Genome Wide Transcriptome Analysis Reveals Complex Regulatory Mechanisms Underlying Phosphate Homeostasis in Soybean Nodules
Abstract
1. Introduction
2. Results
2.1. Growth of Soybean Nodules Is Inhibited by Pi Starvation
2.2. Amino-N Compounds in Nodules Produced in Low P and High P Conditions
2.3. Changes of Transcriptomes in Soybean Nodules Resulting from Pi Starvation
2.4. Analysis of Pi-Responsive Genes Involved in Metabolome
2.5. Identification of Pi-Starvation Responsive Genes Controlling Nodule Pi Homeostasis
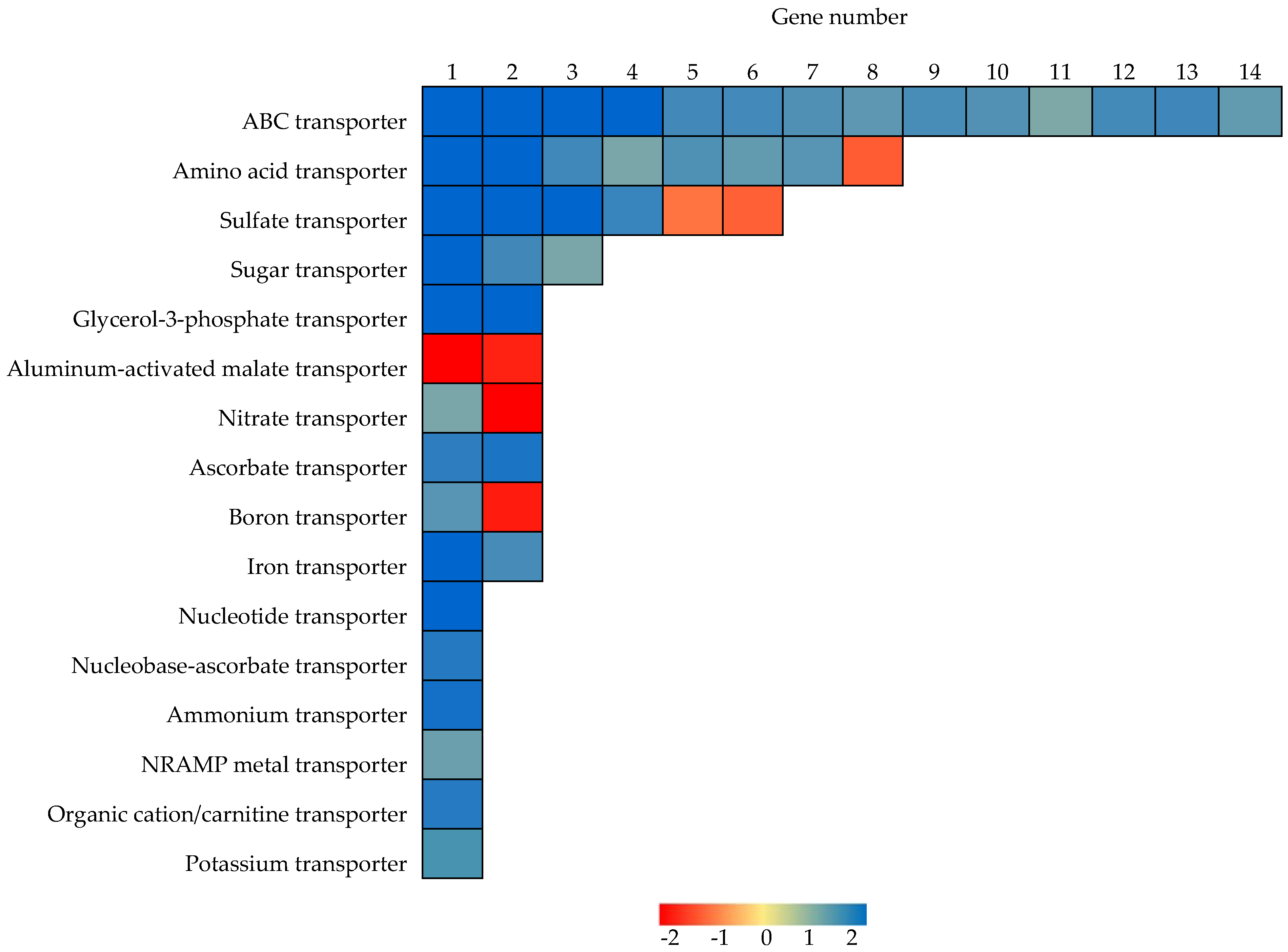
2.6. Identification of Genes Functioning as Transporters
2.7. Genes Involved in Hormonal Signaling Pathways
2.8. Ca2+ Signaling Related Genes in Soybean Nodules Regulated by P Deficiency
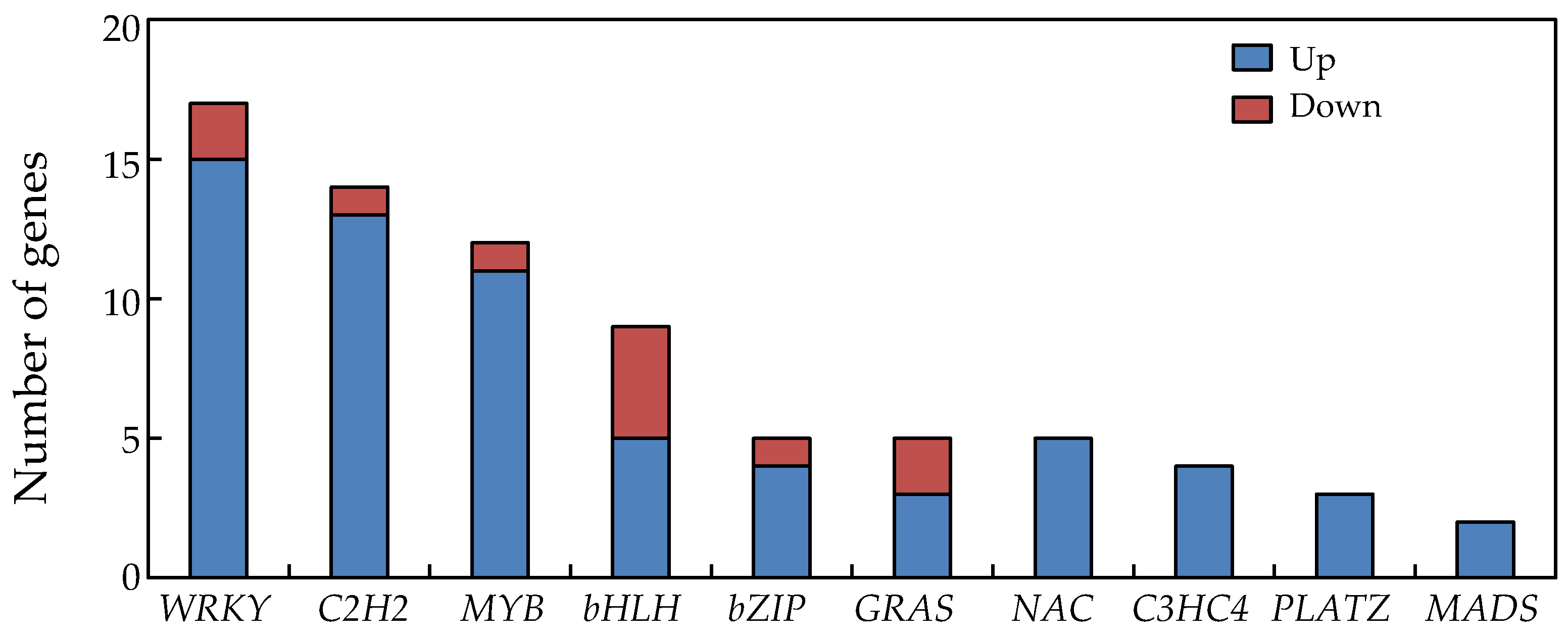
2.9. Transcription Factors in Soybean Nodules Regulated by P Deficiency
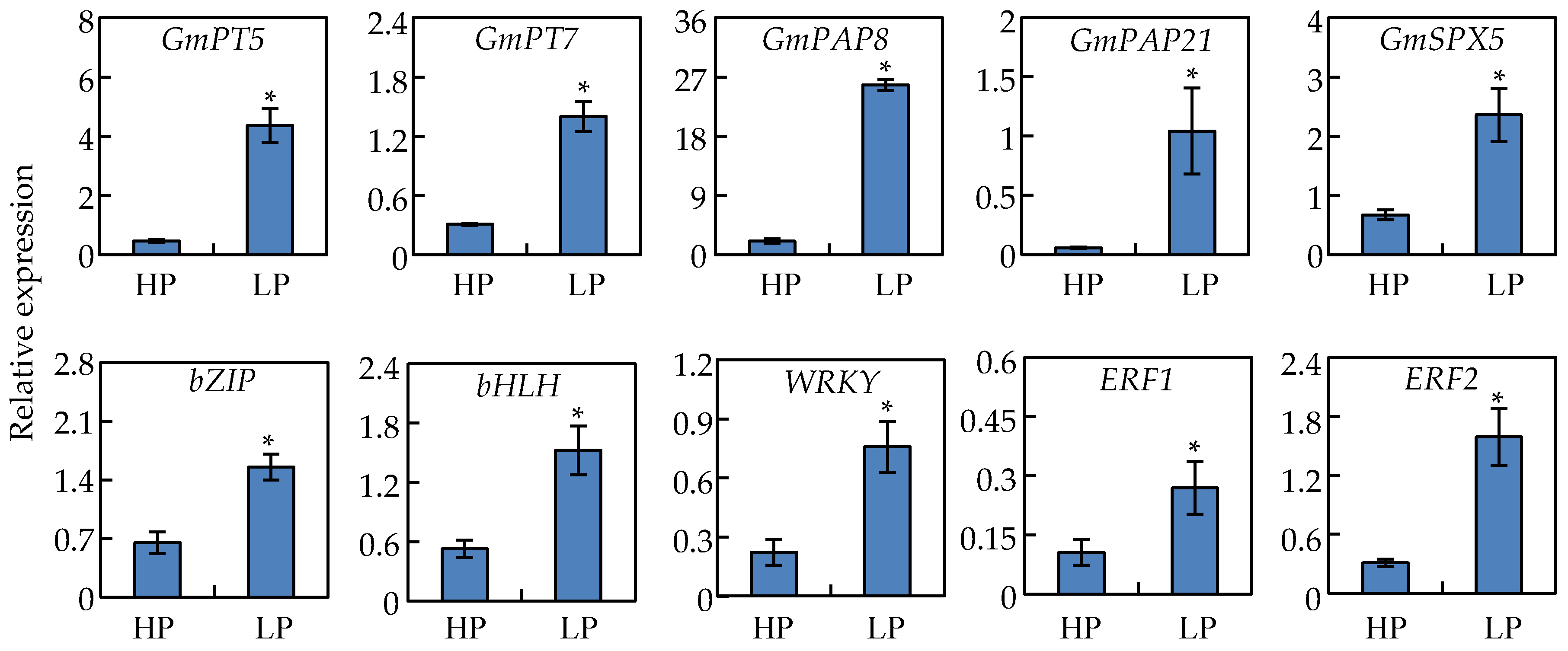
2.10. Analysis of Gene Transcripts Using qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Plant, Rhizobium and Growth Conditions
4.2. Determination of Total P and Soluble Pi Concentrations
4.3. Acid Phosphatase Activity Measurements
4.4. Nodule Nitrogenase Activity Analysis
4.5. Amino Acid Analysis
4.6. cDNA Library Preparation, RNA-Seq and Phylogenetic Analysis
4.7. qRT-PCR Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| SNF | Symbiotic Nitrogen Fixation |
| RNA-seq | RNA sequencing |
| DEGs | Differentially Expressed Genes |
| qRT-PCR | quantitative Real-Time Polymerase Chain Reaction |
| GO | Gene Ontology |
| CK | Cytokinin |
| GA | Gibberellin |
| CHO | Carbohydrate |
References
- Raghothama, K.G. Phosphate acquisition. Annu. Rev. Plant Biol. 1999, 50, 665–693. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.E. Regulating the phosphorus nutrition of plants: Molecular biology meeting agronomic needs. Plant Soil 2009, 322, 17–24. [Google Scholar] [CrossRef]
- Beardsley, T.M. Peak phosphorus. Bioscience 2011, 61, 91. [Google Scholar] [CrossRef]
- Veneklaas, E.J.; Lambers, H.; Bragg, J.; Finnegan, P.M.; Lovelock, C.E.; Plaxton, W.C.; Price, C.A.; Scheible, W.R.; Shane, M.W.; White, P.J.; et al. Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 2012, 195, 306–320. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Liao, H.; Lucas, W. Molecular mechanisms underlying phosphate sensing, signaling and adaptation in plants. J. Integr. Plant Biol. 2014, 56, 192–220. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.P.; Uhde-Stone, C.; Allan, D.L. Phosphorus acquisition and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef]
- Cordell, D.; Drangert, J.O.; White, S. The story of phosphorus: Global food security and food for thought. Glob. Environ. Chang. 2009, 19, 292–305. [Google Scholar] [CrossRef]
- Wang, X.; Yan, X.; Liao, H. Genetic improvement for phosphorus efficiency in soybean: A radical approach. Ann. Bot. 2010, 106, 215–222. [Google Scholar] [CrossRef] [PubMed]
- Chiou, T.J.; Lin, S.I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011, 62, 185–206. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Wang, X.; Tong, Y.; Chen, X.; Liao, H. Bioengineering and management for efficient phosphorus utilization in crops and pastures. Curr. Opin. Biotechnol. 2012, 23, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, J.; Zhao, J.; Tian, J.; Liao, H. Control of phosphate homeostasis through gene regulation in crops. Curr. Opin. Plant Biol. 2014, 21, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Lópezarredondo, D.L.; Leyvagonzález, M.A.; Gonzálezmorales, S.I.; Lópezbucio, J.; Herreraestrella, L. Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Liao, H.; Wang, X.; Yan, X. Phosphorus starvation-induced expression of leaf acid phosphatase isoforms in soybean. Acta Bot. Sin. 2003, 45, 1037–1042. [Google Scholar] [CrossRef]
- Zhao, J.; Fu, J.; Liao, H.; He, Y.; Nian, H.; Hu, Y.; Qiu, L.; Dong, Y.; Yan, X. Characterization of root architecture in an applied core collection for phosphorus efficiency of soybean germplasm. Chin. Sci. Bull. 2004, 49, 1611–1620. [Google Scholar] [CrossRef]
- Liao, H.; Wan, H.; Shaff, J.; Wang, X.; Yan, X.; Kochian, L.V. Phosphorus and aluminum interactions in soybean in relation to aluminum tolerance. Exudation of specific organic acids from different regions of the intact root system. Plant Physiol. 2006, 141, 674–684. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liao, H.; Wang, X.R.; Yan, X.L. Regulation effect of soil P availability on mycorrhizal infection in relation to root architecture and P efficiency of Glycine max. Chin. J. Appl. Ecol. 2008, 19, 564–568. [Google Scholar]
