A Metagenomic-Based Approach for the Characterization of Bacterial Diversity Associated with Spontaneous Malolactic Fermentations in Wine
Abstract
1. Introduction
2. Results
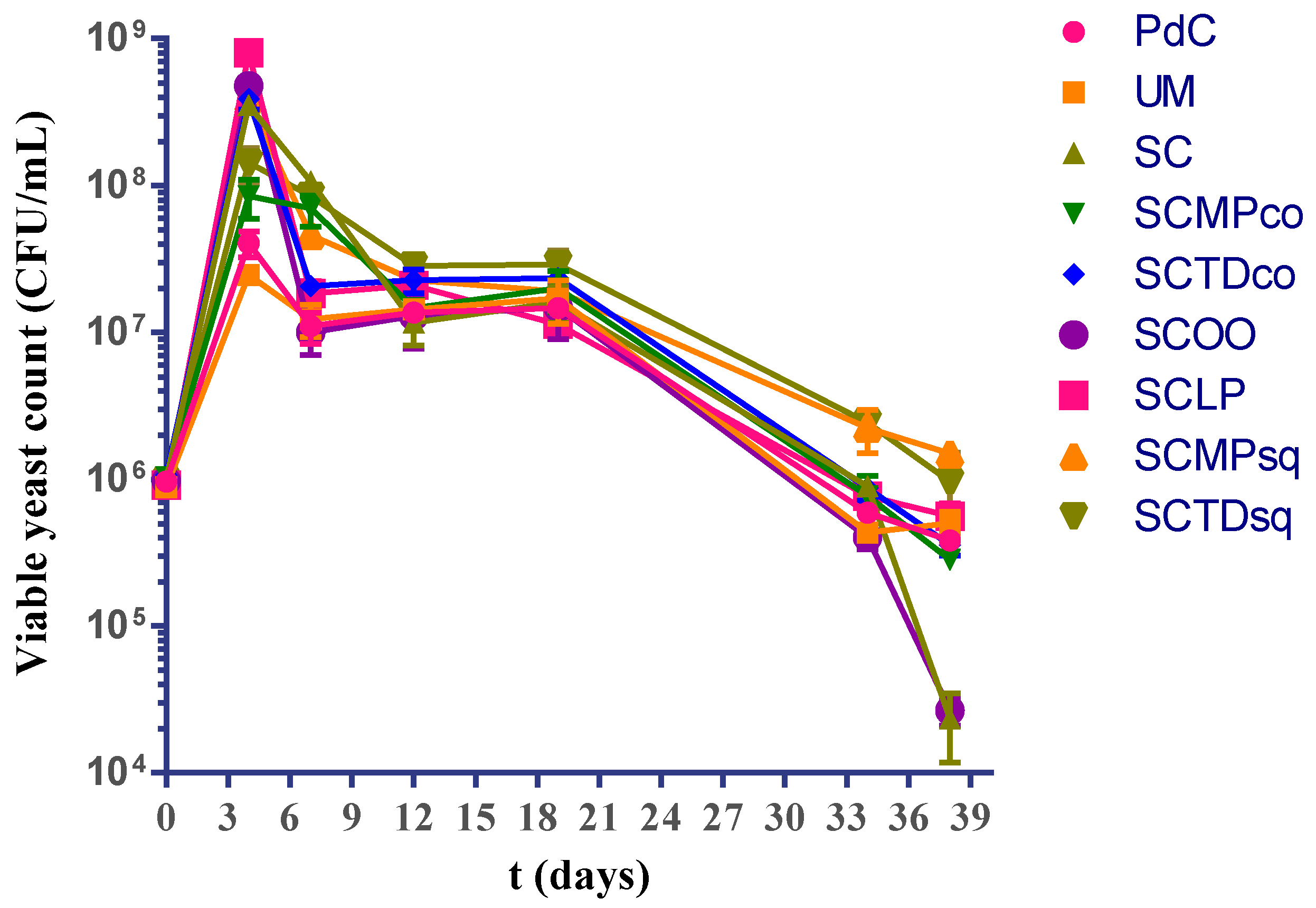
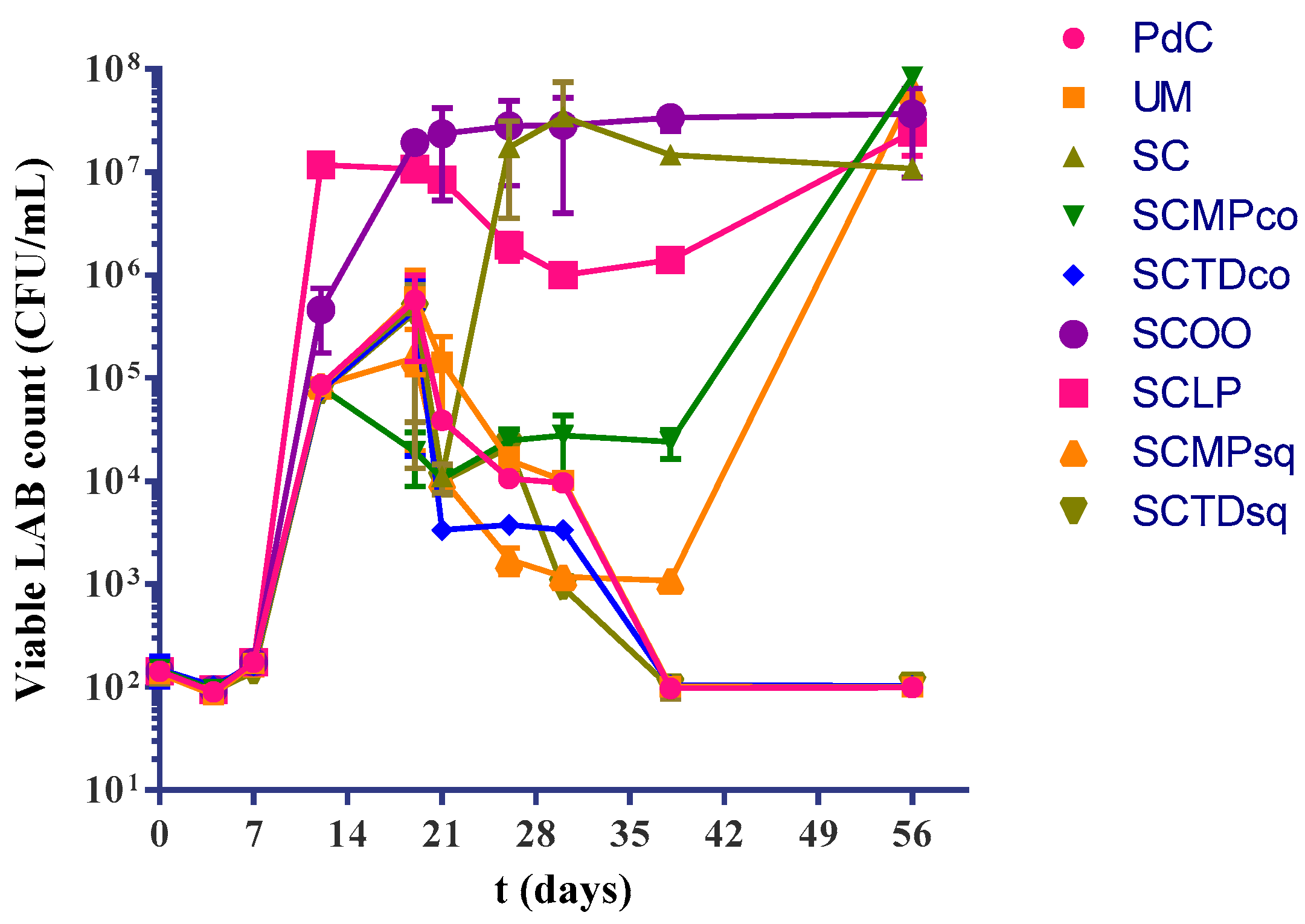
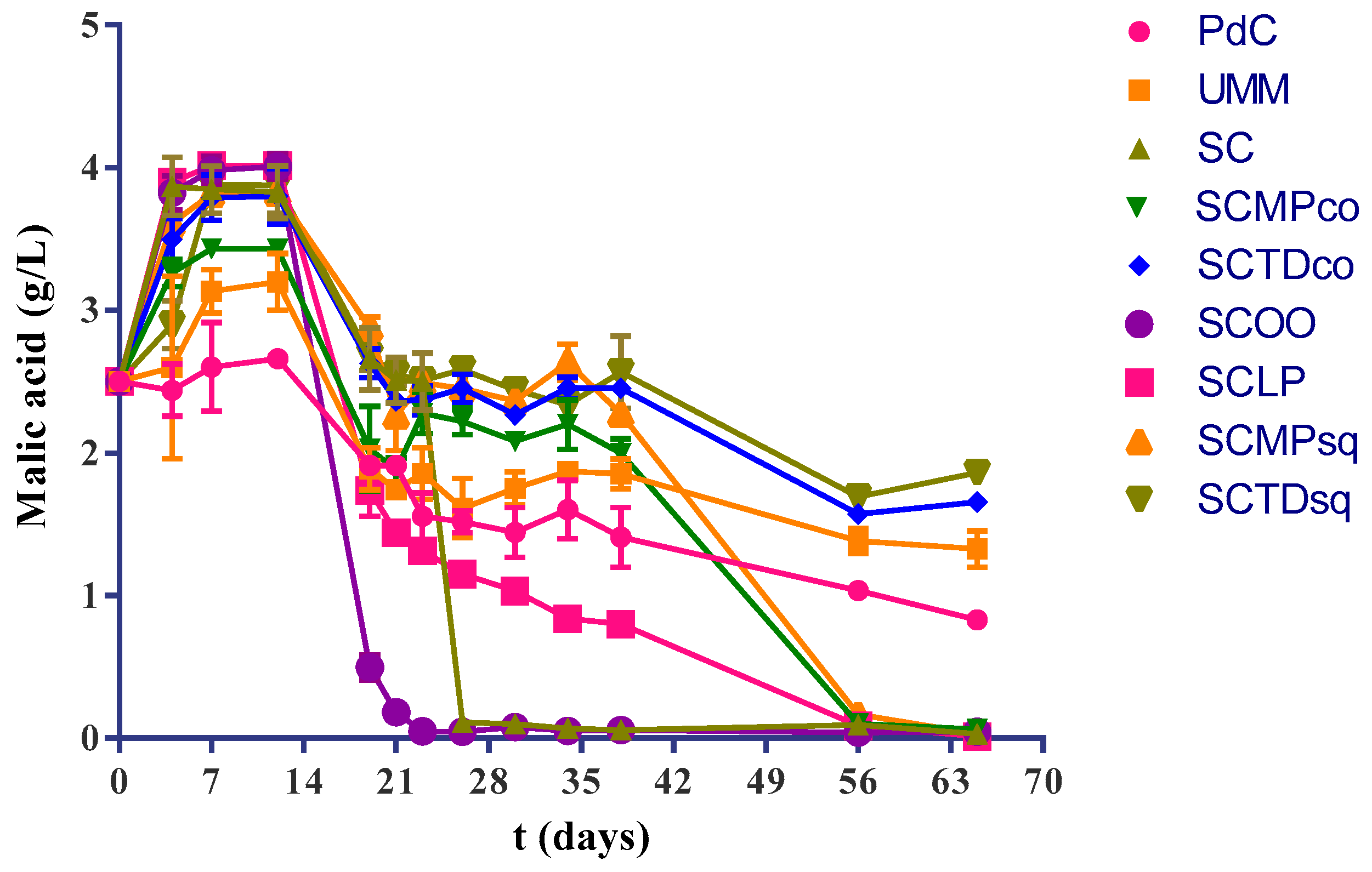
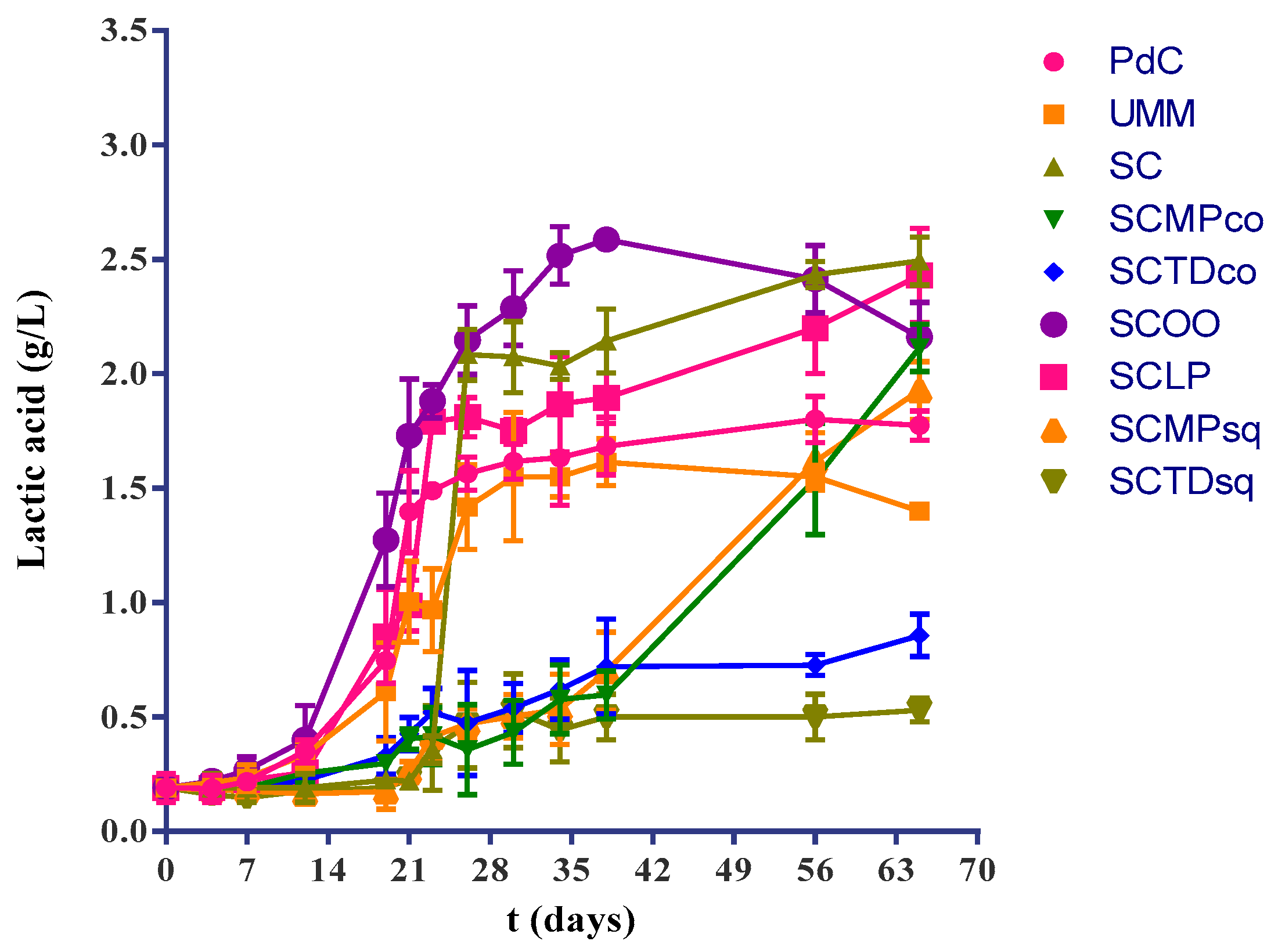
2.1. Experimental Modes and Monitoring of Malolactic Fermentation
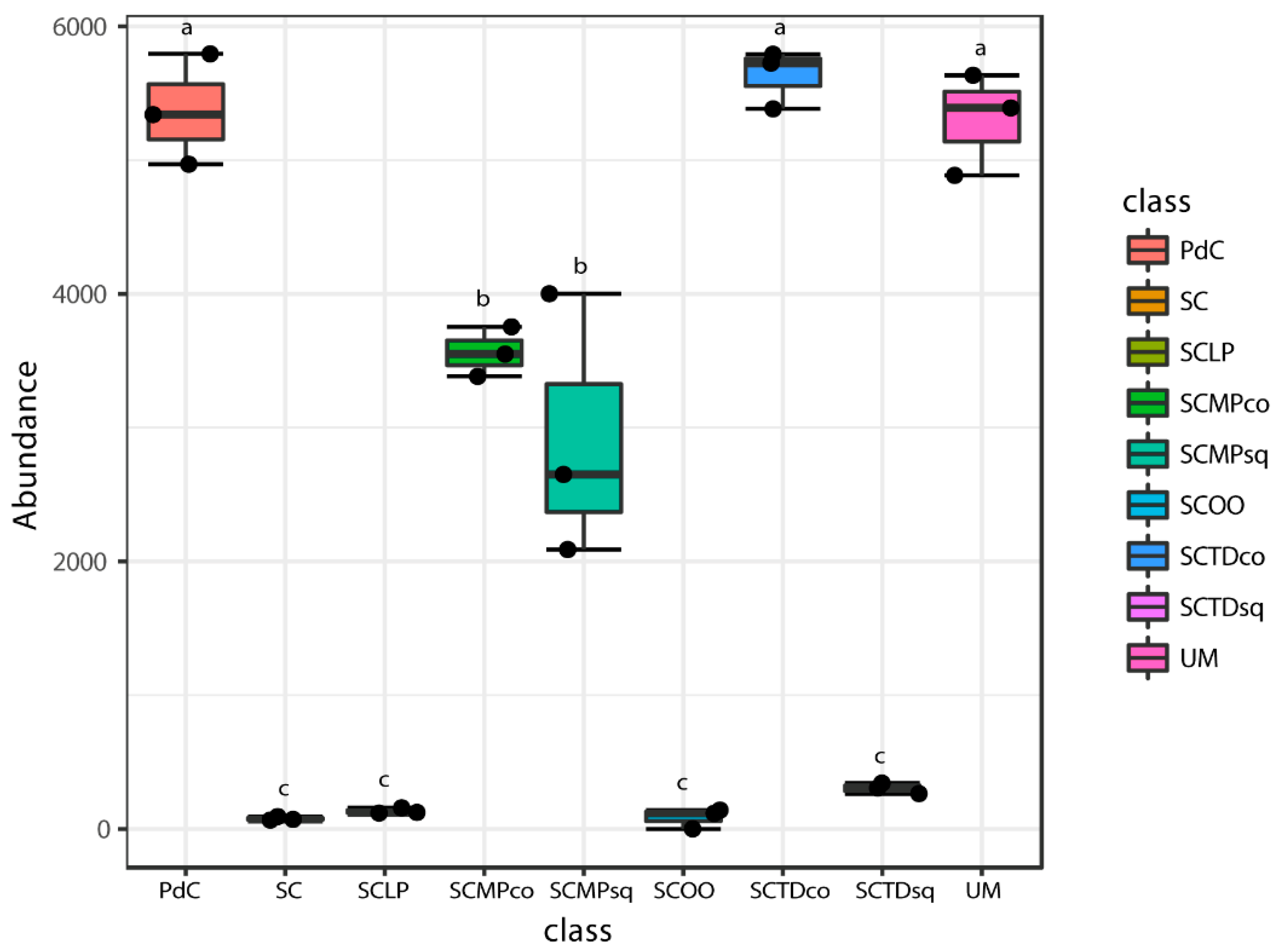
2.2. Malolactic Consortia Monitored using 16S High Throughput Sequencing
3. Discussion
4. Materials and Methods
4.1. Microorganisms
4.2. Vinifications
4.3. Determination of Microbial Population
4.4. Genomic DNA Extraction
4.5. High Throughput Sequencing
4.6. Statistical Analysis and Estimation of Fungal Alpha and Beta Diversity
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AF | Alcoholic Fermentation |
| MLF | Malolactic Fermentation |
| LAB | Lactic Acid Bacteria |
| OTUs | Operational Units |
| NGS | Next Generation Sequencing |
References
- Liu, Y.; Rousseaux, S.; Tourdot-Maréchal, R.; Sadoudi, M.; Gougeon, R.; Schmitt-Kopplin, P.; Alexandre, H. Wine microbiome: A dynamic world of microbial interactions. Crit. Rev. Food Sci. Nutr. 2017, 57, 856–873. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Capece, A.; Comitini, F.; Canonico, L.; Siesto, G.; Romano, P. Yeast Interactions in Inoculated Wine Fermentation. Front. Microbiol. 2016, 7, 555. [Google Scholar] [CrossRef] [PubMed]
- Belda, I.; Ruiz, J.; Esteban-Fernández, A.; Navascués, E.; Marquina, D.; Santos, A.; Moreno-Arribas, M.V. Microbial Contribution to Wine Aroma and Its Intended Use for Wine Quality Improvement. Molecules 2017, 22, 189. [Google Scholar] [CrossRef] [PubMed]
- Bauer, R.; Dicks, L.M.T. Control of Malolactic Fermentation in Wine. A Review. South Afr. J. Enol. Vitic. 2017, 25, 74–88. [Google Scholar] [CrossRef]
- Campbell-Sills, H.; Capozzi, V.; Romano, A.; Cappellin, L.; Spano, G.; Breniaux, M.; Lucas, P.; Biasioli, F. Advances in wine analysis by PTR-ToF-MS: Optimization of the method and discrimination of wines from different geographical origins and fermented with different malolactic starters. Int. J. Mass Spectrom. 2016, 397–398, 42–51. [Google Scholar] [CrossRef]
- Berbegal, C.; Spano, G.; Tristezza, M.; Grieco, F.; Capozzi, V. Microbial Resources and Innovation in the Wine Production Sector. S. Afr. J. Enol. Vitic. 2017, 38, 156–166. [Google Scholar] [CrossRef]
- Garofalo, C.; Arena, M.P.; Laddomada, B.; Cappello, M.S.; Bleve, G.; Grieco, F.; Beneduce, L.; Berbegal, C.; Spano, G.; Capozzi, V. Starter Cultures for Sparkling Wine. Fermentation 2016, 2, 21. [Google Scholar] [CrossRef]
- Petruzzi, L.; Capozzi, V.; Berbegal, C.; Corbo, M.R.; Bevilacqua, A.; Spano, G.; Sinigaglia, M. Microbial Resources and Enological Significance: Opportunities and Benefits. Front. Microbiol. 2017, 8, 995. [Google Scholar] [CrossRef] [PubMed]
- Tempère, S.; Marchal, A.; Barbe, J.-C.; Bely, M.; Masneuf-Pomarede, I.; Marullo, P.; Albertin, W. The complexity of wine: Clarifying the role of microorganisms. Appl. Microbiol. Biotechnol. 2018, 102, 3995–4007. [Google Scholar] [CrossRef]
- Russo, P.; Capozzi, V.; Spano, G.; Corbo, M.R.; Sinigaglia, M.; Bevilacqua, A. Metabolites of Microbial Origin with an Impact on Health: Ochratoxin A and Biogenic Amines. Front. Microbiol. 2016, 7, 482. [Google Scholar] [CrossRef]
- Escribano, R.; González-Arenzana, L.; Portu, J.; Garijo, P.; López-Alfaro, I.; López, R.; Santamaría, P.; Gutiérrez, A.R. Wine aromatic compound production and fermentative behaviour within different non-Saccharomyces species and clones. J. Appl. Microbiol. 2008, 124, 1521–1531. [Google Scholar] [CrossRef] [PubMed]
- Tofalo, R.; Patrignani, F.; Lanciotti, R.; Perpetuini, G.; Schirone, M.; Di Gianvito, P.; Pizzoni, D.; Arfelli, G.; Suzzi, G. Aroma Profile of Montepulciano d’Abruzzo Wine Fermented by Single and Co-culture Starters of Autochthonous Saccharomyces and Non-saccharomyces Yeasts. Front. Microbiol. 2016, 7, 610. [Google Scholar] [CrossRef] [PubMed]
- Chasseriaud, L.; Coulon, J.; Marullo, P.; Albertin, W.; Bely, M. New oenological practice to promote non-Saccharomyces species of interest: Saturating grape juice with carbon dioxide. Appl. Microbiol. Biotechnol. 2018, 102, 3779–3791. [Google Scholar] [CrossRef] [PubMed]
- Canonico, L.; Comitini, F.; Oro, L.; Ciani, M. Sequential Fermentation with Selected Immobilized Non-Saccharomyces Yeast for Reduction of Ethanol Content in Wine. Front. Microbiol. 2016, 7, 278. [Google Scholar] [CrossRef] [PubMed]
