A Novel PL9 Pectate Lyase from Paenibacillus polymyxa KF-1: Cloning, Expression, and Its Application in Pectin Degradation
Abstract
1. Introduction
2. Results and Discussion
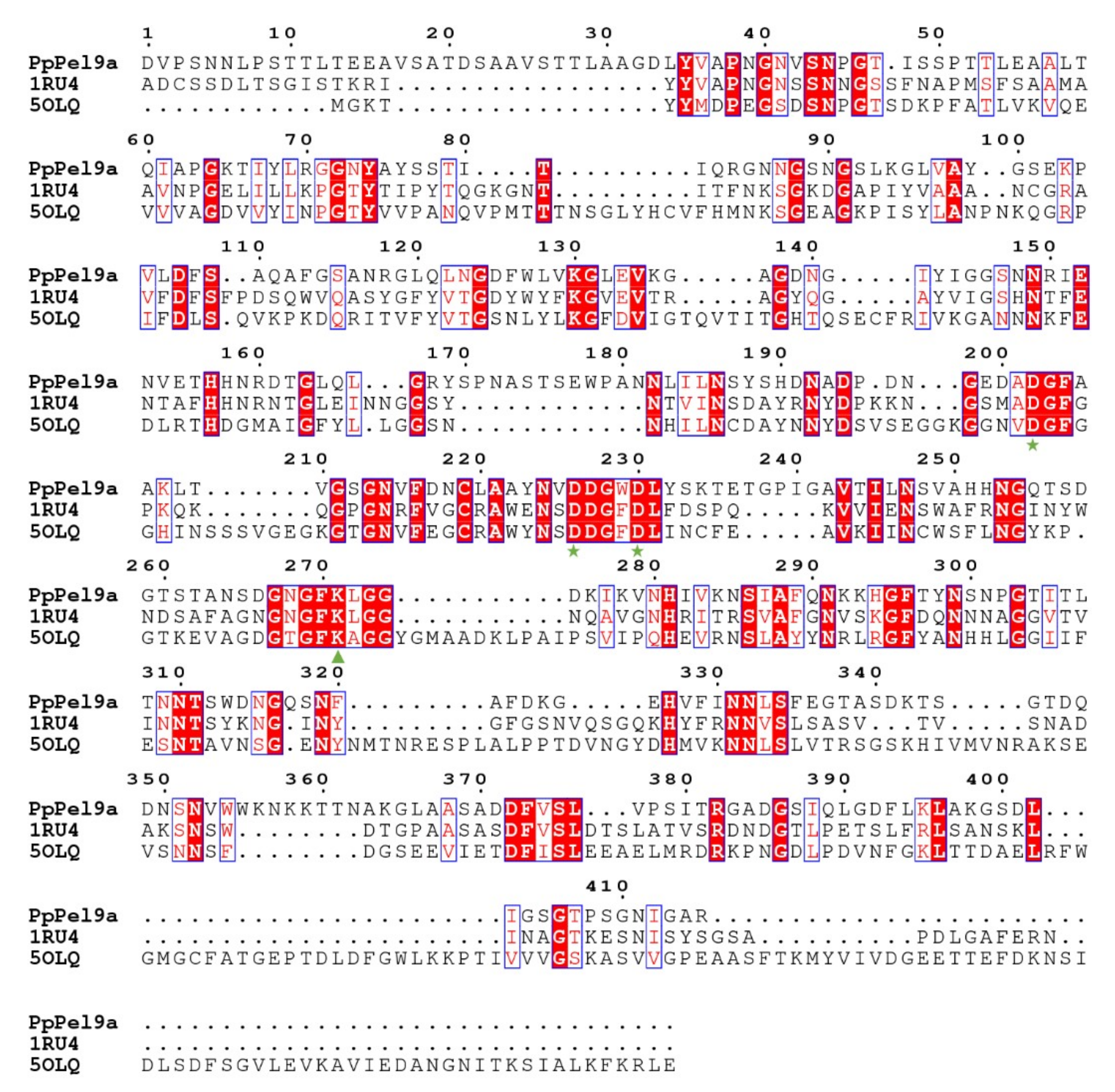
2.1. Gene Cloning and Sequence Analysis of PL9 Pectate Lyase from P. polymyxa KF-1
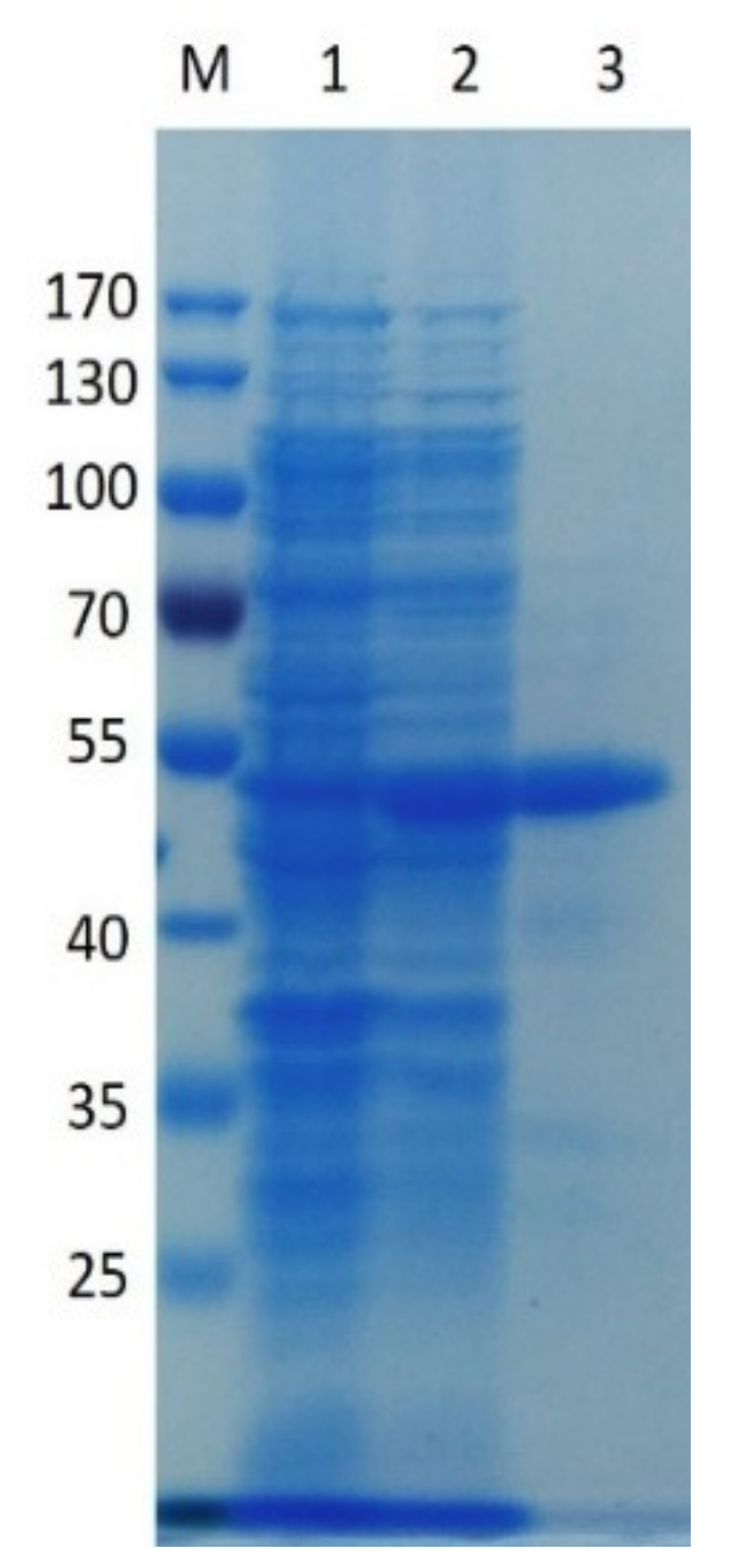
2.2. Expression of Recombinant PpPel9a
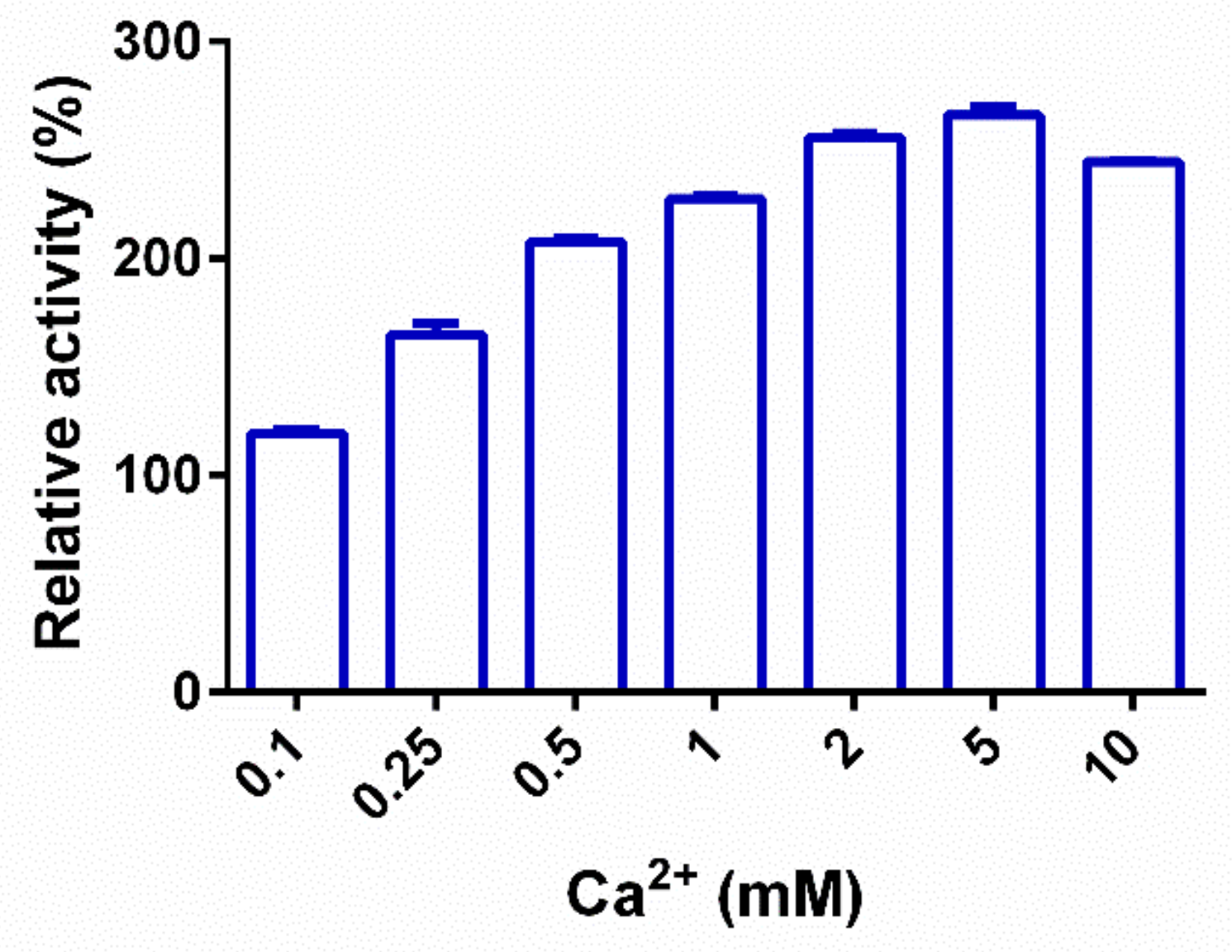
2.3. Biochemical Characterization of PpPel9a
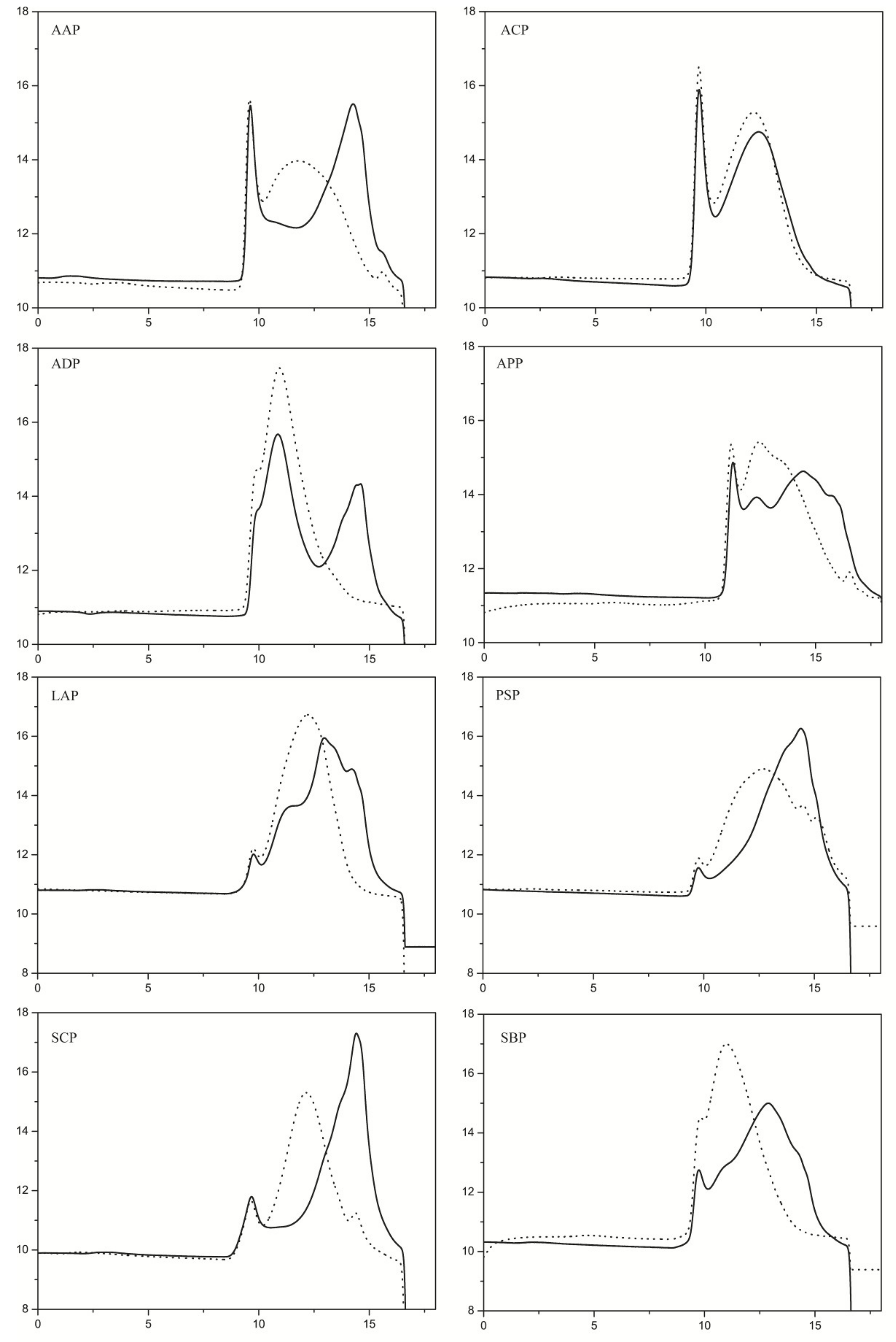
2.4. Substrate Specificity Analysis of PpPel9a
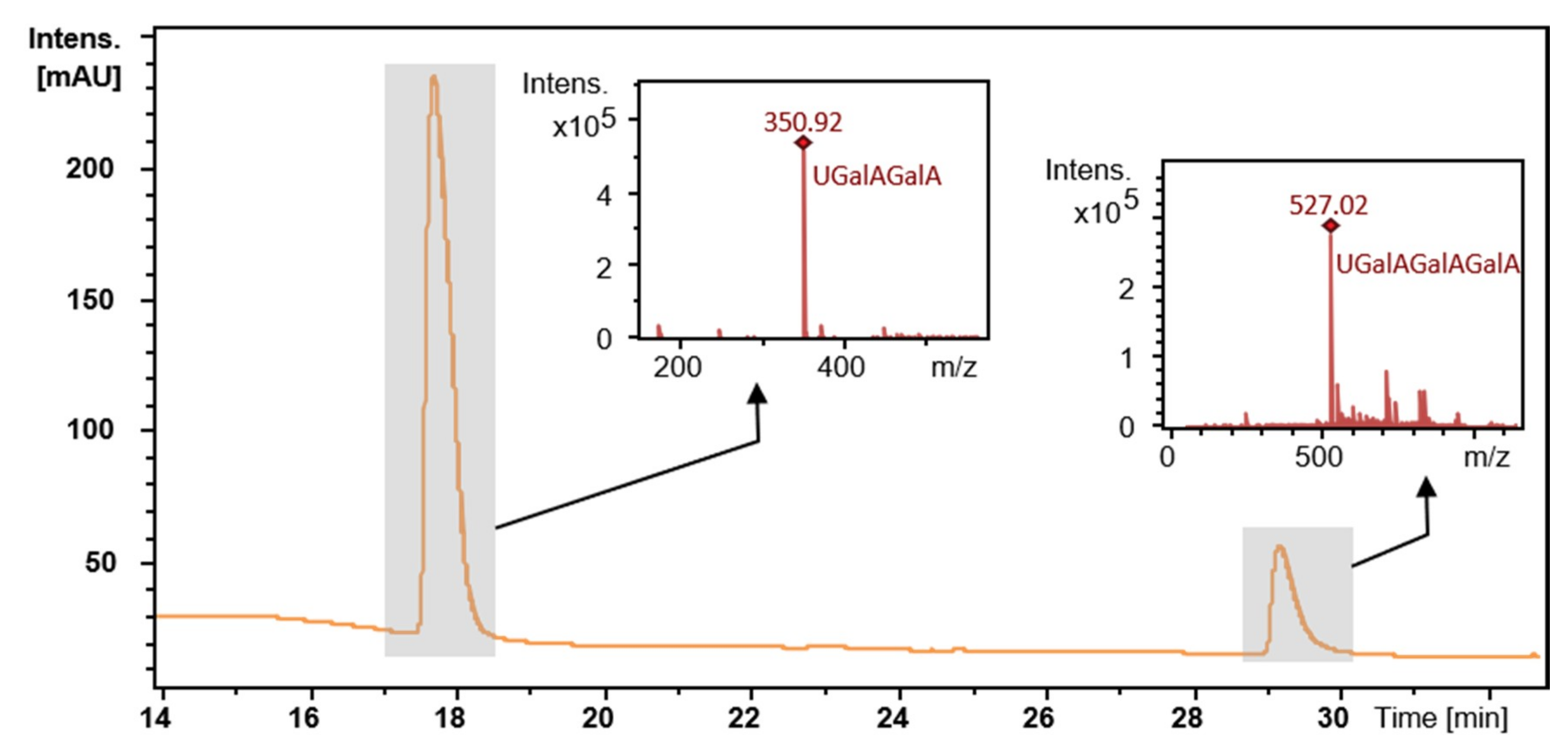
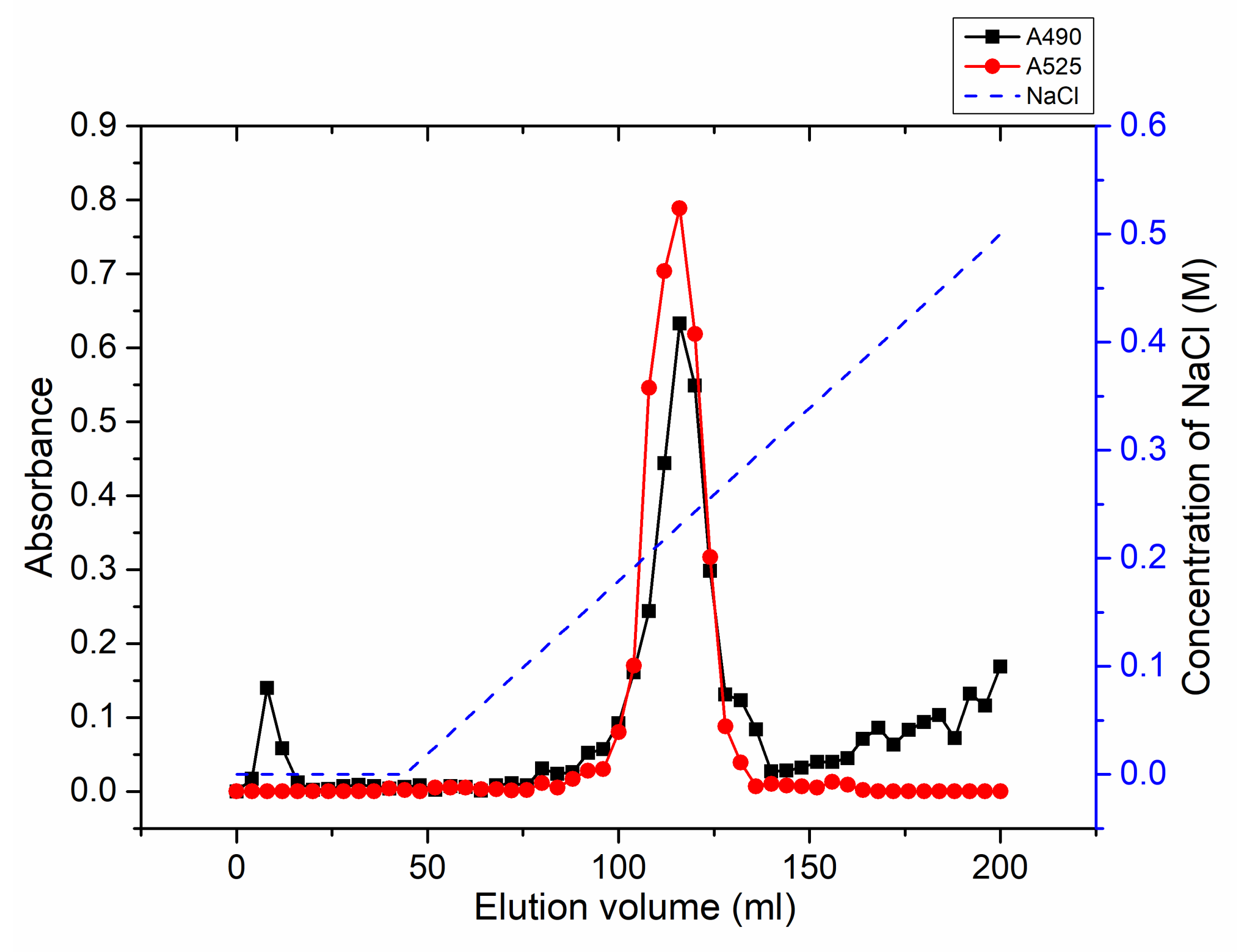
2.5. Degradation of Citrus Pectin and Characterization of the Main Degradation Product
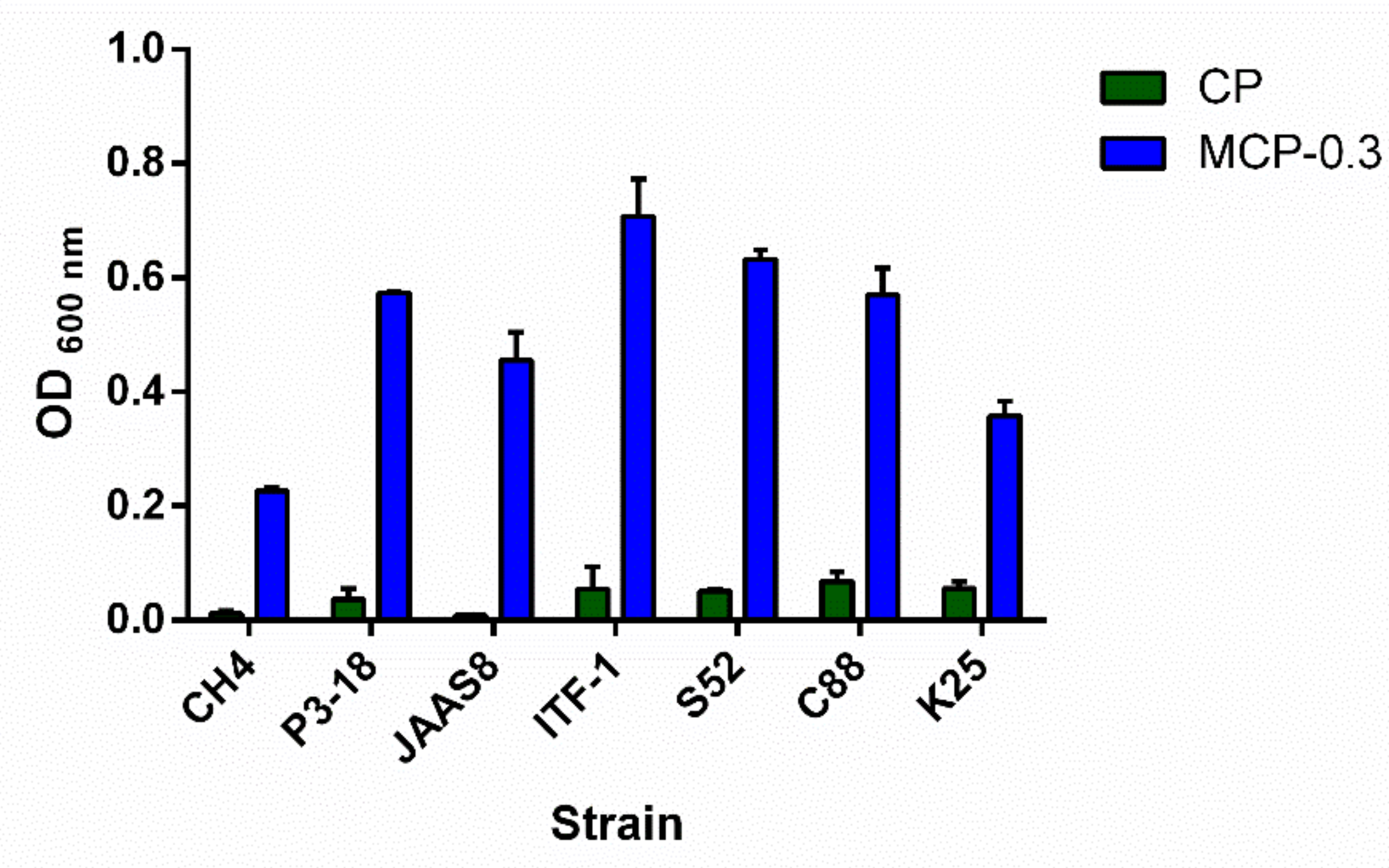
2.6. Growth Effect of MCP-0.3 on Lactobacillus Strains
3. Materials and Methods
3.1. Strains and Reagents
3.2. Substrates
3.3. Analytical Methods
3.4. Analysis of Pectate Lyase Gene
3.5. Cloning of PL9 Pectate Lyase and Its Expression
3.6. Pectate Lyase Activity Assay and Substrate Specificity Analysis of PpPel9a
3.7. Effect of pH, Temperature, and Metal Ions on the Activity and Stability of PpPel9a
3.8. Degradation of CP by PpPel9a and the Preparation of Degradation Product
3.9. Prebiotic Effect of CP and Its Degradation Product MCP-0.3 on Lactobacillus Strains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Christiaens, S.; Van Buggenhout, S.; Houben, K.; Jamsazzadeh Kermani, Z.; Moelants, K.R.N.; Ngouemazong, E.D.; Van Loey, A.; Hendrickx, M.E.G. Process-Structure-Function Relations of Pectin in Food. Crit. Rev. Food Sci. Nutr. 2016, 56, 1021–1042. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.Y.; Choo, W.S.; Young, D.J.; Loh, X.J. Pectin as a rheology modifier: Origin, structure, commercial production and rheology. Carbohydr. Polym. 2017, 161, 118–139. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, S.; Sun, L.; Ji, L.; Zhu, J.; Fan, Y.; Tai, G.; Zhou, Y. Further analysis of the structure and immunological activity of an RG-I type pectin from Panax ginseng. Carbohydr. Polym. 2012, 89, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Minzanova, S.T.; Mironov, V.F.; Arkhipova, D.M.; Khabibullina, A. V.; Mironova, L.G.; Zakirova, Y.M.; Milyukov, V.A. Biological Activity and Pharmacological Application of Pectic Polysaccharides: A Review. Polymers (Basel). 2018, 10, 1407. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dong, M.; Yang, Z.; Pan, S. Anti-diabetic effect of citrus pectin in diabetic rats and potential mechanism via PI3K/Akt signaling pathway. Int. J. Biol. Macromol. 2016, 89, 484–488. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Ye, F.; Zhou, Y.; Zhao, G. Thiolated citrus low-methoxyl pectin: Synthesis, characterization and rheological and oxidation-responsive gelling properties. Carbohydr. Polym. 2018, 181, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Ruvolo, P.P. Galectin 3 as a guardian of the tumor microenvironment. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, P.; Lu, S.-M.; Ling, Z.-Q. Chemoprevention of Low-Molecular-Weight Citrus Pectin (LCP) in Gastrointestinal Cancer Cells. Int. J. Biol. Sci. 2016, 12, 746–756. [Google Scholar] [CrossRef]
- do Prado, S.B.R.; Shiga, T.M.; Harazono, Y.; Hogan, V.A.; Raz, A.; Carpita, N.C.; Fabi, J.P. Migration and proliferation of cancer cells in culture are differentially affected by molecular size of modified citrus pectin. Carbohydr. Polym. 2019, 211, 141–151. [Google Scholar] [CrossRef]
- Liu, M.; Huo, W.; Dai, X.; Dang, Y. Preparation of low-molecular-weight citrus pectin by recombinant Bacillus subtilis pectate lyase and promotion of growth of Bifidobacterium longum. Catal. Commun. 2018, 107, 39–42. [Google Scholar] [CrossRef]
- Chen, J.; Liu, W.; Liu, C.-M.; Li, T.; Liang, R.-H.; Luo, S.-J. Pectin modifications: A review. Crit. Rev. Food Sci. Nutr. 2015, 55, 1684–1698. [Google Scholar] [CrossRef] [PubMed]
