Synthesis and Antimicrobial Evaluation of Chroman-4-One and Homoisoflavonoid Derivatives
Abstract
1. Introduction
2. Results
2.1. Synthesis of Compounds (1–25)
2.2. Antimicrobial Activity of Compounds 1–25
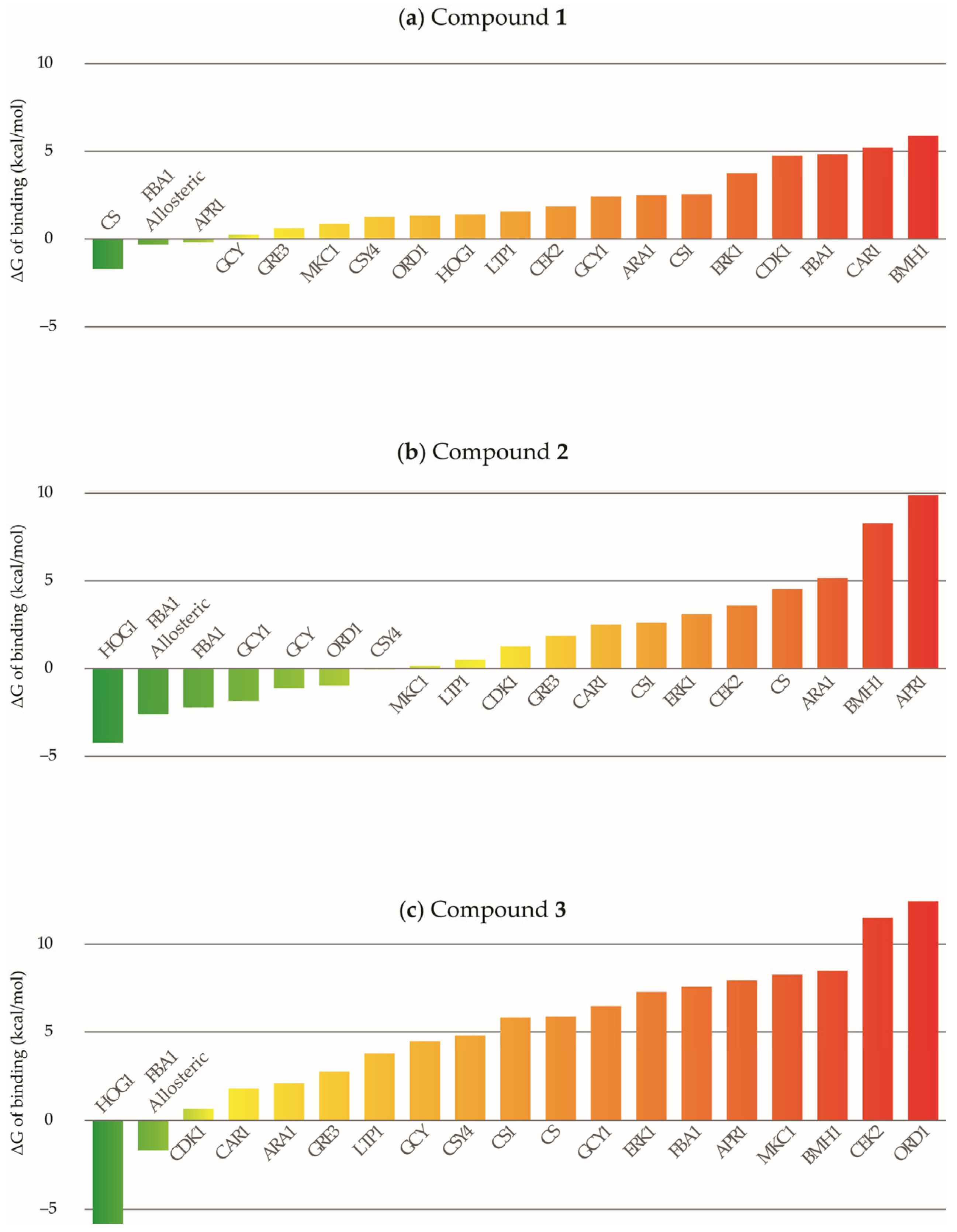
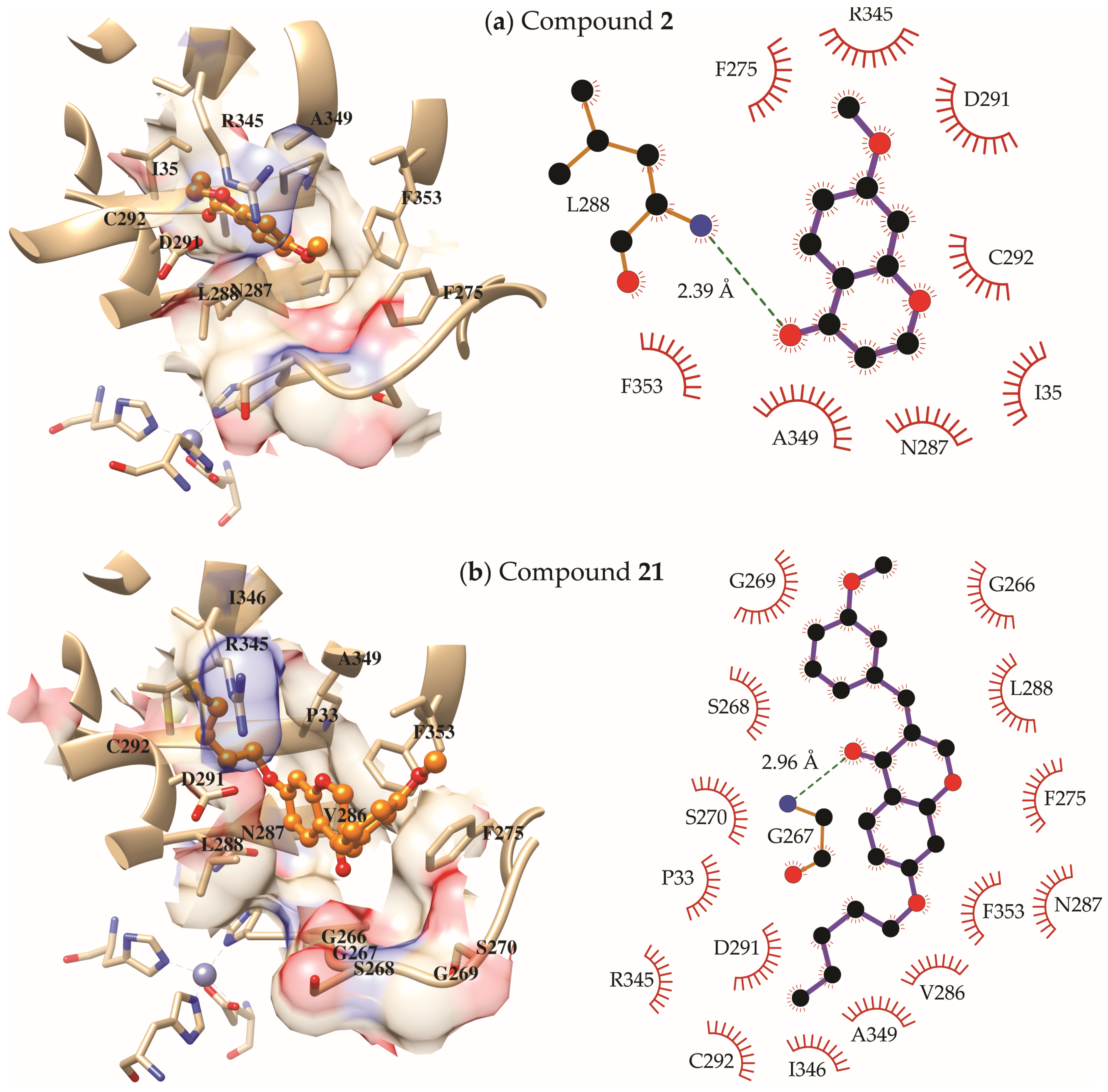
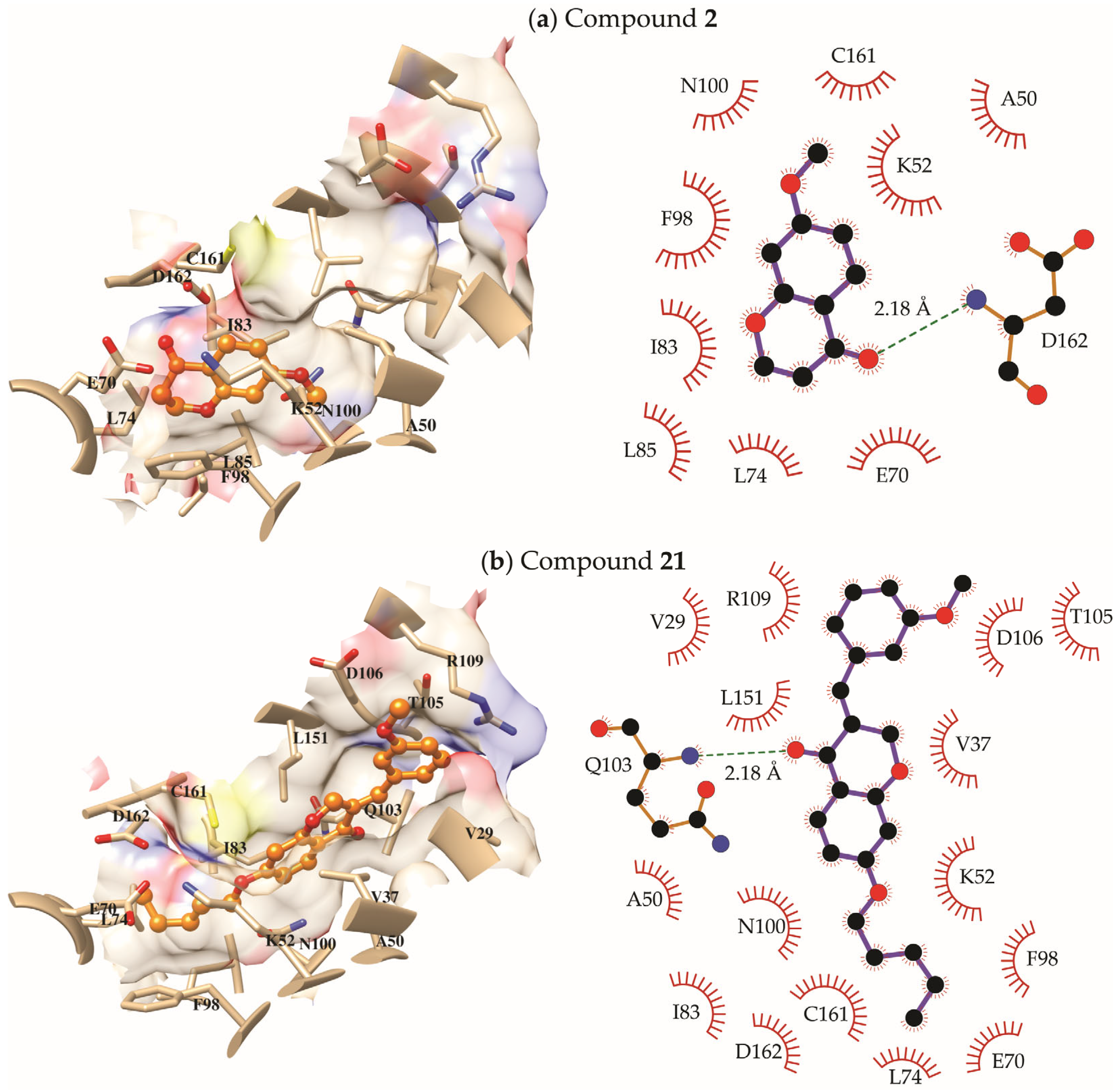
2.3. Molecular Modeling Study
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.1.1. General Procedure for Obtaining 7-Hydroxychroman-4-One (1)
4.1.2. General Procedure for Obtaining Chroman-4-Ones (2–10)
4.1.3. General Procedure for Obtaining Homoisoflavonoids (11–25)
4.2. Biological Evaluation
4.2.1. In Vitro Antimicrobial Assay
4.2.2. Determination of the Minimum Inhibitory Concentration (MIC), Minimum Fungicidal Concentration (MFC), and Minimum Bactericidal Concentration (MBC)
4.3. Molecular Modeling Study
4.3.1. Target Selection
4.3.2. Molecular Docking
4.3.3. Molecular Dynamics Simulations and Free Energies of Binding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huemer, M.; Mairpady Shambat, S.; Brugger, S.D.; Zinkernagel, A.S. Antibiotic Resistance and Persistence—Implications for Human Health and Treatment Perspectives. EMBO Rep 2020, 21, e51034. [Google Scholar] [CrossRef]
- Spellberg, B.; Bartlett, J.G.; Gilbert, D.N. The Future of Antibiotics and Resistance. New Engl. J. Med. 2013, 368, 299–302. [Google Scholar] [CrossRef] [PubMed]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef] [PubMed]
- Samreen; Ahmad, I.; Malak, H.A.; Abulreesh, H.H. Environmental Antimicrobial Resistance and Its Drivers: A Potential Threat to Public Health. J. Glob. Antimicrob. Resist. 2021, 27, 101–111. [Google Scholar] [CrossRef]
- Tang, K.W.K.; Millar, B.C.; Moore, J.E. Antimicrobial Resistance (AMR). Br. J. Biomed. Sci. 2023, 80, 11387. [Google Scholar] [CrossRef]
- Oliveira, M.; Antunes, W.; Mota, S.; Madureira-Carvalho, Á.; Dinis-Oliveira, R.J.; Dias da Silva, D. An Overview of the Recent Advances in Antimicrobial Resistance. Microorganisms 2024, 12, 1920. [Google Scholar] [CrossRef]
- Aslam, B.; Wang, W.; Arshad, M.I.; Khurshid, M.; Muzammil, S.; Rasool, M.H.; Nisar, M.A.; Alvi, R.F.; Aslam, M.A.; Qamar, M.U.; et al. Antibiotic Resistance: A Rundown of a Global Crisis. Infect. Drug Resist. 2018, 11, 1645–1658. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef]
- Ferreira, A.R.; de Sousa, D.P. Chemistry and Antifungal Activity of Homoisoflavonoids. J. Chem. 2023, 2023, 1223690. [Google Scholar] [CrossRef]
- Kamboj, S.; Singh, R. Chromanone-A Prerogative Therapeutic Scaffold: An Overview. Arab. J. Sci. Eng. 2022, 47, 75–111. [Google Scholar] [CrossRef] [PubMed]
- Hegab, M.I. A Review on Chemical and Biological Studies of 4-Chromanone Derivatives. Russ. J. Org. Chem. 2023, 59, 483–497. [Google Scholar] [CrossRef]