- Escribano, R.; González-Arenzana, L.; Garijo, P.; Berlanas, C.; López-Alfaro, I.; López, R.; Gutiérrez, A.R.; Santamaría, P. Screening of enzymatic activities within different enological non-Saccharomyces yeasts. J. Food Sci. Technol. 2017, 54, 1555–1564. [Google Scholar] [CrossRef]
- Roudil, L.; Russo, P.; Berbegal, C.; Albertin, W.; Spano, G.; Capozzi, V. Non-Saccharomyces Commercial Starter Cultures: Scientific Trends, Recent Patents and Innovation in the Wine Sector. Recent Pat. Food Nutr. Agric. 2019. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; Russo, P.; Beneduce, L.; Massa, S.; Spano, G.; Capozzi, V. Non-Saccharomyces biodiversity in wine and the ‘microbial terroir’: A survey on Nero di Troia wine from the Apulian region, Italy. Ann. Microbiol. 2016, 66, 143–150. [Google Scholar] [CrossRef]
- Comitini, F.; Capece, A.; Ciani, M.; Romano, P. New insights on the use of wine yeasts. Curr. Opin. Food Sci. 2017, 13, 44–49. [Google Scholar] [CrossRef]
- Knight, S.; Klaere, S.; Fedrizzi, B.; Goddard, M.R. Regional microbial signatures positively correlate with differential wine phenotypes: Evidence for a microbial aspect to terroir. Sci. Rep. 2015, 5, 14233. [Google Scholar] [CrossRef]
- Bely, M.; Stoeckle, P.; Masneuf-Pomarède, I.; Dubourdieu, D. Impact of mixed Torulaspora delbrueckii–Saccharomyces cerevisiae culture on high-sugar fermentation. Int. J. Food Microbiol. 2008, 122, 312–320. [Google Scholar] [CrossRef]
- Comitini, F.; Gobbi, M.; Domizio, P.; Romani, C.; Lencioni, L.; Mannazzu, I.; Ciani, M. Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 2011, 28, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Mas, A.; Esteve-Zarzoso, B. The Interaction between Saccharomyces cerevisiae and Non-Saccharomyces Yeast during Alcoholic Fermentation Is Species and Strain Specific. Front. Microbiol. 2016, 7, 502. [Google Scholar] [CrossRef] [PubMed]
- Ciani, M.; Comitini, F. Yeast interactions in multi-starter wine fermentation. Curr. Opin. Food Sci. 2015, 1, 1–6. [Google Scholar] [CrossRef]
- Alexandre, H.; Costello, P.J.; Remize, F.; Guzzo, J.; Guilloux-Benatier, M. Saccharomyces cerevisiae–Oenococcus oeni interactions in wine: Current knowledge and perspectives. Int. J. Food Microbiol. 2004, 93, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Nehme, N.; Mathieu, F.; Taillandier, P. Quantitative study of interactions between Saccharomyces cerevisiae and Oenococcus oeni strains. J. Ind. Microbiol. Biotechnol. 2008, 35, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Balmaseda, A.; Bordons, A.; Reguant, C.; Bautista-Gallego, J. Non-Saccharomyces in Wine: Effect Upon Oenococcus oeni and Malolactic Fermentation. Front. Microbiol. 2018, 9, 534. [Google Scholar] [CrossRef] [PubMed]
- Du Plessis, H.; du Toit, M.; Nieuwoudt, H.; van der Rijst, M.; Kidd, M.; Jolly, N. Effect of Saccharomyces, Non-Saccharomyces Yeasts and Malolactic Fermentation Strategies on Fermentation Kinetics and Flavor of Shiraz Wines. Fermentation 2017, 3, 64. [Google Scholar] [CrossRef]
- Du Plessis, H.W.; du Toit, M.; Hoff, J.W.; Hart, R.S.; Ndimba, B.K.; Jolly, N.P. Characterisation of Non-Saccharomyces yeasts using different methodologies and evaluation of their compatibility with malolactic fermentation. S. Afr. J. Enol. Vitic. 2017, 38, 46–63. [Google Scholar] [CrossRef]
- Sumby, K.M.; Bartle, L.; Grbin, P.R.; Jiranek, V. Measures to improve wine malolactic fermentation. Appl. Microbiol. Biotechnol. 2019, 103, 2033–2051. [Google Scholar] [CrossRef]
- Mayo, B.; Rachid, C.T.; Alegría, Á.; Leite, A.M.; Peixoto, R.S.; Delgado, S. Impact of Next Generation Sequencing Techniques in Food Microbiology. Curr. Genomics. 2014, 15, 293–309. [Google Scholar] [CrossRef]
- Morgan, H.H.; du Toit, M.; Setati, M.E. The Grapevine and Wine Microbiome: Insights from High-Throughput Amplicon Sequencing. Front. Microbiol. 2017, 8, 820. [Google Scholar] [CrossRef] [PubMed]
- Leveau, J.H.J.; Tech, J.J. Grapevine microbiomics: Bacterial diversity on grape leaves and berries revealed by high-throughput sequence analysis of 16s rRNA amplicons. Acta Hortic. 2011, 905, 31–42. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Joseph, C.M.L.; Allen, G.; Benson, A.K.; Mills, D.A. Next-Generation Sequencing Reveals Significant Bacterial Diversity of Botrytized Wine. PLoS ONE 2012, 7, e36357. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef] [PubMed]
- Pinto, C.; Pinho, D.; Cardoso, R.; Custódio, V.; Fernandes, J.; Sousa, S.; Pinheiro, M.; Egas, C.; Gomes, A.C. Wine fermentation microbiome: A landscape from different Portuguese wine appellations. Front. Microbiol. 2015, 6, 905. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Swadener, M.; Sakamoto, K.; Mills, D.A.; Bisson, L.F. Sulfur Dioxide Treatment Alters Wine Microbial Diversity and Fermentation Progression in a Dose-Dependent Fashion. Am. J. Enol. Vitic. 2015, 66, 73–79. [Google Scholar] [CrossRef]