- Patidar, M.K.; Nighojkar, S.; Kumar, A.; Nighojkar, A. Pectinolytic enzymes-solid state fermentation, assay methods and applications in fruit juice industries: A review. 3 Biotech 2018, 8, 199. [Google Scholar] [CrossRef] [PubMed]
- Jolie, R.P.; Duvetter, T.; Van Loey, A.M.; Hendrickx, M.E. Pectin methylesterase and its proteinaceous inhibitor: A review. Carbohydr. Res. 2010, 345, 2583–2595. [Google Scholar] [CrossRef] [PubMed]
- Herron, S.R.; Benen, J.A.; Scavetta, R.D.; Visser, J.; Jurnak, F. Structure and function of pectic enzymes: Virulence factors of plant pathogens. Proc. Natl. Acad. Sci. USA 2000, 97, 8762–8769. [Google Scholar] [CrossRef] [PubMed]
- Marin-Rodriguez, M.C.; Orchard, J.; Seymour, G.B. Pectate lyases, cell wall degradation and fruit softening. J. Exp. Bot. 2002, 53, 2115–2119. [Google Scholar] [CrossRef]
- Hugouvieux-Cotte-Pattat, N.; Condemine, G.; Shevchik, V.E. Bacterial pectate lyases, structural and functional diversity. Environ. Microbiol. Rep. 2014, 6, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Hla, S.S.; Kurokawa, J.; Suryani; Kimura, T.; Ohmiya, K.; Sakka, K. A novel thermophilic pectate lyase containing two catalytic modules of Clostridium stercorarium. Biosci. Biotechnol. Biochem. 2005, 69, 2138–2145. [Google Scholar] [CrossRef][Green Version]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Zhou, C.; Xue, Y.; Ma, Y. Cloning, evaluation, and high-level expression of a thermo-alkaline pectate lyase from alkaliphilic Bacillus clausii with potential in ramie degumming. Appl. Microbiol. Biotechnol. 2017, 101, 3663–3676. [Google Scholar] [CrossRef]
- Zhang, C.; Yao, J.; Zhou, C.; Mao, L.; Zhang, G.; Ma, Y. The alkaline pectate lyase PEL168 of Bacillus subtilis heterologously expressed in Pichia pastoris is more stable and efficient for degumming ramie fiber. BMC Biotechnol. 2013, 13, 26. [Google Scholar] [CrossRef]