- Wang, L.; Qin, Y.; Wang, Y.; Zhou, Y.; Liu, B.; Bai, M.; Tong, X.; Fang, R.; Huang, X. Inhibitory Mechanism of Two Homoisoflavonoids from Ophiopogon japonicus on Tyrosinase Activity: Insight from Spectroscopic Analysis and Molecular Docking. RSC. Adv. 2021, 11, 34343–34354. [Google Scholar] [CrossRef]
- Noshita, T.; Fujita, K.; Koga, T.; Ouchi, H.; Tai, A. Synthesis and Biological Activity of (±)-7,3′,4′-Trihydroxyhomoisoflavan and Its Analogs. Bioorg. Med. Chem. Lett. 2021, 31, 127674. [Google Scholar] [CrossRef]
- Diana, E.J.; Kanchana, U.S.; Mathew, T.V. Current Developments in the Synthesis of 4-Chromanone-Derived Compounds. Org. Biomol. Chem. 2021, 19, 7995–8008. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, N.S.; Kushalappa, Y.M.; Sheshappa, S.P.; Nagaraju, H.H. Iron-Catalyzed Crossed-Aldol Condensation for the Synthesis of 3-Benzylidene-4-chromanones: An Efficient Synthesis of Homoisoflavanoids. ChemistrySelect 2019, 4, 13029–13033. [Google Scholar] [CrossRef]
- Perjési, P.; Das, U.; De Clercq, E.; Balzarini, J.; Kawase, M.; Sakagami, H.; Stables, J.P.; Lorand, T.; Rozmer, Z.; Dimmock, J.R. Design, Synthesis and Antiproliferative Activity of Some 3-Benzylidene-2,3-Dihydro-1-Benzopyran-4-Ones Which Display Selective Toxicity for Malignant Cells. Eur. J. Med. Chem. 2008, 43, 839–845. [Google Scholar] [CrossRef]
- Kumar, D.; Sharma, P.; Singh, H.; Nepali, K.; Gupta, G.K.; Jain, S.K.; Ntie-Kang, F. The Value of Pyrans as Anticancer Scaffolds in Medicinal Chemistry. RSC. Adv. 2017, 7, 36977–36999. [Google Scholar] [CrossRef]
- Foroumadi, A.; Samzadeh-Kermani, A.; Emami, S.; Dehghan, G.; Sorkhi, M.; Arabsorkhi, F.; Heidari, M.R.; Abdollahi, M.; Shafiee, A. Synthesis and Antioxidant Properties of Substituted 3-Benzylidene-7-Alkoxychroman-4-Ones. Bioorg. Med. Chem. Lett. 2007, 17, 6764–6769. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Cao, B.; Chen, X.; Xu, M.; Bi, X.; Guan, P.; Jiang, Y.; Xu, J.; Han, L.; Huang, X. Violacin A, a New Chromanone Produced by Streptomyces Violaceoruber and Its Anti-Inflammatory Activity. Bioorg. Med. Chem. Lett. 2018, 28, 947–951. [Google Scholar] [CrossRef]
- Ayati, A.; Falahati, M.; Irannejad, H.; Emami, S. Synthesis, in Vitro Antifungal Evaluation and in Silico Study of 3-Azolyl-4-Chromanone Phenylhydrazones. DARU J. Pharm. Sci. 2012, 20, 46. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.K.; Kumari, N.; Gupta, S.; Pahwa, S.; Nandanwar, H.; Jachak, S.M. 7-Hydroxy-(E)-3-Phenylmethylene-Chroman-4-One Analogues as Efflux Pump Inhibitors against Mycobacterium Smegmatis Mc2 155. Eur. J. Med. Chem. 2013, 66, 499–507. [Google Scholar] [CrossRef]
- Di Pisa, F.; Landi, G.; Dello Iacono, L.; Pozzi, C.; Borsari, C.; Ferrari, S.; Santucci, M.; Santarem, N.; Cordeiro-da-Silva, A.; Moraes, C.; et al. Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity. Molecules 2017, 22, 426. [Google Scholar] [CrossRef]