- Plaxton, W.C.; Tran, H.T. Metabolic adaptations of phosphate starved plants. Plant Physiol. 2011, 156, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Jiang, H.; Tian, J.; Zhao, J.; Liao, H. Rhizobia enhance acquisition of phosphorus from different sources by soybean plants. Plant Soil 2011, 349, 25–36. [Google Scholar] [CrossRef]
- Liang, C.; Tian, J.; Liao, H. Proteomics dissection of plant responses to mineral nutrient deficiency. Proteomics 2013, 13, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Rubio, V.; Linhares, F.; Solano, R.; Martin, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Rus, A.; Sharkhuu, A.; Yokoi, S.; Karthikeyan, A.S.; Raghothama, K.G.; Baek, D.; Koo, Y.D.; Jin, J.B.; Bressan, R.A.; et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. USA 2005, 102, 7760–7765. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Yi, K.; Dang, L.; Huang, H.; Wu, W.; Wu, P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008, 54, 965–975. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, Q.; Kong, Y.H.; Chen, Y.; Duan, J.Y.; Wu, W.H.; Chen, Y.F. Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 2014, 164, 2020–2029. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.F.; Liang, C.Y.; Zhang, Q.; Chen, Z.J.; Xiao, B.X.; Tian, J.; Liao, H. SPX1 is an important component in the phosphorus signaling network of common bean regulating root growth and phosphorus homeostasis. J. Exp. Bot. 2014, 65, 3299–3310. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wang, Z.; Lv, Q.; Shi, J.; Zhong, Y.; Wu, P.; Ma, C. SPX proteins regulate Pi homeostasis and signaling in different subcellular level. Plant Signal. Behav. 2015, 10, e1061163. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhao, J.; Li, X.; Qin, L.; Yan, X.; Liao, H. A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 2011, 66, 541–552. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Gui, S.; Yang, T.; Walk, T.; Wang, X.; Liao, H. Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Ann. Bot. 2012, 109, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Guo, Y.; Chen, L.; Liang, R.; Gu, M.; Xu, G.; Zhao, J.; Walk, T.; Liao, H. Functional characterization of 14 Pht1 family genes in yeast and their expressions in response to nutrient starvation in soybean. PLoS ONE 2012, 7, e47726. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Wang, X.; Hu, R.; Wang, Y.; Xiao, C.; Jiang, Y.; Zhang, X.; Zheng, C.; Fu, Y.F. The pattern of Phosphate transporter 1 genes evolutionary divergence in Glycine max L. BMC Plant Biol. 2013, 13, 48. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, J.; Walk, T.C.; Liao, H. Characterization of soybean β-expansin genes and their expression responses to symbiosis, nutrient deficiency, and hormone treatment. Appl. Microbiol. Biotechnol. 2014, 98, 2805–2817. [Google Scholar] [CrossRef] [PubMed]
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Ai, P.; Sun, S.; Zhao, J.; Xu, G. Regulation and function of Pht1 family phosphate transporters in rice. In Proceedings of the International Plant Nutrition Colloquium XVI; Department of Plant Sciences, UC Davis: Davis, CA, USA, 2009; Available online: http://escholarship.org/uc/item/3657w1q3 (accessed on 2 September 2018).
- Misson, J.; Thibaud, M.C.; Bechtold, N.; Raghothama, K.; Nussaume, L. Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 2004, 55, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Shin, H.S.; Dewbre, G.R.; Harrison, M.J. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004, 39, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Gu, M.; Cao, Y.; Huang, X.; Zhang, X.; Ai, P.; Zhao, J.; Fan, X.; Xu, G. A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 2012, 159, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, Y.; Piñeros, M.A.; Wang, Z.; Wang, W.; Li, C.; Wu, Z.; Kochian, L.V.; Wu, P. Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ. 2014, 37, 1159–1170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Sun, Y.; Pei, W.; Jain, A.; Sun, R.; Cao, Y.; Wu, X.; Jiang, T.; Zhang, L.; Fan, X.; et al. Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J. 2015, 82, 556–569. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, V.K.; Jain, A.; Poling, M.D.; Lewis, A.J.; Raghothama, K.G.; Smith, A.P. Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 2011, 156, 1149–1163. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Zhang, S.; Wang, L.; Yang, Y.; Zhang, H.; Cui, H.; Shao, H.; Xu, G. OsPht1;8, a phosphate transporter, is involved in auxin and phosphate starvation response in rice. J. Exp. Bot. 2017, 68, 5057–5068. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hu, H.; Huang, H.; Duan, K.; Wu, Z.; Wu, P. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J. Integr. Plant Biol. 2009, 51, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Puga, M.I.; Mateos, I.; Charukesi, R.; Wang, Z.; Franco-Zorrilla, J.M.; de Lorenzo, L.; Irigoyen, M.L.; Masiero, S.; Bustos, R.; Rodríguez, J.; et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14947–14952. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Hu, H.; Zhang, K.; Zhang, W.; Yu, Y.; Wu, Z.; Wu, P. The paralogous SPX3 and SPX5 genes redundantly modulate Pi homeostasis in rice. J. Exp. Bot. 2014, 65, 859–870. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ruan, W.; Shi, J.; Zhang, L.; Xiang, D.; Yang, C.; Li, C.; Wu, Z.; Liu, Y.; Yu, Y.; et al. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 14953–14958. [Google Scholar] [CrossRef] [PubMed]
- Wild, R.; Gerasimaite, R.; Jung, J.Y.; Truffault, V.; Pavlovic, I.; Schmidt, A.; Saiardi, A.; Jessen, H.J.; Poirier, Y.; Hothorn, M.; Mayer, A. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 2016, 352, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Shang, W.; Li, C.; Jia, L.; Wang, X.; Xing, G.; Zheng, W. Evolution of the SPX gene family in plants and its role in the response mechanism to phosphorus stress. Open Biol. 2018, 8, 170231. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Chun, H.J.; Yun, D.J.; Kim, M.C. Cross-talk between phosphate starvation and other environmental stress signaling pathways in plants. Mol. Cells 2017, 40, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Andújar, C.; Ruiz-Lozano, J.M.; Dodd, I.C.; Albacete, A.; Pérez-Alfocea, F. Hormonal and nutritional features in contrasting rootstock-mediated tomato growth under low-phosphorus nutrition. Front. Plant Sci. 2017, 11, 533. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Seo, J.S.; Chua, N.H. NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 2014, 26, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Peng, M.; Rothstein, S.J. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in arabidopsis. PLoS Genet. 2011, 7, e1002021. [Google Scholar] [CrossRef] [PubMed]
- Israel, D.W. Investigation of the role of phosphorus in symbiotic dinitrogen fixation. Plant Physiol. 1987, 84, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhao, J.; Tan, Z.; Zeng, R.; Liao, H. GmEXPB2, a cell wall β-expansin, affects soybean nodulation through modifying root architecture and promoting nodule formation and development. Plant Physiol. 2015, 169, 2640–2653. [Google Scholar] [CrossRef] [PubMed]
- Herridge, D.F.; Peoples, M.B.; Boddey, R.M. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 2008, 311, 1–18. [Google Scholar] [CrossRef]
- Peoples, M.B.; Brockwell, J.; Herridge, D.F.; Rochester, I.J.; Alves, B.J.R.; Urquiaga, S.; Boddey, R.M.; Dakora, F.D.; Bhattarai, S.; Maskey, S.L.; et al. The contributions of nitrogen-fixing crop legumes to the productivity of agricultural systems. Symbiosis 2009, 48, 1–17. [Google Scholar] [CrossRef]
- Ferguson, B.J.; Mathesius, U. Phytohormone regulation of legume-rhizobia interactions. J. Chem. Ecol. 2014, 40, 770–790. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, C.; Yang, J.; Yu, N.; Wang, E. Hormone modulation of legume-rhizobial symbiosis. J. Integr. Plant Biol. 2018, 60, 632–648. [Google Scholar] [CrossRef] [PubMed]
- Miyata, K.; Kawaguchi, M.; Nakagawa, T. Two distinct EIN2 genes cooperatively regulate ethylene signaling in Lotus japonicus. Plant Cell Physiol. 2013, 54, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, T.; Maekawa-Yoshikawa, M.; Takeda, N.; Imaizumi-Anraku, H.; Murooka, Y.; Hayashi, M. Gibberellin controls the nodulation signaling pathway in Lotus japonicus. Plant J. 2009, 58, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Fonouni-Farde, C.; Tan, S.; Baudin, M.; Brault, M.; Wen, J.; Mysore, K.S.; Niebel, A.; Frugier, F.; Diet, A. DELLA-mediated gibberellin signalling regulates Nod factor signalling and rhizobial infection. Nat. Commun. 2016, 2, 12636. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Liu, H.; Luo, D.; Yu, N.; Dong, W.; Wang, C.; Zhang, X.; Dai, H.; Yang, J.; Wang, E. DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat. Commun. 2016, 12, 12433. [Google Scholar] [CrossRef] [PubMed]