- van Rensburg, P.; Strauss, M.L.A.; Lambrechts, M.G.; Cordero Otero, R.R.; Pretorius, I.S. The heterologous expression of polysaccharidase-encoding genes with oenological relevance in Saccharomyces cerevisiae. J. Appl. Microbiol. 2007, 103, 2248–2257. [Google Scholar] [CrossRef] [PubMed]
- Willems, J.L.; Low, N.H. Oligosaccharide formation during commercial pear juice processing. Food Chem. 2016, 204, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yuan, Y.; Zhang, X.; Li, Y.; Li, Q.; Zhou, Y.; Gao, J. Screening of a Novel Polysaccharide Lyase Family 10 Pectate Lyase from Paenibacillus polymyxa KF-1: Cloning, Expression and Characterization. Molecules 2018, 23, 2774. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, A.; Sawada, K.; Saito, K.; Hakamada, Y.; Sumitomo, N.; Hatada, Y.; Kobayashi, T.; Ito, S. A new high-alkaline and high-molecular-weight pectate lyase from a Bacillus isolate: Enzymatic properties and cloning of the gene for the enzyme. Biosci. Biotechnol. Biochem. 2000, 64, 1133–1141. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shevchik, V.E.; Kester, H.C.; Benen, J.A.; Visser, J.; Robert-Baudouy, J.; Hugouvieux-Cotte-Pattat, N. Characterization of the exopolygalacturonate lyase PelX of Erwinia chrysanthemi 3937. J. Bacteriol. 1999, 181, 1652–1663. [Google Scholar] [PubMed]
- Brooks, A.D.; He, S.Y.; Gold, S.; Keen, N.T.; Collmer, A.; Hutcheson, S.W. Molecular cloning of the structural gene for exopolygalacturonate lyase from Erwinia chrysanthemi EC16 and characterization of the enzyme product. J. Bacteriol. 1990, 172, 6950–6958. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.; Shevchik, V.E.; Hugouvieux-Cotte-Pattat, N.; Pickersgill, R.W. The crystal structure of pectate lyase Pel9A from Erwinia chrysanthemi. J. Biol. Chem. 2004, 279, 9139–9145. [Google Scholar] [CrossRef]