- Mottaghipisheh, J.; Stuppner, H. A Comprehensive Review on Chemotaxonomic and Phytochemical Aspects of Homoisoflavonoids, as Rare Flavonoid Derivatives. Int. J. Mol. Sci. 2021, 22, 2735. [Google Scholar] [CrossRef]
- Lin, L.-G.; Liu, Q.-Y.; Ye, Y. Naturally Occurring Homoisoflavonoids and Their Pharmacological Activities. Planta Med. 2014, 80, 1053–1066. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fowler, M.I.; Messenger, D.J.; Terry, L.A.; Gu, X.; Zhou, L.; Liu, R.; Su, J.; Shi, S.; Ordaz-Ortiz, J.J.; et al. Homoisoflavonoids Are Potent Glucose Transporter 2 (GLUT2) Inhibitors: A Potential Mechanism for the Glucose-Lowering Properties of Polygonatum odoratum. J. Agric. Food Chem. 2018, 66, 3137–3145. [Google Scholar] [CrossRef] [PubMed]
- Schwikkard, S.; Whitmore, H.; Corson, T.; Sishtla, K.; Langat, M.; Carew, M.; Mulholland, D. Antiangiogenic Activity and Cytotoxicity of Triterpenoids and Homoisoflavonoids from Massonia Pustulata and Massonia Bifolia. Planta. Med. 2018, 84, 638–644. [Google Scholar] [CrossRef]
- Zhao, J.-W.; Chen, D.-S.; Deng, C.-S.; Wang, Q.; Zhu, W.; Lin, L. Evaluation of Anti-Inflammatory Activity of Compounds Isolated from the Rhizome of Ophiopogon japonicas. BMC Complement. Altern. Med. 2017, 17, 1–12. [Google Scholar] [CrossRef]
- Nafisi, S.; Namdar, R. Molecular Aspects on the Specific Interaction of Homoisoflavonoids to DNA. J. Photochem. Photobiol. B. 2012, 117, 207–213. [Google Scholar] [CrossRef]
- Ferreira, A.R.; Alves, D.d.N.; de Castro, R.D.; Perez-Castillo, Y.; de Sousa, D.P. Synthesis of Coumarin and Homoisoflavonoid Derivatives and Analogs: The Search for New Antifungal Agents. Pharmaceuticals 2022, 15, 712. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Patial, V.; Singh, D.; Sharma, U.; Kumar, D. Antimicrobial Homoisoflavonoids from the Rhizomes of Polygonatum verticillatum. Chem. Biodivers. 2018, 15, e1800430. [Google Scholar] [CrossRef]
- O’Donnell, G.; Bucar, F.; Gibbons, S. Phytochemistry and Antimycobacterial Activity of Chlorophytum Inornatum. Phytochemistry 2006, 67, 178–182. [Google Scholar] [CrossRef]
- Rivero-Cruz, J.F. Antimicrobial Compounds Isolated from Haematoxylon Brasiletto. J. Ethnopharmacol. 2008, 119, 99–103. [Google Scholar] [CrossRef]
- Niranjan Reddy, V.L.; Ravikanth, V.; Jansi Lakshmi, V.V.N.S.; Suryanarayan Murty, U.; Venkateswarlu, Y. Inhibitory Activity of Homoisoflavonoids from Caesalpinia Sappan against Beauveria bassiana. Fitoterapia 2003, 74, 600–602. [Google Scholar] [CrossRef]
- Koch, K.; Biggers, M.S. General Preparation of 7-Substituted 4-Chromanones: Synthesis of a Potent Aldose Reductase Inhibitor. J. Org. Chem. 1994, 59, 1216–1218. [Google Scholar] [CrossRef]
