An Overview of Direct Somatic Reprogramming: The Ins and Outs of iPSCs
Abstract
:1. Introduction
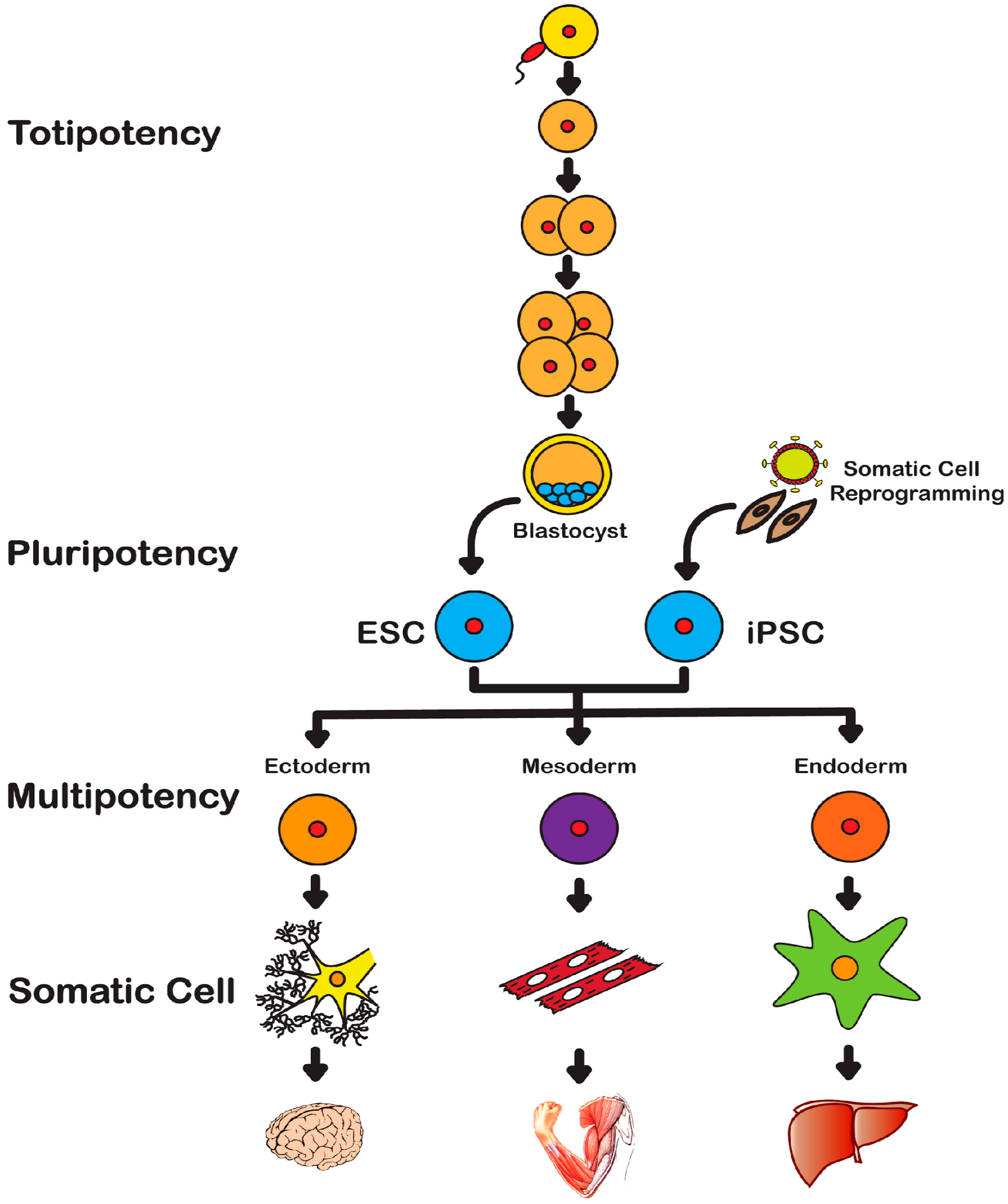
2. History of Stem Cells
2.1. Embryonic Stem Cells
2.2. Induced Pluripotent Stem Cells
3. Reprogramming Techniques
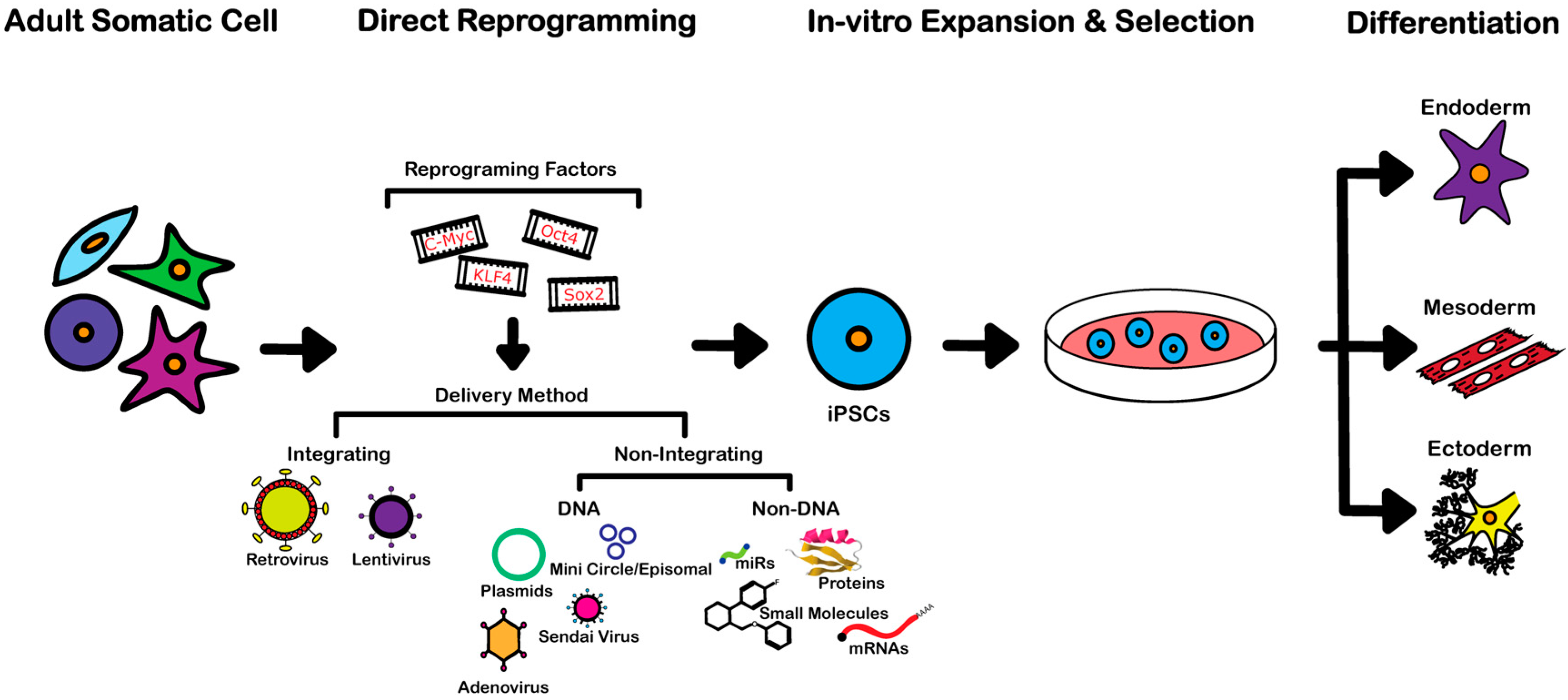
3.1. Integrating Procedures
3.2. Non-Integrating DNA-Based Procedures
3.3. Non-Integrating Non-DNA-Based Procedures
4. Clinical and Therapeutic Application of iPSCs
4.1. Tissue Regeneration and Cell Transplantation Therapy
4.2. Disease Modeling and Drug Screening
| Disease Type | Disease | Cell Line Derived | Reference |
|---|---|---|---|
| Neurological | Huntington’s Disease (HD) | Fibroblasts | [69] |
| Parkinson’s Disease (PD) | Fibroblasts | [70] | |
| Familial Alzheimer's Disease (AD) | Fibroblasts | [71] | |
| Frontotemporal Dementia (FTD) | Fibroblasts | [72] | |
| Amyotrophic Lateral Sclerosis (ALS) | Fibroblasts | [73,74,75] | |
| Spinocerebellar Ataxia Type 2 (SCA 2) | Fibroblasts | [76] | |
| Machado-Joseph Disease (MJD) | Fibroblasts | [77] | |
| Rett Syndrome | Fibroblasts | [78] | |
| Cardiac/Muscular | LEOPARD Syndrome | Fibroblasts | [79] |
| Duchenne Muscular Dystrophy (DMD) | Fibroblasts | [80] | |
| Long QT Syndrome | Fibroblasts | [81] | |
| Pompe Disease | Fibroblasts | [82] | |
| Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC) | Fibroblasts | [83,84] | |
| Dilated Cardiomyopathy (CMD) | Fibroblasts | [85] | |
| Barth Syndrome (BTHS) | Fibroblasts | [86] | |
| Friedreich Ataxia | Fibroblasts | [87] | |
| Metabolic | Alpha-1 Antitrypsin Deficiency (A1ATD) | Fibroblasts | [88] |
| Familial Hypercholesterolemia (FH) | Fibroblasts | [89] | |
| Glycogen Storage Disease Type 1a (GSD1a) | Fibroblasts | [90] | |
| Glycogen Storage Disease Type 1b (GSD1b) | Fibroblasts, Hepatic Cells (nonparenchymal) | [91] | |
| Type 1 Diabetes | Fibroblasts | [92] | |
| Gauchers Disease (GD) | Fibroblasts | [93] | |
| Mitochondrial Encephalomyopathy, Lactic Acidosis, and Stroke-like Episodes (MELAS) | Fibroblasts | [94] | |
| Carnitine Palmitoyltransferase II (CPT II) | Fibroblasts | [95] | |
| Eye/Retina | Usher Syndrome (USH) | Keratinocytes | [96] |
| Retinitis Pigmentosa (RP) | Fibroblasts | [97,98,99] | |
| Leber Congenital Amaurosis (LCA) | Fibroblasts | [100] | |
| Gyrate Atrophy (GA) | Fibroblasts | [101] | |
| Best Vitelliform Macular Dystrophy (BVMD) | Fibroblasts | [102] | |
| Age Related Macular Degeneration (AMD) | T-Cells | [103] | |
| Blood | Shwachman-Diamond Syndrome (SDS) | Fibroblasts | [104] |
| Dyskeratosis Congenita (DKC) | Fibroblasts | [105] | |
| Familial Platelet Disorder (FPD) | Peripheral T Cells | [106] | |
| Sickle Cell Disease | MSCs | [107,108] | |
| β-Thalassemia | Fibroblasts | [109] | |
| Myeloproliferative Disorders (MPDs) | Peripheral Blood CD34+ Cells | [18] | |
| Myelodysplastic Syndromes (MDS) | Hematopoietic Cells | [17] | |
| Skeletal/Bone | Fibrodysplasia Ossificans Progressiva | Fibroblasts | [110] |
| Menkes Disease (MNK) | Fibroblasts | [111] | |
| Skeletal Dysplasia (SD) | Fibroblasts | [112] | |
| Marfan Syndrome (MFS) | Fibroblasts | [113] | |
| Craniometaphyseal Dysplasia (CDM) | Peripheral Blood Mononuclear Cells | [114] | |
| Cancer | Pancreatic ductal Adenocarcinoma (PDAC) | Primary PDAC Cells | [115] |
| Chronic Myeloid Leukemia (CML) | Primary CML Cells | [20,116] | |
| Juvenile Myelomonocytic Leukemia (JML) | Primary JML Cells | [117] | |
| Gastrointestinal Cancer | Gastrointestinal Cancer Cells | [21] | |
| Li-Fraumeni Syndrome (LFS) | Fibroblasts | [118] | |
| Other | Hutchinson Gilford Progeria | Fibroblasts | [119] |
| Primary Ovarian Insufficiency (POI) | Fibroblasts | [120] | |
| Hermansky-Pudlak (HP) | Fibroblasts | [121] | |
| Chediak-Higashi (CH) | Fibroblasts | [121] |
5. Limitations and Concerns of Current iPSCs Technology
5.1. Functionality
5.2. Chromosomal Aberrations and Genetic Modifications
5.3. Cancer and Terotocarcinoma Formation
5.4. Immunogenicity
6. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed]
- Tesar, P.J.; Chenoweth, J.G.; Brook, F.A.; Davies, T.J.; Evans, E.P.; Mack, D.L.; Gardner, R.L.; McKay, R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007, 448, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Ferrer, S.; Scadden, D.T.; Sanchez-Aguilera, A. Bone marrow stem cells: Current and emerging concepts. Ann. N. Y. Acad. Sci. 2015, 1335, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Senarath-Yapa, K.; McArdle, A.; Renda, A.; Longaker, M.T.; Quarto, N. Adipose-derived stem cells: A review of signaling networks governing cell fate and regenerative potential in the context of craniofacial and long bone skeletal repair. Int. J. Mol. Sci. 2014, 15, 9314–9330. [Google Scholar] [CrossRef] [PubMed]
