Exploration of Polysaccharides from Phyllanthus emblica: Isolation, Identification, and Evaluation of Antioxidant and Anti-Glycolipid Metabolism Disorder Activities
Abstract
1. Introduction
2. Results and Discussion
2.1. Preparation and Physicochemical Property Analysis of PEP-1-1
2.2. Monosaccharide Composition
2.3. FT-IR Analysis
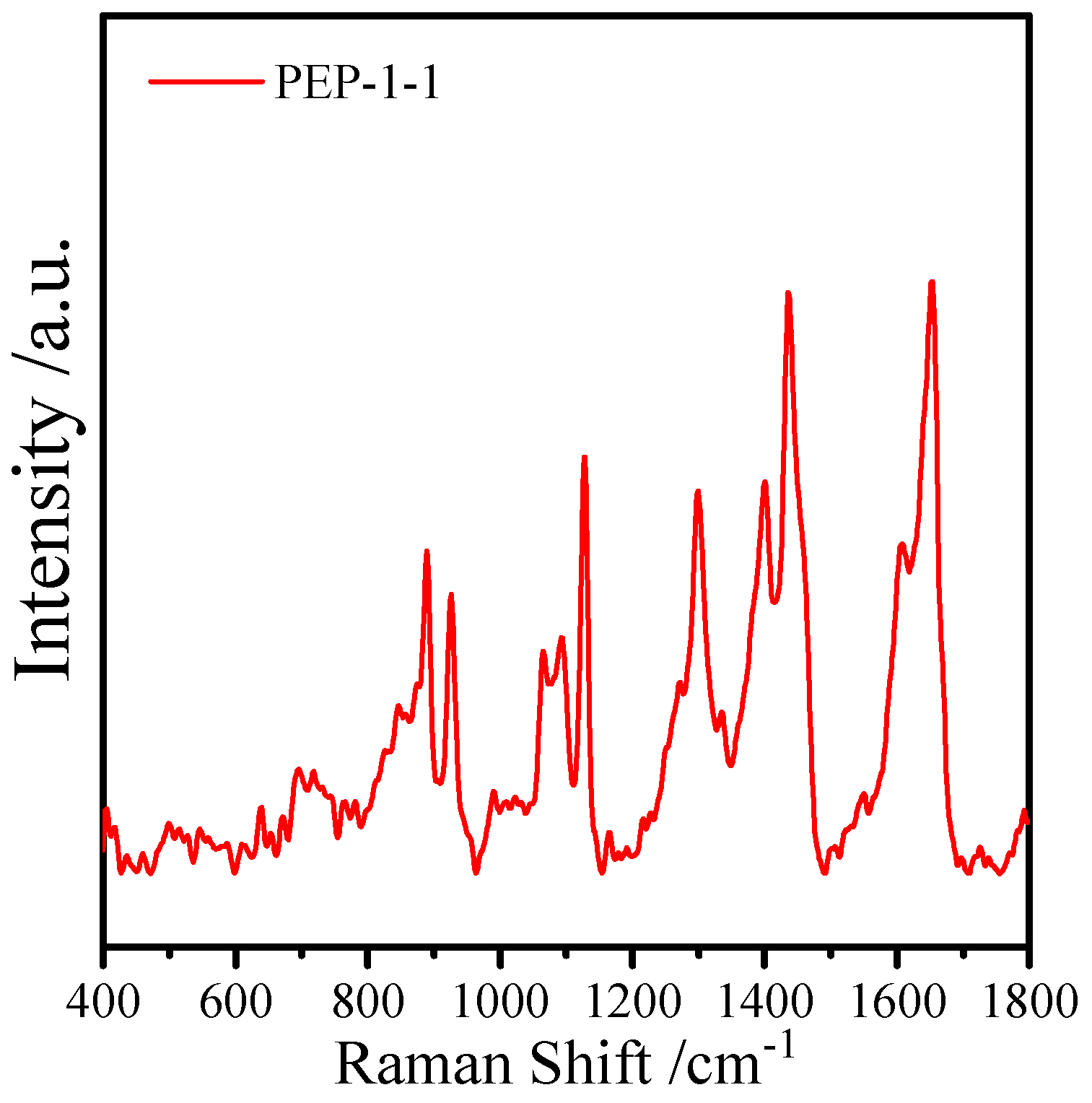
2.4. Surface-Enhanced Raman Spectroscopy Analysis
2.5. Methylation Analysis
2.6. NMR Spectra Analysis
2.7. Congo Red Test Analysis
2.8. SEM Analysis
2.9. Antioxidant Activity of PEP-1-1 In Vitro
2.10. Hypolipidemic Activity and Anti-Glycolipid Metabolism Disorder Activity of PEP-1-1