- Piao, H.; Hawley, E.; Kopf, S.; DeScenzo, R.; Sealock, S.; Henick-Kling, T.; Hess, M. Insights into the bacterial community and its temporal succession during the fermentation of wine grapes. Front. Microbiol. 2015, 6, 809. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations among Wine Grape Microbiome, Metabolome, and Fermentation Behavior Suggest Microbial Contribution to Regional Wine Characteristics. MBio 2016, 7, e00631-16. [Google Scholar] [CrossRef]
- Marzano, M.; Fosso, B.; Manzari, C.; Grieco, F.; Intranuovo, M.; Cozzi, G.; Mulè, G.; Scioscia, G.; Valiente, G.; Tullo, A.; et al. Complexity and Dynamics of the Winemaking Bacterial Communities in Berries, Musts, and Wines from Apulian Grape Cultivars through Time and Space. PLoS ONE 2016, 11, e0157383. [Google Scholar] [CrossRef]
- Salvetti, E.; Campanaro, S.; Campedelli, I.; Fracchetti, F.; Gobbi, A.; Tornielli, G.B.; Torriani, S.; Felis, G.E. Whole-Metagenome-Sequencing-Based Community Profiles of Vitis vinifera L. cv. Corvina Berries Withered in Two Post-harvest Conditions. Front. Microbiol. 2016, 7, 937. [Google Scholar]
- Mezzasalma, V.; Sandionigi, A.; Guzzetti, L.; Galimberti, A.; Grando, M.S.; Tardaguila, J.; Labra, M. Geographical and Cultivar Features Differentiate Grape Microbiota in Northern Italy and Spain Vineyards. Front. Microbiol. 2018, 9, 946. [Google Scholar] [CrossRef] [PubMed]
- Lleixà, J.; Kioroglou, D.; Mas, A.; del Carmen Portillo, M. Microbiome dynamics during spontaneous fermentations of sound grapes in comparison with sour rot and Botrytis infected grapes. Int. J. Food Microbiol. 2018, 281, 36–46. [Google Scholar]
- Vitulo, N.; Lemos, W.J.F.; Calgaro, M.; Confalone, M.; Felis, G.E.; Zapparoli, G.; Nardi, T. Bark and Grape Microbiome of Vitis vinifera: Influence of Geographic Patterns and Agronomic Management on Bacterial Diversity. Front. Microbiol. 2018, 9, 3203. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Chen, J.; Xiao, J.; Cheng, W.; Zheng, X.; Wang, B.; Shi, X. Microbial community composition on grape surface controlled by geographical factors of different wine regions in Xinjiang, China. Food Res. Int. 2019, 122, 348–360. [Google Scholar] [CrossRef] [PubMed]
- Sirén, K.; Mak, S.S.T.; Melkonian, C.; Carøe, C.; Swiegers, J.H.; Molenaar, D.; Fischer, U.; Gilbert, M.T.P. Taxonomic and Functional Characterization of the Microbial Community During Spontaneous in vitro Fermentation of Riesling Must. Front. Microbiol. 2019, 10, 697. [Google Scholar] [CrossRef]
- Clavijo, A.; Calderón, I.L.; Paneque, P. Yeast assessment during alcoholic fermentation inoculated with a natural “pied de cuve” or a commercial yeast strain. World J. Microbiol. Biotechnol. 2011, 27, 1569–1577. [Google Scholar] [CrossRef]
- Mira de Orduña, R. Climate change associated effects on grape and wine quality and production. Food Res. Int. 2010, 43, 1844–1855. [Google Scholar] [CrossRef]
- Capozzi, V.; Russo, P.; Beneduce, L.; Weidmann, S.; Grieco, F.; Guzzo, J.; Spano, G. Technological properties of Oenococcus oeni strains isolated from typical southern Italian wines. Lett. Appl. Microbiol. 2010, 50, 327–334. [Google Scholar] [CrossRef]
- Stefanini, I.; Cavalieri, D. Metagenomic Approaches to Investigate the Contribution of the Vineyard Environment to the Quality of Wine Fermentation: Potentials and Difficulties. Front. Microbiol. 2018. [Google Scholar] [CrossRef]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef]
- Tofalo, R.; Perpetuini, G.; Schirone, M.; Fasoli, G.; Aguzzi, I.; Corsetti, A.; Suzzi, G. Biogeographical characterization of Saccharomyces cerevisiae wine yeast by molecular methods. Front. Microbiol. 2013, 4, 166. [Google Scholar] [CrossRef] [PubMed]
- Capece, A.; Granchi, L.; Guerrini, S.; Mangani, S.; Romaniello, R.; Vincenzini, M.; Romano, P. Diversity of Saccharomyces cerevisiae Strains Isolated from Two Italian Wine-Producing Regions. Front. Microbiol. 2016, 7, 1018. [Google Scholar] [CrossRef] [PubMed]
- Contreras, A.; Curtin, C.; Varela, C. Yeast population dynamics reveal a potential “collaboration” between Metschnikowia pulcherrima and Saccharomyces uvarum for the production of reduced alcohol wines during Shiraz fermentation. Appl. Microbiol. Biotechnol. 2015, 99, 1885–1895. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; El Khoury, M.; Lucas, P.; Bely, M.; Russo, P.; Spano, G.; Capozzi, V. Autochthonous starter cultures and indigenous grape variety for regional wine production. J. Appl. Microbiol. 2015, 118, 1395–1408. [Google Scholar] [CrossRef] [PubMed]
