A Review on Five and Six-Membered Heterocyclic Compounds Targeting the Penicillin-Binding Protein 2 (PBP2A) of Methicillin-Resistant Staphylococcus aureus (MRSA)
Abstract
1. Introduction
History and Emergence of MRSA
2. PBPs, Types of PBP, Their Location, and Contribution in Transpeptidation Reaction
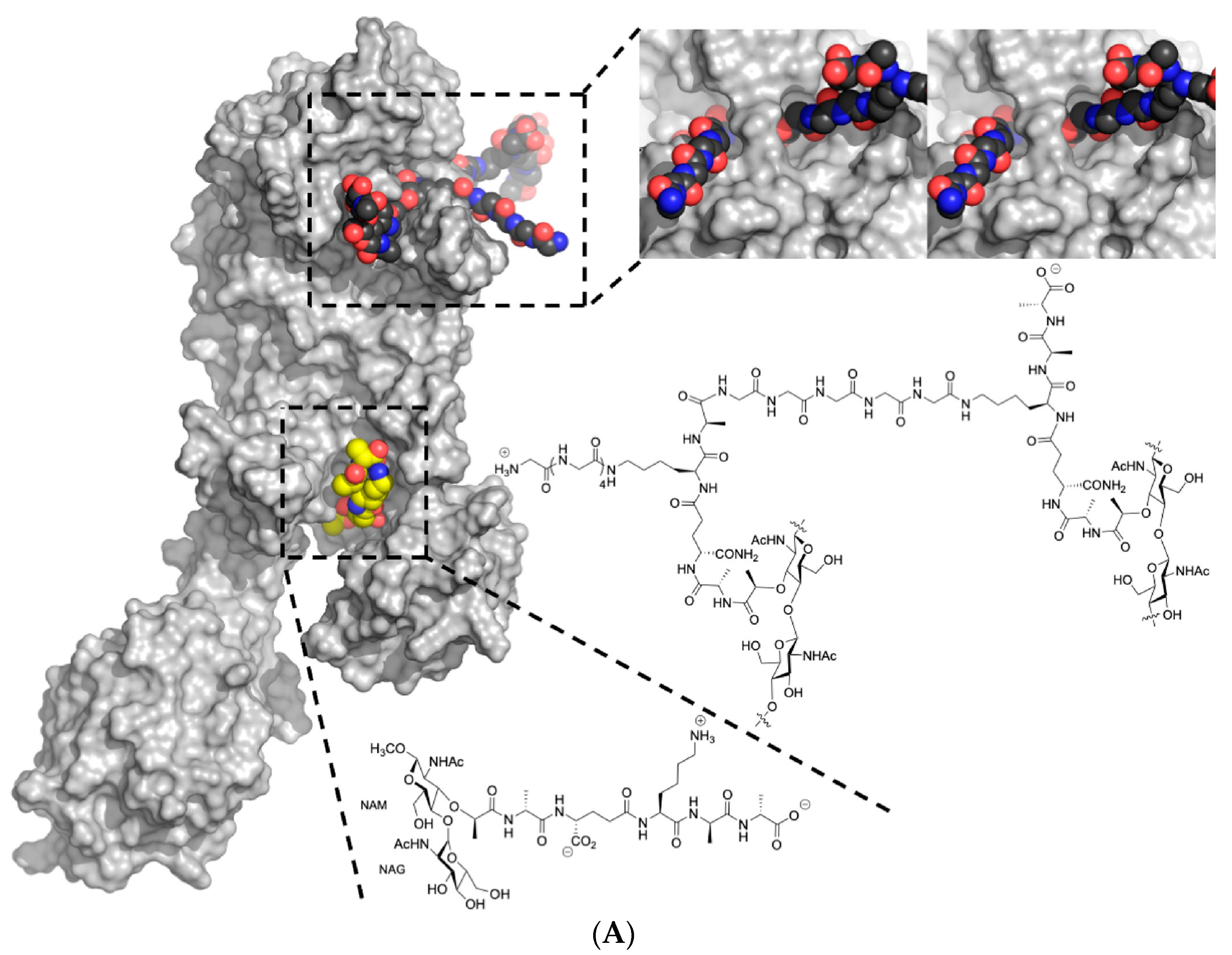
2.1. PBP2a
- PBP2a relies on PBP2′s transglycosylase activity.
- The correct length and sequence of stem peptides are necessary for PBP2a to function effectively.
- The presence of the pentaglycine cross-bridge is essential for PBP2a’s activity [71].
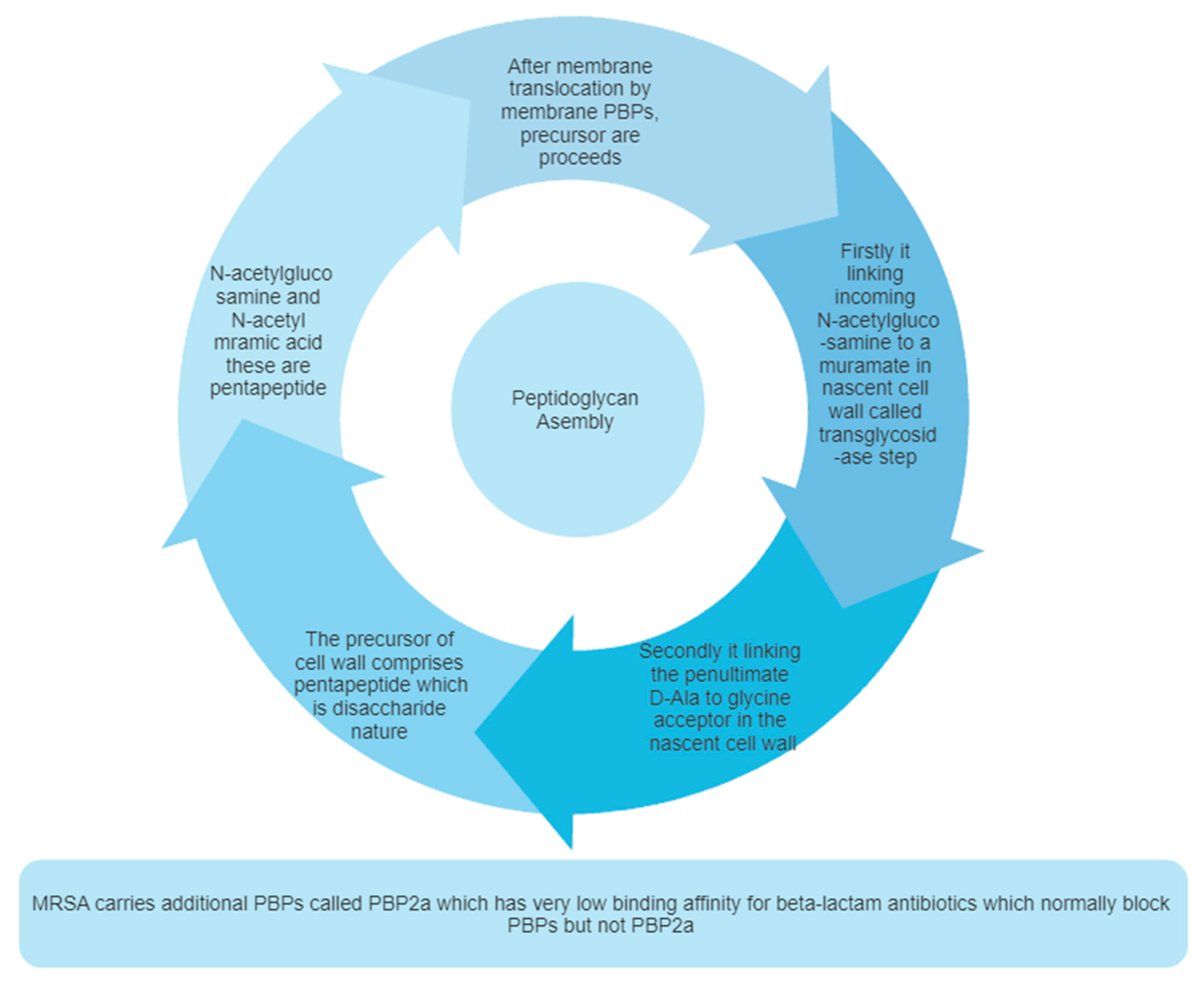
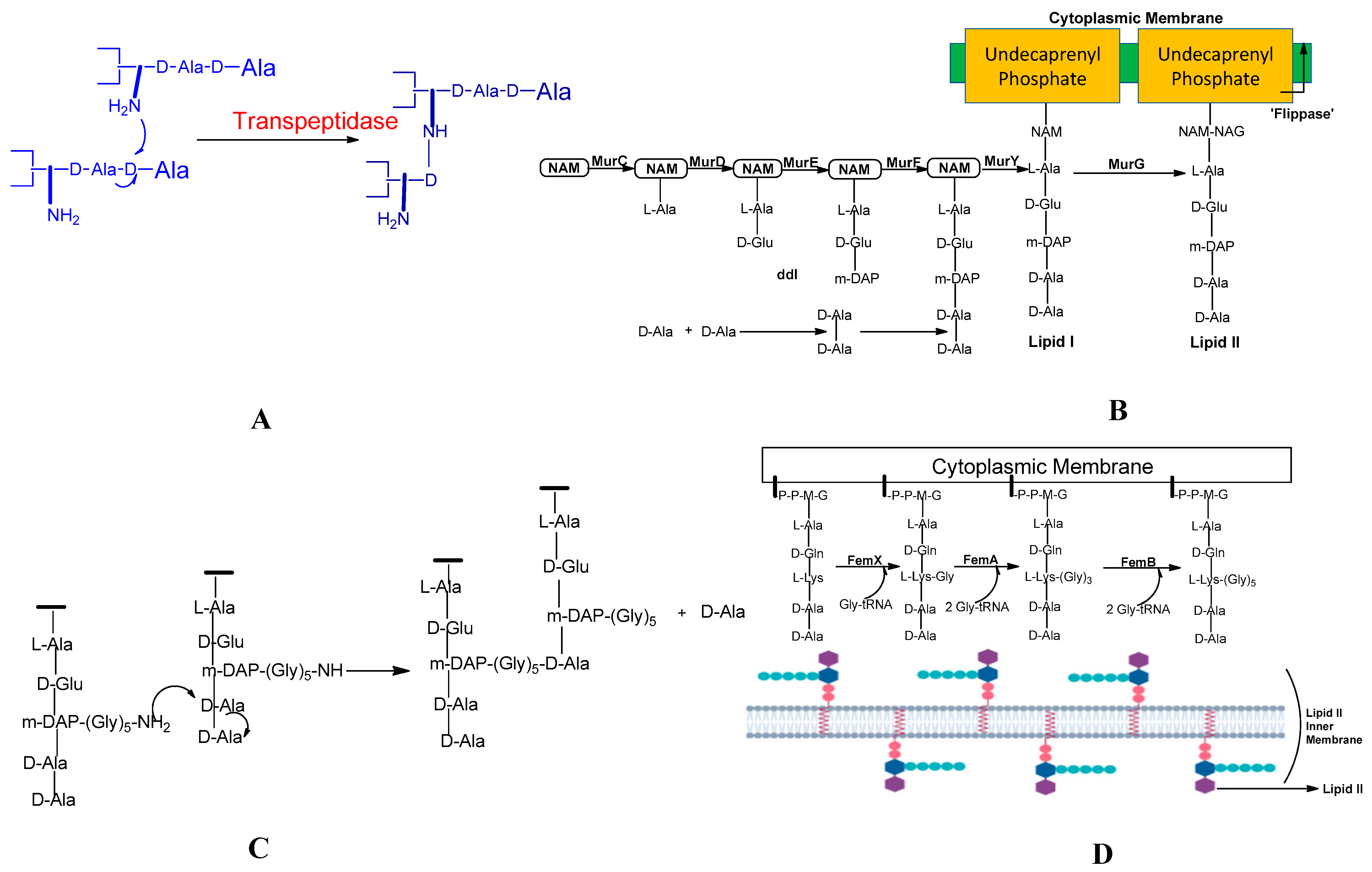
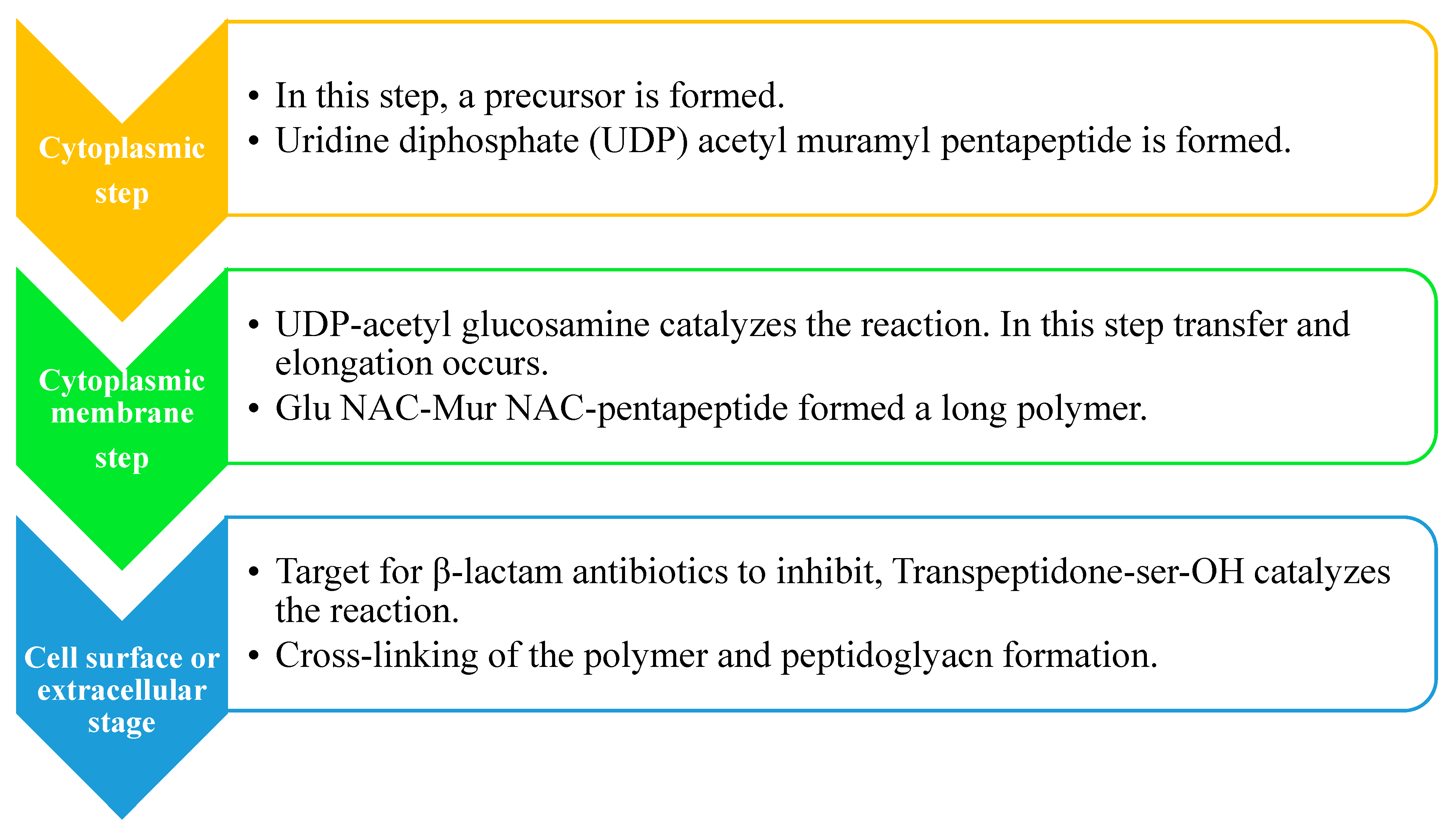
2.2. Peptidoglycan Biosynthesis
| Sr. No. | Name of Mur Enzymes | Role of Enzyme in the Biosynthesis of Peptidoglycan |
|---|---|---|
| 1 | MurA | The enzyme MurA is responsible for catalysing the intracellular stage of peptidoglycan biosynthesis, which is the first stage in peptidoglycan biosynthesis. MurA enzyme brings such a reaction simply by transferring enol pyruvate. This enol pyruvate is produced by conversion from phosphoenolpyruvate to UDP-N-acetylglucosamine. |