- Desideri, N.; Proietti Monaco, L.; Fioravanti, R.; Biava, M.; Yáñez, M.; Alcaro, S.; Ortuso, F. (E)-3-Heteroarylidenechroman-4-Ones as Potent and Selective Monoamine Oxidase-B Inhibitors. Eur. J. Med. Chem. 2016, 117, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Takao, K.; Yamashita, M.; Yashiro, A.; Sugita, Y. Synthesis and Biological Evaluation of 3-Benzylidene-4-Chromanone Derivatives as Free Radical Scavengers and α-Glucosidase Inhibitors. Chem. Pharm. Bull. 2016, 64, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Guevara-Suarez, M.; Sutton, D.A.; Cano-Lira, J.F.; García, D.; Martin-Vicente, A.; Wiederhold, N.; Guarro, J.; Gené, J. Identification and Antifungal Susceptibility of Penicillium-Like Fungi from Clinical Samples in the United States. J. Clin. Microbiol. 2016, 54, 2155–2161. [Google Scholar] [CrossRef][Green Version]
- Pérez-Cantero, A.; López-Fernández, L.; Guarro, J.; Capilla, J. Azole Resistance Mechanisms in Aspergillus: Update and Recent Advances. Int. J. Antimicrob. Agents. 2020, 55, 105807. [Google Scholar] [CrossRef]
- Staniszewska, M. Virulence Factors in Candida Species. Curr. Protein. Pept. Sci. 2020, 21, 313–323. [Google Scholar] [CrossRef]
- Han, J.; Aljahdali, N.; Zhao, S.; Tang, H.; Harbottle, H.; Hoffmann, M.; Frye, J.G.; Foley, S.L. Infection Biology of Salmonella enterica. EcoSal Plus 2024, 12, eesp-0001-2023. [Google Scholar] [CrossRef]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas Aeruginosa: Pathogenesis, Virulence Factors, Antibiotic Resistance, Interaction with Host, Technology Advances and Emerging Therapeutics. Signal Transduct. Target Ther. 2022, 7, 199. [Google Scholar] [CrossRef]
- Burke, Ó.; Zeden, M.S.; O’Gara, J.P. The Pathogenicity and Virulence of the Opportunistic Pathogen Staphylococcus epidermidis. Virulence 2024, 15, 2359483. [Google Scholar] [CrossRef]
- Houghton, P.J.; Howes, M.-J.; Lee, C.C.; Steventon, G. Uses and Abuses of in Vitro Tests in Ethnopharmacology: Visualizing an Elephant. J. Ethnopharmacol. 2007, 110, 391–400. [Google Scholar] [CrossRef]
- Holetz, F.B.; Pessini, G.L.; Sanches, N.R.; Cortez, D.A.G.; Nakamura, C.V.; Dias Filho, B.P. Screening of Some Plants Used in the Brazilian Folk Medicine for the Treatment of Infectious Diseases. Mem. Inst. Oswaldo Cruz 2002, 97, 1027–1031. [Google Scholar] [CrossRef]
- Greenidge, P.A.; Kramer, C.; Mozziconacci, J.-C.; Sherman, W. Improving Docking Results via Reranking of Ensembles of Ligand Poses in Multiple X-Ray Protein Conformations with MM-GBSA. J. Chem. Inf. Model. 2014, 54, 2697–2717. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef]
- Li, D.; She, X.; Calderone, R. Functional Diversity of Complex I Subunits in Candida albicans Mitochondria. Curr. Genet. 2016, 62, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Poli, G.; Granchi, C.; Rizzolio, F.; Tuccinardi, T. Application of MM-PBSA Methods in Virtual Screening. Molecules 2020, 25, 1971. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera?A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Chang, W.; Zhang, M.; Jin, X.; Zhang, H.; Zheng, H.; Zheng, S.; Qiao, Y.; Yu, H.; Sun, B.; Hou, X.; et al. Inhibition of Fungal Pathogenicity by Targeting the H2S-Synthesizing Enzyme Cystathionine β-Synthase. Sci. Adv. 2022, 8, eadd5366. [Google Scholar] [CrossRef]