- Berbegal, C.; Peña, N.; Russo, P.; Grieco, F.; Pardo, I.; Ferrer, S.; Spano, G.; Capozzi, V. Technological properties of Lactobacillus plantarum strains isolated from grape must fermentation. Food Microbiol. 2016, 57, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Lonvaud-Funel, A. Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie Van Leeuwenhoek 1999, 76, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Campbell-Sills, H.; Khoury, M.E.; Gammacurta, M.; Miot-Sertier, C.; Dutilh, L.; Vestner, J.; Capozzi, V.; Sherman, D.; Hubert, C.; Claisse, O.; et al. Two different Oenococcus oeni lineages are associated to either red or white wines in Burgundy: Genomics and metabolomics insights. OENO One 2017, 51, 309. [Google Scholar] [CrossRef]
- Benito, Á.; Calderón, F.; Palomero, F.; Benito, S. Combine Use of Selected Schizosaccharomyces pombe and Lachancea thermotolerans Yeast Strains as an Alternative to the Traditional Malolactic Fermentation in Red Wine Production. Molecules 2015, 20, 9510–9523. [Google Scholar] [CrossRef] [PubMed]
- Lorentzen, M.P.G.; Lucas, P.M. Distribution of Oenococcus oeni populations in natural habitats. Appl. Microbiol. Biotechnol. 2019, 103, 2937–2945. [Google Scholar] [CrossRef] [PubMed]
- Capozzi, V.; Fragasso, M.; Romaniello, R.; Berbegal, C.; Russo, P.; Spano, G. Spontaneous Food Fermentations and Potential Risks for Human Health. Fermentation 2017, 3, 49. [Google Scholar] [CrossRef]
- Spano, G.; Capozzi, V. Food Microbial Biodiversity and Microbes of Protected Origin. Front. Microbiol. 2011, 2, 237. [Google Scholar]
- Gobert, A.; Tourdot-Maréchal, R.; Morge, C.; Sparrow, C.; Liu, Y.; Quintanilla-Casas, B.; Vichi, S.; Alexandre, H. Non-Saccharomyces Yeasts Nitrogen Source Preferences: Impact on Sequential Fermentation and Wine Volatile Compounds Profile. Front. Microbiol. 2017, 8, 2175. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Chua, J.-Y.; Huang, D.; Lee, P.-R.; Liu, S.-Q. Biotransformation of chemical constituents of durian wine with simultaneous alcoholic fermentation by Torulaspora delbrueckii and malolactic fermentation by Oenococcus oeni. Appl. Microbiol. Biotechnol. 2016, 100, 8877–8888. [Google Scholar] [CrossRef] [PubMed]
- Nardi, T.; Panero, L.; Petrozziello, M.; Guaita, M.; Tsolakis, C.; Cassino, C.; Vagnoli, P.; Bosso, A. Managing wine quality using Torulaspora delbrueckii and Oenococcus oeni starters in mixed fermentations of a red Barbera wine. Eur. Food Res. Technol. 2019, 245, 293–307. [Google Scholar] [CrossRef]
- Capozzi, V.; Berbegal, C.; Tufariello, M.; Grieco, F.; Spano, G.; Grieco, F. Impact of co-inoculation of Saccharomyces cerevisiae, Hanseniaspora uvarum and Oenococcus oeni autochthonous strains in controlled multi starter grape must fermentations. LWT 2019, 109, 241–249. [Google Scholar] [CrossRef]
- Padilla, B.; Zulian, L.; Ferreres, À.; Pastor, R.; Esteve-Zarzoso, B.; Beltran, G.; Mas, A. Sequential Inoculation of Native Non-Saccharomyces and Saccharomyces cerevisiae Strains for Wine Making. Front. Microbiol. 2017, 8, 1293. [Google Scholar] [CrossRef] [PubMed]
- Bartowsky, E.J. Oenococcus oeni and malolactic fermentation–Moving into the molecular arena. Aust. J. Grape Wine Res. 2008, 11, 174–187. [Google Scholar] [CrossRef]
- Lucio, O.; Pardo, I.; Heras, J.M.; Krieger-Weber, S.; Ferrer, S. Use of starter cultures of Lactobacillus to induce malolactic fermentation in wine. Aust. J. Grape Wine Res. 2017, 23, 15–21. [Google Scholar] [CrossRef]
- Renault, P.; Gaillardin, C.; Heslot, H. Product of the Lactococcus lactis gene required for malolactic fermentation is homologous to a family of positive regulators. J. Bacteriol. 1989, 171, 3108–3114. [Google Scholar] [CrossRef]
- Bae, S.; Fleet, G.H.; Heard, G.M. Lactic acid bacteria associated with wine grapes from several Australian vineyards. J. Appl. Microbiol. 2006, 100, 712–727. [Google Scholar] [CrossRef]
- Kioroglou, D.; LLeixá, J.; Mas, A.; Del Carmen Portillo, M. Massive Sequencing: A New Tool for the Control of Alcoholic Fermentation in Wine? Fermentation 2018, 4, 7. [Google Scholar] [CrossRef]
- Wolfe, B.E.; Dutton, R.J. Fermented foods as experimentally tractable microbial ecosystems. Cell 2015, 161, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; Berbegal, C.; Grieco, F.; Tufariello, M.; Spano, G.; Capozzi, V. Selection of indigenous yeast strains for the production of sparkling wines from native Apulian grape varieties. Int. J. Food Microbiol. 2018, 285, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 15 September 2018).