| 2 | MurB | Reduce UDP-GlcNAc-enol pyruvate to UDP-acetylmuramic acid. |
| 3 | The ATP-dependent enzyme, Mur ligases enzymes, Enzymes from MurC to MurF | These enzymes are responsible for linking five amino acid residues. These enzymes relate to the UDP-MurNAc and thus help to form the UDP-MurNAc-pentapeptide. |
| 4 | MraY | Produce lipid I. |
| 5 | MurG | MurG adds GlcNAc moiety to lipid I resulting in the formation of lipid II. |
| 6 | MurJ | To export lipid II to the outer leaflet, MurJ switches between inward and outward conformations. Lipid II reversal is involved by MurJ [80,81]. |
2.3. Pentaglycine Bridge and Transpeptidation Reaction in Cell Wall
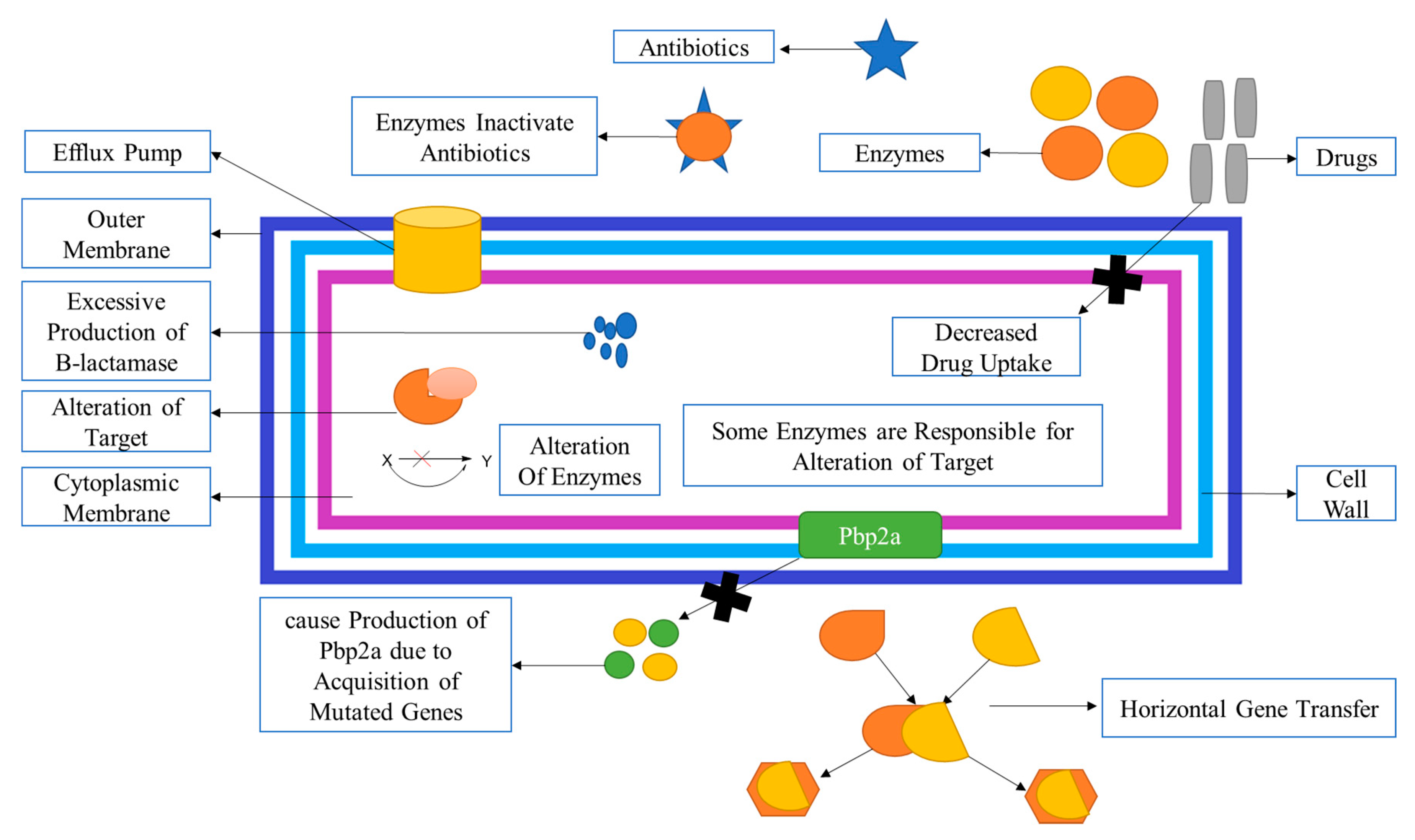
2.4. Mechanism of Methicillin Resistances in S. aureus
2.5. Staphylococcus Cassette Chromosome
- (a)
- mecA Gene Complex: This complex contains the mecA gene, which encodes the penicillin-binding protein PBP2 with reduced affinity for beta-lactam antibiotics.