- Weissman, I.L.; Shizuru, J.A. The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor-specific transplantation tolerance and treat autoimmune diseases. Blood 2008, 112, 3543–3553. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Notarianni, E.; Galli, C.; Laurie, S.; Moor, R.M.; Evans, M.J. Derivation of pluripotent, embryonic cell lines from the pig and sheep. J. Reprod. Fertil. Suppl. 1991, 43, 255–260. [Google Scholar] [PubMed]
- Gurdon, J.B.; Elsdale, T.R.; Fischberg, M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 1958, 182, 64–65. [Google Scholar] [CrossRef] [PubMed]
- Briggs, R.; King, T.J. Transplantation of living nuclei from blastula cells into enucleated frogs’ eggs. Proc. Natl. Acad. Sci. USA 1952, 38, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. A developmental framework for induced pluripotency. Development 2015, 142, 3274–3285. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassady, J.P.; D’Alessio, A.C.; Sarkar, S.; Dani, V.S.; Fan, Z.P.; Ganz, K.; Roessler, R.; Sur, M.; Young, R.A.; Jaenisch, R. Direct lineage conversion of adult mouse liver cells and B lymphocytes to neural stem cells. Stem Cell Rep. 2014, 3, 948–956. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Izpisua Belmonte, J.C. Isolation and cultivation of human keratinocytes from skin or plucked hair for the generation of induced pluripotent stem cells. Nat. Protoc. 2010, 5, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Aasen, T.; Raya, A.; Barrero, M.J.; Garreta, E.; Consiglio, A.; Gonzalez, F.; Vassena, R.; Bilic, J.; Pekarik, V.; Tiscornia, G.; et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 2008, 26, 1276–1284. [Google Scholar] [CrossRef] [PubMed]
- Kotini, A.G.; Chang, C.J.; Boussaad, I.; Delrow, J.J.; Dolezal, E.K.; Nagulapally, A.B.; Perna, F.; Fishbein, G.A.; Klimek, V.M.; Hawkins, R.D.; et al. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat. Biotechnol. 2015, 33, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Zhan, H.; Mali, P.; Dowey, S.; Williams, D.M.; Jang, Y.Y.; Dang, C.V.; Spivak, J.L.; Moliterno, A.R.; Cheng, L. Human-induced pluripotent stem cells from blood cells of healthy donors and patients with acquired blood disorders. Blood 2009, 114, 5473–5480. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Liu, G.H.; Izpisua Belmonte, J.C. Human induced pluripotent stem cells derived hepatocytes: Rising promise for disease modeling, drug development and cell therapy. Protein Cell 2012, 3, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Kumano, K.; Arai, S.; Hosoi, M.; Taoka, K.; Takayama, N.; Otsu, M.; Nagae, G.; Ueda, K.; Nakazaki, K.; Kamikubo, Y.; et al. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood 2012, 119, 6234–6242. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, N.; Ishii, H.; Nagai, K.; Hoshino, H.; Mimori, K.; Tanaka, F.; Nagano, H.; Sekimoto, M.; Doki, Y.; Mori, M. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc. Natl. Acad. Sci. USA 2010, 107, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Ghodsizadeh, A.; Taei, A.; Totonchi, M.; Seifinejad, A.; Gourabi, H.; Pournasr, B.; Aghdami, N.; Malekzadeh, R.; Almadani, N.; Salekdeh, G.H.; et al. Generation of liver disease-specific induced pluripotent stem cells along with efficient differentiation to functional hepatocyte-like cells. Stem Cell Rev. 2010, 6, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, F.; Boue, S.; Izpisua Belmonte, J.C. Methods for making induced pluripotent stem cells: Reprogramming a la carte. Nat. Rev. Genet. 2011, 12, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Seifinejad, A.; Tabebordbar, M.; Baharvand, H.; Boyer, L.A.; Salekdeh, G.H. Progress and promise towards safe induced pluripotent stem cells for therapy. Stem Cell Rev. 2010, 6, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Izpisua Belmonte, J.C. Dynamic pluripotent stem cell states and their applications. Cell Stem Cell 2015, 17, 509–525. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, M.; Koyanagi, M.; Tanabe, K.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Okita, K.; Mochiduki, Y.; Takizawa, N.; Yamanaka, S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008, 26, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Markoulaki, S.; Hanna, J.; Beard, C.; Carey, B.W.; Cheng, A.W.; Lengner, C.J.; Dausman, J.A.; Fu, D.; Gao, Q.; Wu, S.; et al. Transgenic mice with defined combinations of drug-inducible reprogramming factors. Nat. Biotechnol. 2009, 27, 169–171. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Ichisaka, T.; Yamanaka, S. Generation of germline-competent induced pluripotent stem cells. Nature 2007, 448, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Wernig, M.; Meissner, A.; Cassady, J.P.; Jaenisch, R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell 2008, 2, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Aoi, T.; Nakagawa, M.; Ichisaka, T.; Okita, K.; Takahashi, K.; Chiba, T.; Yamanaka, S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 2008, 321, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Stadtfeld, M.; Nagaya, M.; Utikal, J.; Weir, G.; Hochedlinger, K. Induced pluripotent stem cells generated without viral integration. Science 2008, 322, 945–949. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.W.; Lai, Y.S.; Pawlik, K.M.; Liu, K.; Sun, C.W.; Li, C.; Schoeb, T.R.; Townes, T.M. Polycistronic lentiviral vector for “hit and run” reprogramming of adult skin fibroblasts to induced pluripotent stem cells. Stem Cells 2009, 27, 1042–1049. [Google Scholar] [CrossRef] [PubMed]
