In Vitro Evaluation of the Healing Potential and Proteomic Study of Quercus robur L. Leaf Extracts in Human Keratinocytes
Abstract
1. Introduction
2. Results
2.1. Phytochemical Composition and Antioxidant Activity of the Aqueous Extract from Quercus robur Leaves
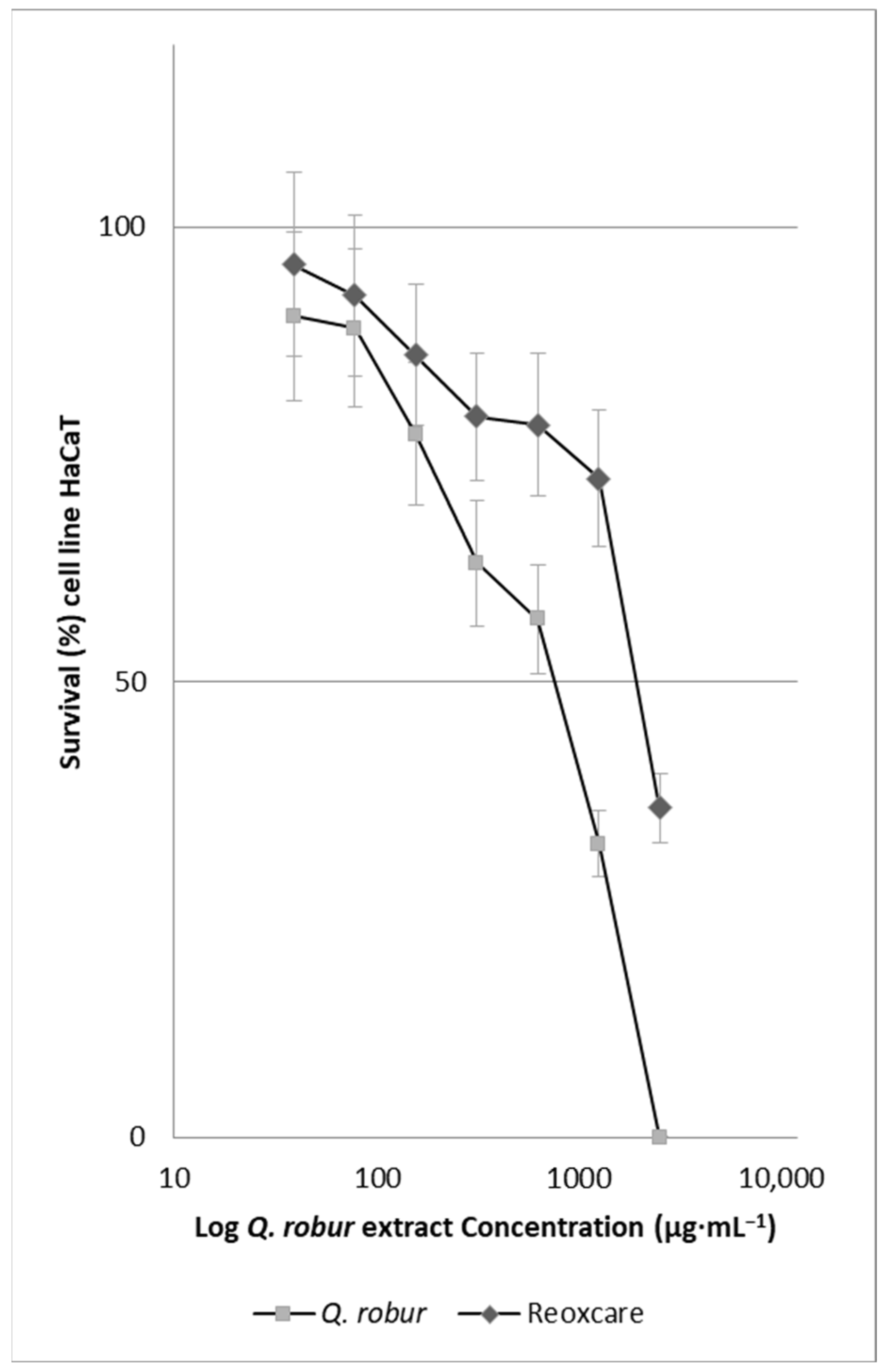
2.2. Effect of Quercus robur Extracts on the Cell Viability
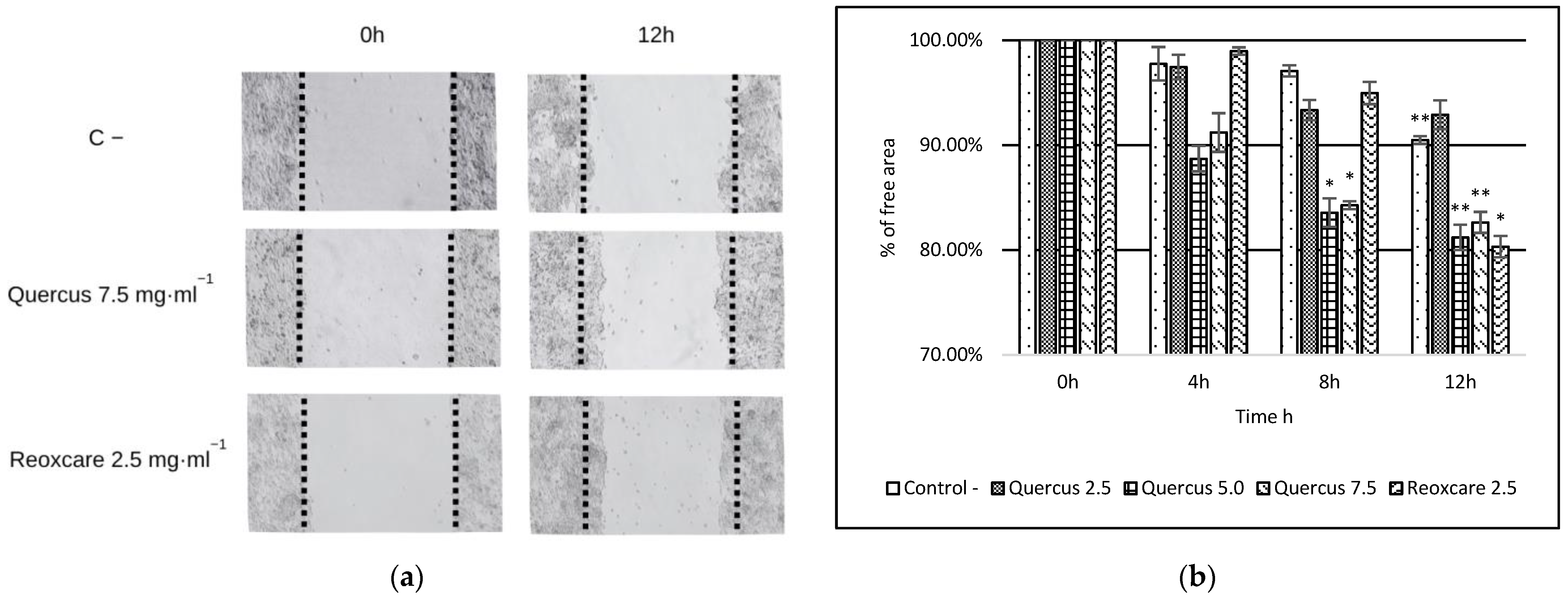
2.3. Effects of Quercus robur Extracts on Cell Migration
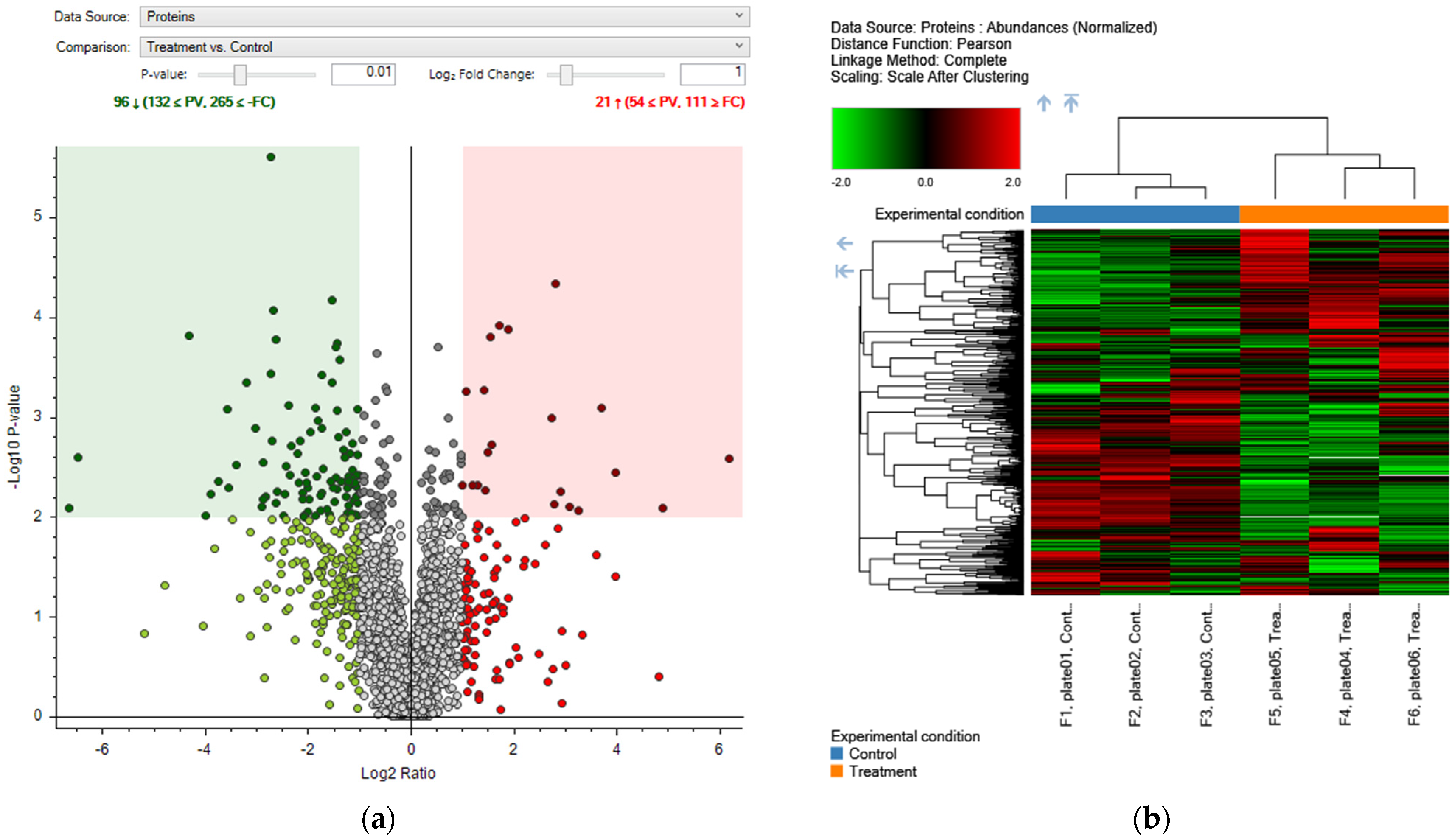
2.4. Proteomic Analysis
2.4.1. Effects of Aqueous Extract of Quercus robur on the Proteomic Profile of Human Keratinocytes
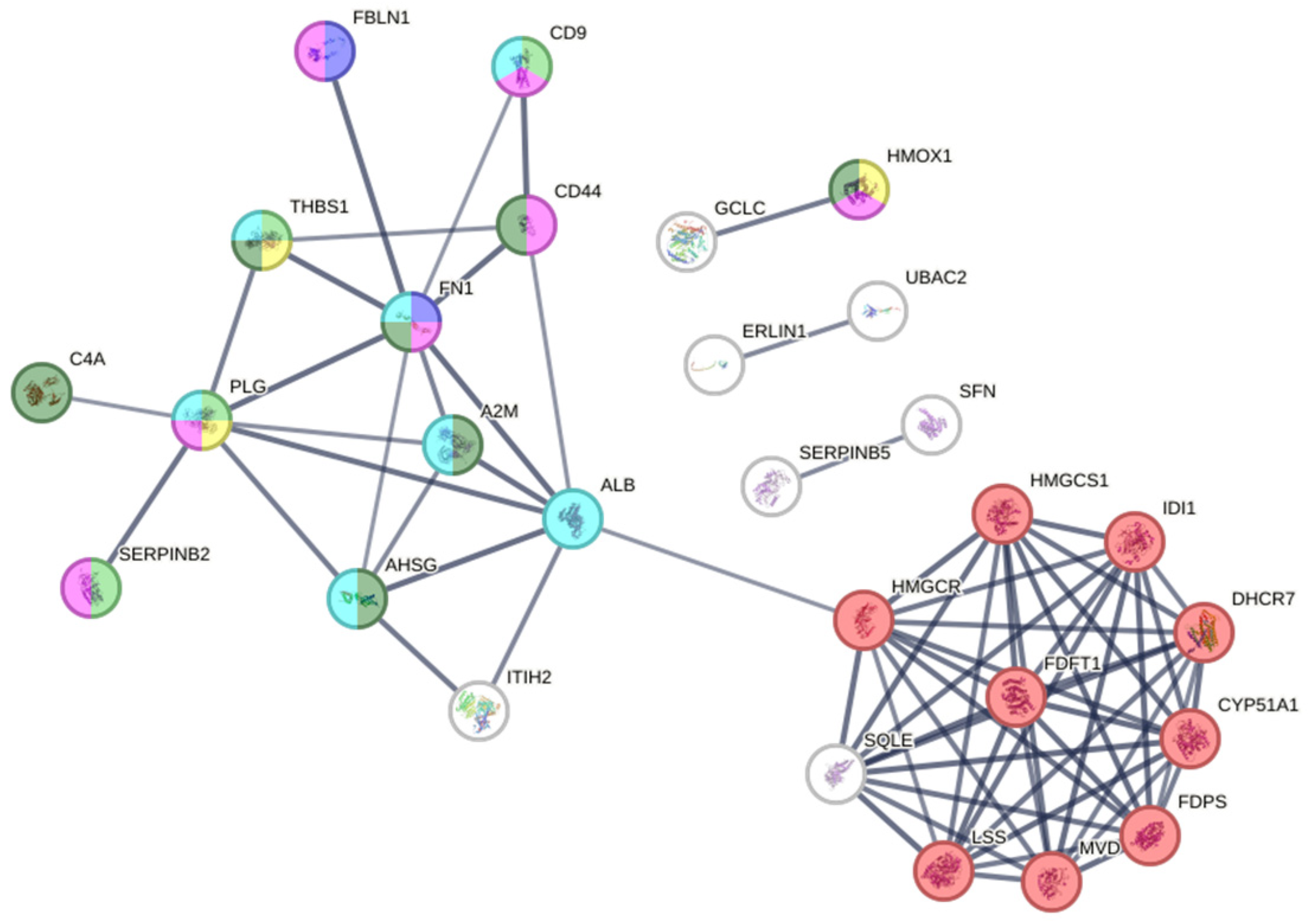
2.4.2. Protein–Protein Interaction (PPI) Network Analysis
2.4.3. Functional Enrichment Analysis
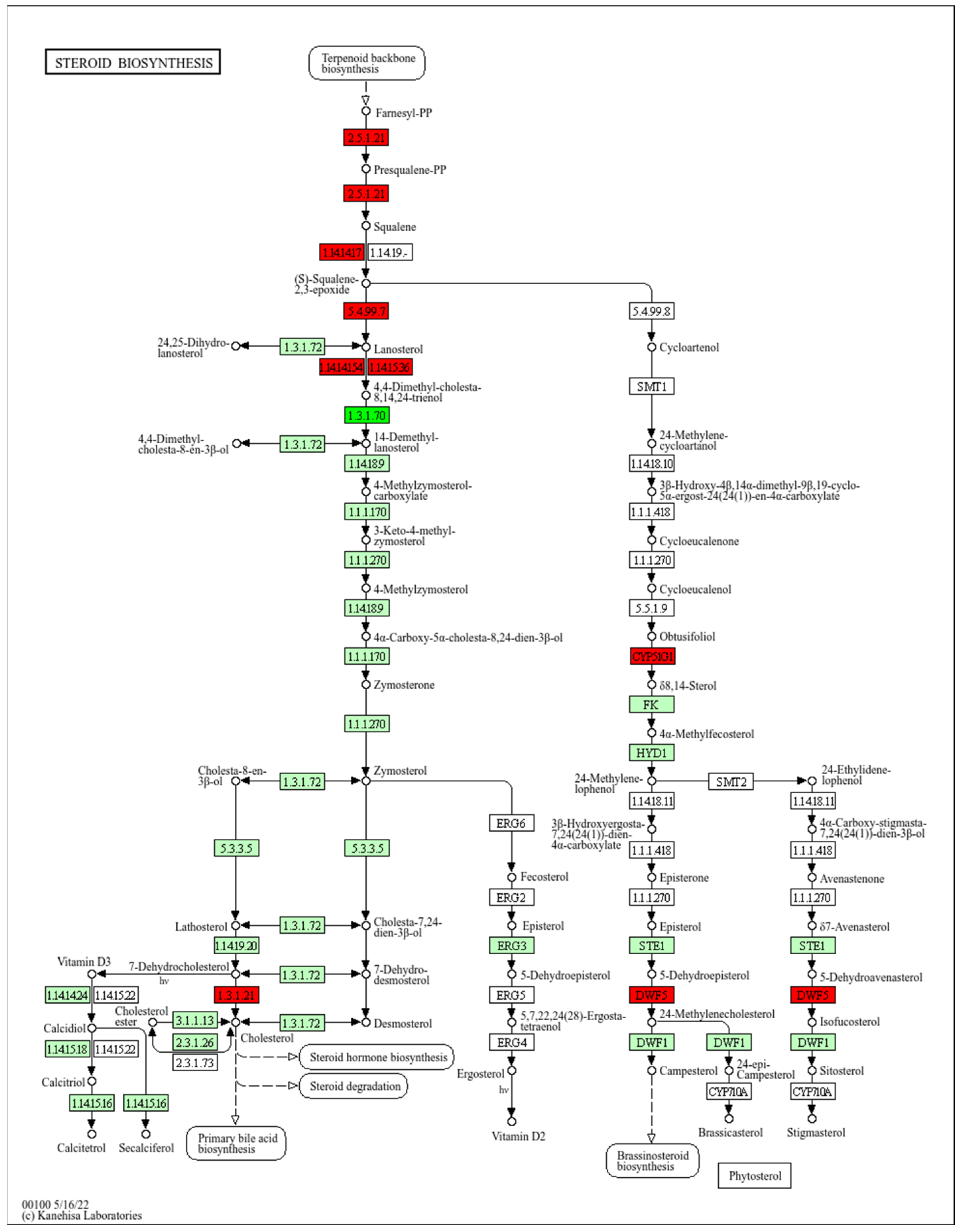
2.4.4. Steroid Biosynthesis Pathway
3. Discussion
3.1. Role of Cholesterol Synthesis in Wound Healing
3.2. Suggested Mechanism of Action
3.3. Future Directions
4. Materials and Methods
4.1. Biological Materials and Extracts from Quercus robur
4.2. Analytical Methods for the Characterization of Compounds and Antioxidant Activity in Quercus robur Leaves
4.3. Cell Culture and Treatments
4.4. Cytotoxicity Assays (MTT Assays)
4.5. Cell Migration Assay
4.6. Proteomic Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MTT | (3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide) assay |
| GAE | Gallic acid equivalent |
| DPPH | 2,2-diphenyl-1-picrylhydrazyl |
| LC-MS | Liquid chromatography-mass spectrometry |
| UHPLC | Ultra-high-performance liquid chromatography |
| FDR | False discovery rate |
| DEPs | Differentially expressed proteins |
| PPI | Protein–protein interaction |
| GO | Gene Ontology |
| DTT | Dithiothreitol |
| BCA | Bicinchoninic acid assay |
Appendix A. Proteins
Overview of Important Differentially Expressed
| Gene Symbol | Protein Name | Fold Change (FC) | FC p-Value | Molecular Function |
|---|---|---|---|---|
| AHSG | Alpha-2-HS-glycoprotein [OS = Homo sapiens] | 72.99 | 0.0026 | enzyme regulator activity |
| FBLN1 | Fibulin-1 [OS = Homo sapiens] | 30.18 | 0.0081 | signal transduction activity or receptor binding; enzyme regulator activity; extracellular structural activity; other molecular function |
| PLG | Plasminogen [OS = Homo sapiens] | 15.82 | 0.0037 | signal transduction activity or receptor binding; other molecular function |
| ALB | Albumin [OS = Homo sapiens] | 13.17 | 0.0008 | nucleic acid binding activity; other molecular function |
| IGFBP2 | Insulin-like growth factor-binding protein 2 [OS = Homo sapiens] | 9.59 | 0.0087 | signal transduction activity or receptor binding; other molecular function |
| IDI1 | Isopentenyl-diphosphate Delta-isomerase 1 [OS = Homo sapiens] | 8.5 | 0.008 | other molecular function |
| HMOX1 | Heme oxygenase 1 [OS = Homo sapiens] | 7.62 | 0.0056 | other molecular function |
| HMGCS1 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic [OS = Homo sapiens] | 7.08 | 0.0 | other molecular function |
| MVD | Diphosphomevalonate decarboxylase [OS = Homo sapiens] | 6.92 | 0.0076 | other molecular function |
| FDFT1 | Squalene synthase [OS = Homo sapiens] | 6.7 | 0.001 | other molecular function |
| CPD | Carboxypeptidase D [OS = Homo sapiens] | 3.71 | 0.0001 | other molecular function |
| GALE | UDP-glucose 4-epimerase [OS = Homo sapiens] | 3.3 | 0.0001 | other molecular function |
| GNAI1 | Guanine nucleotide-binding protein G(i) subunit alpha-1 [OS = Homo sapiens] | 2.97 | 0.0019 | signal transduction activity or receptor binding; other molecular function |
| DHCR7 | 7-dehydrocholesterol reductase [OS = Homo sapiens] | 2.92 | 0.0002 | other molecular function |
| SERPINB5 | Serpin B5 [OS = Homo sapiens] | 2.82 | 0.0023 | enzyme regulator activity |
| POLR2A | DNA-directed RNA polymerase II subunit RPB1 [OS = Homo sapiens] | 2.72 | 0.0054 | nucleic acid binding activity; other molecular function |
| EED | Polycomb protein EED [OS = Homo sapiens] | 2.7 | 0.0005 | enzyme regulator activity; other molecular function |