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Vegan: Community Ecology Package. Ordination methods, Diversity Analysis and Other Functions for Community and Vegetation Ecologists|World Agroforestry Centre. Available online: http://www.worldagroforestry.org/publication/vegan-community-ecology-package-ordination-methods-diversity-analysis-and-other (accessed on 15 September 2018).

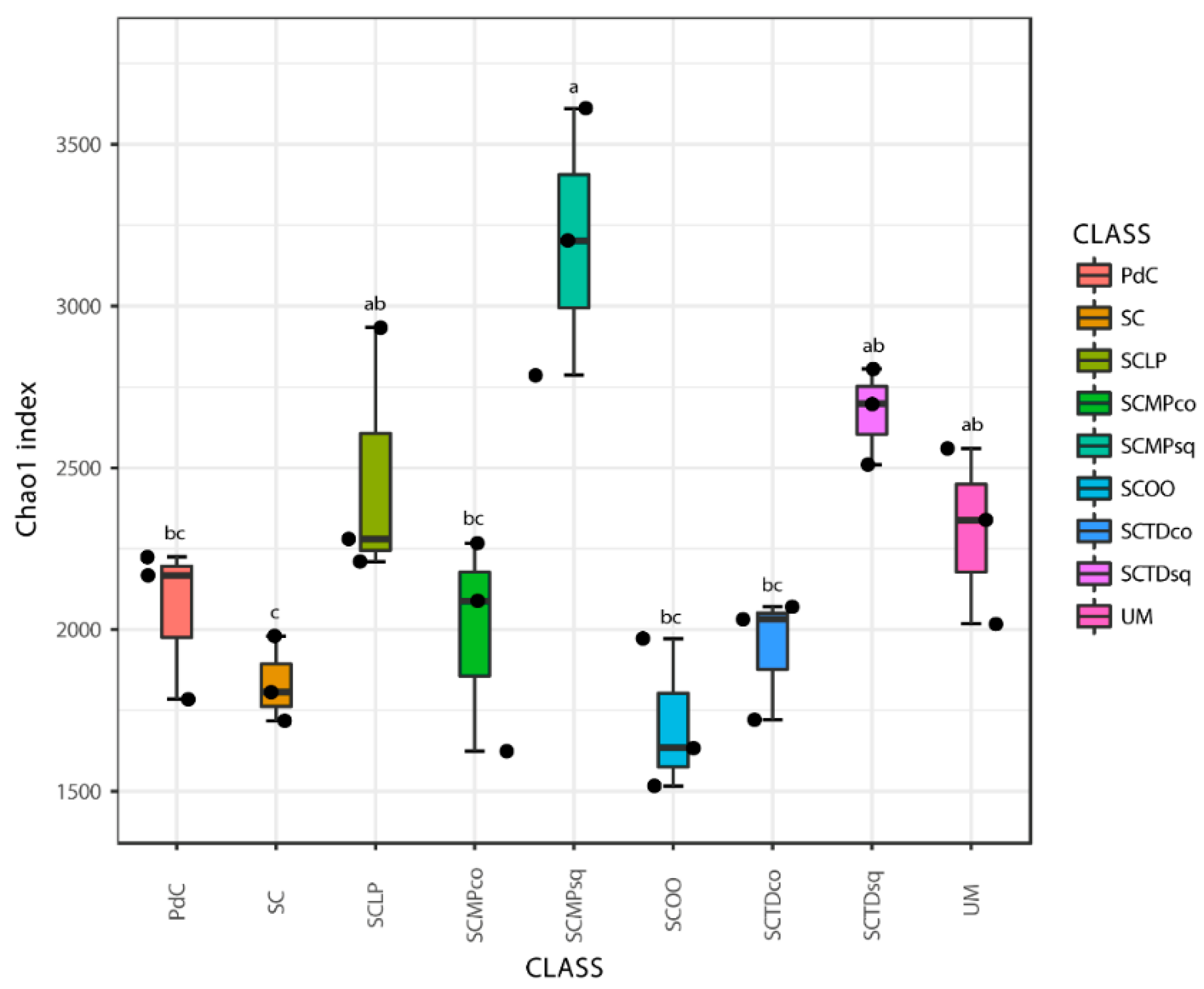
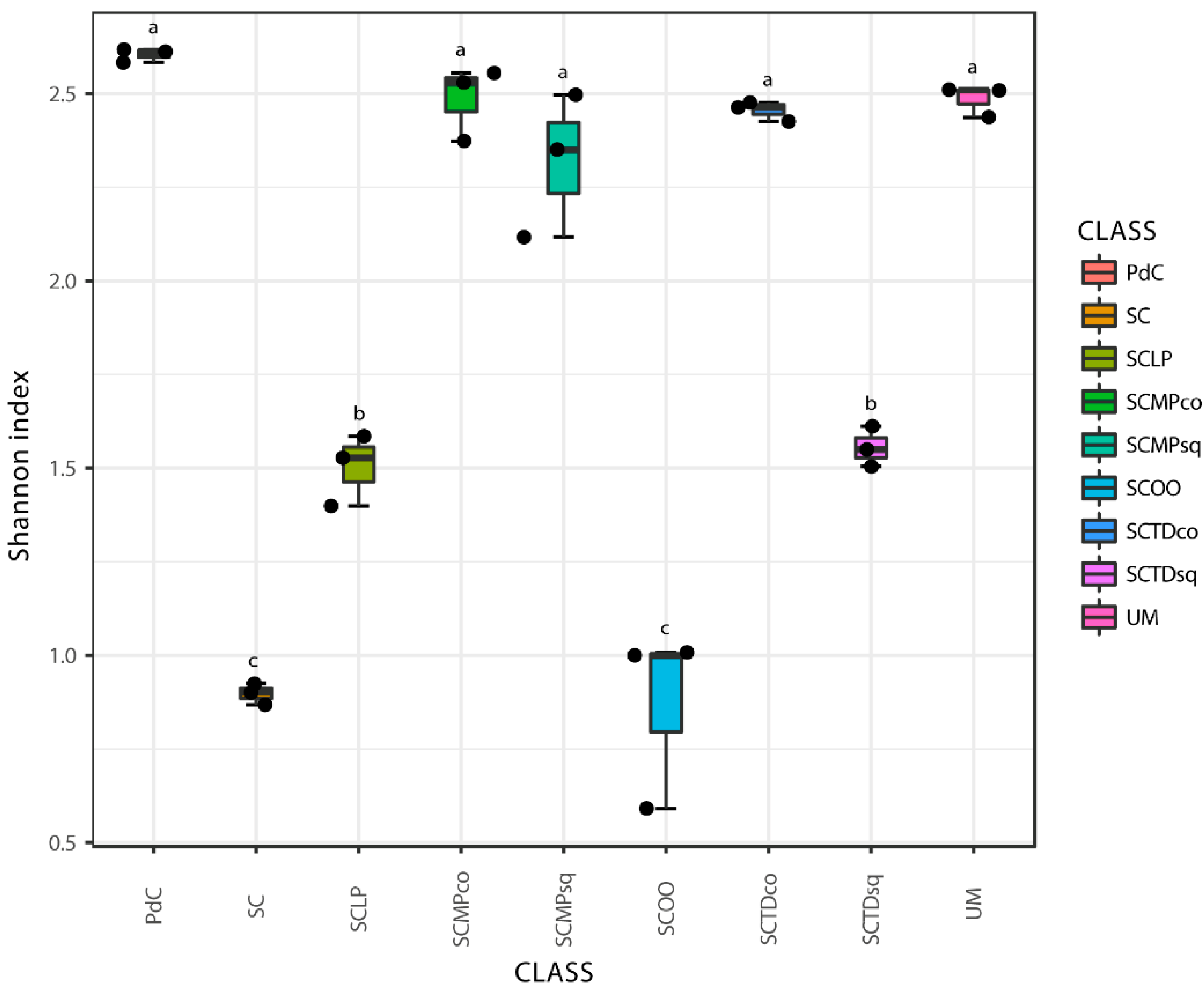
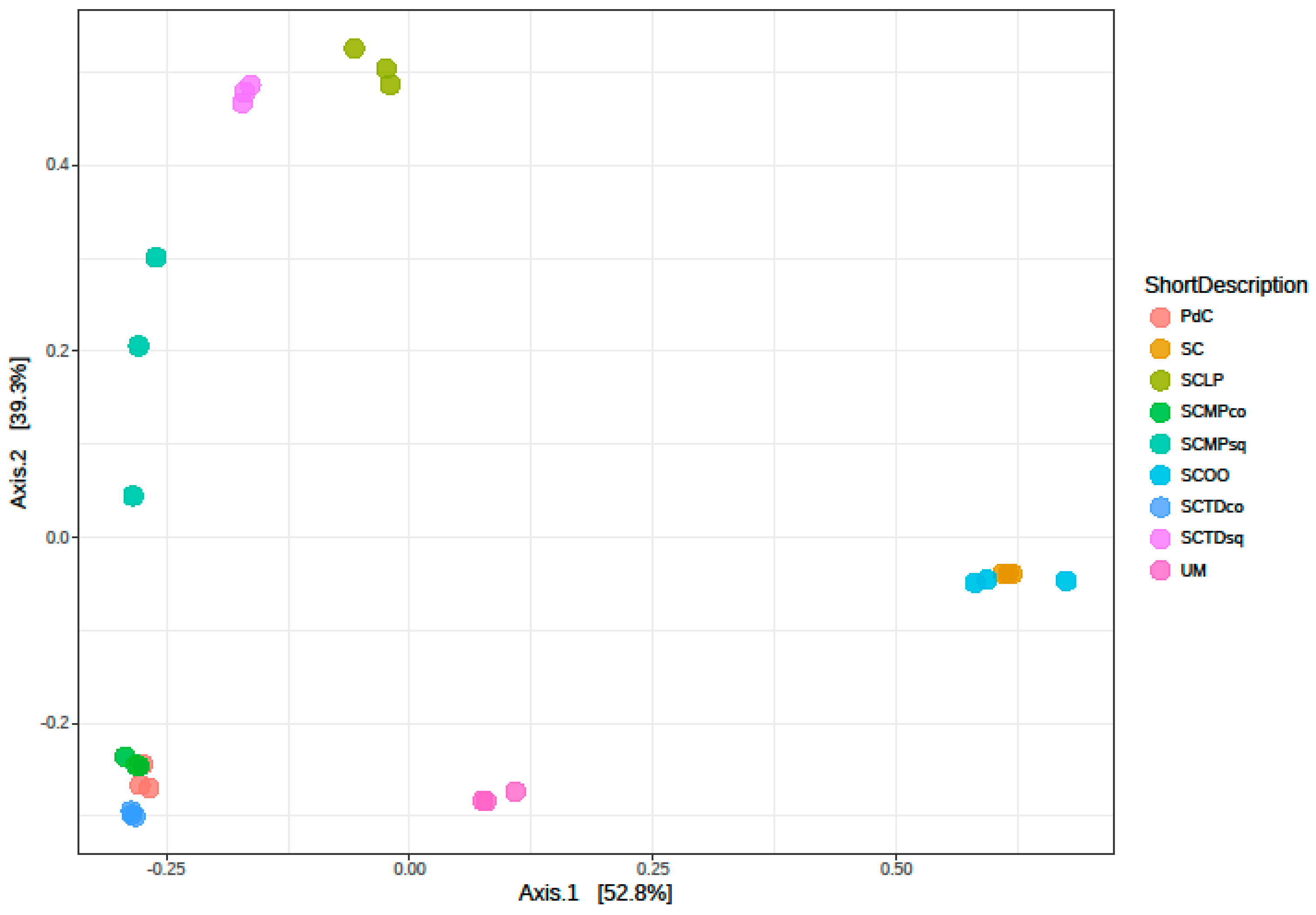
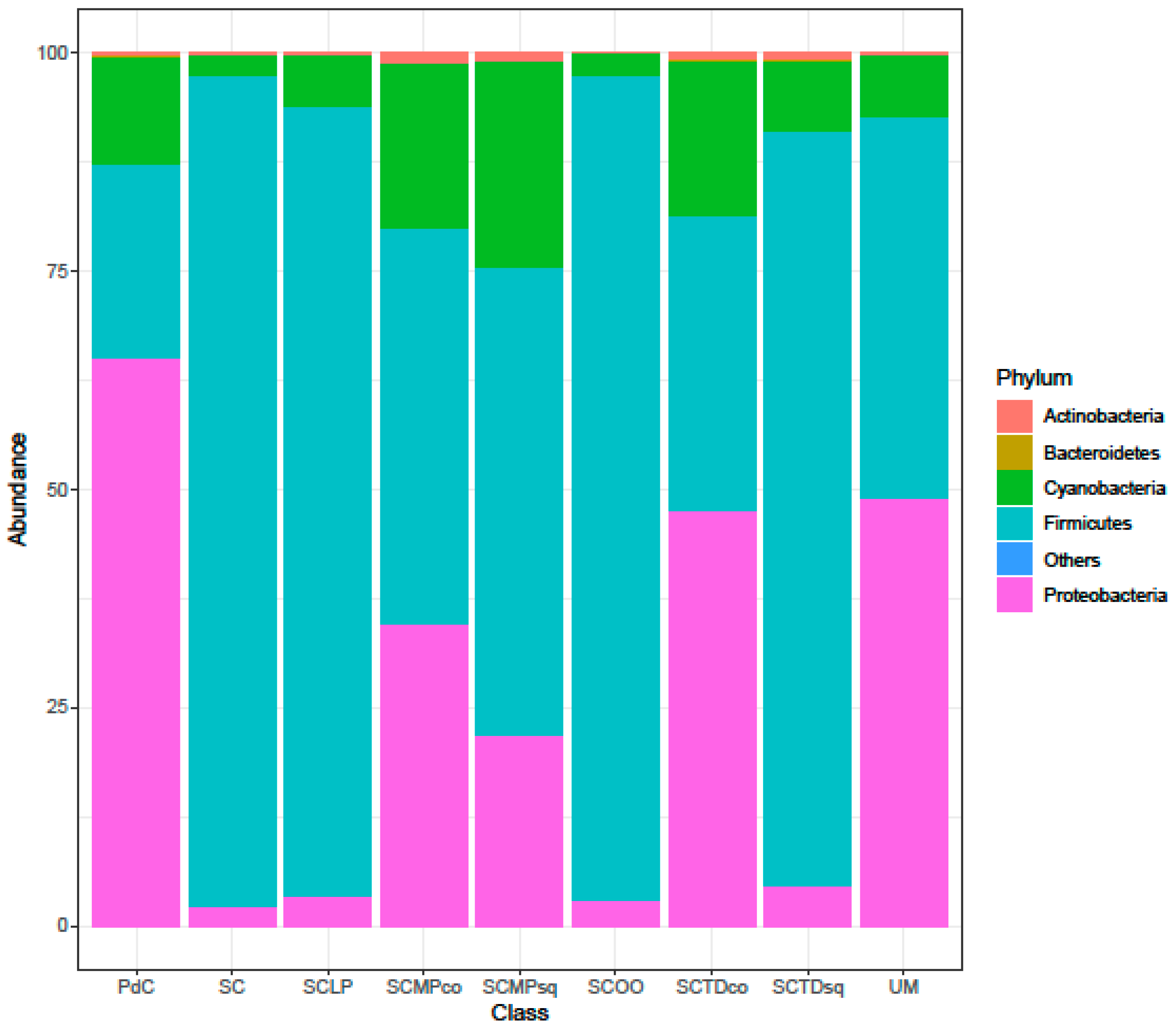
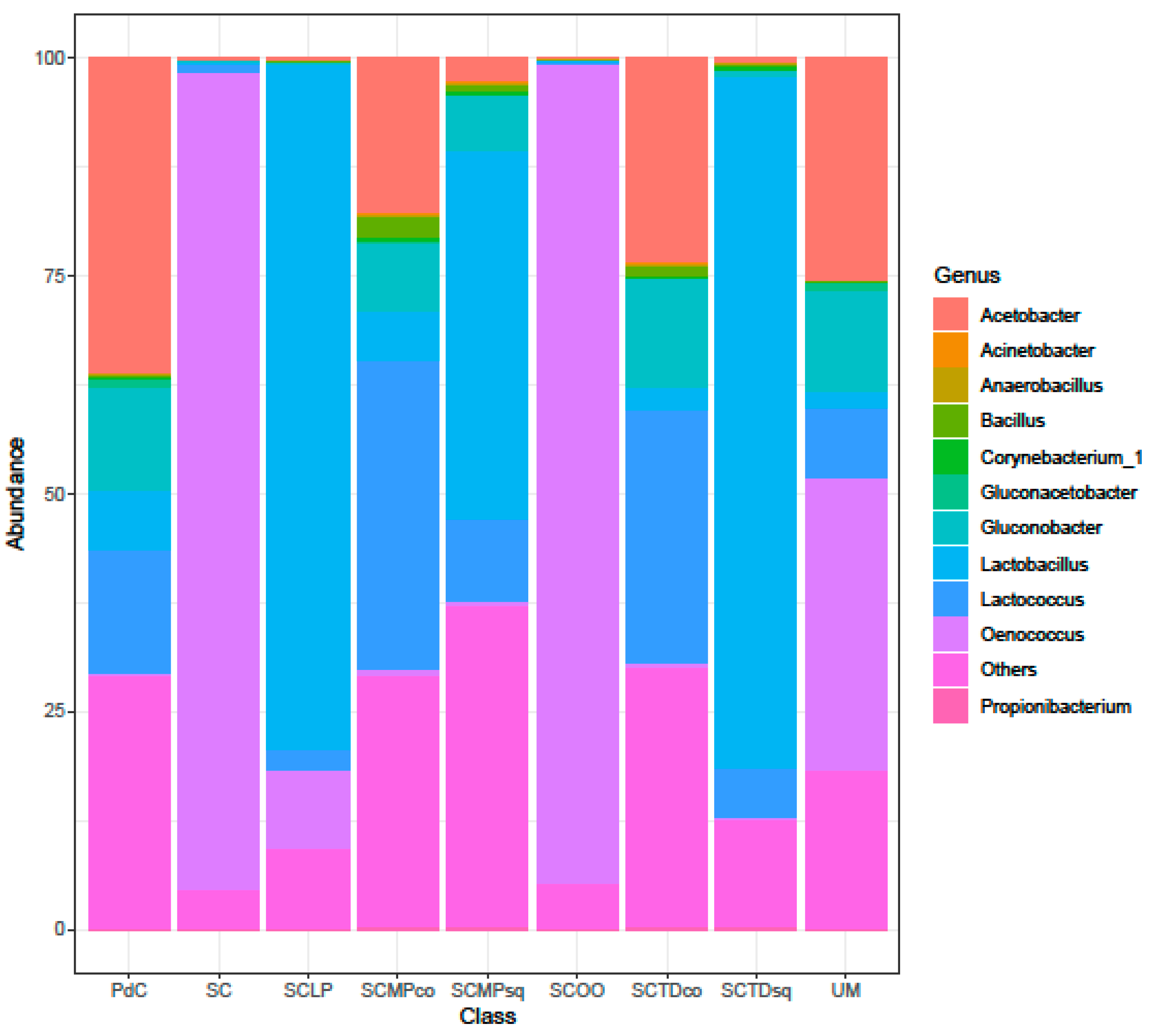
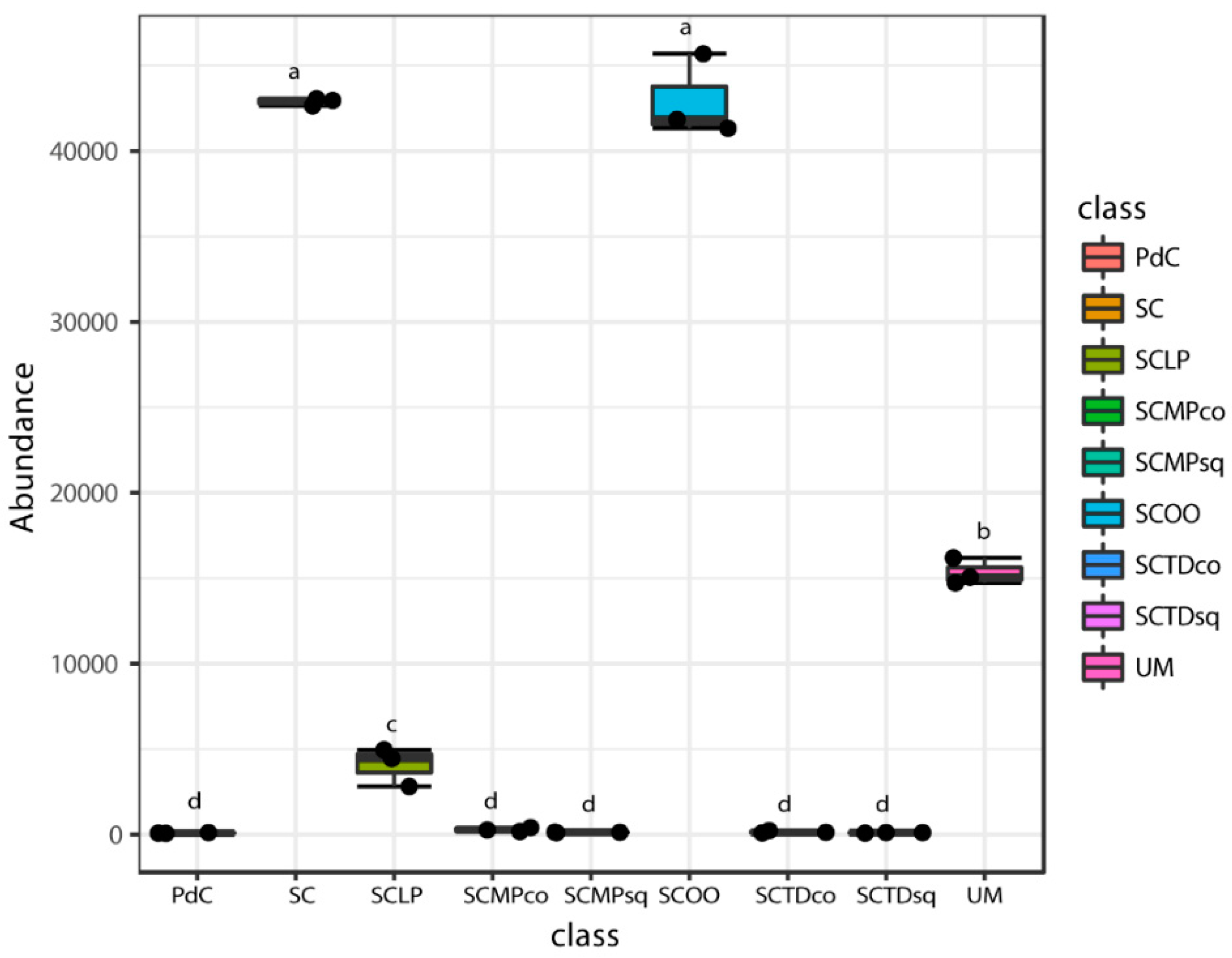
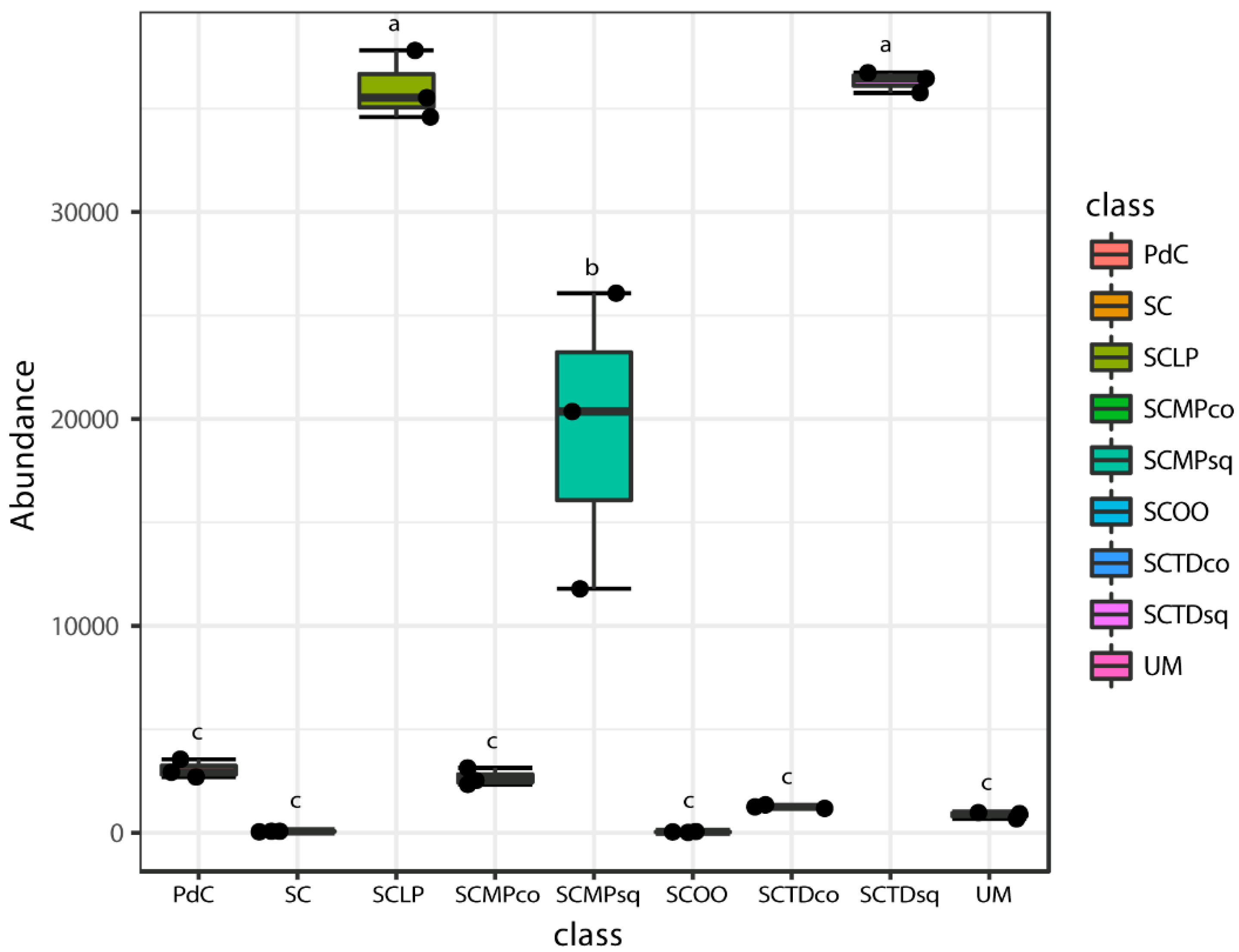
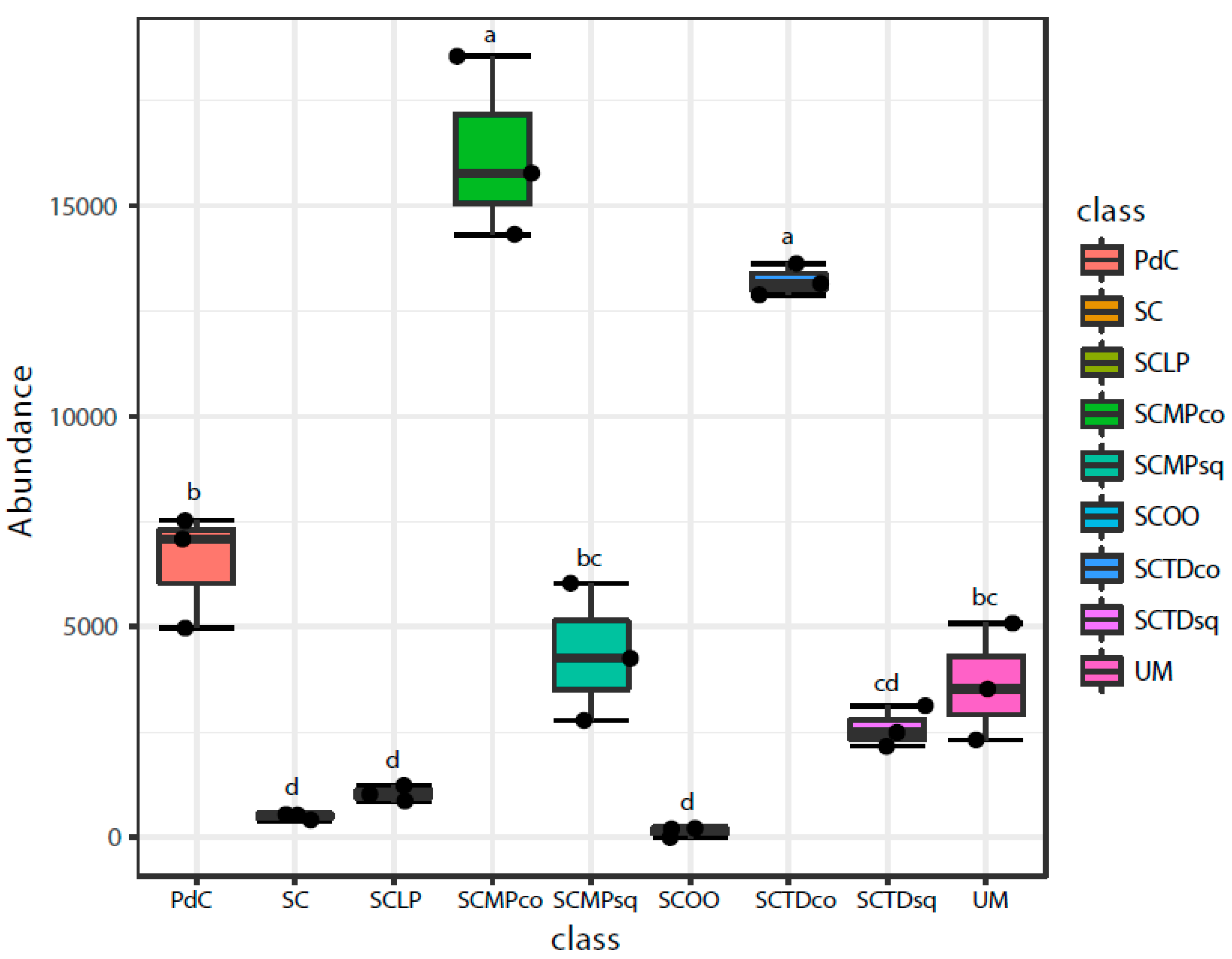

| Year | Subject | References |
|---|---|---|
| 2011 | Study of bacterial communities associated with the leaf and berry surfaces of ‘Chardonnay’ grape | [32] |
| 2012 | Study of bacterial communities associated with botrytized wine fermentations (sampling time compatible with MLF) | [33] |
| 2014 | Grape-associated microbial biogeography from different regions of California | [34] |
| 2015 | Study of bacterial communities associated with spontaneous wine fermentations (from six Portuguese wine appellations) in the initial musts, start and end of alcoholic fermentation | [35] |