- Luis, A.S.; Briggs, J.; Zhang, X.; Farnell, B.; Ndeh, D.; Labourel, A.; Basle, A.; Cartmell, A.; Terrapon, N.; Stott, K.; et al. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat. Microbiol. 2018, 3, 210–219. [Google Scholar] [CrossRef]
- Park, S.R.; Kim, M.K.; Kim, J.O.; Bae, D.W.; Cho, S.J.; Cho, Y.U.; Yun, H.D. Characterization of Erwinia chrysanthemi PY35 cel and pel gene existing in tandem and rapid identification of their gene products. Biochem. Biophys. Res. Commun. 2000, 268, 420–425. [Google Scholar] [CrossRef]
- Zhou, C.; Ye, J.; Xue, Y.; Ma, Y. Directed Evolution and Structural Analysis of Alkaline Pectate Lyase from the Alkaliphilic Bacterium Bacillus sp. Strain N16-5 To Improve Its Thermostability for Efficient Ramie Degumming. Appl. Environ. Microbiol. 2015, 81, 5714–5723. [Google Scholar] [CrossRef]
- Hao, M.; Yuan, X.; Cheng, H.; Xue, H.; Zhang, T.; Zhou, Y.; Tai, G. Comparative studies on the anti-tumor activities of high temperature- and pH-modified citrus pectins. Food Funct. 2013, 4, 960–971. [Google Scholar] [CrossRef]
- Pickersgill, R.; Jenkins, J.; Harris, G.; Nasser, W.; Robert-Baudouy, J. The structure of Bacillus subtilis pectate lyase in complex with calcium. Nat. Struct. Biol. 1994, 1, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, G.; Wang, J.; Hao, Y.; Li, M.; Li, Y.; Hu, B.; Lu, F. Efficient expression of an alkaline pectate lyase gene from Bacillus subtilis and the characterization of the recombinant protein. Biotechnol. Lett. 2012, 34, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.L.; Dreaden, T.M.; Theobald, L.K.; Tran, N.M.; Beal, T.L.; Eid, M.; Gao, M.Y.; Shirley, R.B.; Stoffel, M.T.; Kumar, M.V.; et al. Pectin induces apoptosis in human prostate cancer cells: Correlation of apoptotic function with pectin structure. Glycobiology 2007, 17, 805–819. [Google Scholar] [CrossRef] [PubMed]
- Cipriani, T.R.; Mellinger, C.G.; Bertolini, M.L.C.; Baggio, C.H.; Freitas, C.S.; Marques, M.C.A.; Gorin, P.A.J.; Sassaki, G.L.; Iacomini, M. Gastroprotective effect of a type I arabinogalactan from soybean meal. Food Chem. 2009, 115, 687–690. [Google Scholar] [CrossRef]