| ROCK2 | Rho-associated protein kinase 2 [OS = Homo sapiens] | 2.48 | 0.0048 | cytoskeletal activity; kinase activity; other molecular function |
| GRN | Progranulin [OS = Homo sapiens] | 2.3 | 0.0048 | signal transduction activity or receptor binding; other molecular function |
| SEC23IP | SEC23-interacting protein [OS = Homo sapiens] | 2.12 | 0.0006 | other molecular function |
| ANXA11 | Annexin A11 [OS = Homo sapiens] | 2.02 | 0.0049 | other molecular function |
| PKP3 | Plakophilin-3 [OS = Homo sapiens] | 0.5 | 0.0059 | other molecular function |
| DDX54 | ATP-dependent RNA helicase DDX54 [OS = Homo sapiens] | 0.49 | 0.0038 | signal transduction activity or receptor binding; other molecular function |
| THUMPD1 | THUMP domain-containing protein 1 [OS = Homo sapiens] | 0.49 | 0.0074 | nan |
| CSNK1A1 | Casein kinase I isoform alpha [OS = Homo sapiens] | 0.49 | 0.0008 | kinase activity; other molecular function |
| UHRF1 | E3 ubiquitin-protein ligase UHRF1 [OS = Homo sapiens] | 0.49 | 0.0098 | nucleic acid binding activity; other molecular function |
| DHX16 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [OS = Homo sapiens] | 0.49 | 0.0024 | other molecular function |
| SARNP | SAP domain-containing ribonucleoprotein [OS = Homo sapiens] | 0.48 | 0.0063 | nucleic acid binding activity; other molecular function |
| DNMT1 | DNA (cytosine-5)-methyltransferase 1 [OS = Homo sapiens] | 0.48 | 0.0049 | nucleic acid binding activity; other molecular function |
| DPF2 | Zinc finger protein ubi-d4 [OS = Homo sapiens] | 0.48 | 0.0039 | other molecular function |
| GNL2 | Nucleolar GTP-binding protein 2 [OS = Homo sapiens] | 0.47 | 0.0095 | other molecular function |
| TNKS1BP1 | 182 kDa tankyrase-1-binding protein [OS = Homo sapiens] | 0.47 | 0.0046 | other molecular function |
| MAP2K4 | Dual specificity mitogen-activated protein kinase kinase 4 [OS = Homo sapiens] | 0.47 | 0.0068 | kinase activity; other molecular function |
| CTCF | Transcriptional repressor CTCF [OS = Homo sapiens] | 0.46 | 0.0047 | nucleic acid binding activity; other molecular function |
| TMPO | Lamina-associated polypeptide 2, isoforms beta/gamma [OS = Homo sapiens] | 0.46 | 0.0018 | nucleic acid binding activity; other molecular function |
| SRSF7 | Serine/arginine-rich splicing factor 7 [OS = Homo sapiens] | 0.46 | 0.0034 | nucleic acid binding activity; other molecular function |
| LIMA1 | LIM domain and actin-binding protein 1 [OS = Homo sapiens] | 0.45 | 0.0042 | cytoskeletal activity; other molecular function |
| PPP1R18 | Phostensin [OS = Homo sapiens] | 0.45 | 0.0091 | cytoskeletal activity; other molecular function |
| ATXN2L | Ataxin-2-like protein [OS = Homo sapiens] | 0.44 | 0.0097 | nucleic acid binding activity |
| MARK2 | Serine/threonine-protein kinase MARK2 [OS = Homo sapiens] | 0.44 | 0.0023 | enzyme regulator activity; cytoskeletal activity; kinase activity; other molecular function |
| EPB41L2 | Band 4.1-like protein 2 [OS = Homo sapiens] | 0.44 | 0.0044 | cytoskeletal activity; other molecular function |
| MT1H | Metallothionein-1H [OS = Homo sapiens] | 0.44 | 0.0039 | other molecular function |
| RBM10 | RNA-binding protein 10 [OS = Homo sapiens] | 0.43 | 0.0077 | nucleic acid binding activity; other molecular function |
| PELP1 | Proline-, glutamic acid- and leucine-rich protein 1 [OS = Homo sapiens] | 0.43 | 0.0092 | other molecular function |
| YTHDF3 | YTH domain-containing family protein 3 [OS = Homo sapiens] | 0.43 | 0.0051 | translation activity; nucleic acid binding activity |
| KPNA2 | Importin subunit alpha-1 [OS = Homo sapiens] | 0.42 | 0.0063 | other molecular function |
| PRRC2C | Protein PRRC2C [OS = Homo sapiens] | 0.42 | 0.0045 | nan |
| MTDH | Protein LYRIC [OS = Homo sapiens] | 0.42 | 0.0014 | nucleic acid binding activity; other molecular function |
| RBM14 | RNA-binding protein 14 [OS = Homo sapiens] | 0.42 | 0.0024 | other molecular function |
