The Green Tea Polyphenol Epigallocatechin-Gallate (EGCG) Interferes with Microcin E492 Amyloid Formation
Abstract
:1. Introduction
2. Results
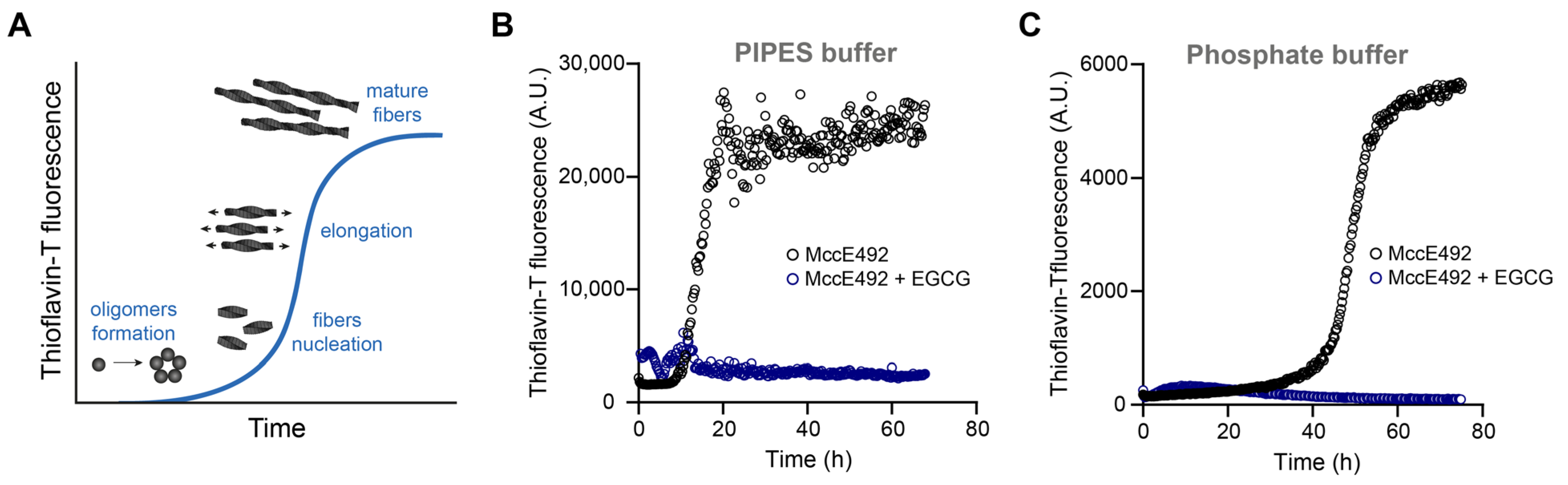
2.1. EGCG Inhibits MccE492 Amyloid Formation Followed by ThT Fluorescence
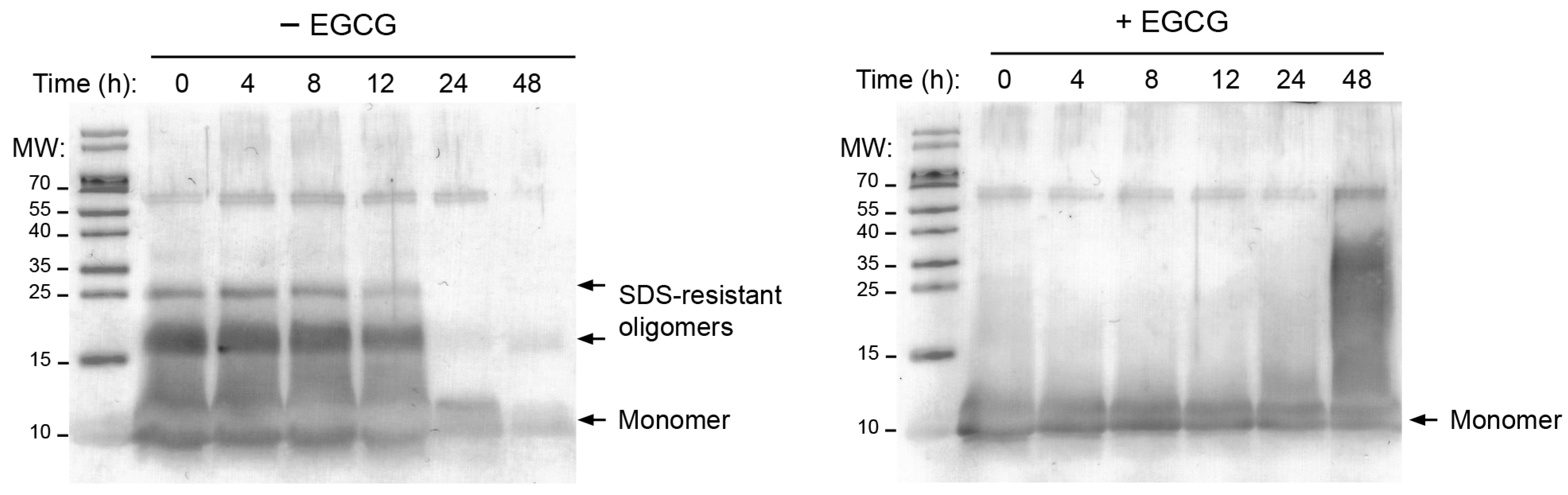
2.2. MccE492 Is Not Incorporated into Higher-Order Aggregates in the Presence of EGCG
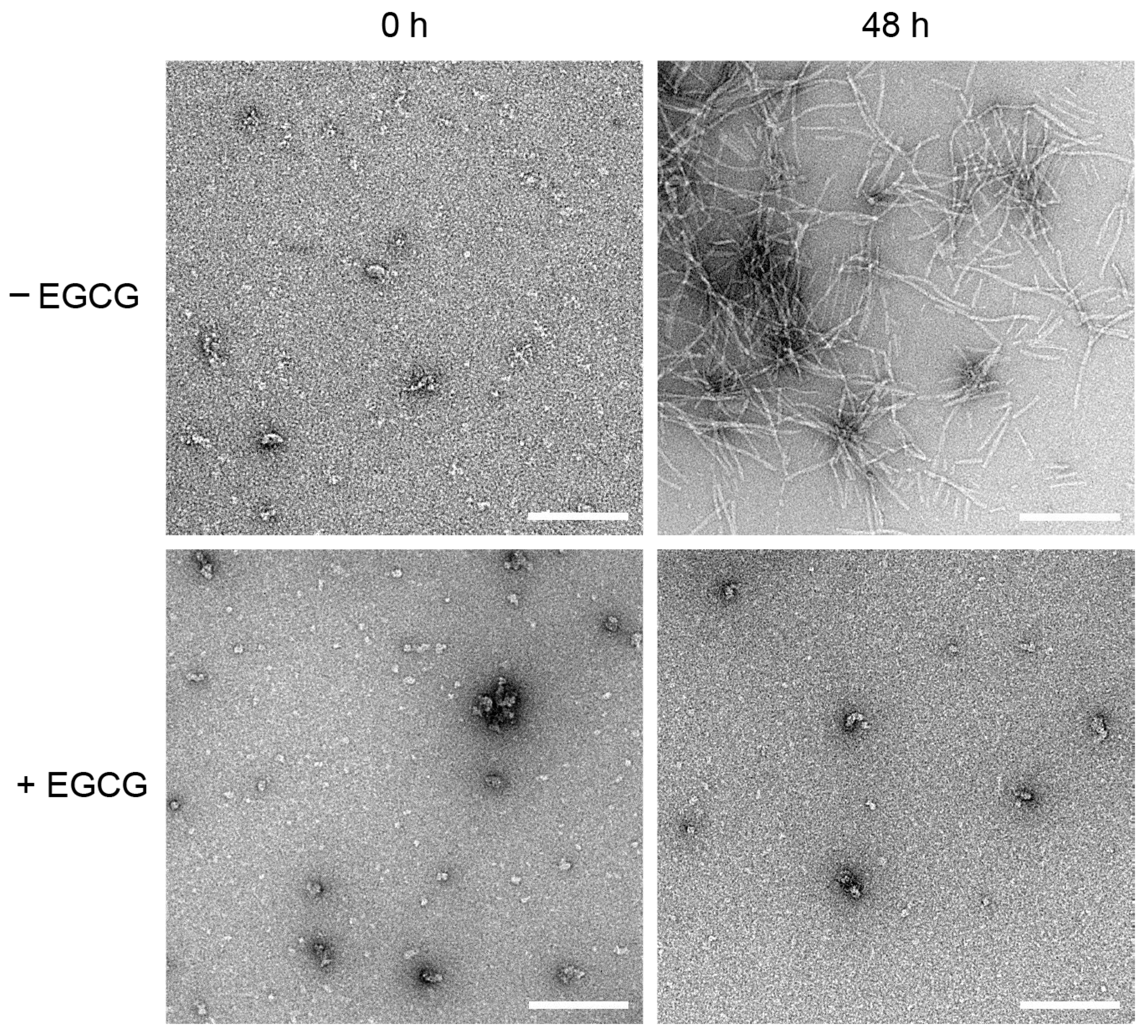
2.3. EGCG Prevents the Formation of MccE492 Amyloid Fibers, as Revealed by Negative-Staining Electron Microscopy
2.4. EGCG Interferes with MccE492 Secondary Structure Transitions to Amyloid-Prone Forms
3. Discussion
4. Materials and Methods
4.1. MccE492 Purification
4.2. MccE492 Reconstitution and Preparation for the Aggregation Assays
4.3. Aggregation Assays Followed by Thioflavin-T Fluorescence
4.4. Determination of Soluble MccE492 during the Aggregation Assay
4.5. SDS-PAGE and Immunoblot for MccE492 Detection
4.6. Negative Staining Electron Microscopy
4.7. Synchrotron Radiation Circular Dichroism (SRCD)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Eisenberg, D.S.; Sawaya, M.R. Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem. 2017, 86, 69–95. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem. 2017, 86, 27–68. [Google Scholar] [CrossRef]
- Fowler, D.M.; Koulov, A.V.; Balch, W.E.; Kelly, J.W. Functional Amyloid—From Bacteria to Humans. Trends Biochem. Sci. 2007, 32, 217–224. [Google Scholar] [CrossRef]
- Van Gerven, N.; Van der Verren, S.E.; Reiter, D.M.; Remaut, H. The Role of Functional Amyloids in Bacterial Virulence. J. Mol. Biol. 2018, 430, 3657–3684. [Google Scholar] [CrossRef] [PubMed]
- Levkovich, S.A.; Gazit, E.; Laor Bar-Yosef, D. Two Decades of Studying Functional Amyloids in Microorganisms. Trends Microbiol. 2021, 29, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.R.; Robinson, L.S.; Pinkner, J.S.; Roth, R.; Heuser, J.; Hammar, M.; Normark, S.; Hultgren, S.J. Role of Escherichia Coli Curli Operons in Directing Amyloid Fiber Formation. Science 2002, 295, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Romero, D.; Aguilar, C.; Losick, R.; Kolter, R. Amyloid Fibers Provide Structural Integrity to Bacillus Subtilis Biofilms. Proc. Natl. Acad. Sci. USA 2010, 107, 2230–2234. [Google Scholar] [CrossRef] [PubMed]