- Schulze, J.; Temple, G.; Temple, S.J.; Beschow, H.; Vance, C.P. Nitrogen fxation by white lupin under phosphorus deficiency. Ann. Bot. 2006, 98, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Cui, Q.; Liang, C.; Sun, L.; Tian, J.; Liao, H. Identification of differentially expressed proteins in soybean nodules under phosphorus deficiency through proteomic analysis. Proteomics 2011, 11, 4648–4659. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Zhao, J.; Tian, J.; Chen, L.; Sun, Z.; Guo, Y.; Lu, X.; Gu, M.; Xu, G.; Liao, H. The high-affinity phosphate transporter GmPT5 regulates phosphate transport to nodules and nodulation in soybean. Plant Physiol. 2012, 159, 1634–1643. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Sui, X.; Wang, F.; Gao, J.; He, X.; Zhang, F.; Yang, J.; Feng, G. Synergistic interactions between Glomus mosseae and Bradyrhizobium japonicum in enhancing proton release from nodules and hyphae. Mycorrhiza 2012, 22, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Hernández, G.; Valdés-López, O.; Ramírez, M.; Goffard, N.; Weiller, G.; Aparicio-Fabre, R.; Fuentes, S.I.; Erban, A.; Kopka, J.; Udvardi, M.K.; et al. Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol. 2009, 151, 1221–1238. [Google Scholar] [CrossRef] [PubMed]
- Cabeza, R.A.; Liese, R.; Lingne, A.; von Stieglitz, I.; Neumann, J.; Salinas-Riester, G.; Pommerenke, C.; Dittert, K.; Schulze, J. RNA-seq transcriptome profiling reveals that Medicago truncatula nodules acclimate N2 fixation before emerging P deficiency reaches the nodules. J. Exp. Bot. 2014, 65, 6035–6048. [Google Scholar] [CrossRef] [PubMed]
- Nasr, E.M.; Inoue, K.; Chu, H.D.; Nguyen, K.H.; Van, H.C.; Watanabe, Y.; Burritt, D.J.; Herrera-Estrella, L.; Mochida, K.; Tran, L.P. Comparative transcriptome analysis of nodules of two mesorhizobium-chickpea associations with differential symbiotic efficiency under phosphate deficiency. Plant J. 2017, 91, 911–926. [Google Scholar] [CrossRef] [PubMed]
- Valentine, A.J.; Benedito, V.A.; Kang, Y. Legume nitrogen fixation and soil abiotic stress: From physiology to genomics and beyond. In Annual Plant Reviews: Nitrogen Metabolism in Plants in the Post-Genomic Era; Foyer, C.H., Zhang, H., Eds.; WileyBlackwell: Oxford, UK, 2010; Volume 42, pp. 207–248. [Google Scholar]
- Amarasinghe, B.H.; de Bruxelles, G.L.; Braddon, M.; Onyeocha, I.; Forde, B.G.; Udvardi, M.K. Regulation of GmNRT2 expression and nitrate transport activity in roots of soybean (Glycine max). Planta 1998, 206, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Misson, J.; Raghothama, K.G.; Jain, A.; Jouhet, J.; Block, M.A.; Bligny, R.; Ortet, P.; Creff, A.; Somerville, S.; Rolland, N. A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. USA 2005, 102, 11934–11939. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.A.; Yang, S.S.; Miller, S.S.; Bucciarelli, B.; Liu, J.; Rydeen, A.; Bozsoki, Z.; Uhde-Stone, C.; Tu, Z.J.; Allan, D.; et al. An RNA-Seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol. 2013, 161, 705–724. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Jabnoune, M.; Walker, H.; Shou, H.; Wu, P.; Poirier, Y.; Whelan, J. Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 2013, 25, 4285–4304. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, J.; Yang, Y.; Du, W.; Zhang, D.; Yu, D.; Cheng, H. A genome-wide expression profile analysis reveals active genes and pathways coping with phosphate starvation in soybean. BMC Genom. 2016, 17, 192. [Google Scholar] [CrossRef] [PubMed]