| RPS4X | 40S ribosomal protein S4, X isoform [OS = Homo sapiens] | 0.41 | 0.0025 | nucleic acid binding activity; other molecular function |
| PDLIM7 | PDZ and LIM domain protein 7 [OS = Homo sapiens] | 0.4 | 0.0022 | cytoskeletal activity; other molecular function |
| H2afy; H2AFY; MACROH2A1 | Core histone macro-H2A.1 [OS = Homo sapiens] | 0.39 | 0.0045 | enzyme regulator activity; nucleic acid binding activity; other molecular function |
| YTHDF1 | YTH domain-containing family protein 1 [OS = Homo sapiens] | 0.39 | 0.0003 | translation activity; nucleic acid binding activity |
| UBAP2L | Ubiquitin-associated protein 2-like [OS = Homo sapiens] | 0.38 | 0.0016 | nan |
| RBM28 | RNA-binding protein 28 [OS = Homo sapiens] | 0.37 | 0.0009 | nan |
| SRRM2 | Serine/arginine repetitive matrix protein 2 [OS = Homo sapiens] | 0.37 | 0.0052 | nucleic acid binding activity; other molecular function |
| TAGLN2 | Transgelin-2 [OS = Homo sapiens] | 0.37 | 0.0002 | cytoskeletal activity |
| RPS16 | 40S ribosomal protein S16 [OS = Homo sapiens] | 0.36 | 0.0002 | nucleic acid binding activity; other molecular function |
| SMU1 | WD40 repeat-containing protein SMU1 [OS = Homo sapiens] | 0.36 | 0.0041 | nan |
| KIF2C | Kinesin-like protein KIF2C [OS = Homo sapiens] | 0.36 | 0.0071 | cytoskeletal activity; nucleic acid binding activity; other molecular function |
| NSA2 | Ribosome biogenesis protein NSA2 homolog [OS = Homo sapiens] | 0.35 | 0.0051 | nan |
| MAP7 | Ensconsin [OS = Homo sapiens] | 0.35 | 0.0005 | signal transduction activity or receptor binding; other molecular function |
| DNTTIP2 | Deoxynucleotidyltransferase terminal-interacting protein 2 [OS = Homo sapiens] | 0.35 | 0.0001 | nan |
| HP1BP3 | Heterochromatin protein 1-binding protein 3 [OS = Homo sapiens] | 0.34 | 0.0045 | nucleic acid binding activity; other molecular function |
| KPNA3 | Importin subunit alpha-4 [OS = Homo sapiens] | 0.34 | 0.0096 | other molecular function |
| UQCRFS1 | Cytochrome b-c1 complex subunit Rieske, mitochondrial [OS = Homo sapiens] | 0.32 | 0.0085 | transporter activity; other molecular function |
| LRRC47 | Leucine-rich repeat-containing protein 47 [OS = Homo sapiens] | 0.31 | 0.0033 | other molecular function |
| RPS11 | 40S ribosomal protein S11 [OS = Homo sapiens] | 0.3 | 0.0004 | nucleic acid binding activity; other molecular function |
| RPL23 | 60S ribosomal protein L23 [OS = Homo sapiens] | 0.3 | 0.0013 | enzyme regulator activity; nucleic acid binding activity; other molecular function |
| nan | Probable ATP-dependent RNA helicase DDX52 [OS = Homo sapiens] | 0.3 | 0.0057 | nan |
| EXOSC6 | Exosome complex component MTR3 [OS = Homo sapiens] | 0.3 | 0.006 | nan |
| MAP1S | Microtubule-associated protein 1S [OS = Homo sapiens] | 0.3 | 0.0086 | cytoskeletal activity; nucleic acid binding activity; other molecular function |
| HMGB2 | High mobility group protein B2 [OS = Homo sapiens] | 0.29 | 0.0011 | signal transduction activity or receptor binding; nucleic acid binding activity; other molecular function |
| TCOF1 | Treacle protein [OS = Homo sapiens] | 0.28 | 0.004 | other molecular function |
| RPL13 | 60S ribosomal protein L13 [OS = Homo sapiens] | 0.28 | 0.0097 | other molecular function |
| MKI67 | Proliferation marker protein Ki-67 [OS = Homo sapiens] | 0.28 | 0.0008 | nucleic acid binding activity; other molecular function |
| RPS14 | 40S ribosomal protein S14 [OS = Homo sapiens] | 0.26 | 0.0014 | nucleic acid binding activity; other molecular function |
| YY1 | Transcriptional repressor protein YY1 [OS = Homo sapiens] | 0.26 | 0.0066 | nucleic acid binding activity; other molecular function |
| RPL37A | 60S ribosomal protein L37a [OS = Homo sapiens] | 0.25 | 0.0051 | other molecular function |
| SAFB | Scaffold attachment factor B1 [OS = Homo sapiens] | 0.25 | 0.0088 | nucleic acid binding activity; other molecular function |
| DDX49 | Probable ATP-dependent RNA helicase DDX49 [OS = Homo sapiens] | 0.24 | 0.0045 | other molecular function |
