, K. Screening and Identification of Novel cGAS Homologues Using a Combination of in Vitro and In Vivo Protein Synthesis. Int. J. Mol. Sci. 2020, 21, 105. https://doi.org/10.3390/ijms21010105
Rolf J, Siedentop R, Lütz S, Rosenthal K. Screening and Identification of Novel cGAS Homologues Using a Combination of in Vitro and In Vivo Protein Synthesis. International Journal of Molecular Sciences. 2020; 21(1):105. https://doi.org/10.3390/ijms21010105
Chicago/Turabian StyleRolf, Jascha, Regine Siedentop, Stephan Lütz, and Katrin Rosenthal. 2020. "Screening and Identification of Novel cGAS Homologues Using a Combination of in Vitro and In Vivo Protein Synthesis" International Journal of Molecular Sciences 21, no. 1: 105. https://doi.org/10.3390/ijms21010105
APA StyleRolf, J., Siedentop, R., Lütz, S., & Rosenthal, K. (2020). Screening and Identification of Novel cGAS Homologues Using a Combination of in Vitro and In Vivo Protein Synthesis. International Journal of Molecular Sciences, 21(1), 105. https://doi.org/10.3390/ijms21010105