- Woodard, L.E.; Wilson, M.H. PiggyBac-ing models and new therapeutic strategies. Trends Biotechnol. 2015, 33, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Woltjen, K.; Michael, I.P.; Mohseni, P.; Desai, R.; Mileikovsky, M.; Hamalainen, R.; Cowling, R.; Wang, W.; Liu, P.; Gertsenstein, M.; et al. PiggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 2009, 458, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Kaji, K.; Norrby, K.; Paca, A.; Mileikovsky, M.; Mohseni, P.; Woltjen, K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature 2009, 458, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Medvedev, S.P.; Shevchenko, A.I.; Zakian, S.M. Induced pluripotent stem cells: Problems and advantages when applying them in regenerative medicine. Acta Naturae 2010, 2, 18–28. [Google Scholar] [PubMed]
- Graham, F.L.; Prevec, L. Adenovirus-based expression vectors and recombinant vaccines. Biotechnology 1992, 20, 363–390. [Google Scholar] [PubMed]
- He, T.C.; Zhou, S.; da Costa, L.T.; Yu, J.; Kinzler, K.W.; Vogelstein, B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 1998, 95, 2509–2514. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Freed, C.R. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells 2009, 27, 2667–2674. [Google Scholar] [CrossRef] [PubMed]
- Narsinh, K.H.; Jia, F.; Robbins, R.C.; Kay, M.A.; Longaker, M.T.; Wu, J.C. Generation of adult human induced pluripotent stem cells using nonviral minicircle DNA vectors. Nat. Protoc. 2011, 6, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Meraviglia, V.; Zanon, A.; Lavdas, A.A.; Schwienbacher, C.; Silipigni, R.; di Segni, M.; Chen, H.S.; Pramstaller, P.P.; Hicks, A.A.; Rossini, A. Generation of induced pluripotent stem cells from frozen buffy coats using non-integrating episomal plasmids. J. Vis. Exp. 2015, e52885. [Google Scholar]
- Hu, W.; He, Y.; Xiong, Y.; Lu, H.; Chen, H.; Hou, L.; Qiu, Z.; Fang, Y.; Zhang, S. Derivation, expansion, and motor neuron differentiation of human-induced pluripotent stem cells with non-integrating episomal vectors and a defined Xenogeneic-free culture system. Mol. Neurobiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Yamakawa, T.; Matsumura, Y.; Sato, Y.; Amano, N.; Watanabe, A.; Goshima, N.; Yamanaka, S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 2013, 31, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Hansen, N.F.; Zhao, L.; Du, Y.; Zou, C.; Donovan, F.X.; Chou, B.K.; Zhou, G.; Li, S.; Dowey, S.N.; et al. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell 2012, 10, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Fusaki, N.; Ban, H.; Nishiyama, A.; Saeki, K.; Hasegawa, M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 348–362. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Verma, P.J. Synthetic mRNA reprogramming of human fibroblast cells. Methods Mol. Biol. 2015, 1330, 17–28. [Google Scholar] [PubMed]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, N.; Ishii, H.; Nagano, H.; Haraguchi, N.; Dewi, D.L.; Kano, Y.; Nishikawa, S.; Tanemura, M.; Mimori, K.; Tanaka, F.; et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 2011, 8, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Sandmaier, S.E.; Telugu, B.P. MicroRNA-mediated reprogramming of somatic cells into induced pluripotent stem cells. Methods Mol. Biol. 2015, 1330, 29–36. [Google Scholar] [PubMed]
- Zhou, H.; Wu, S.; Joo, J.Y.; Zhu, S.; Han, D.W.; Lin, T.; Trauger, S.; Bien, G.; Yao, S.; Zhu, Y.; et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell 2009, 4, 381–384. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, C.H.; Moon, J.I.; Chung, Y.G.; Chang, M.Y.; Han, B.S.; Ko, S.; Yang, E.; Cha, K.Y.; Lanza, R.; et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 2009, 4, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Belting, M.; Sandgren, S.; Wittrup, A. Nuclear delivery of macromolecules: Barriers and carriers. Adv. Drug Deliv. Rev. 2005, 57, 505–527. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.; Futaki, S.; Harashima, H. Delivery of macromolecules using arginine-rich cell-penetrating peptides: Ways to overcome endosomal entrapment. AAPS J. 2009, 11, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, A.; Nervi, P.; Durrenberger, M.; Seelig, J. The cationic cell-penetrating peptide CPP (TAT) derived from the HIV-1 protein TAT is rapidly transported into living fibroblasts: Optical, biophysical, and metabolic evidence. Biochemistry 2005, 44, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Nie, B.; Wang, H.; Laurent, T.; Ding, S. Cellular reprogramming: A small molecule perspective. Curr. Opin. Cell Biol. 2012, 24, 784–792. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, D.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Snitow, M.; Chen, A.E.; Melton, D.A. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 2008, 26, 795–797. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Q.; Yin, X.; Yang, W.; Du, Y.; Hou, P.; Ge, J.; Liu, C.; Zhang, W.; Zhang, X.; et al. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011, 21, 196–204. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Rana, T.M. A kinase inhibitor screen identifies small-molecule enhancers of reprogramming and iPS cell generation. Nat. Commun. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zuo, X.; Jing, J.; Ma, Y.; Wang, J.; Liu, D.; Zhu, J.; Du, X.; Xiong, L.; Du, Y.; et al. Small-molecule-driven direct reprogramming of mouse fibroblasts into functional neurons. Cell Stem Cell 2015, 17, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.L.; Zang, T.; Zou, Y.; Chang, J.C.; Gibson, J.R.; Huber, K.M.; Zhang, C.L. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat. Commun. 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Hu, W.; Qiu, B.; Zhao, J.; Yu, Y.; Guan, W.; Wang, M.; Yang, W.; Pei, G. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 2014, 24, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Ladewig, J.; Mertens, J.; Kesavan, J.; Doerr, J.; Poppe, D.; Glaue, F.; Herms, S.; Wernet, P.; Kogler, G.; Muller, F.J.; et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods 2012, 9, 575–578. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Li, W.; Zhou, H.; Wei, W.; Ambasudhan, R.; Lin, T.; Kim, J.; Zhang, K.; Ding, S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 2010, 7, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wilson, G.F.; Soerens, A.G.; Koonce, C.H.; Yu, J.; Palecek, S.P.; Thomson, J.A.; Kamp, T.J. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ. Res. 2009, 104, e30–e41. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.M.; Huang, Y.J.; Chen, P.; Hsu, Y.C.; Lin, S.W.; Lai, H.; Lee, H.S. Hepatocyte-like cells derived from mouse induced pluripotent stem cells produce functional coagulation factor IX in a hemophilia B mouse model. Cell Transpl. 2015. [Google Scholar] [CrossRef]
- Hanna, J.; Wernig, M.; Markoulaki, S.; Sun, C.W.; Meissner, A.; Cassady, J.P.; Beard, C.; Brambrink, T.; Wu, L.C.; Townes, T.M.; et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 2007, 318, 1920–1923. [Google Scholar] [CrossRef] [PubMed]
- Dimos, J.T.; Rodolfa, K.T.; Niakan, K.K.; Weisenthal, L.M.; Mitsumoto, H.; Chung, W.; Croft, G.F.; Saphier, G.; Leibel, R.; Goland, R.; et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 2008, 321, 1218–1221. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Arora, N.; Huo, H.; Maherali, N.; Ahfeldt, T.; Shimamura, A.; Lensch, M.W.; Cowan, C.; Hochedlinger, K.; Daley, G.Q. Disease-specific induced pluripotent stem cells. Cell 2008, 134, 877–886. [Google Scholar] [CrossRef] [PubMed]
- Consortium, H.D.i. Induced pluripotent stem cells from patients with huntington′s disease show cag-repeat-expansion-associated phenotypes. Cell Stem Cell 2012, 11, 264–278. [Google Scholar]
- Byers, B.; Cord, B.; Nguyen, H.N.; Schule, B.; Fenno, L.; Lee, P.C.; Deisseroth, K.; Langston, J.W.; Pera, R.R.; Palmer, T.D. SNCA triplication Parkinson′s patient′s iPSC-derived Da neurons accumulate α-synuclein and are susceptible to oxidative stress. PLoS ONE 2011, 6, e26159. [Google Scholar] [CrossRef] [PubMed]
- Soldner, F.; Hockemeyer, D.; Beard, C.; Gao, Q.; Bell, G.W.; Cook, E.G.; Hargus, G.; Blak, A.; Cooper, O.; Mitalipova, M.; et al. Parkinson′s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009, 136, 964–977. [Google Scholar] [CrossRef] [PubMed]
- Israel, M.A.; Yuan, S.H.; Bardy, C.; Reyna, S.M.; Mu, Y.; Herrera, C.; Hefferan, M.P.; van Gorp, S.; Nazor, K.L.; Boscolo, F.S.; et al. Probing sporadic and familial Alzheimer′s disease using induced pluripotent stem cells. Nature 2012, 482, 216–220. [Google Scholar] [PubMed]
- Almeida, S.; Gascon, E.; Tran, H.; Chou, H.J.; Gendron, T.F.; Degroot, S.; Tapper, A.R.; Sellier, C.; Charlet-Berguerand, N.; Karydas, A.; et al. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013, 126, 385–399. [Google Scholar] [CrossRef] [PubMed]
- Almeida, S.; Zhang, Z.; Coppola, G.; Mao, W.; Futai, K.; Karydas, A.; Geschwind, M.D.; Tartaglia, M.C.; Gao, F.; Gianni, D.; et al. Induced pluripotent stem cell models of progranulin-deficient frontotemporal dementia uncover specific reversible neuronal defects. Cell Rep. 2012, 2, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Balasubramanian, U.; Cohen, D.; Zhang, P.W.; Mosmiller, E.; Sattler, R.; Maragakis, N.J.; Rothstein, J.D. A comprehensive library of familial human amyotrophic lateral sclerosis induced pluripotent stem cells. PLoS ONE 2015, 10, e0118266. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qian, K.; Du, Z.; Cao, J.; Petersen, A.; Liu, H.; Blackbourn, L.W.T.; Huang, C.L.; Errigo, A.; Yin, Y.; et al. Modeling als with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell 2014, 14, 796–809. [Google Scholar] [CrossRef] [PubMed]