- Yu, K.-W.; Kiyohara, H.; Matsumoto, T.; Yang, H.-C.; Yamada, H. Characterization of pectic polysaccharides having intestinal immune system modulating activity from rhizomes of Atractylodes lancea DC. Carbohydr. Polym. 2001, 46, 125–134. [Google Scholar] [CrossRef]
- Di, R.; Vakkalanka, M.S.; Onumpai, C.; Chau, H.K.; White, A.; Rastall, R.A.; Yam, K.; Hotchkiss, A.T.J. Pectic oligosaccharide structure-function relationships: Prebiotics, inhibitors of Escherichia coli O157:H7 adhesion and reduction of Shiga toxin cytotoxicity in HT29 cells. Food Chem. 2017, 227, 245–254. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, H.; Wang, L.; Liu, F.; Pan, S. Preparation and prebiotic potential of pectin oligosaccharides obtained from citrus peel pectin. Food Chem. 2018, 244, 232–237. [Google Scholar] [CrossRef]
- Zhang, X.; Yu, L.; Bi, H.; Li, X.; Ni, W.; Han, H.; Li, N.; Wang, B.; Zhou, Y.; Tai, G. Total fractionation and characterization of the water-soluble polysaccharides isolated from Panax ginseng C. A. Meyer. Carbohydr. Polym. 2009, 77, 544–552. [Google Scholar] [CrossRef]
- Sun, L.; Ropartz, D.; Cui, L.; Shi, H.; Ralet, M.-C.; Zhou, Y. Structural characterization of rhamnogalacturonan domains from Panax ginseng C. A. Meyer. Carbohydr. Polym. 2019, 203, 119–127. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, L.; Feng, Z.; Wang, H.; Mayo, K.H.; Zhou, Y.; Tai, G. Preparation of individual galactan oligomers, their prebiotic effects, and use in estimating galactan chain length in pectin-derived polysaccharides. Carbohydr. Polym. 2018, 199, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Cui, L.; Yang, G.; Ning, X.; Sun, L.; Zhou, Y. Preparing rhamnogalacturonan II domains from seven plant pectins using Penicillium oxalicum degradation and their structural comparison. Carbohydr. Polym. 2018, 180, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Yang, G.; Fan, X.; Bai, Y.; Ren, X.; Zhou, Y. Controlled methyl-esterification of pectin catalyzed by cation exchange resin. Carbohydr. Polym. 2016, 137, 650–656. [Google Scholar] [CrossRef] [PubMed]