| CDC27 | Cell division cycle protein 27 homolog [OS = Homo sapiens] | 0.24 | 0.0036 | other molecular function |
| CBX8 | Chromobox protein homolog 8 [OS = Homo sapiens] | 0.24 | 0.0099 | enzyme regulator activity; nucleic acid binding activity; other molecular function |
| RPS24 | 40S ribosomal protein S24 [OS = Homo sapiens] | 0.24 | 0.0099 | other molecular function |
| CNN2 | Calponin-2 [OS = Homo sapiens] | 0.23 | 0.0067 | cytoskeletal activity; other molecular function |
| TAF15 | TATA-binding protein-associated factor 2N [OS = Homo sapiens] | 0.23 | 0.0058 | nucleic acid binding activity; other molecular function |
| MARK3 | MAP/microtubule affinity-regulating kinase 3 [OS = Homo sapiens] | 0.22 | 0.0017 | cytoskeletal activity; kinase activity; other molecular function |
| TRIM32 | E3 ubiquitin-protein ligase TRIM32 [OS = Homo sapiens] | 0.22 | 0.0045 | cytoskeletal activity; nucleic acid binding activity; other molecular function |
| MAP4 | Microtubule-associated protein 4 [OS = Homo sapiens] | 0.22 | 0.0023 | cytoskeletal activity; other molecular function |
| NOLC1 | Nucleolar and coiled-body phosphoprotein 1 [OS = Homo sapiens] | 0.2 | 0.002 | nucleic acid binding activity; other molecular function |
| MDC1 | Mediator of DNA damage checkpoint protein 1 [OS = Homo sapiens] | 0.2 | 0.0038 | nan |
| LARP1 | La-related protein 1 [OS = Homo sapiens] | 0.19 | 0.0008 | translation activity; nucleic acid binding activity; other molecular function |
| PRKAR1A | cAMP-dependent protein kinase type I-alpha regulatory subunit [OS = Homo sapiens] | 0.19 | 0.0031 | enzyme regulator activity; other molecular function |
| RPS8 | 40S ribosomal protein S8 [OS = Homo sapiens] | 0.18 | 0.0059 | other molecular function |
| RPL26 | 60S ribosomal protein L26 [OS = Homo sapiens] | 0.18 | 0.0096 | nucleic acid binding activity; other molecular function |
| MINK1 | Misshapen-like kinase 1 [OS = Homo sapiens] | 0.17 | 0.0056 | kinase activity; other molecular function |
| H1-1 | Histone H1.1 [OS = Homo sapiens] | 0.16 | 0.0002 | nucleic acid binding activity; other molecular function |
| RRP1B | Ribosomal RNA processing protein 1 homolog B [OS = Homo sapiens] | 0.16 | 0.0073 | other molecular function |
| SRRM1 | Serine/arginine repetitive matrix protein 1 [OS = Homo sapiens] | 0.16 | 0.0001 | nucleic acid binding activity |
| HNRNPA3 | Heterogeneous nuclear ribonucleoprotein A3 [OS = Homo sapiens] | 0.16 | 0.0018 | transporter activity; nucleic acid binding activity |
| PPME1 | Protein phosphatase methylesterase 1 [OS = Homo sapiens] | 0.15 | 0.0 | enzyme regulator activity; nucleic acid binding activity; other molecular function |
| RPL32 | 60S ribosomal protein L32 [OS = Homo sapiens] | 0.15 | 0.0004 | other molecular function |
| UTP18 | U3 small nucleolar RNA-associated protein 18 homolog [OS = Homo sapiens] | 0.14 | 0.0062 | nan |
| RRM2 | Ribonucleoside-diphosphate reductase subunit M2 [OS = Homo sapiens] | 0.14 | 0.0029 | other molecular function |
| H1-2 | Histone H1.2 [OS = Homo sapiens] | 0.14 | 0.0067 | nucleic acid binding activity; other molecular function |
| RPL36AL | 60S ribosomal protein L36a-like [OS = Homo sapiens] | 0.14 | 0.0081 | other molecular function |
| RPS29 | 40S ribosomal protein S29 [OS = Homo sapiens] | 0.12 | 0.0013 | other molecular function |
| RRN3 | RNA polymerase I-specific transcription initiation factor RRN3 [OS = Homo sapiens] | 0.11 | 0.0005 | nucleic acid binding activity; other molecular function |
| RFC4 | Replication factor C subunit 4 [OS = Homo sapiens] | 0.1 | 0.0031 | nucleic acid binding activity; other molecular function |
| BAZ1B | Tyrosine-protein kinase BAZ1B [OS = Homo sapiens] | 0.09 | 0.0051 | kinase activity; other molecular function |
| EIF4H | Eukaryotic translation initiation factor 4H [OS = Homo sapiens] | 0.08 | 0.0008 | nucleic acid binding activity |
| H1-10 | Histone H1.10 [OS = Homo sapiens] | 0.08 | 0.0045 | nucleic acid binding activity; other molecular function |