| 2015 | Influence of the use of sulfur dioxide (in winemaking) on bacterial diversity | [36] |
| 2015 | Study of the evolution of bacterial communities during the alcoholic fermentation of organically and conventionally produced wines | [37] |
| 2016 | Study of the bacterial communities associated with the main wine fermentation stages including malolactic fermentation | [38] |
| 2016 | Study of the bacterial communities in berries, musts, and wines, including malolactic fermentation | [39] |
| 2016 | Study of the bacterial communities associated with Corvina berries at the end of the withering process performed in two different conditions | [40] |
| 2018 | Correlation between soil- and grape-associated bacterial communities in vineyards | [41] |
| 2018 | Study of bacterial diversity associated with healthy, rotten, botrytized grapes and with the derived musts and wine (up to the alcoholic fermentation) | [42] |
| 2018 | Study of the epiphytic bacterial community of vine bark and its relationships with grape bacterial diversity | [43] |
| 2019 | Study of bacterial diversity associated with the grape surface of samples collected from different wine regions in Xinjiang, China | [44] |
| 2019 | Bacterial communities associated with alcoholic fermentation, including the variation between musts that successfully complete alcoholic fermentation and those that become ‘stuck’ in the process | [45] |
| Codes | Tested Management of The Microbial Resources |
|---|---|
| PdC | ‘Pied-de-cuve’ practice |
| UM | Uninoculated must |
| SC | S. cerevisiae |
| SCMPco | S. cerevisiae + M. pulcherrima (co) |
| SCTDco | S. cerevisiae + T. delbrueckii (co) |
| SCOO | S. cerevisiae + O. oeni |
| SCLP | S. cerevisiae + L. plantarum |
| SCMPsq | S. cerevisiae + M. pulcherrima (sq) |
| SCTDsq | S. cerevisiae + T. delbrueckii (sq) |
| L-malic Acid (g/L) | Lactic Acid (g/L) | |
|---|---|---|
| PdC | 0.830 ± 0.052 a | 1.773 ± 0.064 a |
| UM | 1.327 ± 0.127 b | 1.400 ± 0.001 b |
| SC | 0.030 ± 0.017 c | 2.493 ± 0.105 c |
| SCMPco | 0.062 ± 0.032 c | 2.113 ± 0.102 d |
| SCTDco | 1.657 ± 0.021 d | 0.857 ± 0.093 e |
| SCOO | 0.046 ± 0.020 c | 2.160 ± 0.151 d |
| SCLP | 0.013 ± 0.005 c | 2.430 ± 0.206 cd |
| SCMPsq | 0.300 ± 0.017 c | 1.927 ± 0.127 ad |
| SCTDsq | 1.863 ± 0.025 e | 0.530 ± 0.005 f |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berbegal, C.; Borruso, L.; Fragasso, M.; Tufariello, M.; Russo, P.; Brusetti, L.; Spano, G.; Capozzi, V. A Metagenomic-Based Approach for the Characterization of Bacterial Diversity Associated with Spontaneous Malolactic Fermentations in Wine. Int. J. Mol. Sci. 2019, 20, 3980. https://doi.org/10.3390/ijms20163980