- Dueholm, M.S.; Petersen, S.V.; Sønderkær, M.; Larsen, P.; Christiansen, G.; Hein, K.L.; Enghild, J.J.; Nielsen, J.L.; Nielsen, K.L.; Nielsen, P.H.; et al. Functional Amyloid in Pseudomonas. Mol. Microbiol. 2010, 77, 1009–1020. [Google Scholar] [CrossRef]
- Schwartz, K.; Syed, A.K.; Stephenson, R.E.; Rickard, A.H.; Boles, B.R. Functional Amyloids Composed of Phenol Soluble Modulins Stabilize Staphylococcus Aureus Biofilms. PLoS Pathog. 2012, 8, e1002744. [Google Scholar] [CrossRef]
- Lagos, R.; Tello, M.; Mercado, G.; Garcia, V.; Monasterio, O. Antibacterial and Antitumorigenic Properties of Microcin E492, a Pore- Forming Bacteriocin. Curr. Pharm. Biotechnol. 2009, 10, 74–85. [Google Scholar] [CrossRef]
- Marcoleta, A.E.; Berríos-Pastén, C.; Nuñez, G.; Monasterio, O.; Lagos, R. Klebsiella Pneumoniae Asparagine TDNAs Are Integration Hotspots for Different Genomic Islands Encoding Microcin E492 Production Determinants and Other Putative Virulence Factors Present in Hypervirulent Strains. Front. Microbiol. 2016, 7, 849. [Google Scholar] [CrossRef] [PubMed]
- Lam, M.M.C.; Wyres, K.L.; Duchêne, S.; Wick, R.R.; Judd, L.M.; Gan, Y.H.; Hoh, C.H.; Archuleta, S.; Molton, J.S.; Kalimuddin, S.; et al. Population Genomics of Hypervirulent Klebsiella Pneumoniae Clonal-Group 23 Reveals Early Emergence and Rapid Global Dissemination. Nat. Commun. 2018, 9, 2703. [Google Scholar] [CrossRef] [PubMed]
- Bieler, S.; Estrada, L.; Lagos, R.; Baeza, M.; Castilla, J.; Soto, C. Amyloid Formation Modulates the Biological Activity of a Bacterial Protein. J. Biol. Chem. 2005, 280, 26880–26885. [Google Scholar] [CrossRef]
- Arranz, R.; Mercado, G.; Martín-Benito, J.; Giraldo, R.; Monasterio, O.; Lagos, R.; Valpuesta, J.M. Structural Characterization of Microcin E492 Amyloid Formation: Identification of the Precursors. J. Struct. Biol. 2012, 178, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, P.; Marcoleta, A.; Lobos-Ruiz, P.; Arranz, R.; Valpuesta, J.M.; Monasterio, O.; Lagos, R. Identification of Key Amino Acid Residues Modulating Intracellular and In Vitro Microcin E492 Amyloid Formation. Front. Microbiol. 2016, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Marcoleta, A.; Wien, F.; Arluison, V.; Lagos, R.; Giraldo, R. Bacterial Amyloids. eLS 2019, 1–9. [Google Scholar] [CrossRef]