- Ham, B.K.; Chen, J.; Yan, Y.; Lucas, W.J. Insights into plant phosphate sensing and signaling. Curr. Opin. Biotechnol. 2018, 49, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Tian, J.; Liao, H. Comparative characterization of GmSPX members reveals that GmSPX3 is involved in phosphate homeostasis in soybean. Ann. Bot. 2014, 114, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Delhaize, E.; Taylor, P.; Hocking, P.J.; Simpson, R.J.; Ryan, P.R.; Richardson, A.E. Transgenic barley (Hordeum vulgare L.) expressing the wheat aluminum resistance gene (TaALMT1) shows enhanced phosphorus nutrition and grain production when grown on an acid soil. Plant Biotechnol. J. 2009, 7, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.; Gao, X.; Dong, Z.; Yi, J.; An, L. Improved phosphorus acquisition by tobacco through transgenic expression of mitochondrial malate dehydrogenase from Penicillium oxalicum. Plant Cell Rep. 2012, 31, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Sun, L.; Yao, Z.; Liao, H.; Tian, J. Comparative analysis of PvPAP gene family and their functions in response to phosphorus deficiency in common bean. PLoS ONE 2012, 7, e38106. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhao, X.; Zhang, L.; Lu, W.; Li, X.; Kai, X. TaPht1;4, a high-affinity phosphate transporter gene in wheat (Triticum aestivum), plays an important role in plant phosphate acquisition under phosphorus deprivation. Funct. Plant Biol. 2013, 40, 329–341. [Google Scholar] [CrossRef]
- Wang, L.; Lu, S.; Zhang, Y.; Li, Z.; Du, X.; Liu, D. Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J. Integr. Plant Biol. 2014, 56, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Remy, E.; Cabrito, T.R.; Batista, R.A.; Teixeira, M.C.; Sá-Correia, I.; Duque, P. The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 2012, 195, 356–371. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.D.; Xue, Y.B.; Chen, Z.J.; Liu, G.D.; Tian, J. Characterization of purple acid phosphatases involved in extracellular dNTP utilization in Stylosanthes. J. Exp. Bot. 2016, 67, 4141–4154. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Hurley, B.A.; Plaxton, W.C. Feeding hungry plants: The role of purple acid phosphatases in phosphate nutrition. Plant Sci. 2010, 179, 14–27. [Google Scholar] [CrossRef]
- Tian, J.; Liao, H. The role of intracellular and secreted purple acid phosphatases in plant phosphorus scavenging and recycling. Annu. Plant Rev. 2015, 48, 265–287. [Google Scholar]
- Hegeman, C.E.; Grabau, E.A. A novel phytase with sequence similarity to purple acid phosphatases is expressed in cotyledons of germinating soybean seedlings. Plant Physiol. 2001, 126, 1598–1608. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Wong, F.L.; Phang, T.H.; Cheung, M.Y.; Li, W.Y.; Shao, G.; Yan, X.; Lam, H.M. GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene 2003, 318, 103–111. [Google Scholar] [CrossRef]
- Kong, Y.; Li, X.; Ma, J.; Li, W.; Yan, G.; Zhang, C. GmPAP4, a novel purple acid phosphatase gene isolated from soybean (Glycine max), enhanced extracellular phytate utilization in Arabidopsis thaliana. Plant Cell Rep. 2014, 33, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, C.; Zhang, H.; Liao, H.; Wang, X. The purple acid phosphatase GmPAP21 enhances internal phosphorus utilization and possibly plays a role in symbiosis with rhizobia in soybean. Physiol. Plant 2017, 159, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, X.; Xu, Y.; Yao, M.; Xie, F.; Gai, J.; Li, Y.; Yang, S. Soybean SPX1 is an important component of the response to phosphate deficiency for phosphorus homeostasis. Plant Sci. 2016, 248, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Nasr, E.M.; Kusano, M.; Nguyen, K.H.; Watanabe, Y.; Ha, C.V.; Saito, K.; Sulieman, S.; Herrera-Estrella, L.; Tran, L.S. Adaptation of the symbiotic Mesorhizobium-chickpea relationship to phosphate deficiency relies on reprogramming of whole-plant metabolism. Proc. Natl. Acad. Sci. USA 2016, 113, E4610–E4619. [Google Scholar] [CrossRef] [PubMed]
- Gaufchon, L.; Reisdorf-Cren, M.; Rothstein, S.J.; Chardon, F.; Suzuki, A. Biological functions of asparagine synthetase in plants. Plant Sci. 2010, 179, 141–153. [Google Scholar] [CrossRef]
- Sulieman, S.; Fischinger, S.A.; Gresshoff, P.M.; Schulze, J. Asparagine as a major factor in the N-feedback regulation of N2 fixation in Medicago truncatula. Physiol. Plant 2010, 140, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Sulieman, S.; Tran, L.S.P. Asparagine: An amide of particular distinction in the regulation of symbiotic nitrogen fixation of legumes. Crit. Rev. Biotechnol. 2013, 33, 309–327. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.; Nizampatnam, N.R.; Baron, M.; Coppin, S.; Damodaran, S.; Adhikari, S.; Arunachalam, S.P.; Yu, O.; Subramanian, S. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 2013, 162, 2042–2055. [Google Scholar] [CrossRef] [PubMed]