2.10.1. Effects of PEP-1-1 on Cell Viability
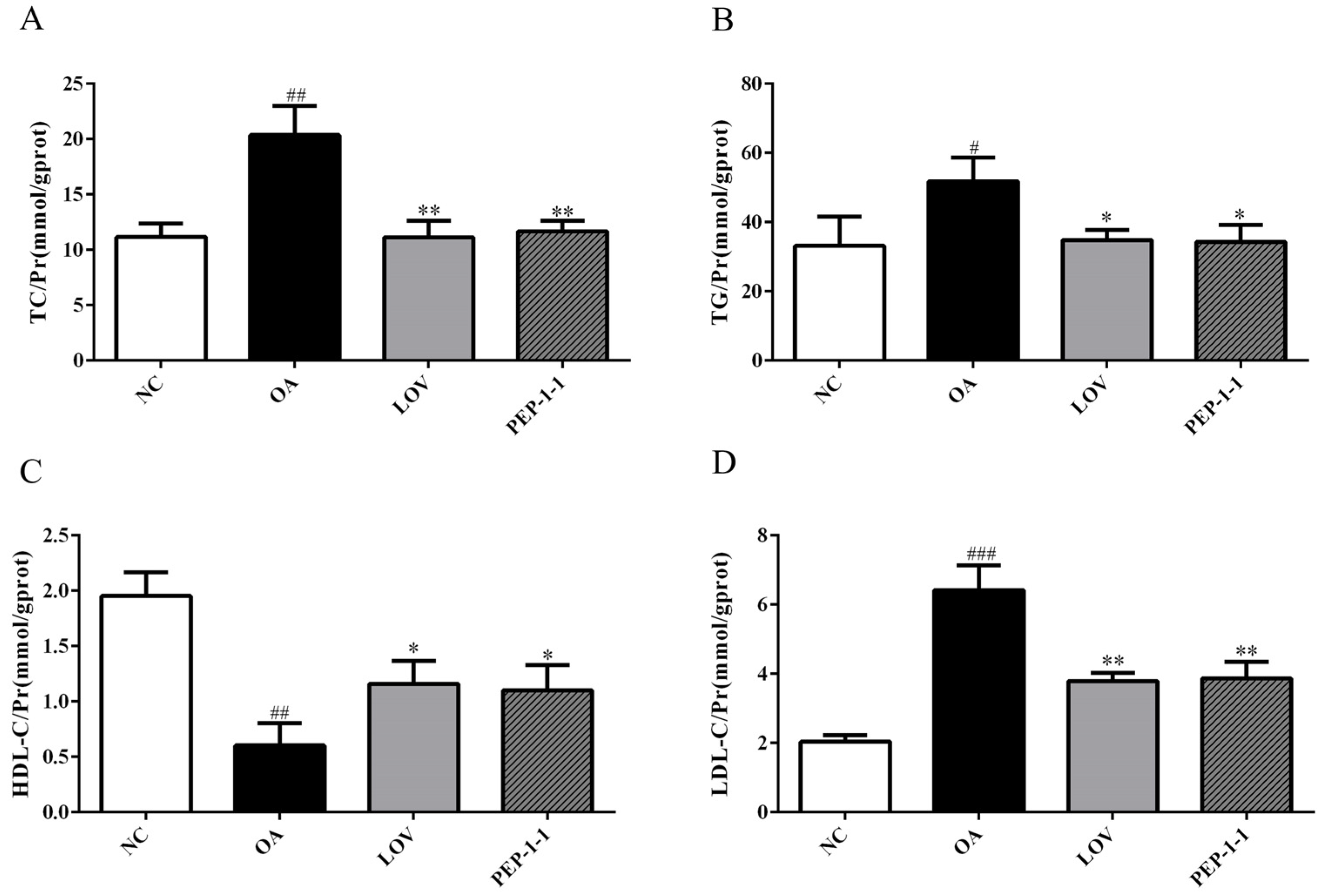
2.10.2. Effects of PEP-1-1 on Lipid Content in High-Fat HepG2 Cell Model Induced by OA
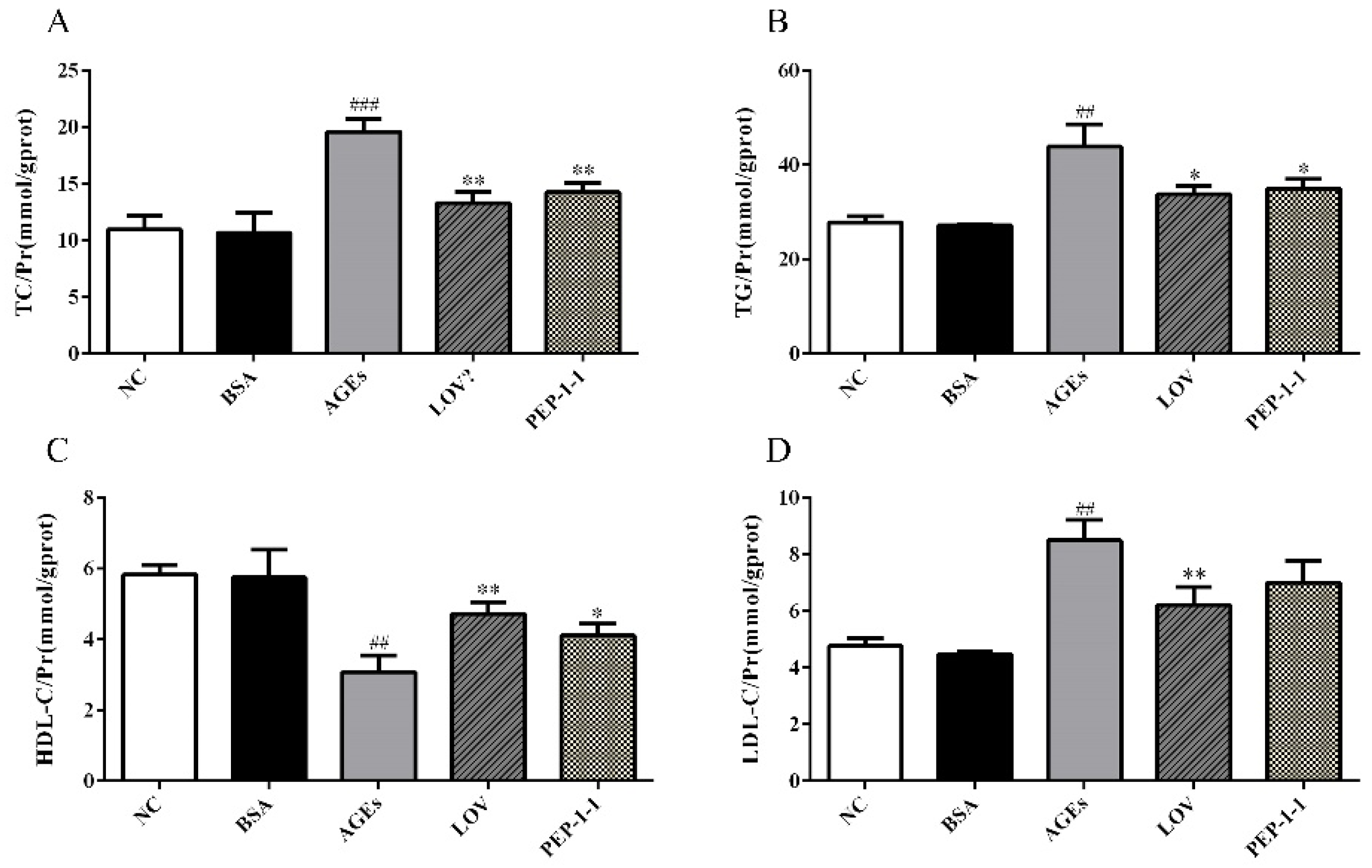
2.10.3. Effects of PEP-1-1 on Lipid Content in High-Fat HepG2 Cell Model Induced by AGEs
3. Materials and Methods
3.1. Materials
3.2. Extraction, Isolation, and Purification of Polysaccharides from Phyllanthus emblica
3.3. Homogeneity and Molecular Weight (Mw)
3.4. Chemical Composition Analysis
3.5. Structural Characteristics of PEP-1-1
3.5.1. Determination of Particle Size and ζ-Potential
3.5.2. Monosaccharide Composition Assay
3.5.3. UV and FT-IR Analysis
3.5.4. Surface Enhanced Raman Spectroscopy (SERS) Analysis
3.5.5. Methylation and GC-MS Analysis
3.5.6. NMR Analysis
3.5.7. Congo Red Test
3.5.8. Scanning Electron Microscopy (SEM) Analysis
3.6. Antioxidant Effects of PEP-1-1 In Vitro
3.6.1. Determination of Scavenging Capacity of Hydroxyl Radicals
3.6.2. Determination of Scavenging Capacity of Superoxide Anion Radicals
3.6.3. Determination of Scavenging Capacity of ABTS Radicals
- A0: Absorption of samples replaced by ultrapure water;
- A1: Absorption of sample groups;
- A2: Absorption of sample groups’ background.