| H1-4 | Histone H1.4 [OS = Homo sapiens] | 0.07 | 0.006 | nucleic acid binding activity; other molecular function |
| RPL39 | 60S ribosomal protein L39 [OS = Homo sapiens] | 0.06 | 0.0096 | nucleic acid binding activity; other molecular function |
| LAD1 | Ladinin-1 [OS = Homo sapiens] | 0.05 | 0.0002 | other molecular function |
| HMGA1 | High mobility group protein HMG-I/HMG-Y [OS = Homo sapiens] | 0.01 | 0.0025 | signal transduction activity or receptor binding; nucleic acid binding activity; other molecular function |
| HMGA2 | High mobility group protein HMGI-C [OS = Homo sapiens] | 0.01 | 0.0081 | nucleic acid binding activity; other molecular function |
References
- Nussbaum, S.R.; Carter, M.J.; Fife, C.E.; DaVanzo, J.; Haught, R.; Nusgart, M.; Cartwright, D. An Economic Evaluation of the Impact, Cost, and Medicare Policy Implications of Chronic Nonhealing Wounds. Value Health 2018, 21, 27–32. [Google Scholar] [CrossRef]
- Cavanagh, P.; Attinger, C.; Abbas, Z.; Bal, A.; Rojas, N.; Xu, Z. Cost of Treating Diabetic Foot Ulcers in Five Different Countries. Diabetes Metab. Res. Rev. 2012, 28, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Vowden, P.; Vowden, K. The Economic Impact of Hard-to- Heal Wounds: Promoting Practice to Address Passivity in wound Management. Wounds Int. 2016, 7, 10–15. [Google Scholar]
- Firlar, I.; Altunbek, M.; McCarthy, C.; Ramalingam, M.; Camci-Unal, G. Functional Hydrogels for Treatment of Chronic Wounds. Gels 2022, 8, 127. [Google Scholar] [CrossRef] [PubMed]
- Sibbald, R.G.; Goodman, L.; Woo, K.Y.; Krasner, D.L.; Smart, H.; Tariq, G.; Ayello, E.A.; Burrell, R.E.; Keast, D.H.; Mayer, D.; et al. Special Considerations in Wound Bed Preparation 2011. Adv. Ski. Wound Care 2011, 24, 415–436. [Google Scholar] [CrossRef]
- Chevallier, A. Encyclopedia of Herbal Medicine; DK Publishing: Londen, UK, 2016. [Google Scholar]
- Ríos, J.L.; Recio, M.C. Medicinal Plants and Antimicrobial Activity. J. Ethnopharmacol. 2005, 100, 80–84. [Google Scholar] [CrossRef]
- Zamani, M.; Kelishadi, M.R.; Ashtary-Larky, D.; Amirani, N.; Goudarzi, K.; Torki, I.A.; Bagheri, R.; Ghanavati, M.; Asbaghi, O. The Effects of Green Tea Supplementation on Cardiovascular Risk Factors: A Systematic Review and Meta-Analysis. Front. Nutr. 2023, 9, 1084455. [Google Scholar] [CrossRef]
- Tanase, C.; Babotă, M.; Nișca, A.; Nicolescu, A.; Ștefănescu, R.; Mocan, A.; Farczadi, L.; Mare, A.D.; Ciurea, C.N.; Man, A. Potential Use of Quercus Dalechampii Ten. and Q. Frainetto Ten. Barks Extracts as Antimicrobial, Enzyme Inhibitory, Antioxidant and Cytotoxic Agents. Pharmaceutics 2023, 15, 343. [Google Scholar] [CrossRef]
- Panahi, Y.; Fazlolahzadeh, O.; Atkin, S.L.; Majeed, M.; Butler, A.E.; Johnston, T.P.; Sahebkar, A. Evidence of Curcumin and Curcumin Analogue Effects in Skin Diseases: A Narrative Review. J. Cell. Physiol. 2019, 234, 1165–1178. [Google Scholar] [CrossRef]
- Karioti, A.; Bilia, A.R.; Gabbiani, C.; Messori, L.; Skaltsa, H. Proanthocyanidin Glycosides from the Leaves of Quercus Ilex L. (Fagaceae). Tetrahedron Lett. 2009, 50, 1771–1776. [Google Scholar] [CrossRef]
- Kazmi, S.T.B.; Majid, M.; Maryam, S.; Rahat, A.; Ahmed, M.; Khan, M.R.; ul Haq, I. Quercus Dilatata Lindl. Ex Royle Ameliorates BPA Induced Hepatotoxicity in Sprague Dawley Rats. Biomed. Pharmacother. 2018, 102, 728–738. [Google Scholar] [CrossRef] [PubMed]
- Şöhretoğlu, D.; Renda, G. The Polyphenolic Profile of Oak (Quercus) Species: A Phytochemical and Pharmacological Overview. Phytochem. Rev. 2020, 19, 1379–1426. [Google Scholar] [CrossRef]
- Yang, J.; Zhou, Y.; Liu, H.; Wang, J.; Hu, J. MCI Extraction from Turkish Galls Played Protective Roles against X-Ray-Induced Damage in AHH-1 Cells. Int. J. Clin. Exp. Pathol. 2015, 8, 8122–8128. [Google Scholar]