- (b)
| Sr. No. | Name of Genes | Role in MRSA |
|---|---|---|
| s1 | Mec (mecA, mecR1-mecIs) | The close relationship between two regulatory elements, one is mec and the second one is bla. BlaR2 or a close relative may be participating in mecA activation here too. |
| 2 | Chromosomal gene | “FEM” stands for factors essential for methicillin resistance. Its inactivation reduces the methicillin-resistant and aux (auxiliary) factors usually present in staphylococcal aureus. The majority of elements are engaged in cell wall biosynthesis and some turnover of bacteria. |
| 3 | The PBP2a operon in normal S. aureus contributes to resistance | The mecA is responsible for the production of PBP2a, which helps in the biosynthesis of bacteria’s cell walls. However, its role in the resistance of S. aureus is still unclear. |
| 4 | Fmt | Reduce methicillin resistance. |
| 5 | femX, femAB | When reduction of the length of the glycine side chain occurs, it is because of the inactivation of the femAB factor. The inactivation and reduced length lead to impairment in the growth of peptidoglycan and the turnover of cell-wall being reduced. All these reactions are responsible for lowering peptidoglycan cross-linking and hypersusceptibility to all β-lactam antibiotics and other antibiotics, which leads to the decrease in methicillin resistance. |
| 6 | glnRA (femC) (Mutated gene) | The precursors of muropeptide use an amino acid which is a non-amidated D-glutamate amino acid, and it takes part in the stem peptide of the transpeptidation reactions less readily, which leads to a reduction of methicillin resistance |
| 7 | glmM (femD, femR) | When the rate of precursor formation for peptidoglycan is reduced, it reduces methicillin resistance. It increases teicoplanin susceptibility and thus decreases methicillin resistance in MRSA. |
| 8 | Lytic enzymes (Hydrolysis Enzyme) | Murein, which is present in Staphylococcus aureus, causes hydrolysis. It is then needed for the growth of peptidoglycan, which is a precious cell wall content. This plays one of the leading roles in peptidoglycan growth. |
| 9 | murE (femF) | In the formation of peptidoglycan precursors, MurE necessitates the presence of this factor. This is because the abnormal precursors of peptidoglycan are present, which