- York, W.S.; Darvill, A.G.; McNeil, M.; Albersheim, P. 3-deoxy-d-manno-2-octulosonic acid (KDO) is a component of rhamnogalacturonan II, a pectic polysaccharide in the primary cell walls of plants. Carbohydr. Res. 1985, 138, 109–126. [Google Scholar] [CrossRef]
- Sayers, E.W.; Barrett, T.; Benson, D.A.; Bolton, E.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; DiCuccio, M.; Federhen, S.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011, 39, D38–D51. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [PubMed]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinforma. 2014, 48, 3–13. [Google Scholar]
- Gouet, P.; Robert, X.; Courcelle, E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003, 31, 3320–3323. [Google Scholar] [CrossRef]
- Mooers, B.H.M. Simplifying and enhancing the use of PyMOL with horizontal scripts. Protein Sci. 2016, 25, 1873–1882. [Google Scholar] [CrossRef] [PubMed]
- Schagger, H. Tricine-SDS-PAGE. Nat. Protoc. 2006, 1, 16–22. [Google Scholar] [CrossRef] [PubMed]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric Method for Determination of Sugars and Related Substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Blumenkrantz, N.; Asboe-Hansen, G. New method for quantitative determination of uronic acids. Anal. Biochem. 1973, 54, 484–489. [Google Scholar] [CrossRef]


| Metal Ions or Chemicals (5 mM) | Relative Activity (%) 1 |
|---|---|
| NaCl | 86.7 ± 3.3 |
| KCl | 79.0 ± 0.6 |
| CaCl2 | 266.5 ± 7.0 |
| MgCl2 | 94.5 ± 4.6 |
| FeSO4 | 54.5 ± 0.7 |
| EDTA | 63.1 ± 0.5 |
| ZnCl2 | 88.0 ± 1.6 |
| AlCl3 | 106.1 ± 6.1 |
| CoCl2 | 51.4 ± 2.4 |
| SDS | 37.7 ± 0.2 |
| Tween-20 | 122.5 ± 0.6 |
| Tween-40 | 51.4 ± 2.1 |