- Natella, F.; Leoni, G.; Maldini, M.; Natarelli, L.; Comitato, R.; Schonlau, F.; Virgili, F.; Canali, R. Absorption, Metabolism, and Effects at Transcriptome Level of a Standardized French Oak Wood Extract, Robuvit, in Healthy Volunteers: Pilot Study. J. Agric. Food Chem. 2014, 62, 443–453. [Google Scholar] [CrossRef]
- Országhová, Z.; Waczulíková, I.; Burki, C.; Rohdewald, P.; Ďuračková, Z. An Effect of Oak-Wood Extract (Robuvit®) on Energy State of Healthy Adults—A Pilot Study. Phytother. Res. 2015, 29, 1219–1224. [Google Scholar] [CrossRef]
- Panchal, S.K.; Brown, L. Cardioprotective and Hepatoprotective Effects of Ellagitannins from European Oak Bark (Quercus petraea L.) Extract in Rats. Eur. J. Nutr. 2013, 52, 397–408. [Google Scholar] [CrossRef]
- Popović, B.M.; Štajner, D.; Ždero, R.; Orlović, S.; Galić, Z. Antioxidant Characterization of Oak Extracts Combining Spectrophotometric Assays and Chemometrics. Sci. World J. 2013, 2013, 134656. [Google Scholar] [CrossRef]
- Uyar, A.; Jhangir, G.M.; Keleş, Ö.F.; Yener, Z. The Effects of Quercus (Oak) Acorn on Cutaneous Wound Healing in Rats. Int. J. Plant Based Pharm. 2023, 3, 148–155. [Google Scholar] [CrossRef]
- Tuzlaci, E.; Erol, M.K. Turkish Folk Medicinal Plants. Part II: Eğirdir (Isparta). Fitoterapia 1999, 70, 593–610. [Google Scholar] [CrossRef]
- Banc, R.; Rusu, M.E.; Filip, L.; Popa, D.S. Phytochemical Profiling and Biological Activities of Quercus sp. Galls (Oak Galls): A Systematic Review of Studies Published in the Last 5 Years. Plants 2023, 12, 3873. [Google Scholar] [CrossRef]
- Anlas, C.; Bakirel, T.; Ustun-Alkan, F.; Celik, B.; Yuzbasioglu Baran, M.; Ustuner, O.; Kuruuzum-Uz, A. In Vitro Evaluation of the Therapeutic Potential of Anatolian Kermes Oak (Quercus coccifera L.) as an Alternative Wound Healing Agent. Ind. Crop. Prod. 2019, 137, 24–32. [Google Scholar] [CrossRef]
- Dardmah, F.; Farahpour, M.R. Quercus Infectoria Gall Extract Aids Wound Healing in a Streptozocin-Induced Diabetic Mouse Model. J. Wound Care 2021, 30, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Morton, L.M.; Phillips, T.J. Wound Healing and Treating Wounds. J. Am. Acad. Dermatol. 2016, 74, 589–605. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Xie, Z.; Gao, W.; Pu, L.; Wei, J.; Guo, C. Quercetin Regulates Hepatic Cholesterol Metabolism by Promoting Cholesterol-to-Bile Acid Conversion and Cholesterol Efflux in Rats. Nutr. Res. 2016, 36, 271–279. [Google Scholar] [CrossRef]
- Luu, W.; Sharpe, L.J.; Capell-Hattam, I.; Gelissen, I.C.; Brown, A.J. Oxysterols: Old Tale, New Twists. Annu. Rev. Pharmacol. Toxicol. 2016, 56, 447–467. [Google Scholar] [CrossRef]
- Luo, J.; Yang, H.; Song, B.-L. Mechanisms and Regulation of Cholesterol Homeostasis. Nat. Rev. Mol. Cell. Biol. 2020, 21, 225–245. [Google Scholar] [CrossRef]
- Porter, J.A.; Young, K.E.; Beachy, P.A. Cholesterol Modification of Hedgehog Signaling Proteins in Animal Development. Science 1996, 274, 255–259. [Google Scholar] [CrossRef]
- Kuzu, O.F.; Noory, M.A.; Robertson, G.P. The Role of Cholesterol in Cancer. Cancer Res. 2016, 76, 2063–2070. [Google Scholar] [CrossRef]
- Qin, P.; Zhou, P.; Huang, Y.; Long, B.; Gao, R.; Zhang, S.; Zhu, B.; Li, Y.-Q.; Li, Q. Upregulation of Rate-Limiting Enzymes in Cholesterol Metabolism by PKCδ Mediates Endothelial Apoptosis in Diabetic Wound Healing. Cell Death Discov. 2024, 10, 263. [Google Scholar] [CrossRef]
- Velnar, T.; Bailey, T.; Smrkolj, V. The Wound Healing Process: An Overview of the Cellular and Molecular Mechanisms. J. Int. Med. Res. 2009, 37, 1528–1542. [Google Scholar] [CrossRef]
- Chokpaisarn, J.; Chusri, S.; Amnuaikit, T.; Udomuksorn, W.; Voravuthikunchai, S.P. Potential Wound Healing Activity of Quercus Infectoria Formulation in Diabetic Rats. PeerJ 2017, 2017, e3608. [Google Scholar] [CrossRef] [PubMed]
- Zangeneh, M.M.; Pooyanmehr, M.; Zangeneh, A. Biochemical, Histopathological, and Pharmacological Evaluations of Cutaneous Wound Healing Properties of Quercus Brantii Ethanolic Extract Ointment in Male Rats. Comp. Clin. Path. 2019, 28, 1483–1493. [Google Scholar] [CrossRef]