causes a decline in methicillin resistance; it is also possible that there may be abnormal shortening precursors involved in the peptidoglycan biosynthesis. It is still unclear. |
| 10 | Llm gene | The llm gene encodes lipophilic membrane proteins, affecting methicillin resistance levels and necessarily causing bacterial cell lysis rate reduction. Its functions, however, are still unclear. |
| 11 | Global regulators: sar, agr, sigB | The global regulators, like sar and agr, control cell density-dependent synthesis of cell wall factors that are cell wall-associated, and extracellular virulence factors. In heterogeneous MRSA, this global regulator appears to have just a slight effect on methicillin resistance in MRSA. |
| 12 | ctaA | Reduces the resistance to methicillin in MRSA |
| 13 | Blaz | The blaZ gene has been mutated in Staphylococcus aureus, and it is responsible for the breakdown of the β-lactam ring of mainly penicillin antibiotics. |
| 14 | blaI | blaI is a repressor protein that is responsible for preventing the transcription of mutated genes like blaZ or mecA. |
| 15 | blaR1 | blaR1 is a transmembrane protein that signals blaZ or mecA transcription. |
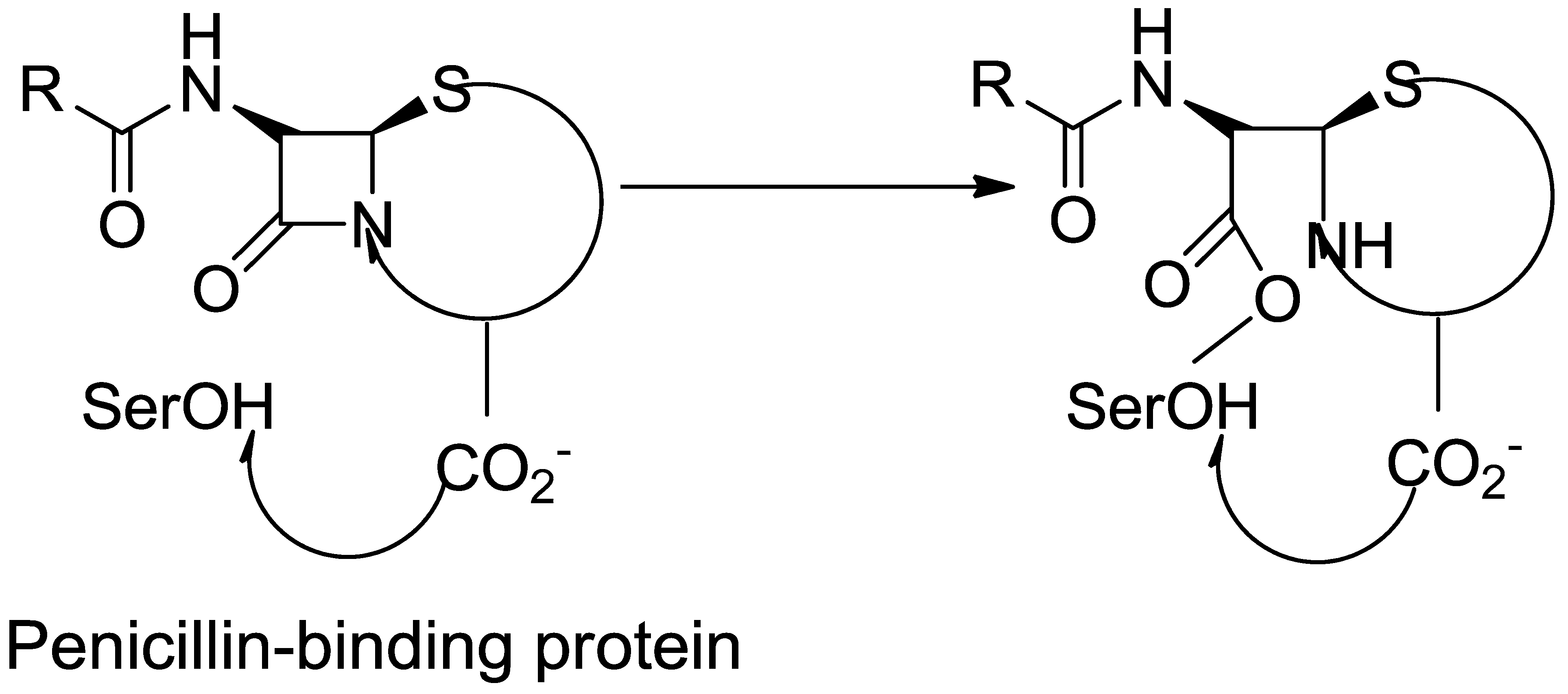
3. Structure Basis for PBP2A
3.1. β-Lactam as Substrate Analogues
3.2. About Crystal Structures for PBP2a
4. Development of PBP2a Inhibitors
4.1. Penicillin
4.2. Cephalosporins


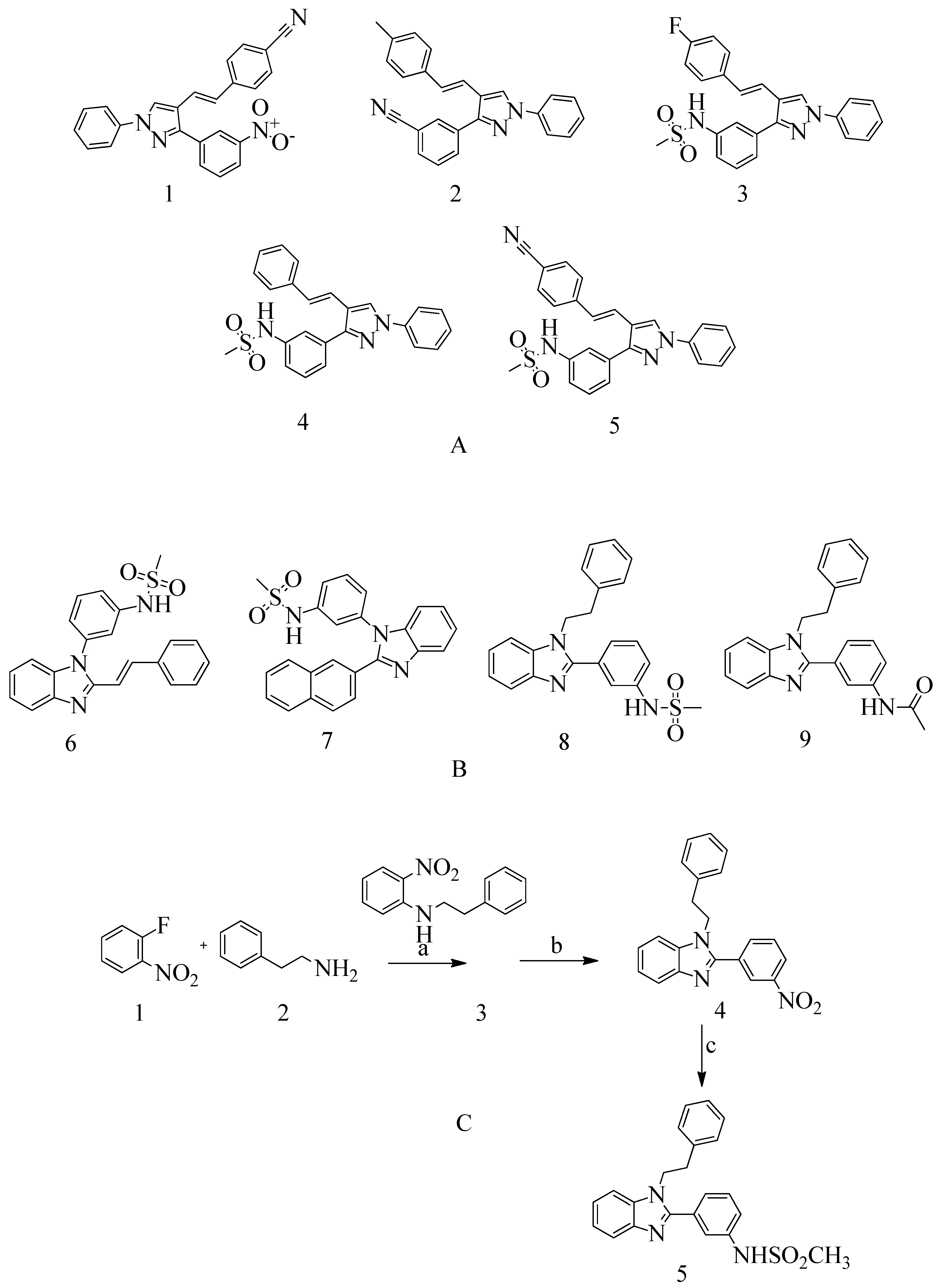
4.3. Novel Pyrazole-Benzimidazole Based Derivatives
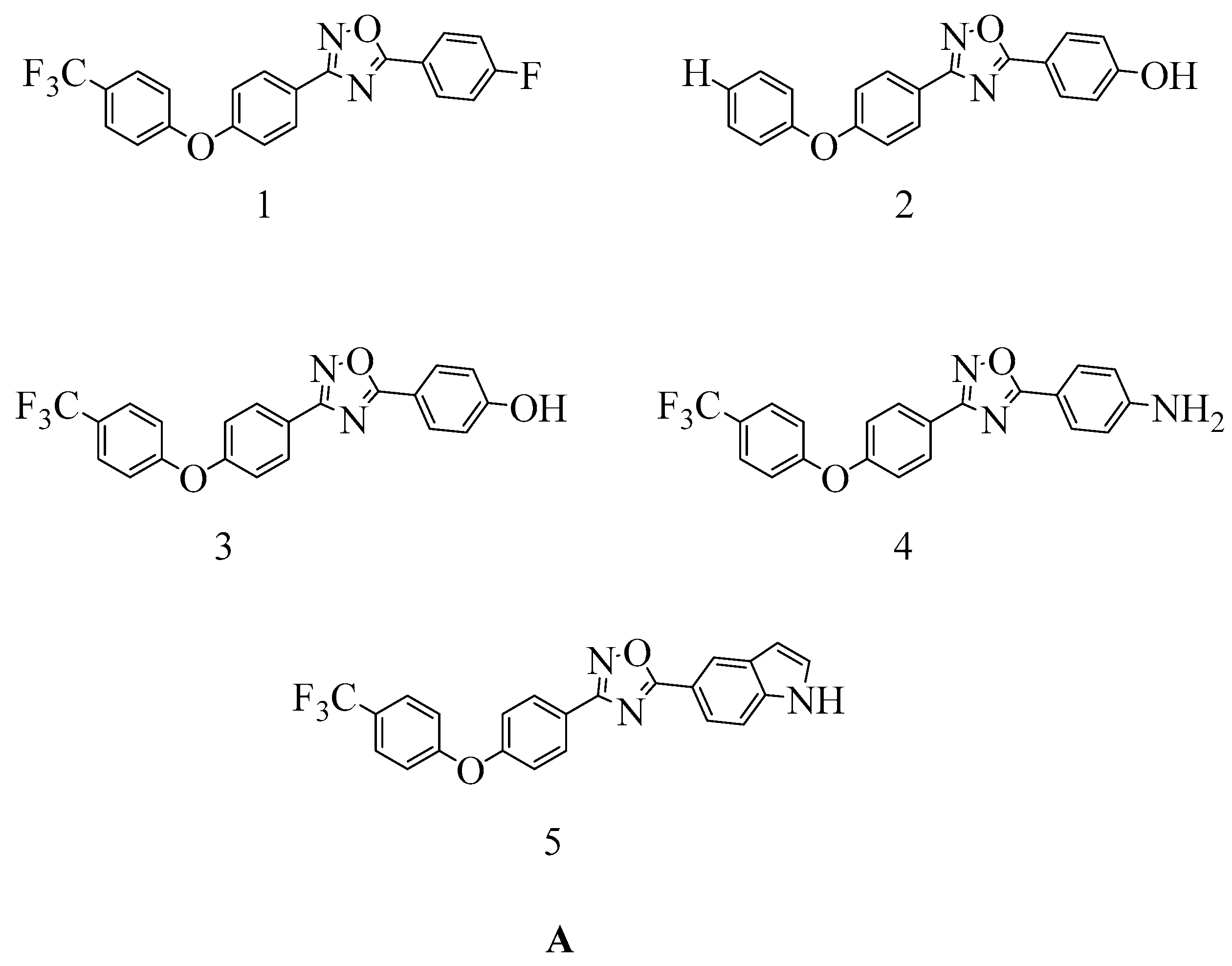
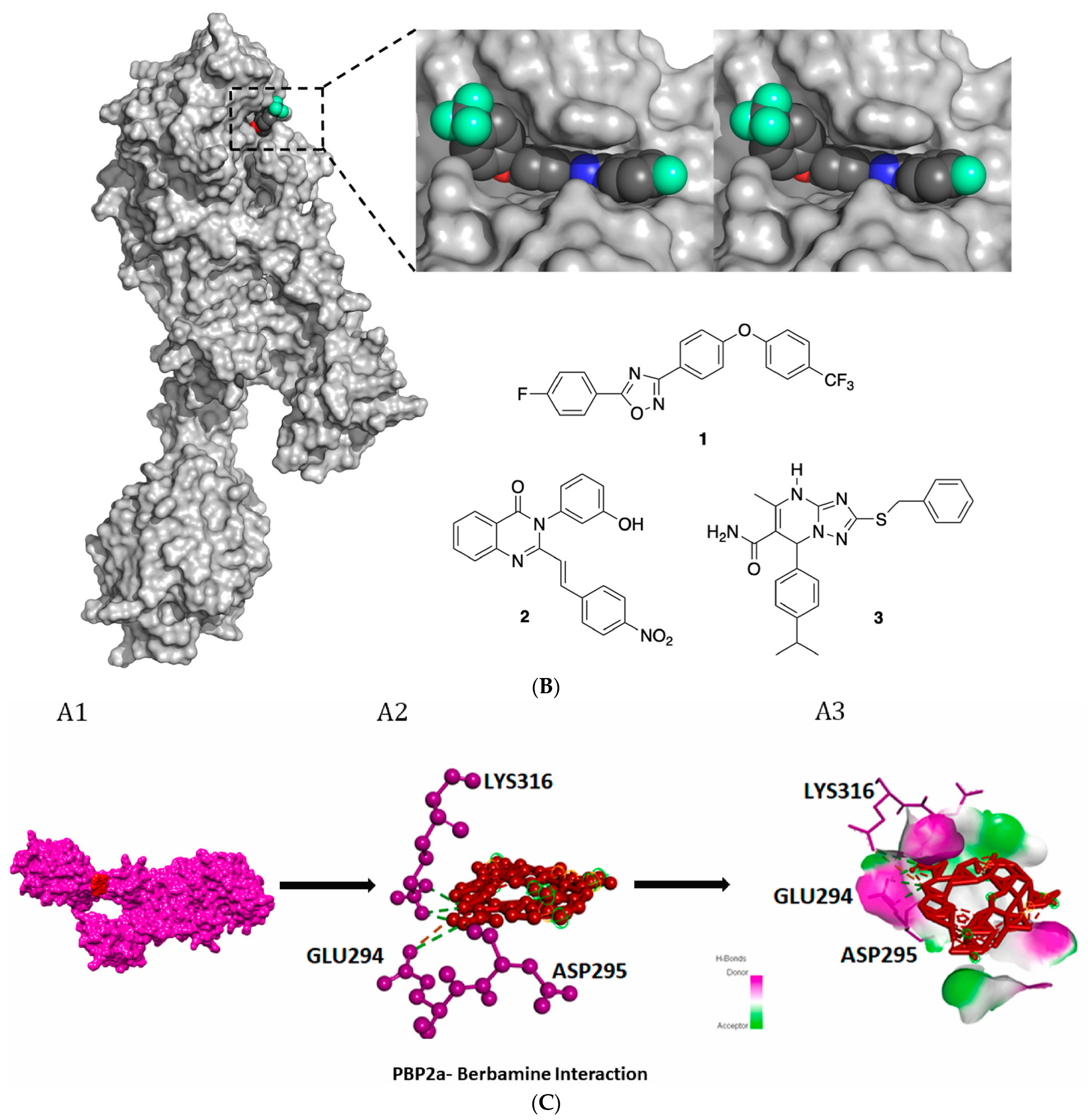
4.4. Novel 1,2,4-Oxadiazole-Containing Derivatives
4.5. Non-β-Lactam Allosteric Inhibitors
4.6. 4-(3H)-Quinazolinones
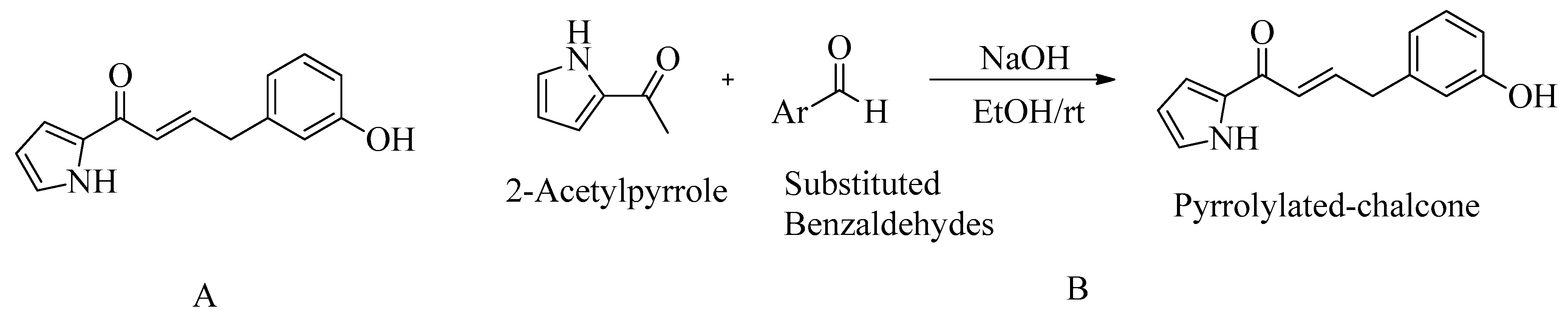
4.7. Pyrrolylated-Chalcones
4.8. Bis-2-oxoazetidinyl Macrocycle (β-Lactams with 1,3-Bridges)
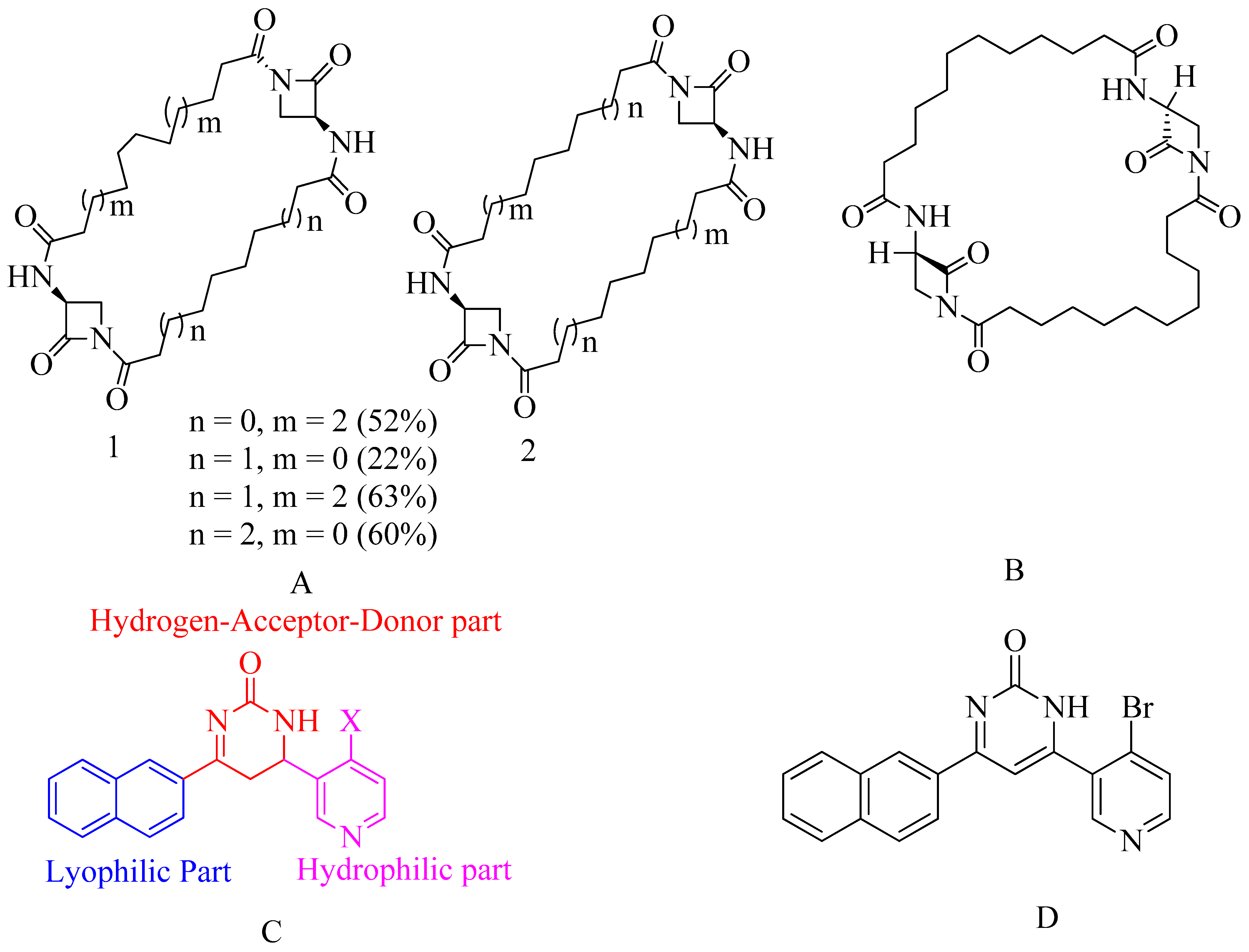
4.9. Macrocycle-Embedded β-Lactams as Novel Inhibitors
4.10. Pyridine-Coupled Pyrimidinones/Pyrimidinthiones
- Naphthyl ring–Lyophilic unit
- Pyridine ring–Hydrophilic unit
- Pyrimidinone ring–Hydrogen-Donor-Acceptor (HAD) unit.
4.11. Novel Naphthalimide Corbelled Aminothiazoximes
4.12. Non-Covalent Inhibitors
4.13. Investigational β-Lactam Antibiotics
4.14. Carbapenem
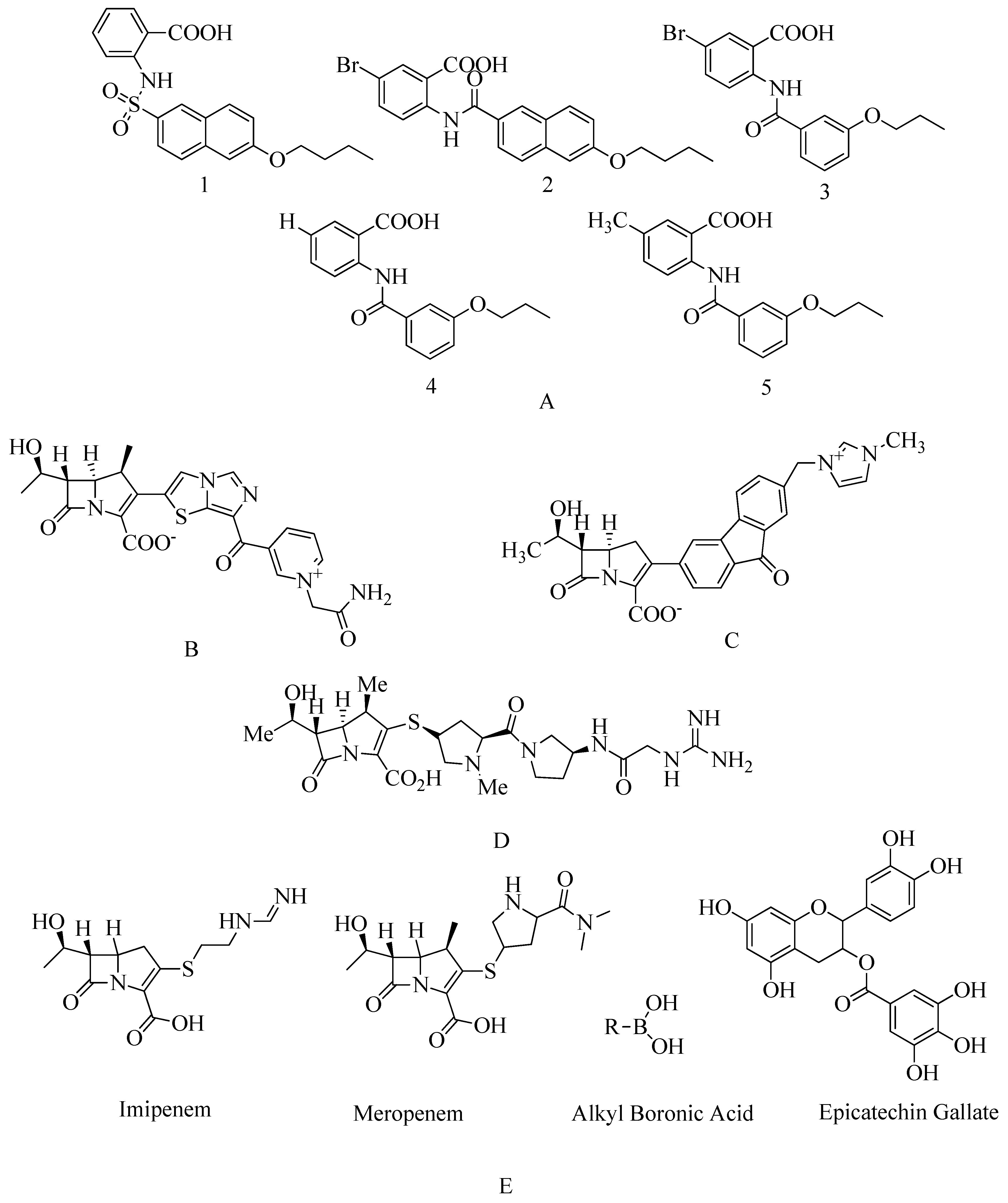