| Tween-60 | 40.0 ± 0.6 |
| Tween-80 | 2.0 ± 0.1 |
| TritonX-100 | 4.3 ± 0.3 |
| Type of Pectin | Substrate 1 | Specific Activity (U/mg) |
|---|---|---|
| Pectin | PGP | 48.1 ± 4.9 |
| RGP | 59.1 ± 0.1 | |
| CP | 107.0 ± 0.2 | |
| RG-I | PGP-RGI | 2.1 ± 0.9 |
| RGP-RGI | 21.0 ± 0.3 | |
| HG | PGP-HG | 106.8 ± 0.6 |
| RGP-HG | 68.9 ± 0.5 | |
| Completely esterified HG | CP-CeHG | 47.1 ± 0.1 |
| De-esterified HG | CP-DeHG | 710.7 ± 3.3 |
| Monosaccharide (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Man | GlcA | Rha | GalA | Glc | Gal | Xyl | Ara | Fuc | |
| CP | 1.20 | 0.57 | 3.62 | 62.18 | 4.12 | 13.73 | - 1 | 14.42 | 0.16 |
| MCP-0.3 | 1.12 | 1.66 | 3.68 | 86.97 | 1.02 | 2.06 | - | 2.49 | 1.00 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Y.; Zhang, X.-Y.; Zhao, Y.; Zhang, H.; Zhou, Y.-F.; Gao, J. A Novel PL9 Pectate Lyase from Paenibacillus polymyxa KF-1: Cloning, Expression, and Its Application in Pectin Degradation. Int. J. Mol. Sci. 2019, 20, 3060. https://doi.org/10.3390/ijms20123060
Yuan Y, Zhang X-Y, Zhao Y, Zhang H, Zhou Y-F, Gao J. A Novel PL9 Pectate Lyase from Paenibacillus polymyxa KF-1: Cloning, Expression, and Its Application in Pectin Degradation. International Journal of Molecular Sciences. 2019; 20(12):3060. https://doi.org/10.3390/ijms20123060
Chicago/Turabian StyleYuan, Ye, Xin-Yu Zhang, Yan Zhao, Han Zhang, Yi-Fa Zhou, and Juan Gao. 2019. "A Novel PL9 Pectate Lyase from Paenibacillus polymyxa KF-1: Cloning, Expression, and Its Application in Pectin Degradation" International Journal of Molecular Sciences 20, no. 12: 3060. https://doi.org/10.3390/ijms20123060
APA StyleYuan, Y., Zhang, X.-Y., Zhao, Y., Zhang, H., Zhou, Y.-F., & Gao, J. (2019). A Novel PL9 Pectate Lyase from Paenibacillus polymyxa KF-1: Cloning, Expression, and Its Application in Pectin Degradation. International Journal of Molecular Sciences, 20(12), 3060. https://doi.org/10.3390/ijms20123060