- Visakorpi, K.; Riutta, T.; Malhi, Y.; Salminen, J.-P.; Salinas, N.; Gripenberg, S. Changes in Oak (Quercus robur) Photosynthesis after Winter Moth (Operophtera brumata) Herbivory Are Not Explained by Changes in Chemical or Structural Leaf Traits. PLoS ONE 2020, 15, e0228157. [Google Scholar] [CrossRef] [PubMed]
- Rotowa, O.J.; Małek, S.; Jasik, M.; Staszel-Szlachta, K. Effect of Innovative Peat-Free Organic Growing Media and Fertilizer on Nutrient Allocation in Pedunculate Oak (Quercus robur L.) and European Beech (Fagus sylvatica L.) Seedlings. New For. 2025, 56, 17. [Google Scholar] [CrossRef]
- Tuyen, P.T.; Khang, D.T.; Thu Ha, P.T.; Hai, T.N.; Elzaawely, A.A.; Xuan, T.D. Antioxidant Capacity and Phenolic Contents of Three Quercus Species. Int. Lett. Nat. Sci. 2016, 54, 85–99. [Google Scholar] [CrossRef]
- Jong, J.K.; Bimal, K.G.; Hyeun, C.S.; Kyung, J.L.; Ki, S.S.; Young, S.C.; Taek, S.Y.; Ye Ji, L.; Eun Hye, K.; Ill Min, C. Comparison of Phenolic Compounds Content in Indeciduous Quercus Species. J. Med. Plants Res. 2012, 6, 5228–5239. [Google Scholar] [CrossRef]
- Coyotl-Martinez, E.; Hernández-Rivera, J.A.; Parra-Suarez, J.L.A.; Reyes-Carmona, S.R.; Carrasco-Carballo, A. Phytochemical Profile, Antioxidant and Antimicrobial Activity of Two Species of Oak: Quercus Sartorii and Quercus Rysophylla. Appl. Biosci. 2025, 4, 13. [Google Scholar] [CrossRef]
- Hong, J.-A.; Bae, D.; Oh, K.-N.; Oh, D.-R.; Kim, Y.; Kim, Y.; Jeong Im, S.; Choi, E.; Lee, S.; Kim, M.; et al. Protective Effects of Quercus Acuta Thunb. Fruit Extract against UVB-Induced Photoaging through ERK/AP-1 Signaling Modulation in Human Keratinocytes. BMC Complement. Med. Ther. 2022, 22, 6. [Google Scholar] [CrossRef]
- Wunnoo, S.; Sermwittayawong, D.; Praparatana, R.; Voravuthikunchai, S.P.; Jakkawanpitak, C. Quercus Infectoria Gall Ethanolic Extract Accelerates Wound Healing through Attenuating Inflammation and Oxidative Injuries in Skin Fibroblasts. Antioxidants 2024, 13, 1094. [Google Scholar] [CrossRef]
- Wang, Y.; Ren, J.; Xia, K.; Wang, S.; Yin, T.; Xie, D.; Li, L. Effect of Mitomycin on Normal Dermal Fibroblast and HaCat Cell: An in Vitro Study. J. Zhejiang Univ. Sci. B 2012, 13, 997–1005. [Google Scholar] [CrossRef]
- Berniak, K.; Moradi, A.; Lichawska-Cieslar, A.; Szukala, W.; Jura, J.; Stachewicz, U. Controlled Therapeutic Cholesterol Delivery to Cells for the Proliferation and Differentiation of Keratinocytes. J. Mater. Chem. B 2024, 12, 11110–11122. [Google Scholar] [CrossRef] [PubMed]
- McCarty, K.D.; Sullivan, M.E.; Tateishi, Y.; Hargrove, T.Y.; Lepesheva, G.I.; Guengerich, F.P. Processive Kinetics in the Three-Step Lanosterol 14α-Demethylation Reaction Catalyzed by Human Cytochrome P450 51A1. J. Biol. Chem. 2023, 299, 104841. [Google Scholar] [CrossRef] [PubMed]
- Lepesheva, G.I.; Waterman, M.R. CYP51—The Omnipotent P450. Mol. Cell. Endocrinol. 2004, 215, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Belter, A.; Skupinska, M.; Giel-Pietraszuk, M.; Grabarkiewicz, T.; Rychlewski, L.; Barciszewski, J. Squalene Monooxygenase—A Target for Hypercholesterolemic Therapy. Biol. Chem. 2011, 392, 1053–1075. [Google Scholar] [CrossRef]
- Yoshioka, H.; Coates, H.W.; Chua, N.K.; Hashimoto, Y.; Brown, A.J.; Ohgane, K. A Key Mammalian Cholesterol Synthesis Enzyme, Squalene Monooxygenase, Is Allosterically Stabilized by Its Substrate. Proc. Natl. Acad. Sci. USA 2020, 117, 7150–7158. [Google Scholar] [CrossRef]
- Taib, M.; Rezzak, Y.; Bouyazza, L.; Lyoussi, B. Medicinal Uses, Phytochemistry, and Pharmacological Activities of Quercus Species. Evid.-Based Complement. Altern. Med. 2020, 2020, 1920683. [Google Scholar] [CrossRef]
- Linnaeus, C. Species Plantarum; Laurentii Salvii: Stockholm, Sweden, 1753; Volume 2. [Google Scholar]
- Histocell. Reoxcare: Productos Para el Cuidado de la Piel y las Heridas: Línea Reoxcare [Brochure]. Reoxcare. 2022. Available online: https://www.reoxcare.com/wp-content/uploads/2022/08/reoxcare-linea.pdf (accessed on 13 May 2025).