- Miwa, H.; Sun, J.; Oldroyd, G.E.D.; Downie, J.A. Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J. 2006, 48, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Kudla, J.; Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 2010, 61, 593–620. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Parniske, M. Activation of calcium- and calmodulin—Dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr. Opin. Plant Biol. 2012, 15, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Chu, X.; Wang, C.; Xiao, A.; Zhu, H.; Yuan, S.; Yang, Z.; Ke, D.; Xiao, S.; Hong, Z.; et al. A MYB coiled-coil transcription factor interacts with NSP2 and is involved in nodulation in Lotus japonicus. New Phytol. 2014, 201, 837–849. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, S.; Kim, J.; Munoz, A.; Heckmann, A.B.; Downie, J.A.; Oldroyd, G.E. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 2009, 21, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.; Riley, J. A modifed single solution method for the determination of phosphate in natural water. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Vadez, V.; Beck, D.P.; Lasso, J.H.; Drevon, J.J. Utilization of the acetylene reduction assay to screen for tolerance of symbiotic N2, fixation to limiting P nutrition in common bean. Physiol. Plant 1997, 99, 227–232. [Google Scholar] [CrossRef]
- Wang, M.; Ma, H.; Tian, C.; Liu, S.; Ye, X.; Zhou, D.; Li, Y.; Hui, N.; Li, X. Bioassay-guided isolation of glycoprotein SPG-56 from sweet potato Zhongshu-1 and its anti-colon cancer activity in vitro and in vivo. J. Funct. Foods 2017, 35, 315–324. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Feng, Z.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MapMan: A user-driven tool to display genomic data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Paszkowski, U.; Kroken, S.; Roux, C.; Briggs, S.P. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2002, 99, 13324–13329. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Versaw, W.K.; Pumplin, N.; Gomez, S.K.; Blaylock, L.A.; Harrison, M.J. Closely related members of the Medicago truncatula PHT1 phosphate transporter gene family encode phosphate transporters with distinct biochemical activities. J. Biol. Chem. 2008, 283, 24673–24681. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhu, H.; Liu, K.; Liu, X.; Leggewie, G.; Udvardi, M.; Wang, D. Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J. Biol. Chem. 2002, 277, 27772–27781. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, C.; Tian, J.; Li, K.; Shou, H. Identification of rice purple acid phosphatases related to phosphate starvation signalling. Plant Biol. 2011, 13, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Katagi, H.; Harrison, M.; Wang, Z.Y. Improved phosphorus acquisition and biomass production in Arabidopsis, by transgenic expression of a purple acid phosphatase gene from m. truncatula. Plant Sci. 2006, 170, 191–202. [Google Scholar] [CrossRef]
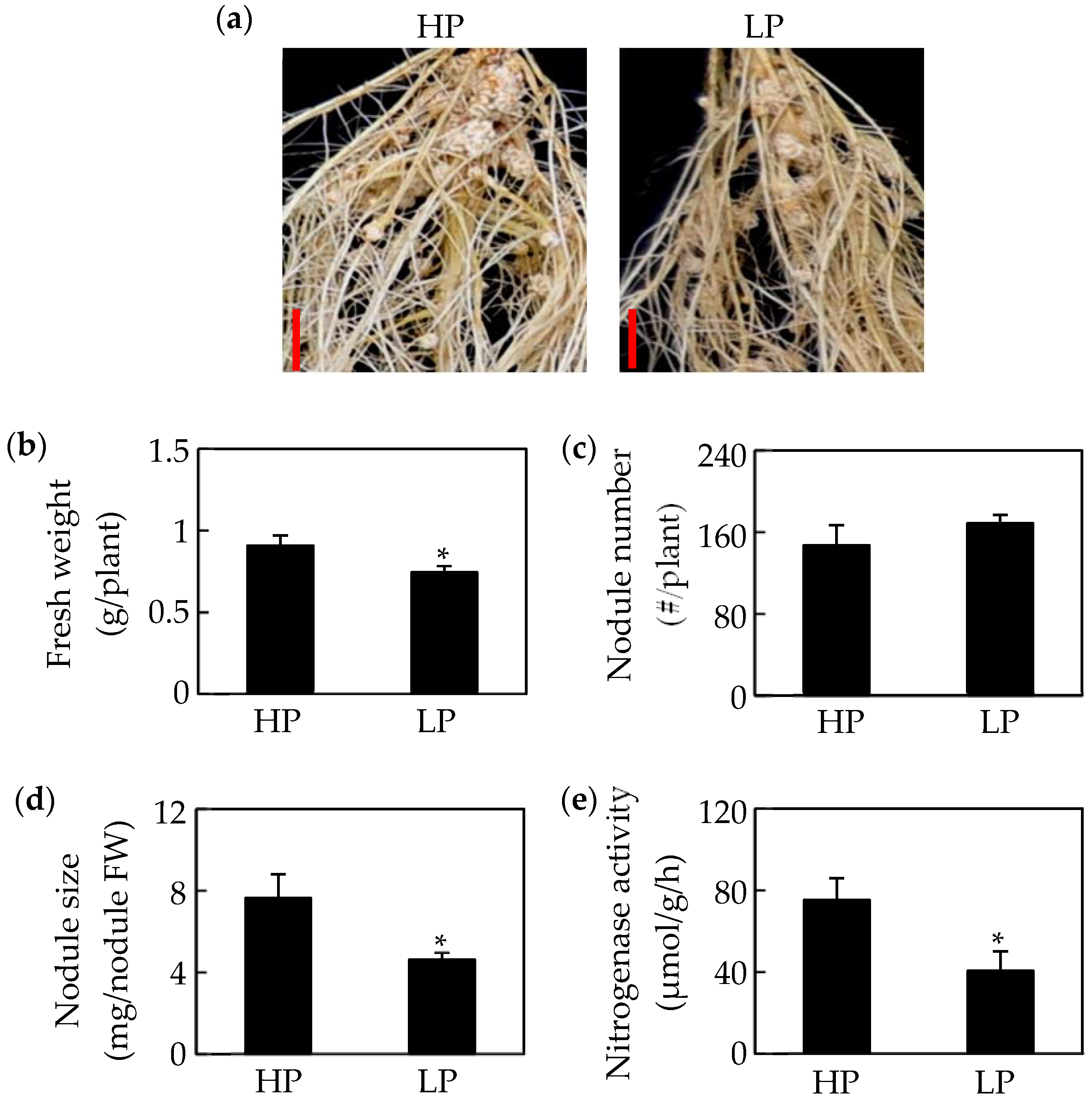
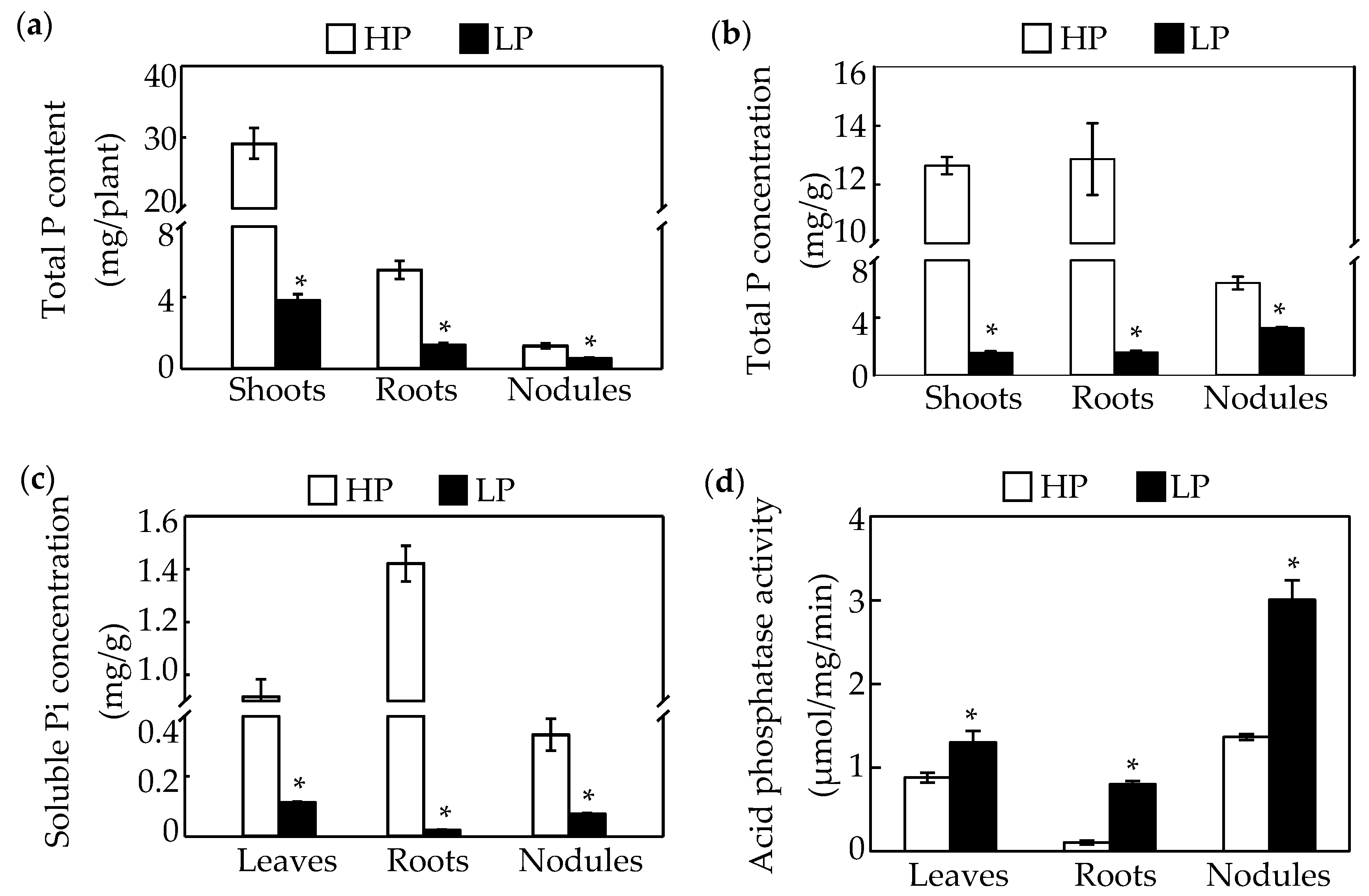
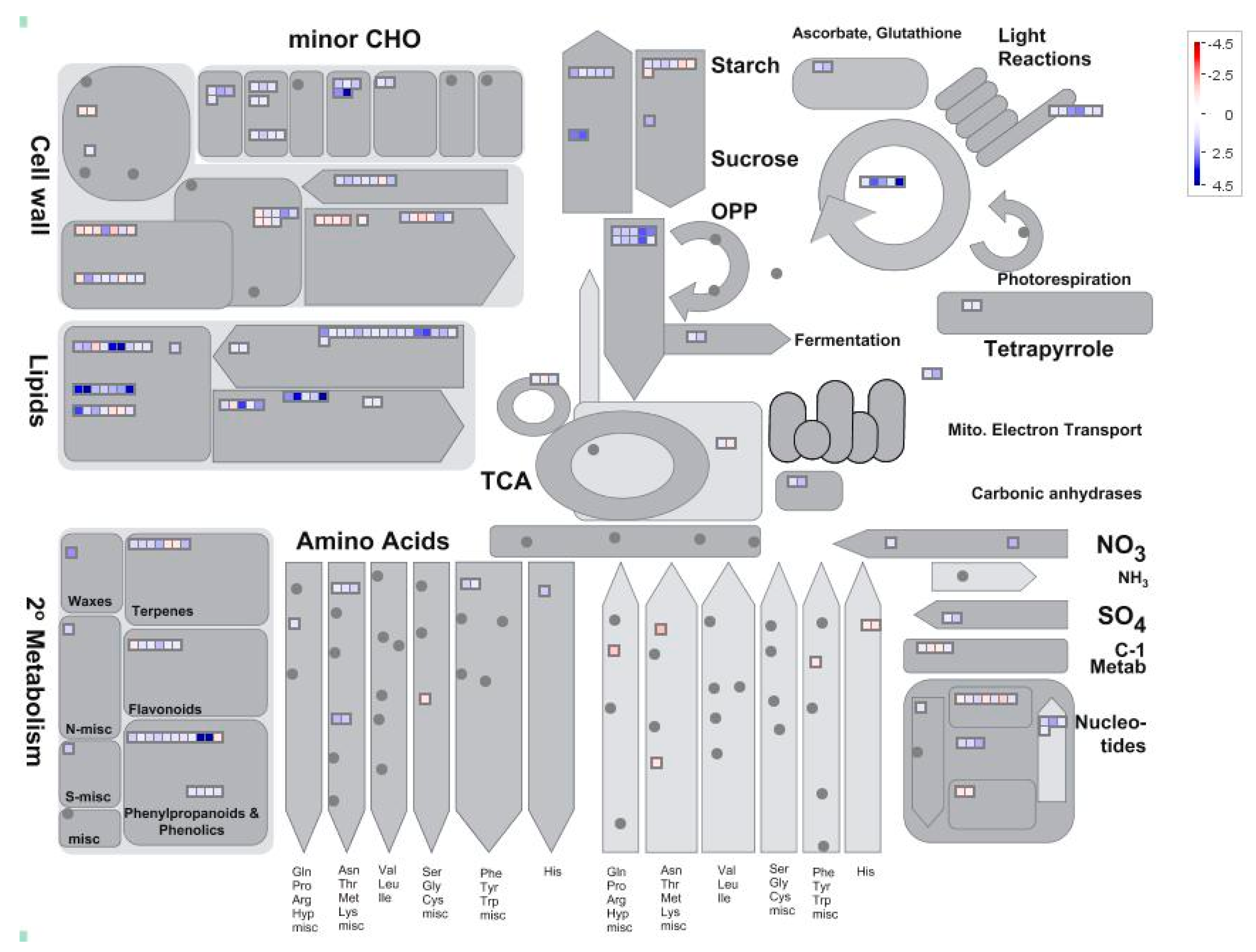
| HP | LP | Log2(LP/HP) | |||
|---|---|---|---|---|---|
| µg/g FW | % | µg/g FW | % | ||
| Asparagine | 3225.00 ± 570.99 | 69.47 | 5039.99 ± 720.71 | 79.83 | 0.64 * |
| Glutamic acid | 619.90 ± 45.82 | 13.35 | 285.56 ± 32.07 | 4.52 | −1.12 * |
| γ-Aminobutyric acid | 148.83 ± 9.96 | 3.21 | 134.97 ± 23.90 | 2.14 | −0.14 |
| Serine | 118.56 ± 9.95 | 2.55 | 117.30 ± 0.88 | 1.86 | −0.02 |
| Aspartic acid | 107.75 ± 8.35 | 2.32 | 68.12 ± 8.58 | 1.08 | −0.66 * |
| Alanine | 62.91 ± 4.19 | 1.36 | 53.39 ± 22.40 | 0.85 | −0.24 |
| Tryptophane | 51.22 ± 3.52 | 1.10 | 42.19 ± 5.36 | 0.67 | −0.28 |
| Glutamine | 43.98 ± 53.87 | 0.95 | 84.27 ± 33.24 | 1.33 | 0.94 |
| Arginine | 43.30 ± 4.81 | 0.93 | 155.11 ± 12.52 | 2.46 | 1.84 * |
| β-Alanine | 39.17 ± 4.00 | 0.84 | 30.68 ± 2.15 | 0.49 | −0.35 * |
| Phosphoserine | 34.65 ± 1.88 | 0.75 | 33.11 ± 2.49 | 0.52 | −0.07 |
| Histidine | 30.23 ± 3.20 | 0.65 | 135.99 ± 7.34 | 2.15 | 2.17 * |
| Threonine | 23.54 ± 3.23 | 0.51 | 22.33 ± 2.22 | 0.35 | −0.08 |
| Phenylalanine | 18.79 ± 2.42 | 0.40 | 17.25 ± 1.68 | 0.27 | −0.12 |
| Tyrosine | 12.91 ± 4.17 | 0.28 | 11.63 ± 1.44 | 0.18 | −0.15 |