- Li, D.-D.; Wang, Y.; Dai, B.-D.; Li, X.-X.; Zhao, L.-X.; Cao, Y.-B.; Yan, L.; Jiang, Y.-Y. ECM17-Dependent Methionine/Cysteine Biosynthesis Contributes to Biofilm Formation in Candida albicans. Fungal Genet. Biol. 2013, 51, 50–59. [Google Scholar] [CrossRef]
- Yadav, A.K.; Desai, P.R.; Rai, M.N.; Kaur, R.; Ganesan, K.; Bachhawat, A.K. Glutathione Biosynthesis in the Yeast Pathogens Candida glabrata and Candida albicans: Essential in C. glabrata, and Essential for Virulence in C. albicans. Microbiology 2011, 157, 484–495. [Google Scholar] [CrossRef]
- Rodaki, A.; Young, T.; Brown, A.J.P. Effects of Depleting the Essential Central Metabolic Enzyme Fructose-1,6-Bisphosphate Aldolase on the Growth and Viability of Candida albicans: Implications for Antifungal Drug Target Discovery. Eukaryot. Cell 2006, 5, 1371–1377. [Google Scholar] [CrossRef]
- Wen, W.; Cao, H.; Huang, Y.; Tu, J.; Wan, C.; Wan, J.; Han, X.; Chen, H.; Liu, J.; Rao, L.; et al. Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis. J. Med. Chem. 2022, 65, 2656–2674. [Google Scholar] [CrossRef]
- Wang, X.-R.; Zhong, H.; Ma, S.-S.; Huang, Y.-H.; Xu, W.-H.; Wang, Y. Discovery of Petroselinic Acid with in Vitro and in Vivo Antifungal Activity by Targeting Fructose-1,6-Bisphosphate Aldolase. Phytomedicine 2024, 133, 155948. [Google Scholar] [CrossRef] [PubMed]
- Cheetham, J.; MacCallum, D.M.; Doris, K.S.; da Silva Dantas, A.; Scorfield, S.; Odds, F.; Smith, D.A.; Quinn, J. MAPKKK-Independent Regulation of the Hog1 Stress-Activated Protein Kinase in Candida albicans. J. Biol. Chem. 2011, 286, 42002–42016. [Google Scholar] [CrossRef] [PubMed]
- Dinér, P.; Veide Vilg, J.; Kjellén, J.; Migdal, I.; Andersson, T.; Gebbia, M.; Giaever, G.; Nislow, C.; Hohmann, S.; Wysocki, R.; et al. Design, Synthesis, and Characterization of a Highly Effective Hog1 Inhibitor: A Powerful Tool for Analyzing MAP Kinase Signaling in Yeast. PLoS ONE 2011, 6, e20012. [Google Scholar] [CrossRef] [PubMed]
- Correia, I.; Wilson, D.; Hube, B.; Pla, J. Characterization of a Candida albicans Mutant Defective in All MAPKs Highlights the Major Role of Hog1 in the MAPK Signaling Network. J. Fungi 2020, 6, 230. [Google Scholar] [CrossRef]
- Hameed, S. Protein Kinases as Potential Anticandidal Drug Targets. Front. Biosci. 2020, 25, 4862. [Google Scholar] [CrossRef]
- Emami, S.; Ghanbarimasir, Z. Recent Advances of Chroman-4-One Derivatives: Synthetic Approaches and Bioactivities. Eur. J. Med. Chem. 2015, 93, 539–563. [Google Scholar] [CrossRef]
- Sartoratto, A.; Machado, A.L.M.; Delarmelina, C.; Figueira, G.M.; Duarte, M.C.T.; Rehder, V.L.G. Composition and Antimicrobial Activity of Essential Oils from Aromatic Plants Used in Brazil. Braz. J. Microbiol. 2004, 35, 275–280. [Google Scholar] [CrossRef]