3.7. Hypolipidemic Activity and Anti-Glycolipid Metabolism Disorder Activity of PEP-1-1
3.7.1. Preparation of AGEs
3.7.2. Cell Culture
3.7.3. Cell Viability Assay
3.7.4. Effect of PEP-1-1 on Lipid Content in High-Fat HepG2 Cell Model Induced by OA
3.7.5. Effect of PEP-1-1 on Lipid Content in High-Fat HepG2 Cell Model Induced by AGEs
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sheng-Shou, H. Report on cardiovascular health and diseases in china 2021: An updated summary. J. Geriatr. Cardiol. 2023, 20, 573–603. [Google Scholar]
- Townsend, N.; Kazakiewicz, D.; Lucy Wright, F.; Timmis, A.; Huculeci, R.; Torbica, A.; Gale, C.P.; Achenbach, S.; Weidinger, F.; Vardas, P. Epidemiology of cardiovascular disease in Europe. Nat. Rev. Cardiol. 2022, 19, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Kaminsky, L.A.; German, C.; Imboden, M.; Ozemek, C.; Peterman, J.E.; Brubaker, P.H. The importance of healthy lifestyle behaviors in the prevention of cardiovascular disease. Prog. Cardiovasc. Dis. 2022, 70, 8–15. [Google Scholar] [CrossRef]
- Hamedi, A.; Sakhteman, A.; Moheimani, S.M. An in silico approach towards investigation of possible effects of essential oils constituents on receptors involved in cardiovascular diseases (CVD) and associated risk factors (Diabetes Mellitus and Hyperlipidemia). Cardiovasc. Hematol. Agents Med. Chem. (Former. Curr. Med. Chem.-Cardiovasc. Hematol. Agents) 2021, 19, 32–42. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, B.J. Effect of low-carbohydrate diets on cardiometabolic risk, insulin resistance, and metabolic syndrome. Curr. Opin. Endocrinol. Diabetes Obes. 2020, 27, 301–307. [Google Scholar] [CrossRef]
- Zhou, Y.-J.; Xu, N.; Zhang, X.-C.; Zhu, Y.-Y.; Liu, S.-W.; Chang, Y.-N. Chrysin improves glucose and lipid metabolism disorders by regulating the AMPK/PI3K/AKT signaling pathway in insulin-resistant HepG2 cells and HFD/STZ-induced C57BL/6J mice. J. Agric. Food Chem. 2021, 69, 5618–5627. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Yin, X.; Liu, Z.; Wang, J. Non-alcoholic fatty liver disease (NAFLD) pathogenesis and natural products for prevention and treatment. Int. J. Mol. Sci. 2022, 23, 15489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Jiang, L.; Li, G.; Wu, J.; Tian, P.; Zhang, D.; Qin, Y.; Shi, Z.; Gao, Z.; Zhang, N. Advanced Glycosylation End Products Induced Synaptic Deficits and Cognitive Decline Through ROS-JNK-p53/miR-34c/SYT1 Axis in Diabetic Encephalopathy. J. Alzheimer’s Dis. 2022, 87, 843–861. [Google Scholar] [CrossRef] [PubMed]
- Huo, S.; Wang, Q.; Shi, W.; Peng, L.; Jiang, Y.; Zhu, M.; Guo, J.; Peng, D.; Wang, M.; Men, L. ATF3/SPI1/SLC31A1 signaling promotes cuproptosis induced by advanced glycosylation end products in diabetic myocardial injury. Int. J. Mol. Sci. 2023, 24, 1667. [Google Scholar] [CrossRef] [PubMed]
- Mengstie, M.A.; Chekol Abebe, E.; Behaile Teklemariam, A.; Tilahun Mulu, A.; Agidew, M.M.; Teshome Azezew, M.; Zewde, E.A.; Agegnehu Teshome, A. Endogenous advanced glycation end products in the pathogenesis of chronic diabetic complications. Front. Mol. Biosci. 2022, 9, 1002710. [Google Scholar] [CrossRef]
- Abdul-Rahman, T.; Bukhari, S.M.A.; Herrera, E.C.; Awuah, W.A.; Lawrence, J.; de Andrade, H.; Patel, N.; Shah, R.; Shaikh, R.; Capriles, C.A.A. Lipid lowering therapy: An era beyond statins. Curr. Probl. Cardiol. 2022, 47, 101342. [Google Scholar] [CrossRef]
- Sirtori, C.R. The pharmacology of statins. Pharmacol. Res. 2014, 88, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Sharma, M.; Singh, S.; Goyal, A. Recent advances of α-glucosidase inhibitors: A comprehensive review. Curr. Top. Med. Chem. 2022, 22, 2069–2086. [Google Scholar] [CrossRef] [PubMed]
- Triggle, C.R.; Mohammed, I.; Bshesh, K.; Marei, I.; Ye, K.; Ding, H.; MacDonald, R.; Hollenberg, M.D.; Hill, M.A. Metformin: Is it a drug for all reasons and diseases? Metabolism 2022, 133, 155223. [Google Scholar] [CrossRef] [PubMed]