- Egawa, N.; Kitaoka, S.; Tsukita, K.; Naitoh, M.; Takahashi, K.; Yamamoto, T.; Adachi, F.; Kondo, T.; Okita, K.; Asaka, I.; et al. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med. 2012, 4, 145ra104. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.; Santostefano, K.; Hamazaki, T.; Liu, J.; Subramony, S.H.; Terada, N.; Ashizawa, T. Generation of human-induced pluripotent stem cells to model spinocerebellar ataxia type 2 in vitro. J. Mol. Neurosci. 2013, 51, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Koch, P.; Breuer, P.; Peitz, M.; Jungverdorben, J.; Kesavan, J.; Poppe, D.; Doerr, J.; Ladewig, J.; Mertens, J.; Tuting, T.; et al. Excitation-induced ataxin-3 aggregation in neurons from patients with machado-joseph disease. Nature 2011, 480, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Djuric, U.; Cheung, A.Y.; Zhang, W.; Mok, R.S.; Lai, W.; Piekna, A.; Hendry, J.A.; Ross, P.J.; Pasceri, P.; Kim, D.S.; et al. MECP2E1 isoform mutation affects the form and function of neurons derived from rett syndrome patient ips cells. Neurobiol. Dis. 2015, 76, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Carvajal-Vergara, X.; Sevilla, A.; D′Souza, S.L.; Ang, Y.S.; Schaniel, C.; Lee, D.F.; Yang, L.; Kaplan, A.D.; Adler, E.D.; Rozov, R.; et al. Patient-specific induced pluripotent stem-cell-derived models of leopard syndrome. Nature 2010, 465, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Shoji, E.; Sakurai, H.; Nishino, T.; Nakahata, T.; Heike, T.; Awaya, T.; Fujii, N.; Manabe, Y.; Matsuo, M.; Sehara-Fujisawa, A. Early pathogenesis of duchenne muscular dystrophy modelled in patient-derived human induced pluripotent stem cells. Sci. Rep. 2015, 5, 12831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moretti, A.; Bellin, M.; Welling, A.; Jung, C.B.; Lam, J.T.; Bott-Flugel, L.; Dorn, T.; Goedel, A.; Hohnke, C.; Hofmann, F.; et al. Patient-specific induced pluripotent stem-cell models for long-qt syndrome. N. Engl. J. Med. 2010, 363, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.P.; Chen, P.H.; Hwu, W.L.; Chuang, C.Y.; Chien, Y.H.; Stone, L.; Chien, C.L.; Li, L.T.; Chiang, S.C.; Chen, H.F.; et al. Human pompe disease-induced pluripotent stem cells for pathogenesis modeling, drug testing and disease marker identification. Hum. Mol. Genet. 2011, 20, 4851–4864. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Wong, J.; Wen, J.; Wang, S.; Wang, C.; Spiering, S.; Kan, N.G.; Forcales, S.; Puri, P.L.; Leone, T.C.; et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature 2013, 494, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Caspi, O.; Huber, I.; Gepstein, A.; Arbel, G.; Maizels, L.; Boulos, M.; Gepstein, L. Modeling of arrhythmogenic right ventricular cardiomyopathy with human induced pluripotent stem cells. Circ. Cardiovasc. Genet. 2013, 6, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Siu, C.W.; Lee, Y.K.; Ho, J.C.; Lai, W.H.; Chan, Y.C.; Ng, K.M.; Wong, L.Y.; Au, K.W.; Lau, Y.M.; Zhang, J.; et al. Modeling of lamin A/C mutation premature cardiac aging using patient-specific induced pluripotent stem cells. Aging 2012, 4, 803–822. [Google Scholar] [PubMed]
- Dudek, J.; Cheng, I.F.; Balleininger, M.; Vaz, F.M.; Streckfuss-Bomeke, K.; Hubscher, D.; Vukotic, M.; Wanders, R.J.; Rehling, P.; Guan, K. Cardiolipin deficiency affects respiratory chain function and organization in an induced pluripotent stem cell model of barth syndrome. Stem Cell Res. 2013, 11, 806–819. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Verma, P.J.; Evans-Galea, M.V.; Delatycki, M.B.; Michalska, A.; Leung, J.; Crombie, D.; Sarsero, J.P.; Williamson, R.; Dottori, M.; et al. Generation of induced pluripotent stem cell lines from friedreich ataxia patients. Stem Cell Rev. 2011, 7, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Rashid, S.T.; Corbineau, S.; Hannan, N.; Marciniak, S.J.; Miranda, E.; Alexander, G.; Huang-Doran, I.; Griffin, J.; Ahrlund-Richter, L.; Skepper, J.; et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J. Clin. Investig. 2010, 120, 3127–3136. [Google Scholar] [CrossRef] [PubMed]
- Satoh, D.; Maeda, T.; Ito, T.; Nakajima, Y.; Ohte, M.; Ukai, A.; Nakamura, K.; Enosawa, S.; Toyota, M.; Miyagawa, Y.; et al. Establishment and directed differentiation of induced pluripotent stem cells from glycogen storage disease type ib patient. Genes Cells 2013, 18, 1053–1069. [Google Scholar] [CrossRef] [PubMed]
- Maehr, R.; Chen, S.; Snitow, M.; Ludwig, T.; Yagasaki, L.; Goland, R.; Leibel, R.L.; Melton, D.A. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc. Natl. Acad. Sci. USA 2009, 106, 15768–15773. [Google Scholar] [CrossRef] [PubMed]
- Panicker, L.M.; Miller, D.; Park, T.S.; Patel, B.; Azevedo, J.L.; Awad, O.; Masood, M.A.; Veenstra, T.D.; Goldin, E.; Stubblefield, B.K.; et al. Induced pluripotent stem cell model recapitulates pathologic hallmarks of gaucher disease. Proc. Natl. Acad. Sci. USA 2012, 109, 18054–18059. [Google Scholar] [CrossRef] [PubMed]