- Xie, Y.; Yang, W.; Tang, F.; Chen, X.; Ren, L. Antibacterial Activities of Flavonoids: Structure-Activity Relationship and Mechanism. Curr. Med. Chem. 2014, 22, 132–149. [Google Scholar] [CrossRef]
- Albrecht, U.; Lalk, M.; Langer, P. Synthesis and Structure–Activity Relationships of 2-Vinylchroman-4-Ones as Potent Antibiotic Agents. Bioorg. Med. Chem. 2005, 13, 1531–1536. [Google Scholar] [CrossRef]
- Cleeland, L.; Squires, E. Evaluation of New Antimicrobials in Vitro and in Experimental Animal Infections. In Antibiotics in Laboratory Medicine, 3rd ed.; Lorian, V.M.D., Ed.; Williams & Wilkins: Baltimore, MD, USA, 1991; pp. 739–788. [Google Scholar]
- Antunes, R.M.P.; Lima, E.O.; Pereira, M.S.V.; Camara, C.A.; Arruda, T.A.; Catão, R.M.R.; Barbosa, T.P.; Nunes, X.P.; Dias, C.S.; Silva, T.M.S. Atividade Antimicrobiana “in Vitro” e Determinação Da Concentração Inibitória Mínina (CIM) de Fitoconstituintes e Produtos Sintéticos Sobre Bactérias e Fungos Leveduriformes. Rev. Bras. Farmacogn. 2006, 16, 517–524. [Google Scholar] [CrossRef]
- Freire, I.C.M.; Pérez, A.L.A.L.; Cardoso, A.M.R.; Mariz, B.A.L.A.; Almeida, L.F.D.; Cavalcanti, Y.W.; Padilha, W.W.N. Atividade Antibacteriana de Óleos Essenciais Sobre Streptococcus Mutans e Staphylococcus Aureus. Rev. Bras. Plantas Med. 2014, 16, 372–377. [Google Scholar] [CrossRef]
- Hafidh, R.R. Inhibition of Growth of Highly Resistant Bacterial and Fungal Pathogens by a Natural Product. Open Microbiol. J. 2011, 5, 96–106. [Google Scholar] [CrossRef] [PubMed]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in Vitro Evaluating Antimicrobial Activity: A Review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef]
- Ncube, N.S.; Afolayan, A.J.; Okoh, A.I. Assessment Techniques of Antimicrobial Properties of Natural Compounds of Plant Origin: Current Methods and Future Trends. Afr. J. Biotechnol. 2008, 7, 1797–1806. [Google Scholar] [CrossRef]
- de Morais, M.C.; de Oliveira Lima, E.; Perez-Castillo, Y.; de Sousa, D.P. Synthetic Cinnamides and Cinnamates: Antimicrobial Activity, Mechanism of Action, and In Silico Study. Molecules 2023, 28, 1918. [Google Scholar] [CrossRef]
- Keiser, M.J.; Roth, B.L.; Armbruster, B.N.; Ernsberger, P.; Irwin, J.J.; Shoichet, B.K. Relating Protein Pharmacology by Ligand Chemistry. Nat. Biotechnol. 2007, 25, 197–206. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. Conformer Generation with OMEGA: Algorithm and Validation Using High Quality Structures from the Protein Databank and Cambridge Structural Database. J. Chem. Inf. Model. 2010, 50, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, P.C.D.; Skillman, A.G.; Warren, G.L.; Ellingson, B.A.; Stahl, M.T. OMEGA [Internet]; OpenEye Scientific Software: Santa Fe, NM, USA. Available online: http://www.eyesopen.com (accessed on 28 April 2025).