- Ward, N.C.; Watts, G.F.; Eckel, R.H. Statin toxicity: Mechanistic insights and clinical implications. Circ. Res. 2019, 124, 328–350. [Google Scholar] [CrossRef] [PubMed]
- Kores, K.; Konc, J.; Bren, U. Mechanistic insights into side effects of troglitazone and rosiglitazone using a novel inverse molecular docking protocol. Pharmaceutics 2021, 13, 315. [Google Scholar] [CrossRef] [PubMed]
- Silva, M.L.; Bernardo, M.A.; Singh, J.; de Mesquita, M.F. Cinnamon as a complementary therapeutic approach for dysglycemia and dyslipidemia control in type 2 diabetes mellitus and its molecular mechanism of action: A review. Nutrients 2022, 14, 2773. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Yu, Q.; Zhang, D.-K.; Li, M.; Yu, J.; Yu, X.; Xia, C.; Lin, J.-Z.; Han, L.; Huang, H.-Z. Phyllanthus emblica fruits: A polyphenol-rich fruit with potential benefits for oral management. Food Funct. 2023, 14, 7738–7759. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Liu, M.; Wang, F.; Cai, J.; Luo, Q.; Li, S.; Zhu, J.; Tang, Z.; Fang, Z.; Wang, C. The inhibition mechanism of polyphenols from Phyllanthus emblica Linn. fruit on acetylcholinesterase: A interaction, kinetic, spectroscopic, and molecular simulation study. Food Res. Int. 2022, 158, 111497. [Google Scholar] [CrossRef]
- Jaisamut, P.; Tohlang, C.; Wanna, S.; Thanakun, A.; Srisuwan, T.; Limsuwan, S.; Rattanachai, W.; Suwannachot, J.; Chusri, S. Clinical Evaluation of a Novel Tablet Formulation of Traditional Thai Polyherbal Medicine Named Nawametho in Comparison with Its Decoction in the Treatment of Hyperlipidemia. Evid.-Based Complement. Altern. Med. 2022, 2022, 2530266. [Google Scholar] [CrossRef]
- Saini, R.; Sharma, N.; Oladeji, O.S.; Sourirajan, A.; Dev, K.; Zengin, G.; El-Shazly, M.; Kumar, V. Traditional uses, bioactive composition, pharmacology, and toxicology of Phyllanthus emblica fruits: A comprehensive review. J. Ethnopharmacol. 2022, 282, 114570. [Google Scholar] [CrossRef] [PubMed]
- Mahata, S.; Pandey, A.; Shukla, S.; Tyagi, A.; Husain, S.A.; Das, B.C.; Bharti, A.C. Anticancer activity of Phyllanthus emblica Linn.(Indian gooseberry): Inhibition of transcription factor AP-1 and HPV gene expression in cervical cancer cells. Nutr. Cancer 2013, 65, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Ngamkitidechakul, C.; Jaijoy, K.; Hansakul, P.; Soonthornchareonnon, N.; Sireeratawong, S. Antitumour effects of Phyllanthus emblica L.: Induction of cancer cell apoptosis and inhibition of in vivo tumour promotion and in vitro invasion of human cancer cells. Phytother. Res. 2010, 24, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Panich, S.; Amatatongchai, M. A non-toxic approach to assess total antioxidant capacity (TAC) of exotic tropical fruits from Thailand. J. Food Sci. Technol. 2019, 56, 3547–3552. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, X.; Chen, R.; Li, Y.; Miao, J.; Liu, G.; Lan, Y.; Chen, Y.; Cao, Y. HPLC fingerprint analysis of Phyllanthus emblica ethanol extract and their antioxidant and anti-inflammatory properties. J. Ethnopharmacol. 2020, 254, 112740. [Google Scholar] [CrossRef] [PubMed]
- Pedro-Botet, J.; Climent, E.; Benaiges, D. Atherosclerosis and inflammation. New therapeutic approaches. Med. Clínica (Engl. Ed.) 2020, 155, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Gan, J.; Zhang, X.; Ma, C.; Sun, L.; Feng, Y.; He, Z.; Zhang, H. Purification of polyphenols from Phyllanthus emblica L. pomace using macroporous resins: Antioxidant activity and potential anti-Alzheimer’s effects. J. Food Sci. 2022, 87, 1244–1256. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, J.; Cao, L.; Li, L.; Wang, F.; Liao, Z.; Chen, J.; Wu, S.; Zhang, L. Characterization of a novel polysaccharide isolated from Phyllanthus emblica L. and analysis of its antioxidant activities. J. Food Sci. Technol. 2018, 55, 2758–2764. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Xia, X.; Liao, S.; Wu, T.; Wang, L.; Chen, Q.; Wei, S.; Gu, X.; Zhu, Z. Physicochemical characterization and antioxidant and hypolipidaemic activities of a polysaccharide from the fruit of Kadsura coccinea (Lem.) AC Smith. Front. Nutr. 2022, 9, 903218. [Google Scholar] [CrossRef]