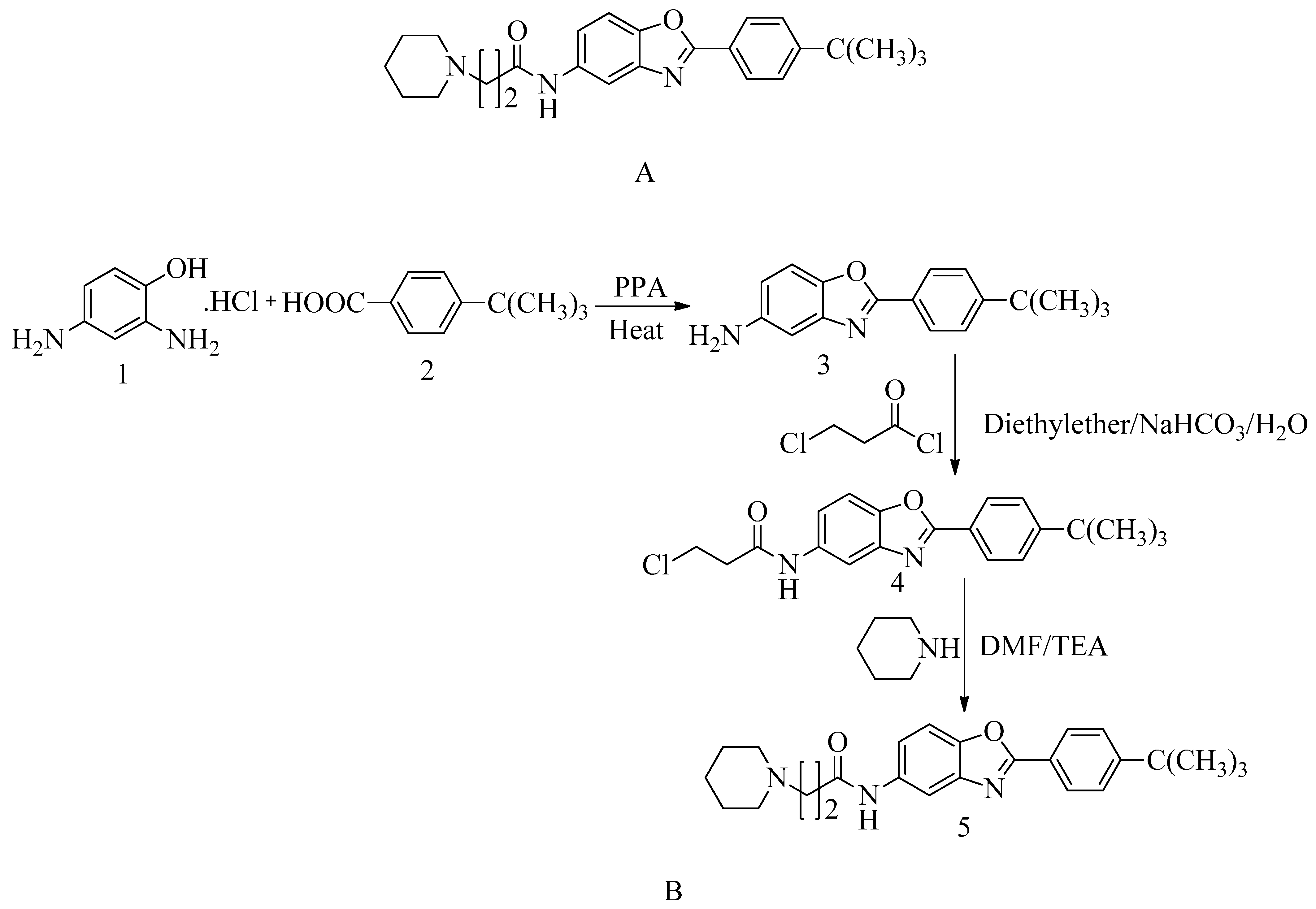
4.15. Novel Benzoxazole Derivatives
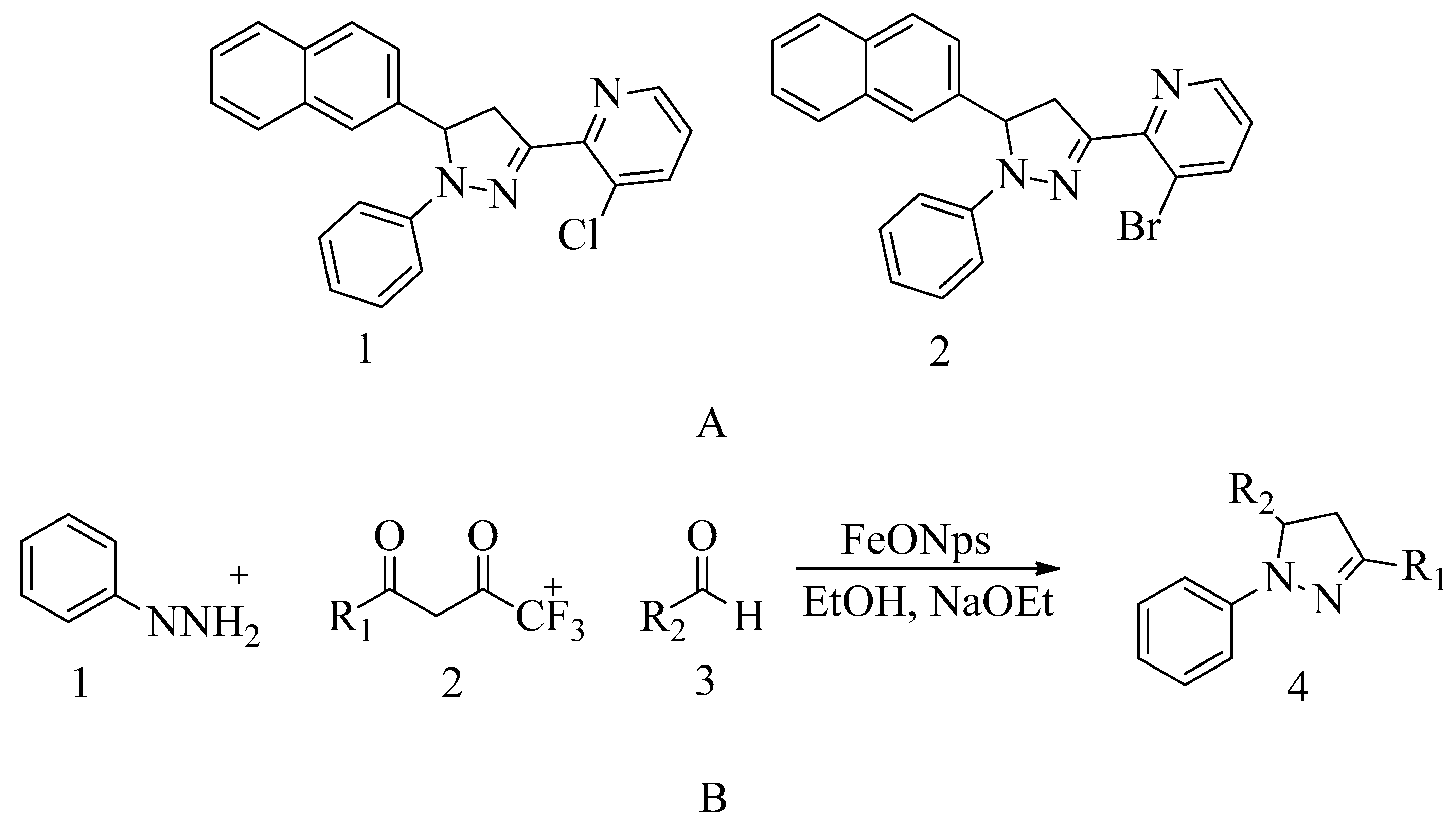
4.16. Pyrazolylpyridine Analogues
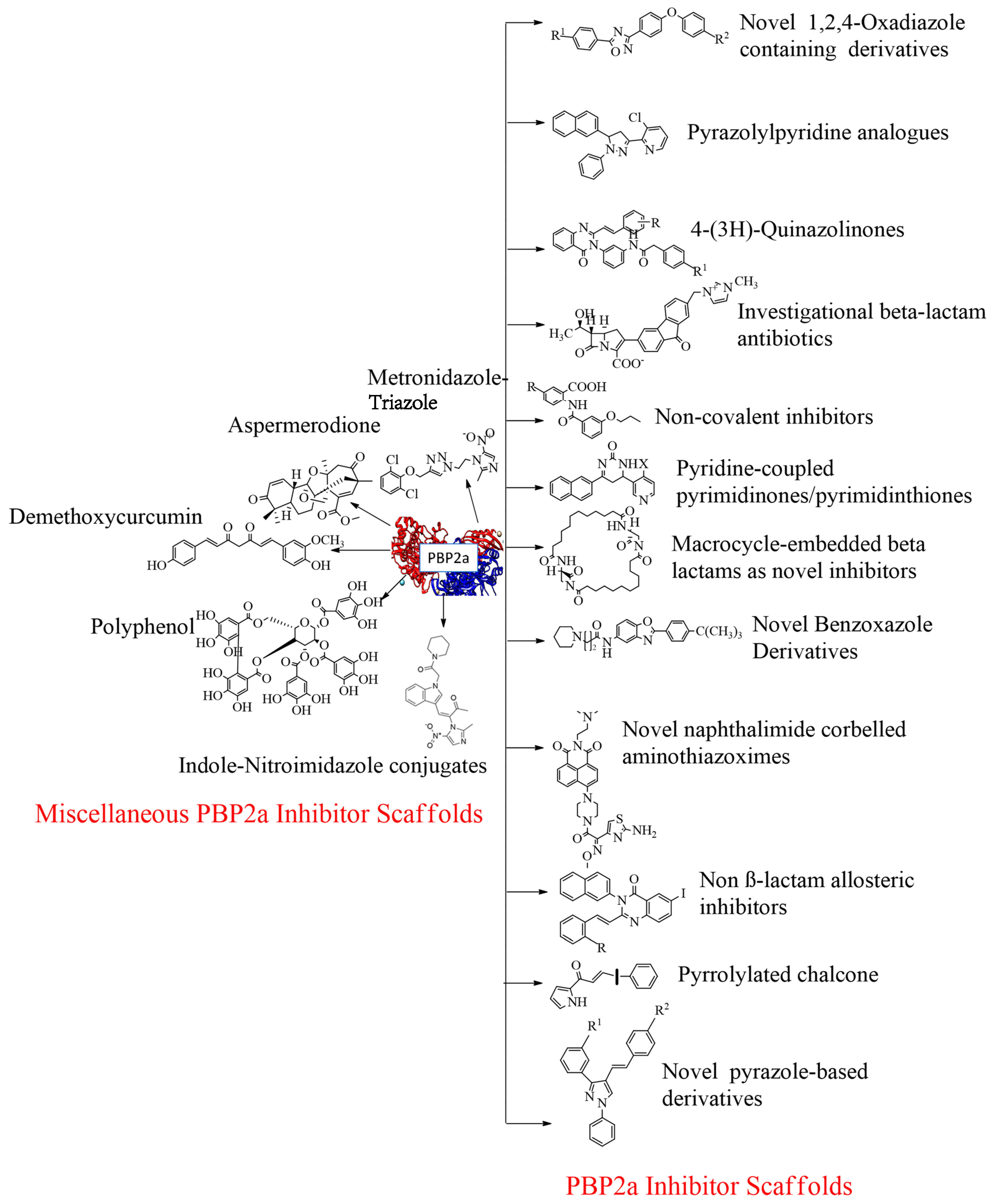
4.17. Miscellaneous PBP2a Inhibitors
4.17.1. Flavonoids
4.17.2. Demethoxycurcumin
4.17.3. Quercetin 3-o-Rutinoside
4.17.4. Ursolic Acid 3-O-α-l-arabinopyranoside with Oxacillin
4.17.5. Thioridazine
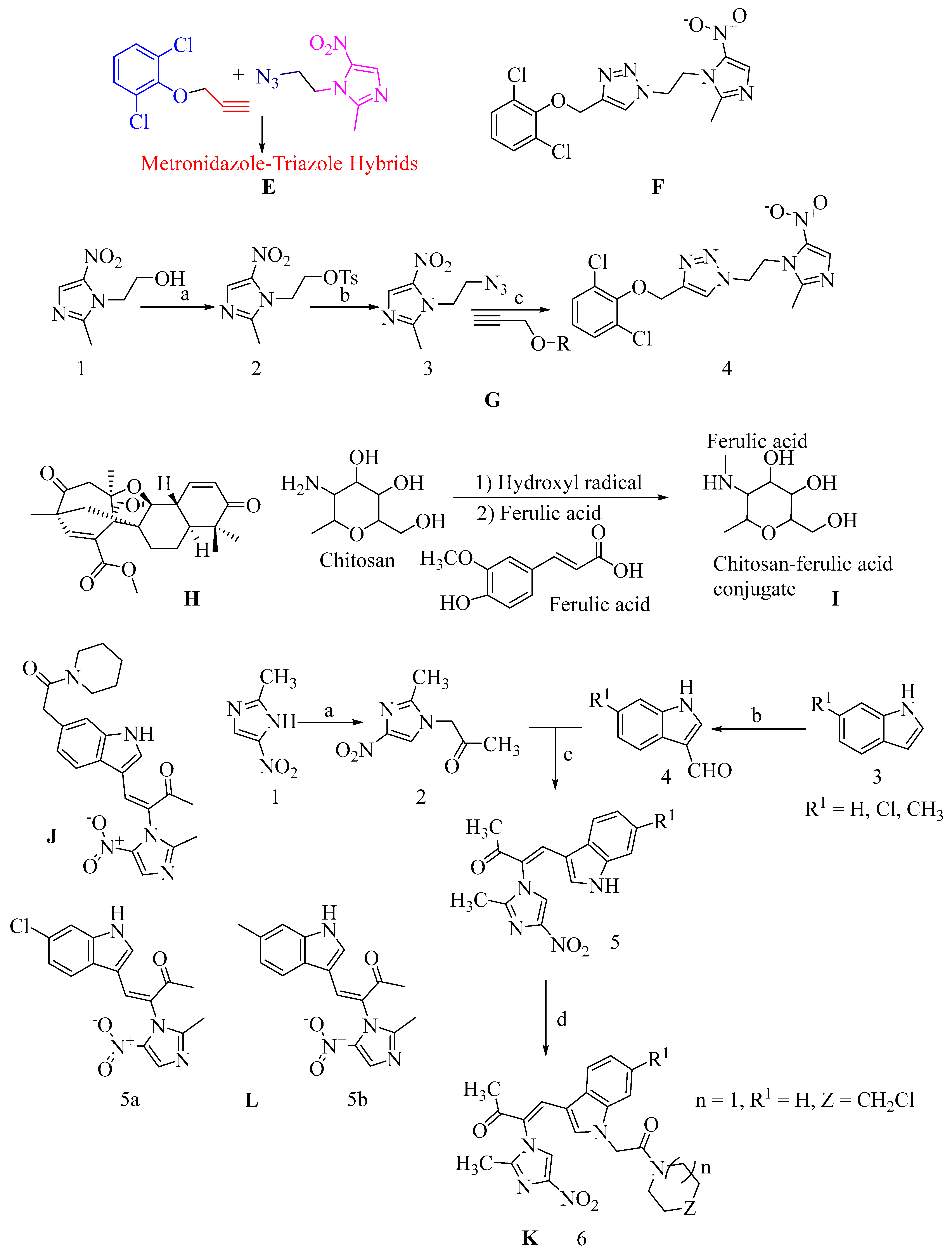
4.17.6. Metronidazole-Triazole Hybrids
4.17.7. Aspermerodione
4.17.8. Chitosan-Ferulic Acid Conjugates
4.17.9. Indole-Nitroimidazole Conjugates
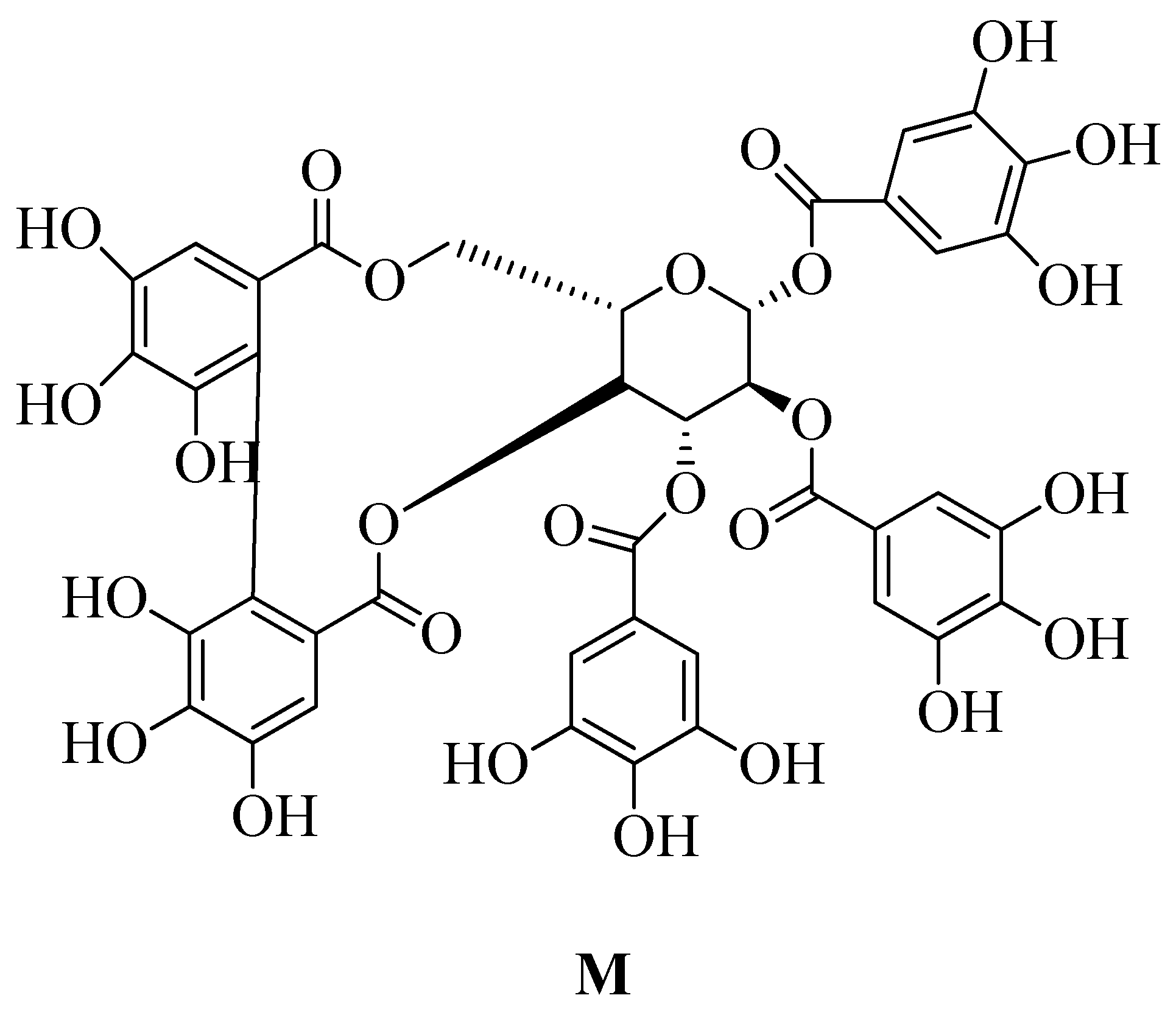
4.17.10. Polyphenol

5. Screening Technologies
5.1. High throughput Screening
5.2. Computer-Aided Screening
5.3. In Silico Docking Screening
5.4. Multiple Virtual Screening Techniques
5.5. X-ray Crystallography
5.6. Miscellaneous Method of Screening
5.6.1. Inverse Virtual Screening
5.6.2. Reverse Virtual Screening
5.6.3. Microtiter Plate-Based Assay
5.6.4. Ab Initio Methods of Screening
6. Conclusions
7. The Outlook
- The generation of peptidoglycan precursors is a critical process in methicillin-resistant Staphylococcus aureus (MRSA).