- Shahnawaz, M.; Park, K.W.; Mukherjee, A.; Diaz-Espinoza, R.; Soto, C. Prion-like Characteristics of the Bacterial Protein Microcin E492. Sci. Rep. 2017, 7, 45720. [Google Scholar] [CrossRef]
- Marcoleta, A.; Marín, M.; Mercado, G.; Valpuesta, J.M.; Monasterio, O.; Lagos, R. Microcin E492 Amyloid Formation Is Retarded by Posttranslational Modification. J. Bacteriol. 2013, 195, 3995–4004. [Google Scholar] [CrossRef]
- Porat, Y.; Abramowitz, A.; Gazit, E. Inhibition of Amyloid Fibril Formation by Polyphenols: Structural Similarity and Aromatic Interactions as a Common Inhibition Mechanism. Chem. Biol. Drug Des. 2006, 67, 27–37. [Google Scholar] [CrossRef]
- Ehrnhoefer, D.E.; Duennwald, M.; Markovic, P.; Wacker, J.L.; Engemann, S.; Roark, M.; Legleiter, J.; Marsh, J.L.; Thompson, L.M.; Lindquist, S.; et al. Green Tea (-)-Epigallocatechin-Gallate Modulates Early Events in Huntingtin Misfolding and Reduces Toxicity in Huntington’s Disease Models. Hum. Mol. Genet. 2006, 15, 2743–2751. [Google Scholar] [CrossRef]
- Bieschke, J.; Russ, J.; Friedrich, R.P.; Ehrnhoefer, D.E.; Wobst, H.; Neugebauer, K.; Wanker, E.E. EGCG Remodels Mature α-Synuclein and Amyloid-β Fibrils and Reduces Cellular Toxicity. Proc. Natl. Acad. Sci. USA 2010, 107, 7710–7715. [Google Scholar] [CrossRef] [PubMed]
- Marinelli, P.; Pallares, I.; Navarro, S.; Ventura, S. Dissecting the Contribution of Staphylococcus Aureus α-Phenol-Soluble Modulins to Biofilm Amyloid Structure. Sci. Rep. 2016, 6, 34552. [Google Scholar] [CrossRef] [PubMed]
- Stenvang, M.; Dueholm, M.S.; Vad, B.S.; Seviour, T.; Zeng, G.; Geifman-Shochat, S.; Søndergaard, M.T.; Christiansen, G.; Meyer, R.L.; Kjelleberg, S.; et al. Epigallocatechin Gallate Remodels Overexpressed Functional Amyloids in Pseudomonas Aeruginosa and Increases Biofilm Susceptibility to Antibiotic Treatment. J. Biol. Chem. 2016, 291, 26540–26553. [Google Scholar] [CrossRef] [PubMed]
- Serra, D.O.; Mika, F.; Richter, A.M.; Hengge, R. The Green Tea Polyphenol EGCG Inhibits E. Coli Biofilm Formation by Impairing Amyloid Curli Fibre Assembly and Downregulating the Biofilm Regulator CsgD via the ΣE-Dependent SRNA RybB. Mol. Microbiol. 2016, 101, 136–151. [Google Scholar] [CrossRef] [PubMed]