- Chen, F.; Huang, G.; Yang, Z.; Hou, Y. Antioxidant activity of Momordica charantia polysaccharide and its derivatives. Int. J. Biol. Macromol. 2019, 138, 673–680. [Google Scholar] [CrossRef]
- Li, F.; Wei, Y.; Liang, L.; Huang, L.; Yu, G.; Li, Q. A novel low-molecular-mass pumpkin polysaccharide: Structural characterization, antioxidant activity, and hypoglycemic potential. Carbohydr. Polym. 2021, 251, 117090. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, H.; Liu, Y.; Nan, J.; Park, H.J.; Chen, Y.; Yang, L. The chemical structure and immunomodulatory activity of an exopolysaccharide produced by Morchella esculenta under submerged fermentation. Food Funct. 2021, 12, 9327–9338. [Google Scholar] [CrossRef] [PubMed]
- Özbalci, B.; Boyaci, İ.H.; Topcu, A.; Kadılar, C.; Tamer, U. Rapid analysis of sugars in honey by processing Raman spectrum using chemometric methods and artificial neural networks. Food Chem. 2013, 136, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Söderholm, S.; Roos, Y.H.; Meinander, N.; Hotokka, M. Raman spectra of fructose and glucose in the amorphous and crystalline states. J. Raman Spectrosc. 1999, 30, 1009–1018. [Google Scholar] [CrossRef]
- Tahir, M.; Majeed, M.I.; Nawaz, H.; Ali, S.; Rashid, N.; Kashif, M.; Ashfaq, I.; Ahmad, W.; Ghauri, K.; Sattar, F. Raman spectroscopy for the analysis of different exo-polysaccharides produced by bacteria. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 237, 118408. [Google Scholar] [CrossRef] [PubMed]
- Synytsya, A.; Čopíková, J.; Matějka, P.; Machovič, V. Fourier transform Raman and infrared spectroscopy of pectins. Carbohydr. Polym. 2003, 54, 97–106. [Google Scholar] [CrossRef]
- Vasko, P.; Blackwell, J.; Koenig, J. Infrared and raman spectroscopy of carbohydrates.: Part II: Normal coordinate analysis of α-D-glucose. Carbohydr. Res. 1972, 23, 407–416. [Google Scholar] [CrossRef]
- Peng, Q.; Liu, H.; Lei, H.; Wang, X. Relationship between structure and immunological activity of an arabinogalactan from Lycium ruthenicum. Food Chem. 2016, 194, 595–600. [Google Scholar] [CrossRef]
- Huang, W.; Zhao, M.; Wang, X.; Tian, Y.; Wang, C.; Sun, J.; Wang, Z.; Gong, G.; Huang, L. Revisiting the structure of arabinogalactan from Lycium barbarum and the impact of its side chain on anti-ageing activity. Carbohydr. Polym. 2022, 286, 119282. [Google Scholar] [CrossRef]
- Huang, H.; Yang, X.; Li, W.; Han, Q.; Xu, Z.; Xia, W.; Wu, M.; Zhang, W. Structural characterization and immunomodulatory activity of an arabinogalactan from Jasminum sambac (L.) Aiton tea processing waste. Int. J. Biol. Macromol. 2023, 235, 123816. [Google Scholar] [CrossRef]
- Kim, M.; Kim, S.-R.; Park, J.; Mun, S.-H.; Kwak, M.; Ko, H.-J.; Baek, S.-H. Structure and antiviral activity of a pectic polysaccharide from the root of Sanguisorba officinalis against enterovirus 71 in vitro/vivo. Carbohydr. Polym. 2022, 281, 119057. [Google Scholar] [CrossRef] [PubMed]
- Golovchenko, V.; Popov, S.; Smirnov, V.; Khlopin, V.; Vityazev, F.; Naranmandakh, S.; Dmitrenok, A.S.; Shashkov, A.S. Polysaccharides of Salsola passerina: Extraction, Structural Characterization and Antioxidant Activity. Int. J. Mol. Sci. 2022, 23, 13175. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, J.; Xu, X.; Luo, Z.; Sun, J.; Wang, H.; Liu, C.; Ni, X.; Sun, J.; Xu, J. Separation and Structural Characterization of a Novel Exopolysaccharide from Rhizopus nigricans. Molecules 2022, 27, 7756. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Gong, Z.; Liu, H.; Wang, J.; Wang, D.; Yang, Y.; Zhong, S. Structural characterization and anti-tumor activity in vitro of a water-soluble polysaccharide from dark brick tea. Int. J. Biol. Macromol. 2022, 205, 615–625. [Google Scholar] [CrossRef]