- Hamalainen, R.H.; Manninen, T.; Koivumaki, H.; Kislin, M.; Otonkoski, T.; Suomalainen, A. Tissue- and cell-type-specific manifestations of heteroplasmic mtDNA 3243a>g mutation in human induced pluripotent stem cell-derived disease model. Proc. Natl. Acad. Sci. USA 2013, 110, E3622–E3630. [Google Scholar] [CrossRef] [PubMed]
- Yasuno, T.; Osafune, K.; Sakurai, H.; Asaka, I.; Tanaka, A.; Yamaguchi, S.; Yamada, K.; Hitomi, H.; Arai, S.; Kurose, Y.; et al. Functional analysis of iPSC-derived myocytes from a patient with carnitine palmitoyltransferase ii deficiency. Biochem. Biophys. Res. Commun. 2014, 448, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Tucker, B.A.; Mullins, R.F.; Streb, L.M.; Anfinson, K.; Eyestone, M.E.; Kaalberg, E.; Riker, M.J.; Drack, A.V.; Braun, T.A.; Stone, E.M. Patient-specific iPSC-derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. Elife 2013, 2, e00824. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, N.; Carr, A.J.; Lane, A.; Moeller, F.; Chen, L.L.; Aguila, M.; Nommiste, B.; Muthiah, M.N.; Kanuga, N.; Wolfrum, U.; et al. Translational read-through of the RP2 Arg120stop mutation in patient iPSC-derived retinal pigment epithelium cells. Hum. Mol. Genet. 2015, 24, 972–986. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Ozawa, Y.; Suzuki, K.; Yuki, K.; Ohyama, M.; Akamatsu, W.; Matsuzaki, Y.; Shimmura, S.; Mitani, K.; Tsubota, K.; et al. The use of induced pluripotent stem cells to reveal pathogenic gene mutations and explore treatments for retinitis pigmentosa. Mol. Brain 2014, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.B.; Okamoto, S.; Osakada, F.; Homma, K.; Assawachananont, J.; Hirami, Y.; Iwata, T.; Takahashi, M. Modeling retinal degeneration using patient-specific induced pluripotent stem cells. PLoS ONE 2011, 6, e17084. [Google Scholar] [CrossRef] [PubMed]
- Lustremant, C.; Habeler, W.; Plancheron, A.; Goureau, O.; Grenot, L.; de la Grange, P.; Audo, I.; Nandrot, E.F.; Monville, C. Human induced pluripotent stem cells as a tool to model a form of leber congenital amaurosis. Cell. Reprogram. 2013, 15, 233–246. [Google Scholar] [PubMed]
- Meyer, J.S.; Howden, S.E.; Wallace, K.A.; Verhoeven, A.D.; Wright, L.S.; Capowski, E.E.; Pinilla, I.; Martin, J.M.; Tian, S.; Stewart, R.; et al. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells 2011, 29, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Shen, W.; Kuai, D.; Martin, J.M.; Guo, X.; Smith, M.A.; Perez, E.T.; Phillips, M.J.; Simonett, J.M.; Wallace, K.A.; et al. IPS cell modeling of best disease: Insights into the pathophysiology of an inherited macular degeneration. Hum. Mol. Genet. 2013, 22, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Chang, W.C.; Hung, K.H.; Yang, D.M.; Cheng, Y.H.; Liao, Y.W.; Woung, L.C.; Tsai, C.Y.; Hsu, C.C.; Lin, T.C.; et al. The generation of induced pluripotent stem cells for macular degeneration as a drug screening platform: Identification of curcumin as a protective agent for retinal pigment epithelial cells against oxidative stress. Front. Aging Neurosci. 2014, 6, 191. [Google Scholar] [CrossRef] [PubMed]
- Tulpule, A.; Kelley, J.M.; Lensch, M.W.; McPherson, J.; Park, I.H.; Hartung, O.; Nakamura, T.; Schlaeger, T.M.; Shimamura, A.; Daley, G.Q. Pluripotent stem cell models of shwachman-diamond syndrome reveal a common mechanism for pancreatic and hematopoietic dysfunction. Cell Stem Cell 2013, 12, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Batista, L.F.; Pech, M.F.; Zhong, F.L.; Nguyen, H.N.; Xie, K.T.; Zaug, A.J.; Crary, S.M.; Choi, J.; Sebastiano, V.; Cherry, A.; et al. Telomere shortening and loss of self-renewal in dyskeratosis congenita induced pluripotent stem cells. Nature 2011, 474, 399–402. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, M.; Kunimoto, H.; Watanabe, N.; Fukuchi, Y.; Yuasa, S.; Yamazaki, S.; Nishimura, T.; Sadahira, K.; Fukuda, K.; Okano, H.; et al. Impaired hematopoietic differentiation of RUNX1-mutated induced pluripotent stem cells derived from FPD/AML patients. Leukemia 2014, 28, 2344–2354. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Mali, P.; Huang, X.; Dowey, S.N.; Cheng, L. Site-specific gene correction of a point mutation in human ips cells derived from an adult patient with sickle cell disease. Blood 2011, 118, 4599–4608. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Chou, B.K.; Yen, J.; Ye, Z.; Zou, J.; Dowey, S.; Brodsky, R.A.; Ohm, J.E.; Yu, W.; Baylin, S.B.; et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells 2010, 28, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Chang, J.C.; Lin, C.; Sun, X.; Yu, J.; Kan, Y.W. Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc. Natl. Acad. Sci. USA 2009, 106, 9826–9830. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Hayashi, Y.; Schlieve, C.R.; Ikeya, M.; Kim, H.; Nguyen, T.D.; Sami, S.; Baba, S.; Barruet, E.; Nasu, A.; et al. Induced pluripotent stem cells from patients with human fibrodysplasia ossificans progressiva show increased mineralization and cartilage formation. Orphanet J. Rare Dis. 2013, 8, 190. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Choi, J.; Han, K.M.; Lee, B.H.; Choi, J.H.; Yoo, H.W.; Han, Y.M. Impaired osteogenesis in menkes disease-derived induced pluripotent stem cells. Stem Cell Res. Ther. 2015, 6, 160. [Google Scholar] [CrossRef] [PubMed]