| Lysine | 12.10 ± 1.71 | 0.26 | 13.32 ± 1.72 | 0.21 | 0.14 |
| Citrulline | 10.17 ± 4.09 | 0.22 | 7.28 ± 1.69 | 0.12 | −0.48 |
| Glycine | 8.31 ± 1.29 | 0.18 | 8.86 ± 0.66 | 0.14 | 0.09 |
| Isoleucine | 5.92 ± 0.55 | 0.13 | 10.24 ± 0.38 | 0.16 | 0.79 * |
| Cystine | 5.89 ± 0.75 | 0.13 | 3.90 ± 1.28 | 0.06 | −0.59 * |
| Leucine | 4.89 ± 1.48 | 0.11 | 5.49 ± 0.94 | 0.09 | 0.17 |
| Valine | 4.71 ± 0.83 | 0.10 | 3.48 ± 0.33 | 0.06 | −0.44 |
| Ornithine | 3.54 ± 1.20 | 0.08 | 2.52 ± 0.78 | 0.04 | −0.49 |
| Methionine | 3.18 ± 0.09 | 0.07 | 2.25 ± 1.18 | 0.04 | −0.5 |
| Phosphorylethanolamine | 2.64 ± 1.13 | 0.06 | 24.30 ± 0.56 | 0.38 | 3.20 * |
| Total | 4642.09 | 100 | 6313.53 | 100 |
| Total Expressed Genes | Up-Regulated | Down-Regulated | |
|---|---|---|---|
| HP | 38,813 | ||
| LP | 38,874 | ||
| DEGs * | 2055 | 1431 | 624 |
| Accession No. | Name/Description | Log2(LP/HP) | Q Value |
|---|---|---|---|
| Glyma.02G005800 | GmPT1 | 1.15 | 3.0 × 10−2 |
| Glyma.03G162800 | GmPT2 | 1.41 | 1.34 × 10−15 |
| Glyma.10G006700 | GmPT4 | 3.99 | 1.12 × 10−45 |
| Glyma.10G036800 | GmPT5 | 4.04 | 2.84 × 10−166 |
| Glyma.10G186400 | GmPT6 | 4.01 | 2.56 × 10−30 |
| Glyma.10G186500 | GmPT7 | 2.28 | 5.85 × 10−50 |
| Glyma.20G204000 | GmPT13 | 1.82 | 1.3 × 10−13 |
| Glyma.20G204100 | GmPT14 | 3.67 | 1.02 × 10−21 |
| Glyma.02G117000 | GmPAP1 | 2.15 | 3.94 × 10−15 |
| Glyma.05G138400 | GmPAP8 | 3.03 | 3.05 × 10−43 |
| Glyma.05G247900 | GmPAP9 | 2.45 | 5.49 × 10−93 |
| Glyma.05G247800 | GmPAP10 | 2.28 | 4.15 × 10−29 |
| Glyma.06G028200 | GmPAP11 | 5.41 | 1.90 × 10−239 |
| Glyma.08G056400 | GmPAP13 | 2.43 | 1.11 × 10−84 |
| Glyma.08G093500 | GmPAP15 | 2.19 | 2.92 × 10−46 |
| Glyma.08G093600 | GmPAP16 | 2.29 | 8.75 × 10−47 |
| Glyma.08G291600 | GmPAP17 | 1.70 | 2.33 × 10−19 |
| Glyma.09G229200 | GmPAP20 | 3.70 | 3.36 × 10−162 |
| Glyma.10G071000 | GmPAP21 | 4.61 | 9.92 × 10−78 |
| Glyma.12G007500 | GmPAP23 | 3.14 | 3.07 × 10−110 |
| Glyma.18G132500 | GmPAP30 | 1.29 | 3.01 × 10−13 |
| Glyma.19G026600 | GmPAP31 | 4.86 | 1.32 × 10−73 |
| Glyma.19G193900 | GmPAP32 | 1.81 | 2.24 × 10−4 |
| Glyma.20G026800 | GmPAP35 | 1.66 | 1.02 × 10−35 |
| Glyma.01G135500 | GmSPX1 | 6.35 | 0 |
| Glyma.04G067400 | GmSPX2 | 2.09 | 1.39 × 10−5 |
| Glyma.04G147600 | GmSPX3 | 4.55 | 9.01 × 10−105 |
| Glyma.06G069000 | GmSPX4 | 5.89 | 5.27 × 10−124 |
| Glyma.10G261900 | GmSPX5 | 1.99 | 2.31 × 10−14 |
| Glyma.13G166800 | GmSPX7 | 3.41 | 4.79 × 10−103 |
| Glyma.17G114700 | GmSPX8 | 3.52 | 1.03 × 10−73 |
| Glyma.20G129000 | GmSPX9 | 7.40 | 2.6 × 10−118 |
| Hormone | Accession No. | Name/Description | Log2(LP/HP) | Q Value |
|---|---|---|---|---|
| Auxin | Glyma.02G142400 | AUX/IAA family auxin-responsive protein | 2.25 | 3.56 × 10−8 |
| Glyma.02G142500 | AUX/IAA family auxin-responsive protein | 2.28 | 5.80 × 10−16 | |
| Glyma.04G025300 | AUX/IAA family auxin-responsive protein | 1.91 | 3.46 × 10−24 | |
| Glyma.05G196300 | AUX/IAA family auxin-responsive protein | 1.58 | 4.80 × 10−7 | |
| Glyma.09G193000 | AUX/IAA family auxin-responsive protein | 1.06 | 3.69 × 10−2 | |
| Glyma.10G031900 | SAUR family auxin-responsive protein | 1.86 | 7.77 × 10−6 | |
| Glyma.10G056200 | SAUR family auxin-responsive protein | −1.07 | 9.92 × 10−4 | |
| Glyma.12G035700 | SAUR family auxin-responsive protein | 1.84 | 5.57 × 10−5 | |
| Glyma.13G361200 | SAUR family auxin-responsive protein | 1.06 | 2.09 × 10−6 | |
| Glyma.16G020800 | SAUR family auxin-responsive protein | −1.27 | 1.70 × 10−2 | |
| Glyma.19G206100 | Auxin response factor | 2.16 | 7.42 × 10−6 | |
| Cytokinin | Glyma.04G055600 | Cytokinin dehydrogenase | −1.25 | 2.96 × 10−3 |
| Glyma.20G159600 | Cytokinin hydroxylase | 1.56 | 1.95 × 10−3 | |
| Ethylene | Glyma.01G206600 | AP2-like ethylene response factor | 1.18 | 3 × 10−2 |
| Glyma.02G132500 | AP2-like ethylene response factor | 1.90 | 2.06 × 10−5 | |
| Glyma.05G063500 | AP2-like ethylene response factor | 1.12 | 3.03 × 10−4 | |
| Glyma.06G236400 | AP2-like ethylene response factor | 1.09 | 6.61 × 10−6 | |
| Glyma.07G113800 | AP2-like ethylene response factor | 1.28 | 4.46 × 10−3 | |
| Glyma.07G212400 | AP2-like ethylene response factor | 2.60 | 1.05 × 10−18 | |
| Glyma.09G072000 | AP2-like ethylene response factor | 2.30 | 2.54 × 10−10 | |
| Glyma.10G118900 | AP2-like ethylene response factor | 1.01 | 7.27 × 10−6 | |