- Wei, X.; Yao, J.; Wang, F.; Wu, D.; Zhang, R. Extraction, isolation, structural characterization, and antioxidant activity of polysaccharides from elderberry fruit. Front. Nutr. 2022, 9, 947706. [Google Scholar] [CrossRef]
- Makarova, E.N.; Shakhmatov, E.G.; Belyy, V.A. Structural studies of water-extractable pectic polysaccharides and arabinogalactan proteins from Picea abies greenery. Carbohydr. Polym. 2018, 195, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, S.; Feng, W.; Zhang, Z.; Li, H. Structural characterization and immunomodulatory activities of two polysaccharides from Rehmanniae Radix Praeparata. Int. J. Biol. Macromol. 2021, 186, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Kang, J.; Xu, Z.; Guo, Q.; Zhang, L.; Ning, H.; Cui, S.W. Triple-helix polysaccharides: Formation mechanisms and analytical methods. Carbohydr. Polym. 2021, 262, 117962. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Huang, L.; Zhang, C.; Xie, P.; Cheng, J.; Wang, X.; Liu, L. Novel polysaccharide from Chaenomeles speciosa seeds: Structural characterization, α-amylase and α-glucosidase inhibitory activity evaluation. Int. J. Biol. Macromol. 2020, 153, 755–766. [Google Scholar] [CrossRef]
- Chen, X.; Zuo, X.; Xu, A.a.; Xu, P.; Wang, Y. Comparative Study on the Physicochemical Characteristics and Antioxidant Activities of Polysaccharides in Different Tea Cultivars. J. Tea Sci. 2022, 42, 806–818. [Google Scholar]
- Pu, X.; Ma, X.; Liu, L.; Ren, J.; Li, H.; Li, X.; Yu, S.; Zhang, W.; Fan, W. Structural characterization and antioxidant activity in vitro of polysaccharides from angelica and astragalus. Carbohydr. Polym. 2016, 137, 154–164. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, H.-M.; Qin, G.-Y. Structure characterization and antioxidant activity of polysaccharides from Chinese quince seed meal. Food Chem. 2017, 234, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Zheng, Y.; Tang, S.; Gu, L.; Zhu, Y.; Ying, R.; Liu, Y.; Ma, J.; Guo, R.; Gao, P. Radical oxygen species: An important breakthrough point for botanical drugs to regulate oxidative stress and treat the disorder of glycolipid metabolism. Front. Pharmacol. 2023, 14, 1166178. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, B. Hydroxyl radical and its scavengers in health and disease. Oxidative Med. Cell. Longev. 2011, 2011, 809696. [Google Scholar] [CrossRef] [PubMed]
- Olech, M.; Nowacka-Jechalke, N.; Masłyk, M.; Martyna, A.; Pietrzak, W.; Kubiński, K.; Załuski, D.; Nowak, R. Polysaccharide-rich fractions from Rosa rugosa Thunb.—Composition and chemopreventive potential. Molecules 2019, 24, 1354. [Google Scholar] [CrossRef] [PubMed]
- Nowotny, K.; Jung, T.; Höhn, A.; Weber, D.; Grune, T. Advanced glycation end products and oxidative stress in type 2 diabetes mellitus. Biomolecules 2015, 5, 194–222. [Google Scholar] [CrossRef]
- Kuzan, A. Toxicity of advanced glycation end products. Biomed. Rep. 2021, 14, 46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xing, N.; Peng, D.; Wang, Q.; Kuang, H. Study on Isolation, Purification and in vitro Antioxidant Activity of the Polysaccharides from Cucumis satiuus. China Pharm. 2021, 32, 432–438. [Google Scholar]
- Tian, W.; Dai, L.; Lu, S.; Luo, Z.; Qiu, Z.; Li, J.; Li, P.; Du, B. Effect of Bacillus sp. DU-106 fermentation on Dendrobium officinale polysaccharide: Structure and immunoregulatory activities. Int. J. Biol. Macromol. 2019, 135, 1034–1042. [Google Scholar] [CrossRef]