- Partouche, D.; Turbant, F.; Hamoui, O.E.; Campidelli, C.; Bombled, M.; Trépout, S.; Wien, F.; Arluison, V. Epigallocatechin Gallate Remodelling of Hfq Amyloid-like Region Affects Escherichia Coli Survival. Pathogens 2018, 7, 95. [Google Scholar] [CrossRef]
- Busi, F.; Turbant, F.; Waeytens, J.; Hamoui, O.E.; Wien, F.; Arluison, V. Evaluation of Amyloid Inhibitor Efficiency to Block Bacterial Survival. Methods Mol. Biol. 2022, 2538, 145–163. [Google Scholar] [CrossRef]
- Micsonai, A.; Wien, F.; Bulyáki, É.; Kun, J.; Moussong, É.; Lee, Y.H.; Goto, Y.; Réfrégiers, M.; Kardos, J. BeStSel: A Web Server for Accurate Protein Secondary Structure Prediction and Fold Recognition from the Circular Dichroism Spectra. Nucleic Acids Res. 2018, 46, W315–W322. [Google Scholar] [CrossRef]
- Lorenzo, V. De Isolation and Characterization of Microcin E 492 from Klebsiella Pneumoniae. Arch. Microbiol. 1984, 139, 72–75. [Google Scholar] [CrossRef]
- Xu, Y.Q.; Yu, P.; Zhou, W. Combined Effect of PH and Temperature on the Stability and Antioxidant Capacity of Epigallocatechin Gallate (EGCG) in Aqueous System. J. Food Eng. 2019, 250, 46–54. [Google Scholar] [CrossRef]
- Waeytens, J.; Turbant, F.; Arluison, V.; Raussens, V.; Wien, F. Analysis of Bacterial Amyloid Interaction with Lipidic Membrane by Orientated Circular Dichroism and Infrared Spectroscopies. In Bacterial Amyloids: Methods and Protocols; Arluison, V., Wien, F., Marcoleta, A., Eds.; Springer: New York, NY, USA, 2022; pp. 217–234. ISBN 978-1-0716-2529-3. [Google Scholar]
- Giuliani, A.; Jamme, F.; Rouam, V.; Wien, F.; Giorgetta, J.L.; Lagarde, B.; Chubar, O.; Bac, S.; Yao, I.; Rey, S.; et al. DISCO: A Low-Energy Multipurpose Beamline at Synchrotron SOLEIL. J. Synchrotron Radiat. 2009, 16, 835–841. [Google Scholar] [CrossRef]
- Huang, K.; Zeng, J.; Liu, X.; Jiang, T.; Wang, J. Structure of the Mannose Phosphotransferase System (Man-PTS) Complexed with Microcin E492, a Pore-Forming Bacteriocin. Cell Discov. 2021, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Meade, R.M.; Fairlie, D.P.; Mason, J.M. Alpha-Synuclein Structure and Parkinson’s Disease. Mol. Neurodegener. 