| Glyma.10G194200 | AP2-like ethylene response factor | 1.23 | 6.20 × 10−3 | |
| Glyma.12G110400 | AP2-like ethylene response factor | 1.04 | 4 × 10−2 | |
| Glyma.12G203100 | AP2-like ethylene response factor | 1.32 | 3.03 × 10−4 | |
| Glyma.13G040400 | AP2-like ethylene response factor | −1.13 | 2 × 10−2 | |
| Glyma.13G112400 | AP2-like ethylene response factor | 2.16 | 1.95 × 10−6 | |
| Glyma.15G180000 | AP2-like ethylene response factor | 2.87 | 3.81 × 10−16 | |
| Glyma.17G047300 | AP2-like ethylene response factor | 1.20 | 3 × 10−2 | |
| Glyma.17G070800 | AP2-like ethylene response factor | 1.25 | 6.47 × 10−3 | |
| Glyma.17G145300 | AP2-like ethylene response factor | 1.26 | 1.54 × 10−5 | |
| Glyma.18G281400 | AP2-like ethylene response factor | −1.35 | 3.86 × 10-4 | |
| Glyma.19G256800 | AP2-like ethylene response factor | 1.27 | 2 × 10−2 | |
| Glyma.20G070100 | AP2-like ethylene response factor | 1.93 | 2.56 × 10−9 | |
| Glyma.20G172800 | AP2-like ethylene response factor | −1.18 | 2 × 10−2 | |
| Glyma.14G041500 | EIN3-like ethylene response factor | 1.32 | 8.33 × 10−19 | |
| Gibberellin | Glyma.12G137700 | Gibberellin-responsive protein | 1.44 | 7.13 × 10−4 |
| Glyma.13G285400 | Gibberellin-responsive protein | 1.49 | 2.82 × 10−8 | |
| Glyma.19G013000 | Gibberellin-regulated protein | 1.06 | 3 × 10−2 |
| Accession No. | Name/Description | Log2(LP/HP) | Q Value |
|---|---|---|---|
| Glyma.05G085200 | Annexin | 1.86 | 8.71 × 10−9 |
| Glyma.05G178200 | Annexin | 1.18 | 7.92 × 10−16 |
| Glyma.10G002200 | Annexin-like | −1.29 | 4.65 × 10−14 |
| Glyma.01G166100 | Calmodulin-binding transcription activator | 1.76 | 2.47 × 10−17 |
| Glyma.02G059200 | Calcium-transporting ATPase | 1.66 | 4.21 × 10−13 |
| Glyma.02G245700 | Calcium-transporting ATPase | 1.44 | 4.46 × 10−13 |
| Glyma.03G138000 | Calmodulin-like protein | −1.03 | 3.38 × 10−4 |
| Glyma.04G064800 | Calmodulin-like protein | 1.43 | 3.68 × 10−5 |
| Glyma.04G136200 | Calcium-binding protein | 1.32 | 1.40 × 10−4 |
| Glyma.05G047100 | Calcium uptake protein | 1.15 | 2 × 10−2 |
| Glyma.05G199400 | Calcium-binding protein | 1.47 | 5.09 × 10−4 |
| Glyma.06G171100 | Calcium-binding protein | 1.06 | 4 × 10−2 |
| Glyma.07G229500 | Calcium-binding protein | 1.56 | 8.46 × 10−6 |
| Glyma.08G006900 | Calcium-binding protein | −1.19 | 5.63 × 10−3 |
| Glyma.11G048300 | Calcium-binding protein | 1.50 | 2.93 × 10−4 |
| Glyma.11G077300 | Calcium-binding protein | 1.16 | 3.34 × 10-3 |
| Glyma.11G157100 | Calcium-binding protein | 1.11 | 2 × 10−2 |
| Glyma.12G217700 | Calcium-binding protein | 2.23 | 1.37 × 10−9 |
| Glyma.14G156300 | Calcium-binding protein | −1.48 | 2.72 × 10−3 |
| Glyma.14G222000 | Calcium-binding protein | 1.58 | 2.06 × 10−20 |
| Glyma.16G142100 | Calcium-binding protein | 1.12 | 1.60 × 10−9 |
| Glyma.17G128900 | Calcium-dependent protein kinase | 1.40 | 1.65 × 10−6 |
| Glyma.20G034200 | Calcium-dependent protein kinase | 1.06 | 1.73 × 10−4 |
| Glyma.20G066800 | Calcium-dependent protein kinase | 1.55 | 9.09 × 10−16 |
| Glyma.05G248000 | Calcium-dependent protein kinase | −1.08 | 2.08 × 10−4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, Y.; Zhuang, Q.; Zhu, S.; Xiao, B.; Liang, C.; Liao, H.; Tian, J. Genome Wide Transcriptome Analysis Reveals Complex Regulatory Mechanisms Underlying Phosphate Homeostasis in Soybean Nodules. Int. J. Mol. Sci. 2018, 19, 2924. https://doi.org/10.3390/ijms19102924
Xue Y, Zhuang Q, Zhu S, Xiao B, Liang C, Liao H, Tian J. Genome Wide Transcriptome Analysis Reveals Complex Regulatory Mechanisms Underlying Phosphate Homeostasis in Soybean Nodules. International Journal of Molecular Sciences. 2018; 19(10):2924. https://doi.org/10.3390/ijms19102924
Chicago/Turabian StyleXue, Yingbin, Qingli Zhuang, Shengnan Zhu, Bixian Xiao, Cuiyue Liang, Hong Liao, and Jiang Tian. 2018. "Genome Wide Transcriptome Analysis Reveals Complex Regulatory Mechanisms Underlying Phosphate Homeostasis in Soybean Nodules" International Journal of Molecular Sciences 19, no. 10: 2924. https://doi.org/10.3390/ijms19102924
APA StyleXue, Y., Zhuang, Q., Zhu, S., Xiao, B., Liang, C., Liao, H., & Tian, J. (2018). Genome Wide Transcriptome Analysis Reveals Complex Regulatory Mechanisms Underlying Phosphate Homeostasis in Soybean Nodules. International Journal of Molecular Sciences, 19(10), 2924. https://doi.org/10.3390/ijms19102924