- Bremner, J.M. Total Nitrogen. In Methods of Soil Analysis; Norman, A.G., Ed.; American Society of Agronomy, Inc.: Madison, WI, USA, 1965; pp. 1149–1178. ISBN 9780891182047. [Google Scholar]
- 4500-NH3 nitrogen (ammonia). In Standard Methods For the Examination of Water and Wastewater; Amer Public Health Assn: Washington, DC, USA, 2005.
- Murphy, J.; Riley, J.P. A Modified Single Solution Method for the Determination of Phosphate in Natural Waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- 4500-P phosphorus. In Standard Methods for the Examination of Water and Wastewater; Amer Public Health Assn: Washington, DC, USA, 2005.
- Krul, E.S. Calculation of Nitrogen-to-Protein Conversion Factors: A Review with a Focus on Soy Protein. J. Am. Oil. Chem. Soc. 2019, 96, 339–364. [Google Scholar] [CrossRef]
- Abdala-Díaz, R.T.; Cabello-Pasini, A.; Pérez-Rodríguez, E.; Álvarez, R.M.C.; Figueroa, F.L. Daily and Seasonal Variations of Optimum Quantum Yield and Phenolic Compounds in Cystoseira Tamariscifolia (Phaeophyta). Mar. Biol. 2006, 148, 459–465. [Google Scholar] [CrossRef]
- Maaloul, A.; Pérez Manríquez, C.; Decara, J.; Marí-Beffa, M.; Álvarez-Torres, D.; Latorre Redoli, S.; Martínez-Albardonedo, B.; Araya-Rojas, M.; Fajardo, V.; Abdala Díaz, R.T. Biological Effects of Polysaccharides from Bovistella Utriformis as Cytotoxic, Antioxidant, and Antihyperglycemic Agents: In Vitro and In Vivo Studies. Pharmaceutics 2025, 17, 335. [Google Scholar] [CrossRef]
- Brand-Williams, W.; Cuvelier, M.E.; Berset, C. Use of a Free Radical Method to Evaluate Antioxidant Activity. LWT—Food Sci. Technol. 1995, 28, 25–30. [Google Scholar] [CrossRef]
- Vijayabaskar, P.; Vaseela, N. In Vitro Antioxidant Properties of Sulfated Polysaccharide from Brown Marine Algae Sargassum Tenerrimum. Asian Pac. J. Trop. Dis. 2012, 2, S890–S896. [Google Scholar] [CrossRef]
- García-Márquez, J.; Moreira, B.R.; Valverde-Guillén, P.; Latorre-Redoli, S.; Caneda-Santiago, C.T.; Acién, G.; Martínez-Manzanares, E.; Marí-Beffa, M.; Abdala-Díaz, R.T. In Vitro and In Vivo Effects of Ulvan Polysaccharides from Ulva Rigida. Pharmaceuticals 2023, 16, 660. [Google Scholar] [CrossRef] [PubMed]
- Abdala Díaz, R.T.; Chabrillón, M.; Cabello-Pasini, A.; Gómez-Pinchetti, J.L.; Figueroa, F.L. Characterization of Polysaccharides from Hypnea Spinella (Gigartinales) and Halopithys Incurva (Ceramiales) and Their Effect on RAW 264.7 Macrophage Activity. J. Appl. Phycol. 2011, 23, 523–528. [Google Scholar] [CrossRef]
- © ibidi GmbH. Ibidi Application Guide: Wound Healing and Migration Assays; Guide; ibidi GmbH: Gräfelfing, Germany, 2019. [Google Scholar]
- Casas-Arrojo, V.; Arrojo Agudo, M.d.l.Á.; Cárdenas García, C.; Carrillo, P.; Pérez Manríquez, C.; Martínez-Manzanares, E.; Abdala Díaz, R.T. Antioxidant, Immunomodulatory and Potential Anticancer Capacity of Polysaccharides (Glucans) from Euglena gracilis G.A. Klebs. Pharmaceuticals 2022, 15, 1379. [Google Scholar] [CrossRef]
| Compound/Element | Amount |
|---|---|
| Total Nitrogen (N) | 2.38 ± 0.2% |
| Phosphorus (P) | 0.09 ± 0.001% |
| Total protein | 14.87 ± 0.9% |
| Total phenolic | 3.20 ± 0.15 mg·g−1 DW |
| GO-Term | Description | Count in Network | Strength | False Discovery Rate (FDR) |
|---|---|---|---|---|
| GO:0010647 | Regulation of substrate-dependent cell migration, cell adhesion | 2 | 2.42 | 0.021 |
| GO:0006695 | Cholesterol biosynthetic process | 2 | 1.97 | 7.95 × 10−4 |
| GO:0007596 | Blood coagulation | 9 | 1.52 | 3.12 × 10−4 |
| GO:0002040 | Positive regulation of endothelial cell migration | 7 | 0.77 | 0.0099 |
| GO:0022617 | Wound healing | 8 | 0.77 | 0.028 |
| GO:0006954 | Inflammatory response | 8 | 0.77 | 0.0077 |
| Pathway | Description | Count in Network | Strength | False Discovery Rate (FDR) |
|---|---|---|---|---|
| R-HSA-114608 | Platelet degranulation | 7 | 1.34 | 1.44 × 10−5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojas-Velis, N.; Cárdenas-García, C.; Pérez, E.; Toledo, J.R.; Medina, M.Á.; Astuya-Villalón, A.; Abdala-Díaz, R.T. In Vitro Evaluation of the Healing Potential and Proteomic Study of Quercus robur L. Leaf Extracts in Human Keratinocytes. Molecules 2025, 30, 2152. https://doi.org/10.3390/molecules30102152
Rojas-Velis N, Cárdenas-García C, Pérez E, Toledo JR, Medina MÁ, Astuya-Villalón A, Abdala-Díaz RT. In Vitro Evaluation of the Healing Potential and Proteomic Study of Quercus robur L. Leaf Extracts in Human Keratinocytes. Molecules. 2025; 30(10):2152. https://doi.org/10.3390/molecules30102152
Chicago/Turabian StyleRojas-Velis, Nelson, Casimiro Cárdenas-García, Erik Pérez, Jorge R. Toledo, Miguel Ángel Medina, Allisson Astuya-Villalón, and Roberto T. Abdala-Díaz. 2025. "In Vitro Evaluation of the Healing Potential and Proteomic Study of Quercus robur L. Leaf Extracts in Human Keratinocytes" Molecules 30, no. 10: 2152. https://doi.org/10.3390/molecules30102152
APA StyleRojas-Velis, N., Cárdenas-García, C., Pérez, E., Toledo, J. R., Medina, M. Á., Astuya-Villalón, A., & Abdala-Díaz, R. T. (2025). In Vitro Evaluation of the Healing Potential and Proteomic Study of Quercus robur L. Leaf Extracts in Human Keratinocytes. Molecules, 30(10), 2152. https://doi.org/10.3390/molecules30102152