2019, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Singh, Y.; Sharpe, P.C.; Hoang, H.N.; Lucke, A.J.; McDowall, A.W.; Bottomley, S.P.; Fairlie, D.P. Amyloid Formation from an α-Helix Peptide Bundle Is Seeded by 3 10-Helix Aggregates. Chemistry 2011, 17, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Ferreira, R.; Taylor, N.M.I.; Mona, D.; Ringler, P.; Lauer, M.E.; Riek, R.; Britschgi, M.; Stahlberg, H. Cryo-EM Structure of Alpha-Synuclein Fibrils. eLife 2018, 7, e36402. [Google Scholar] [CrossRef]
- Mokra, D.; Joskova, M.; Mokry, J. Therapeutic Effects of Green Tea Polyphenol (−)-Epigallocatechin-3-Gallate (EGCG) in Relation to Molecular Pathways Controlling Inflammation, Oxidative Stress, and Apoptosis. Int. J. Mol. Sci. 2023, 24, 340. [Google Scholar] [CrossRef]
- Almatrood, S.A.; Almatroudi, A.; Khan, A.A.; Alhumaydh, F.A.; Alsahl, M.A.; Rahmani, A.H. Potential Therapeutic Targets of Epigallocatechin Gallate (EGCG), the Most Abundant Catechin in Green Tea, and Its Role in the Therapy of Various Types of Cancer. Molecules 2020, 25, 3146. [Google Scholar] [CrossRef]
- Romano, A.; Martel, F. The Role of EGCG in Breast Cancer Prevention and Therapy. Mini-Rev. Med. Chem. 2020, 21, 883–898. [Google Scholar] [CrossRef]
- Naujokat, C.; McKee, D.L. The “Big Five” Phytochemicals Targeting Cancer Stem Cells: Curcumin, EGCG, Sulforaphane, Resveratrol and Genistein. Curr. Med. Chem. 2020, 28, 4321–4342. [Google Scholar] [CrossRef]
- Payne, A.; Nahashon, S.; Taka, E.; Adinew, G.M.; Soliman, K.F.A. Epigallocatechin-3-Gallate (EGCG): New Therapeutic Perspectives for Neuroprotection, Aging, and Neuroinflammation for the Modern Age. Biomolecules 2022, 12, 371. [Google Scholar] [CrossRef]
- Palhano, F.L.; Lee, J.; Grimster, N.P.; Kelly, J.W. Toward the Molecular Mechanism(s) by Which EGCG Treatment Remodels Mature Amyloid Fibrils. J. Am. Chem. Soc. 2013, 135, 7503–7510. [Google Scholar] [CrossRef]
- Sønderby, T.V.; Louros, N.N.; Khodaparast, L.; Khodaparast, L.; Madsen, D.J.; Olsen, W.P.; Moonen, N.; Nagaraj, M.; Sereikaite, V.; Strømgaard, K.; et al. Sequence-Targeted Peptides Divert Functional Bacterial Amyloid Towards Destabilized Aggregates and Reduce Biofilm Formation. J. Mol. Biol. 2023, 435, 168039. [Google Scholar] [CrossRef] [PubMed]