Berbegal C, Borruso L, Fragasso M, Tufariello M, Russo P, Brusetti L, Spano G, Capozzi V. A Metagenomic-Based Approach for the Characterization of Bacterial Diversity Associated with Spontaneous Malolactic Fermentations in Wine. International Journal of Molecular Sciences. 2019; 20(16):3980. https://doi.org/10.3390/ijms20163980
Chicago/Turabian StyleBerbegal, Carmen, Luigimaria Borruso, Mariagiovanna Fragasso, Maria Tufariello, Pasquale Russo, Lorenzo Brusetti, Giuseppe Spano, and Vittorio Capozzi. 2019. "A Metagenomic-Based Approach for the Characterization of Bacterial Diversity Associated with Spontaneous Malolactic Fermentations in Wine" International Journal of Molecular Sciences 20, no. 16: 3980. https://doi.org/10.3390/ijms20163980
APA StyleBerbegal, C., Borruso, L., Fragasso, M., Tufariello, M., Russo, P., Brusetti, L., Spano, G., & Capozzi, V. (2019). A Metagenomic-Based Approach for the Characterization of Bacterial Diversity Associated with Spontaneous Malolactic Fermentations in Wine. International Journal of Molecular Sciences, 20(16), 3980. https://doi.org/10.3390/ijms20163980