- QUACPAC, 2.2.5.1; OpenEye Scientific Software: Santa Fe, NM, USA, 2025.
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic. Acids. Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology Modelling of Protein Structures and Complexes. Nucleic. Acids. Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and Validation of a Genetic Algorithm for Flexible Docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Perez-Castillo, Y.; Montes, R.C.; da Silva, C.R.; Neto, J.B.d.A.; Dias, C.d.S.; Brunna Sucupira Duarte, A.; Júnior, H.V.N.; de Sousa, D.P. Antifungal Activity of N-(4-Halobenzyl)Amides against Candida Spp. and Molecular Modeling Studies. Int. J. Mol. Sci. 2021, 23, 419. [Google Scholar] [CrossRef]
- Case, D.A.; Aktulga, H.M.; Belfon, K.; Ben-Shalom, I.Y.; Berryman, J.T.; Brozell, S.R.; Carvahol, F.S.; Cerutti, D.S.; Cheatham, T.E.; Cisneros, G.A.; et al. AMBER, 2024; University of California: San Francisco, CA, USA, 2025.
- Lopes, S.P.; Castillo, Y.P.; Monteiro, M.L.; Menezes, R.R.P.P.B.d.; Almeida, R.N.; Martins, A.M.C.; de Sousa, D.P. Trypanocidal Mechanism of Action and in Silico Studies of P-Coumaric Acid Derivatives. Int. J. Mol. Sci. 2019, 20, 5916. [Google Scholar] [CrossRef]
- Machado, M.R.; Pantano, S. Split the Charge Difference in Two! A Rule of Thumb for Adding Proper Amounts of Ions in MD Simulations. J. Chem. Theory Comput. 2020, 16, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Merz, K.M. MCPB.Py: A Python Based Metal Center Parameter Builder. J. Chem. Inf. Model. 2016, 56, 599–604. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Pifferi, G.; Da Re, P.; Valenti, P.; Bisi, A. Synthesis of the 3-Homologue of Ipriflavone. Il Farm. (Pavia) 1996, 51, 689–691. [Google Scholar]
- Cloete, S.J.; N’Da, C.I.; Legoabe, L.J.; Petzer, A.; Petzer, J.P. The Evaluation of 1-Tetralone and 4-Chromanone Derivatives as Inhibitors of Monoamine Oxidase. Mol. Divers. 2021, 25, 491–507. [Google Scholar] [CrossRef]



| Compound | S. epidermidis ATCC-12228 | P. aeruginosa ATCC-25853 | S. enteritidis ATCC-6017 | MBC |
|---|---|---|---|---|
| 1 | 128 (779.7) | 128 (779.7) | 256 (1559.5) | 512 (3119.0) |
| 2 | 128 (718.40) | 128 (718.40) | 256 (1436.8) | 512 (2873.6) |
| 3 | 256 (1241.3) | 512 (2482.6) | 512 (2482.6) | 1024 (4965.2) |
| 4 | + | + | + | X |
| 5 | + | + | + | X |
| 6 | + | + | + | X |
| 7 | + | + | + | X |
| 8 | 256 (906.7) | 256 (906.7) | 256 (906.7) | 512 (1813.4) |
| 9 | + | + | + | X |
| 10 | 512 (1588.7) | 512 (1588.7) | 512 (1588.7) | X |
| 11 | 512 (1922.7) | 512 (1922.7) | 512 (1922.7) | X |
| 12 | 256 (869.7) | 256 (869.7) | 256 (869.7) | 1024 (3478.8) |
| 13 | 1024 (3478.8) | 1024 (3478.8) | 1024 (3478.8) | X |
| 14 | 512 (1588.1) | 512 (1588.1) | 512 (1588.1) | X |
| 15 | + | + | + | X |
| 16 | + | + | + | X |
| 17 | + | + | + | X |
| 18 | 512 (1161.7) | 512 (1161.7) | 512 (1161.7) | X |
| 19 | + | + | + | X |
| 20 | 128 (394.6) | 256 (789.2) | 256 (789.2) | 512 (1578.4) |
| 21 | 128 (363.2) | 128 (363.2) | 128 (363.2) | 512 (1452.8) |
| 22 | 256 (567.2) | 512 (1134.4) | 512 (1134.4) | 512 (1134.4) |
| 23 | + | + | + | X |
| 24 | + | + | + | X |
| 25 | + | + | + | X |
| Control: Culture medium | - | - | - | X |
| Control: Microorganism * | + | + | + | X |
| Control: Gentamicin | 128 (268.0) | 256 (536.0) | 256 (536.0) | X |
| Compound | C. albicans ATCC-76445 | C. albicans LM-92 | C. tropicalis ATCC-13803 | N. glabratus ATCC-90030 | A. flavus ATCC-13013 | P. citrinum ATCC-4001 | MFC |
|---|---|---|---|---|---|---|---|
| 1 | 64 (389.9) | 64 (389.9) | 64 (389.9) | 64 (389.9) | 256 (1559.5) | 256 (1559.5) | 256 (1559.5) |
| 2 | 64 (359.2) | 64 (359.2) | 64 (359.2) | 64 (359.2) | 512 (2873.6) | 512 (2873.6) | 256 (1436.8) |
| 3 | 128 (620.6) | 128 (620.6) | 256 (1241.3) | 256 (1241.3) | 512 (2482.6) | 512 (2482.6) | 512 (2482.6) |
| 4 | + | + | + | + | + | + | X |
| 5 | + | + | + | + | + | + | X |