- Arita-Morioka, K.I.; Yamanaka, K.; Mizunoe, Y.; Tanaka, Y.; Ogura, T.; Sugimoto, S. Inhibitory Effects of Myricetin Derivatives on Curli-Dependent Biofilm Formation in Escherichia Coli. Sci. Rep. 2018, 8, 8452. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, P.; Marín, J.; Lagos, R.; Marcoleta, A. Identification of Aggregation-Prone and Gatekeeper Residues in Bacterial Amyloids Using Site-Directed Mutagenesis and Flow Cytometry. Methods Mol. Biol. 2022, 2538, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H.; von Jagow, G. Tricine-Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis for the Separation of Proteins in the Range from 1 to 100 KDa. Anal. Biochem. 1987, 166, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Miles, A.J.; Wallace, B.A. CDtoolX, a Downloadable Software Package for Processing and Analyses of Circular Dichroism Spectroscopic Data. Protein Sci. 2018, 27, 1717–1722. [Google Scholar] [CrossRef]
- Anthis, N.J.; Clore, G.M. Sequence-Specific Determination of Protein and Peptide Concentrations by Absorbance at 205 Nm. Protein Sci. 2013, 22, 851–858. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguilera, P.; Berríos-Pastén, C.; Veloso, M.; Gálvez-Silva, M.; Turbant, F.; Lagos, R.; Wien, F.; Arluison, V.; Marcoleta, A.E. The Green Tea Polyphenol Epigallocatechin-Gallate (EGCG) Interferes with Microcin E492 Amyloid Formation. Molecules 2023, 28, 7262. https://doi.org/10.3390/molecules28217262
Aguilera P, Berríos-Pastén C, Veloso M, Gálvez-Silva M, Turbant F, Lagos R, Wien F, Arluison V, Marcoleta AE. The Green Tea Polyphenol Epigallocatechin-Gallate (EGCG) Interferes with Microcin E492 Amyloid Formation. Molecules. 2023; 28(21):7262. https://doi.org/10.3390/molecules28217262
Chicago/Turabian StyleAguilera, Paulina, Camilo Berríos-Pastén, Marcelo Veloso, Matías Gálvez-Silva, Florian Turbant, Rosalba Lagos, Frank Wien, Veronique Arluison, and Andrés E. Marcoleta. 2023. "The Green Tea Polyphenol Epigallocatechin-Gallate (EGCG) Interferes with Microcin E492 Amyloid Formation" Molecules 28, no. 21: 7262. https://doi.org/10.3390/molecules28217262
APA StyleAguilera, P., Berríos-Pastén, C., Veloso, M., Gálvez-Silva, M., Turbant, F., Lagos, R., Wien, F., Arluison, V., & Marcoleta, A. E. (2023). The Green Tea Polyphenol Epigallocatechin-Gallate (EGCG) Interferes with Microcin E492 Amyloid Formation. Molecules, 28(21), 7262. https://doi.org/10.3390/molecules28217262