- DuBois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.t.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Filisetti-Cozzi, T.M.; Carpita, N.C. Measurement of uronic acids without interference from neutral sugars. Anal. Biochem. 1991, 197, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Sun, Y.; Li, X.; Zeng, B.; Chen, D. Extraction, structural characterization, and antioxidant and immunomodulatory activities of a polysaccharide from Notarchus leachii freeri eggs. Bioorg. Chem. 2021, 116, 105275. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Dong, X.; Ma, R.; Ji, H.; Yu, J.; Liu, A. Characterization of a polysaccharide from Polygala tenuifolia willd. with immune activity via activation MAPKs pathway. Bioorg. Chem. 2023, 130, 106214. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Jia, R.-B.; Ou, Z.-R.; Li, Z.-R.; Zhao, M.; Luo, D.; Lin, L. Comparative study on the structural characterization and α-glucosidase inhibitory activity of polysaccharide fractions extracted from Sargassum fusiforme at different pH conditions. Int. J. Biol. Macromol. 2022, 194, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Wu, Y.-T.; Chang, H.-J.; Huang, C.-C.; Cheng, K.-C.; Hsu, H.-Y.; Hsieh, C.-W. Anti-Inflammatory and Anti-Oxidative Effects of Polysaccharides Extracted from Unripe Carica papaya L. Fruit. Antioxidants 2023, 12, 1506. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Chen, P.; Zhao, Y.; Zeng, Q.; Ou, S.; Zhang, Y.; Wang, P.; Chen, N.; Ou, J. Isolation, structural characterization and anti-oxidant activity of a novel polysaccharide from garlic bolt. Carbohydr. Polym. 2021, 267, 118194. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, L.-S.; Dong, L.-L. Inhibitory effect of polysaccharides from pumpkin on advanced glycation end-products formation and aldose reductase activity. Food Chem. 2012, 130, 821–825. [Google Scholar] [CrossRef]


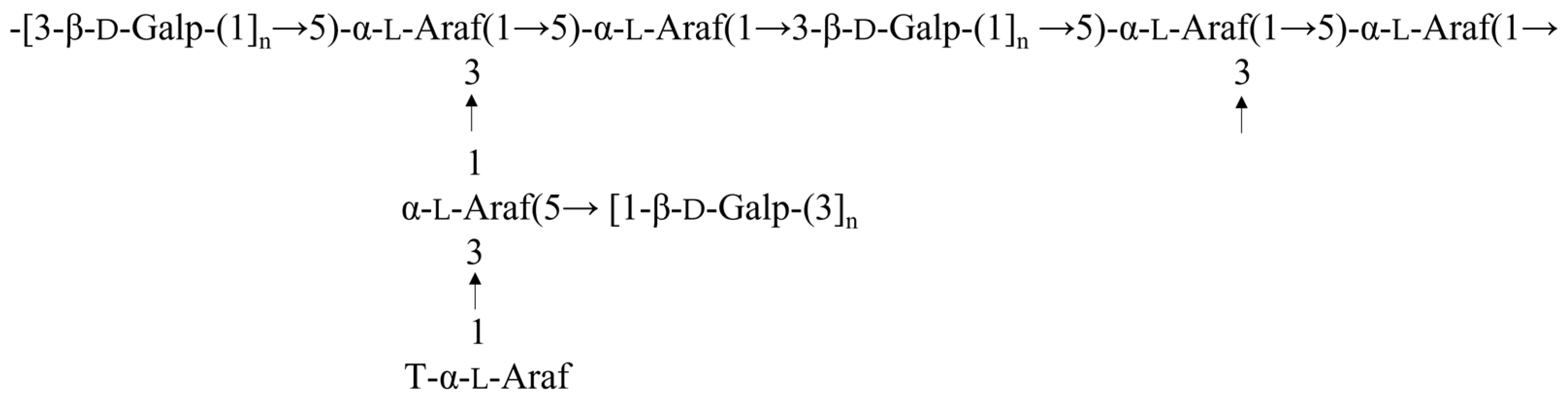

| Polysaccharide | Chemical Composition | Molecular Properties | |||
|---|---|---|---|---|---|
| Total Sugar | Protein | Uronic Acid | Particle Size | ζ-Potential | |
| PEP-1-1 | 91.23% | 0.35% | 6.22% | 1093 nm | −4.22 mV |
| Code | Residues | Chemical Shifts (ppm) | |||||
|---|---|---|---|---|---|---|---|
| H1/C1 | H2/C2 | H3/C3 | H4/C4 | H5/C5 | H6/C6 | ||
| A | T-α-l-Araf-(1- | 5.03/107.31 | 4.07/81.03 | 3.96/76.34 | 4.15/82.07 | 3.75/60.99 | |
| B | -5-α-l-Araf-(1- | 5.17/106.35 | 4.24/81.34 | 3.89/76.68 | 3.98/83.83 | 3.54/72.04 | |
| C | -3,5)-α-l-Araf-(1- | 5.10/106.90 | 4.22/79.10 | 4.03/82.06 | 4.24/84.27 | 3.82/69.26 | |
| D | -3-β-d-Galp-(1- | 4.60/104.50 | 3.62/71.72 | 4.12/77.50 | 3.75/71.50 | 3.72/73.21 | 3.65/60.68 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, P.; Chen, M.; Wang, W.; Li, Q.; Chen, X.; Liang, J.; He, Y.; Wu, Y. Exploration of Polysaccharides from Phyllanthus emblica: Isolation, Identification, and Evaluation of Antioxidant and Anti-Glycolipid Metabolism Disorder Activities. Molecules 2024, 29, 1751. https://doi.org/10.3390/molecules29081751
Guo P, Chen M, Wang W, Li Q, Chen X, Liang J, He Y, Wu Y. Exploration of Polysaccharides from Phyllanthus emblica: Isolation, Identification, and Evaluation of Antioxidant and Anti-Glycolipid Metabolism Disorder Activities. Molecules. 2024; 29(8):1751. https://doi.org/10.3390/molecules29081751
Chicago/Turabian StyleGuo, Peng, Meng Chen, Wenzhao Wang, Qiuyun Li, Xinyu Chen, Jiayue Liang, Yiyang He, and Yanli Wu. 2024. "Exploration of Polysaccharides from Phyllanthus emblica: Isolation, Identification, and Evaluation of Antioxidant and Anti-Glycolipid Metabolism Disorder Activities" Molecules 29, no. 8: 1751. https://doi.org/10.3390/molecules29081751
APA StyleGuo, P., Chen, M., Wang, W., Li, Q., Chen, X., Liang, J., He, Y., & Wu, Y. (2024). Exploration of Polysaccharides from Phyllanthus emblica: Isolation, Identification, and Evaluation of Antioxidant and Anti-Glycolipid Metabolism Disorder Activities. Molecules, 29(8), 1751. https://doi.org/10.3390/molecules29081751