| 6 | + | + | + | + | + | + | X |
| 7 | + | + | + | + | + | + | X |
| 8 | 256 (906.7) | 256 (906.7) | 256 (906.7) | 256 (906.7) | 512 (1813.4) | 512 (1813.4) | 512 (1813.4) |
| 9 | + | + | + | + | + | + | X |
| 10 | 512 (1588.7) | 512 (1588.7) | 512 (1588.7) | 512 (1588.7) | 1024 (3177.4) | 1024 (3177.4) | X |
| 11 | 512 (1922.7) | 512 (1922.7) | 512 (1922.7) | 512 (1922.7) | 1024 (3845.4) | 1024 (3845.4) | X |
| 12 | 256 (869.7) | 256 (869.7) | 256 (869.7) | 256 (869.7) | 1024 (3478.8) | 1024 (3478.8) | 1024 (3478.8) |
| 13 | 1024 (3478.8) | 1024 (3478.8) | 1024 (3478.8) | 1024 (3478.8) | 1024 (3478.8) | 1024 (3478.8) | X |
| 14 | 512 (1588.1) | 512 (1588.1) | 512 (1588.1) | 512 (1588.1) | 1024 (3176.2) | 1024 (3176.2) | X |
| 15 | + | + | + | + | + | + | X |
| 16 | + | + | + | + | + | + | X |
| 17 | + | + | + | + | + | + | X |
| 18 | 512 (1161.7) | 512 (1161.7) | 512 (1161.7) | 512 (1161.7) | 512 (1161.7) | 512 (1161.7) | X |
| 19 | + | + | + | + | + | + | X |
| 20 | 256 (789.2) | 256 (789.2) | 256 (789.2) | 256 (789.2) | 512 (1578.4) | 512 (1578.4) | 512 (1578.4) |
| 21 | 128 (363.2) | 128 (363.2) | 128 (363.2) | 128 (363.2) | 512 (1452.8) | 512 (1452.8) | 512 (1452.8) |
| 22 | 256 (567.2) | 256 (567.2) | 256 (567.2) | 256 (567.2) | 512 (1134.4) | 512 (1134.4) | 512 (1134.4) |
| 23 | + | + | + | + | + | + | X |
| 24 | + | + | + | + | + | + | X |
| 25 | + | + | + | + | + | + | X |
| Control: Culture medium | - | - | - | - | - | - | X |
| Control: Microorganism * | + | + | + | + | + | + | X |
| Control: Fluconazole | 128 (417.9) | 128 (417.9) | 128 (417.9) | 128 (417.9) | 256 (835.8) | 256 (835.8) | X |
| UniProt Accession | Description | ID | Compound (a) |
|---|---|---|---|
| A0A1D8PE91 | Tyrosine protein phosphatase | LTP1 | 1 |
| A0A1D8PP00 | Arginase | CAR1 | 1 |
| O42766 | 14-3-3 protein | BMH1 | 1 |
| A0A1D8PI24 | D-arabinose 1-dehydrogenase | ARA1 | 2, 21 |
| A0A1D8PNK3 | D-xylose reductase | GRE3 | 2, 21 |
| A0A1D8PQ13 | Cysteine synthase | CS | 2 |
| C4YT51 | Cysteine synthase | CS1 | 2 |
| P78599 | Ornithine decarboxylase | ORD1 | 2 |
| Q59T95 | Cystathionine beta-synthase | CSY4 | 2 |
| Q59U59 | Proteinase A | APR1 | 2 |
| Q5ADT3 | Glycerol dehydrogenase | GCY | 2, 21 |
| Q5ADT4 | Glycerol 2-dehydrogenase | GCY1 | 2, 21 |
| Q9URB4 | Fructose-bisphosphate aldolase | FBA1 | 2 |
| A0A1D8PT38 | Mitogen-activated protein kinase | CEK2 | 21 |
| P43063 | Cyclin-dependent kinase 1 | CDK1 | 21 |
| Q5A1D3 | Extracellular signal-regulated kinase 1 | ERK1 | 21 |
| Q5AAG6 | Mitogen-activated protein kinase | MKC1 | 21 |
| Q92207 | Mitogen-activated protein kinase | HOG1 | 21 |
| Q9B8D6 | NADH-ubiquinone oxidoreductase | NAD1 | 21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filho, C.d.S.M.B.; Galvão, J.L.F.M.; Lima, E.O.; Perez-Castillo, Y.; Velásquez-López, Y.; de Sousa, D.P. Synthesis and Antimicrobial Evaluation of Chroman-4-One and Homoisoflavonoid Derivatives. Molecules 2025, 30, 3575. https://doi.org/10.3390/molecules30173575
Filho CdSMB, Galvão JLFM, Lima EO, Perez-Castillo Y, Velásquez-López Y, de Sousa DP. Synthesis and Antimicrobial Evaluation of Chroman-4-One and Homoisoflavonoid Derivatives. Molecules. 2025; 30(17):3575. https://doi.org/10.3390/molecules30173575
Chicago/Turabian StyleFilho, Carlos d. S. M. Bezerra, José L. F. M. Galvão, Edeltrudes O. Lima, Yunierkis Perez-Castillo, Yendrek Velásquez-López, and Damião P. de Sousa. 2025. "Synthesis and Antimicrobial Evaluation of Chroman-4-One and Homoisoflavonoid Derivatives" Molecules 30, no. 17: 3575. https://doi.org/10.3390/molecules30173575
APA StyleFilho, C. d. S. M. B., Galvão, J. L. F. M., Lima, E. O., Perez-Castillo, Y., Velásquez-López, Y., & de Sousa, D. P. (2025). Synthesis and Antimicrobial Evaluation of Chroman-4-One and Homoisoflavonoid Derivatives. Molecules, 30(17), 3575. https://doi.org/10.3390/molecules30173575