- Saitta, B.; Passarini, J.; Sareen, D.; Ornelas, L.; Sahabian, A.; Argade, S.; Krakow, D.; Cohn, D.H.; Svendsen, C.N.; Rimoin, D.L. Patient-derived skeletal dysplasia induced pluripotent stem cells display abnormal chondrogenic marker expression and regulation by BMP2 and TGFBETA1. Stem Cells Dev. 2014, 23, 1464–1478. [Google Scholar] [CrossRef] [PubMed]
- Quarto, N.; Leonard, B.; Li, S.; Marchand, M.; Anderson, E.; Behr, B.; Francke, U.; Reijo-Pera, R.; Chiao, E.; Longaker, M.T. Skeletogenic phenotype of human marfan embryonic stem cells faithfully phenocopied by patient-specific induced-pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2012, 109, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.P.; Fukuda, K.; Fusaki, N.; Iida, A.; Hasegawa, M.; Lichtler, A.; Reichenberger, E.J. Induced pluripotent stem cell reprogramming by integration-free sendai virus vectors from peripheral blood of patients with craniometaphyseal dysplasia. Cell. Reprogram. 2013, 15, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Hoffman, J.P.; Alpaugh, R.K.; Rhim, A.D.; Reichert, M.; Stanger, B.Z.; Furth, E.E.; Sepulveda, A.R.; Yuan, C.X.; Won, K.J.; et al. An iPSC line from human pancreatic ductal adenocarcinoma undergoes early to invasive stages of pancreatic cancer progression. Cell. Rep. 2013, 3, 2088–2099. [Google Scholar] [CrossRef] [PubMed]
- Carette, J.E.; Pruszak, J.; Varadarajan, M.; Blomen, V.A.; Gokhale, S.; Camargo, F.D.; Wernig, M.; Jaenisch, R.; Brummelkamp, T.R. Generation of iPSCs from cultured human malignant cells. Blood 2010, 115, 4039–4042. [Google Scholar] [CrossRef] [PubMed]
- Gandre-Babbe, S.; Paluru, P.; Aribeana, C.; Chou, S.T.; Bresolin, S.; Lu, L.; Sullivan, S.K.; Tasian, S.K.; Weng, J.; Favre, H.; et al. Patient-derived induced pluripotent stem cells recapitulate hematopoietic abnormalities of juvenile myelomonocytic leukemia. Blood 2013, 121, 4925–4929. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.F.; Su, J.; Kim, H.S.; Chang, B.; Papatsenko, D.; Zhao, R.; Yuan, Y.; Gingold, J.; Xia, W.; Darr, H.; et al. Modeling familial cancer with induced pluripotent stem cells. Cell 2015, 161, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lian, Q.; Zhu, G.; Zhou, F.; Sui, L.; Tan, C.; Mutalif, R.A.; Navasankari, R.; Zhang, Y.; Tse, H.F.; et al. A human iPSC model of hutchinson gilford progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell 2011, 8, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Leng, L.; Tan, Y.; Gong, F.; Hu, L.; Ouyang, Q.; Zhao, Y.; Lu, G.; Lin, G. Differentiation of primordial germ cells from induced pluripotent stem cells of primary ovarian insufficiency. Hum. Reprod. 2015, 30, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Mica, Y.; Lee, G.; Chambers, S.M.; Tomishima, M.J.; Studer, L. Modeling neural crest induction, melanocyte specification, and disease-related pigmentation defects in hescs and patient-specific iPSCs. Cell Rep. 2013, 3, 1140–1152. [Google Scholar] [CrossRef] [PubMed]
- Singleton, A.B.; Farrer, M.; Johnson, J.; Singleton, A.; Hague, S.; Kachergus, J.; Hulihan, M.; Peuralinna, T.; Dutra, A.; Nussbaum, R.; et al. α-Synuclein locus triplication causes Parkinson’s disease. Science 2003, 302, 841. [Google Scholar] [CrossRef] [PubMed]
- Chartier-Harlin, M.C.; Kachergus, J.; Roumier, C.; Mouroux, V.; Douay, X.; Lincoln, S.; Levecque, C.; Larvor, L.; Andrieux, J.; Hulihan, M.; et al. α-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 2004, 364, 1167–1169. [Google Scholar] [CrossRef]
- Quarto, N.; Li, S.; Renda, A.; Longaker, M.T. Exogenous activation of BMP-2 signaling overcomes TGFβ-mediated inhibition of osteogenesis in Marfan embryonic stem cells and Marfan patient-specific induced pluripotent stem cells. Stem Cells 2012, 30, 2709–2719. [Google Scholar] [CrossRef] [PubMed]
- Drachtman, R.A.; Alter, B.P. Dyskeratosis congenita. Dermatol. Clin. 1995, 13, 33–39. [Google Scholar] [PubMed]
- Agarwal, S.; Loh, Y.H.; McLoughlin, E.M.; Huang, J.; Park, I.H.; Miller, J.D.; Huo, H.; Okuka, M.; Dos Reis, R.M.; Loewer, S.; et al. Telomere elongation in induced pluripotent stem cells from dyskeratosis congenita patients. Nature 2010, 464, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.Y.; Weick, J.P.; Yu, J.; Ma, L.X.; Zhang, X.Q.; Thomson, J.A.; Zhang, S.C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA 2010, 107, 4335–4340. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Freire, V.; Lee, A.S.; Hu, S.; Abilez, O.J.; Liang, P.; Lan, F.; Huber, B.C.; Ong, S.G.; Hong, W.X.; Huang, M.; et al. Effect of human donor cell source on differentiation and function of cardiac induced pluripotent stem cells. J. Am. Coll. Cardiol. 2014, 64, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Chin, M.H.; Mason, M.J.; Xie, W.; Volinia, S.; Singer, M.; Peterson, C.; Ambartsumyan, G.; Aimiuwu, O.; Richter, L.; Zhang, J.; et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 2009, 5, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Shoemaker, R.; Xie, B.; Gore, A.; LeProust, E.M.; Antosiewicz-Bourget, J.; Egli, D.; Maherali, N.; Park, I.H.; Yu, J.; et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 2009, 27, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Laurent, L.C.; Ulitsky, I.; Slavin, I.; Tran, H.; Schork, A.; Morey, R.; Lynch, C.; Harness, J.V.; Lee, S.; Barrero, M.J.; et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 2011, 8, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Mayshar, Y.; Ben-David, U.; Lavon, N.; Biancotti, J.C.; Yakir, B.; Clark, A.T.; Plath, K.; Lowry, W.E.; Benvenisty, N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 2010, 7, 521–531. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Iskow, R.; Yang, L.; Gokcumen, O.; Haseley, P.; Luquette, L.J., 3rd; Lohr, J.G.; Harris, C.C.; Ding, L.; Wilson, R.K.; et al. Landscape of somatic retrotransposition in human cancers. Science 2012, 337, 967–971. [Google Scholar] [CrossRef] [PubMed]
- Hamdorf, M.; Idica, A.; Zisoulis, D.G.; Gamelin, L.; Martin, C.; Sanders, K.J.; Pedersen, I.M. miR-128 represses L1 retrotransposition by binding directly to L1 RNA. Nat. Struct. Mol. Biol. 2015, 22, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Zhang, Z.N.; Rong, Z.; Xu, Y. Immunogenicity of induced pluripotent stem cells. Nature 2011, 474, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Doi, A.; Park, I.H.; Wen, B.; Murakami, P.; Aryee, M.J.; Irizarry, R.; Herb, B.; Ladd-Acosta, C.; Rho, J.; Loewer, S.; et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009, 41, 1350–1353. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Doi, A.; Wen, B.; Ng, K.; Zhao, R.; Cahan, P.; Kim, J.; Aryee, M.J.; Ji, H.; Ehrlich, L.I.; et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010, 467, 285–290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polo, J.M.; Liu, S.; Figueroa, M.E.; Kulalert, W.; Eminli, S.; Tan, K.Y.; Apostolou, E.; Stadtfeld, M.; Li, Y.; Shioda, T.; et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010, 28, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Drukker, M. Potential barriers to therapeutics utilizing pluripotent cell derivatives: Intrinsic immunogenicity of in vitro maintained and matured populations. Semin. Immunopathol. 2011, 33, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Muotri, A.; Gage, F.; Varki, A. Human embryonic stem cells express an immunogenic nonhuman sialic acid. Nat. Med. 2005, 11, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Yu, M.; Shen, C.; Chen, X.; Feng, T.; Yao, Y.; Li, J.; Li, H.; Tu, W. Negligible immunogenicity of induced pluripotent stem cells derived from human skin fibroblasts. PLoS ONE 2014, 9, e114949. [Google Scholar]
- Guha, P.; Morgan, J.W.; Mostoslavsky, G.; Rodrigues, N.P.; Boyd, A.S. Lack of immune response to differentiated cells derived from syngeneic induced pluripotent stem cells. Cell Stem Cell 2013, 12, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Morizane, A.; Doi, D.; Kikuchi, T.; Okita, K.; Hotta, A.; Kawasaki, T.; Hayashi, T.; Onoe, H.; Shiina, T.; Yamanaka, S.; et al. Direct comparison of autologous and allogeneic transplantation of iPSC-derived neural cells in the brain of a non-human primate. Stem Cell Rep. 2013, 1, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Todorova, D.; Kim, J.; Hamzeinejad, S.; He, J.; Xu, Y. Brief Report: Immune microenvironment determines the immunogenicity of induced pluripotent stem cell derivatives. Stem Cells 2015. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menon, S.; Shailendra, S.; Renda, A.; Longaker, M.; Quarto, N. An Overview of Direct Somatic Reprogramming: The Ins and Outs of iPSCs. Int. J. Mol. Sci. 2016, 17, 141. https://doi.org/10.3390/ijms17010141
Menon S, Shailendra S, Renda A, Longaker M, Quarto N. An Overview of Direct Somatic Reprogramming: The Ins and Outs of iPSCs. International Journal of Molecular Sciences. 2016; 17(1):141. https://doi.org/10.3390/ijms17010141
Chicago/Turabian StyleMenon, Siddharth, Siny Shailendra, Andrea Renda, Michael Longaker, and Natalina Quarto. 2016. "An Overview of Direct Somatic Reprogramming: The Ins and Outs of iPSCs" International Journal of Molecular Sciences 17, no. 1: 141. https://doi.org/10.3390/ijms17010141
APA StyleMenon, S., Shailendra, S., Renda, A., Longaker, M., & Quarto, N. (2016). An Overview of Direct Somatic Reprogramming: The Ins and Outs of iPSCs. International Journal of Molecular Sciences, 17(1), 141. https://doi.org/10.3390/ijms17010141