- Discovering novel medicines against MRSA is often better and more convenient using a ligand-based method.
- The main challenges in creating novel anti-MRSA agents include: (a) How to leverage existing knowledge to develop anti-virulence medications. (b) How to integrate these findings into the evolving field of bacterial interactions within the human microbiome.
- Utilizing knowledge from various articles is crucial for discovering new antibiotics to combat MRSA infections. As antibiotic resistance poses a significant threat to public health, it is essential to improve existing antibiotic classes or explore entirely new classes of antibiotics (Figure 25) [183].
Author Contributions
Funding
Conflicts of Interest
References
- Tansey, E.M. Superbugs and Superdrugs: A History of MRSA; The Transcript of a Witness Seminar Held by the Wellcome Trust Centre for the History of Medicine at UCL: London, UK, 2008; Volume 32. [Google Scholar]
- Campbell, J.; Singh, A.K.; Santa, J.P.M., Jr.; Kim, Y.; Swoboda, J.G.; Mylonakis, E.; Wilkinson, B.J.; Walker, S. Synthetic Lethal Compound Combinations Reveal a Fundamental Connection between Wall Teichoic Acid and Peptidoglycan Biosyntheses in Staphylococcus aureus. ACS Chem. Biol. 2012, 6, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Guo, Y.; Song, G.; Sun, M.; Wang, J.; Wang, Y.; Wang, Y. Prevalence and Therapies of Antibiotic-Resistance in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2020, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, J.F.; de Azevedo, Í.M.; Rocha, K.B.F.; Medeiros, A.C.; da Silva Carriço, A. Oxacillin Magnetically Targeted for the Treatment of Methicillin-Resistant S. aureus infection in rats. Acta Cir. Bras. 2017, 32, 46–55. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lowy, F.D.; Lowy, F.D. Antimicrobial Resistance: The Example of Staphylococcus aureus Find the Latest Version: Antimicrobial Resistance: The Example of Staphylococcus aureus. J. Clin. Investig. 2003, 111, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Malachowa, N.; DeLeo, F.R. Mobile genetic elements of Staphylococcus aureus. Cell. Mol. Life Sci. 2010, 67, 3057–3071. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.J.; Foster, T.J. The Staphylococcus aureus “Superbug”. J. Clin. Investig. 2004, 114, 1693–1696. [Google Scholar] [CrossRef] [PubMed]
- Saraiva, F.B.; de Araújo, A.C.C.; de Araújo, A.É.V.; Senna, J.P.M. Monoclonal Antibody Anti-PBP2a Protects Mice against MRSA (Methicillin-Resistant Staphylococcus aureus) Infections. PLoS ONE 2019, 14, e0225752. [Google Scholar] [CrossRef]
- Deng, J.Z. Methicillin/per-6-(4-Methoxylbenzyl)-Amino-6-Deoxy-β-Cyclodextrin 1:1 Complex and Its Potentiation In Vitro against Methicillin-Resistant Staphylococcus aureus. J. Antibiot. 2013, 66, 517–521. [Google Scholar] [CrossRef]
- Vernozy-rozand, C.; Mazuy, C.; Lasne, Y.; Fiedler, F.; Etienne, J. Staphylococcus fleurettii sp. Nov., Isolated from Goat’s Milk Cheeses. Int. J. Syst. Evol. Microbiol. 2000, 50, 1521–1527. [Google Scholar] [CrossRef]
- Berndsen, R.; Cunningham, T.; Kaelin, L.; Callender, M.; Boldog, W.D.; Viering, B.; King, A.; Labban, N.; Pollock, J.A.; Miller, H.B.; et al. Identi Fi Cation and Evaluation of Brominated Carbazoles as a Novel Antibiotic Adjuvant Sca Ff Old in MRSA. ACS Med. Chem. Lett. 2022, 13, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Dai, M.; Wang, Y.; Huang, L.; Yuan, Z. Key Genetic Elements and Regulation Systems in Methicillin-Resistant Staphylococcus aureus. Future Microbiol. 2012, 7, 1315–1329. [Google Scholar] [CrossRef] [PubMed]
- Barber, M.; Rozwadowska-Dowzenko, M. Infection by penicillin-resistant staphylococci. Lancet 1948, 23, 641–644. [Google Scholar] [CrossRef] [PubMed]
- Harkins, C.P.; Pichon, B.; Doumith, M.; Parkhill, J.; Westh, H.; Tomasz, A.; de Lencastre, H.; Bentley, S.D.; Kearns, A.M.; Holden, M.T.G. Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice. Genome Biol. 2017, 20, 130. [Google Scholar] [CrossRef] [PubMed]
- Lyont, B.R.; Skurray, R.O.N. Antimicrobial Resistance of Staphylococcus aureus: Genetic Basis. Microbiol. Rev. 1987, 51, 88–134. [Google Scholar]
- Vizdi, Z.; AmraHuki, M. Staphylococcus aureus (MRSA) as a cause of nosocomial wound. Bosn J Basic Med. Sci. 2010, 10, 32–37. [Google Scholar]
- Grundmann, H.; Aires-de-sousa, M.; Boyce, J.; Tiemersma, E. Emergence and Resurgence of Meticillin-Resistant Staphylococcus aureus as a Public-Health Threat. Lancet 2006, 368, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Liscano, Y.; Amú, A.; González, A.; Oñate-Garzón, J.; Salamanca, C.H. In Silico Characterization of the Interaction between the Pbp2a “Decoy” Protein of Resistant Staphylococcus aureus and the Monomeric Units of Eudragit e-100 and Poly(Maleic Acid-Alt-Octadecene) Polymers. Polymers 2021, 13, 2320. [Google Scholar] [CrossRef]
- Moellering, R.C., Jr. MRSA: The First Half Century. J. Antimicrob. Chemother. 2012, 67, 4–11. [Google Scholar] [CrossRef]
- Oehler, R.L. MRSA: Historical Perspective. 2000, 1961. (Table 1). Available online: http://www.antimicrobe.org/h04c.files/history/MRSA-Hx%20Perspective.asp (accessed on 4 October 2023).
- Andrade, M.M.; Luiz, W.B.; Oliveira, S.; Amorim, J.H. The History of Methicillin-Resistant Staphylococcus aureus in Brazil. Can. J. Infect. Dis. Med. Microbiol. 2020, 2020, 1721936. [Google Scholar] [CrossRef]
- Haamann, F.; Dulon, M.; Nienhaus, A. MRSA as an Occupational Disease: A Case Series. Int. Arch. Occup. Environ. Health 2011, 84, 259–266. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Diep, B.A.; Carleton, H.A.; Chang, R.F.; Sensabaugh, G.F. Roles of 34 Virulence Genes in the Evolution of Hospital- and Community-Associated Strains of Methicillin-Resistant Staphylococcus aureus. J. Infect. Dis. 2006, 94110, 1495–1503. [Google Scholar] [CrossRef]
- Enright, M.C.; Robinson, D.A.; Randle, G.; Feil, E.J.; Grundmann, H.; Spratt, B.G. The Evolutionary History of Methicillin-Resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 2002, 99, 7687–7692. [Google Scholar] [CrossRef] [PubMed]
- Stefani, S.; Varaldo, P.E. Epidemiology of Methicillin-Resistant Staphylococci in Europe. Clin. Microbiol. Infect. 2003, 9, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Janout, V.; Matouskova, I. Current knowledge of methicillin-resistant Staphylococcus aureus and community-associated methicillin-resistant Staphylococcus aureus. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2008, 152, 191–202. [Google Scholar]
- Majed, H.; Johnston, T.; Kelso, C.; Monachino, E.; Jergic, S.; Dixon, N.E.; Mylonakis, E.; Kelso, M.J. Structure-Activity Relationships of Pyrazole-4-Carbodithioates as Antibacterials against Methicillin-Resistant Staphylococcus aureus. Bioorg. Med. Chem. Lett. 2018, 28, 3526–3528. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Ippolito, G.; Leone, S.; Lauria, F.N.; Nicastri, E.; Wenzel, R.P. International Journal of Infectious Diseases Methicillin-Resistant Staphylococcus aureus: The Superbug. Int. J. Infect. Dis. 2010, 4, 7–11. [Google Scholar] [CrossRef]
- Pantosti, A.; Venditti, M. What is MRSA? Eur. Respir. J. 2009, 34, 1190–1196. [Google Scholar] [CrossRef]
- Wong, S.C.Y.; Tse, H.; Chen, J.H.K. Multidrug-Resistant Staphylococcus aureus, Colistin-Resistant Enterobacteriaceae Carrying the Mcr-1 Gene among Patients in Hong Kong. Emerg. Infect. Dis. 2016, 22, 2015–2016. [Google Scholar] [CrossRef]
- Mehta, Y.; Hegde, A.; Pande, R.; Zirpe, K.G.; Gupta, V.; Ahdal, J.; Qamra, A.; Motlekar, S. Methicillin-Resistant Staphylococcus aureus in Intensive Care Unit Setting of India: A Review of Clinical Burden, Patterns of Prevalence, Preventive Measures, and Future Strategies. Indian J. Crit. Care Med. Peer-Rev. Off. Publ. Indian Soc. Crit. Care Med. 2020, 24, 55. [Google Scholar]
- Jindal, N.; Malhotra, R.; Grover, P.; Singh, S.; Bansal, R.; Kaur, S. Methicillin resistant Staphylococcus aureus (MRSA) in Malwa region of Punjab (North-West India). Indian J. Med. Res. 2016, 143, 371–372. [Google Scholar] [CrossRef] [PubMed]
- Ld, I.M.; David, M.G.; Esposito, S.; Garau, J.; Lina, G.; Mazzei, T.; Peters, G. International Journal of Antimicrobial Agents New Insights into Meticillin-Resistant Staphylococcus aureus (MRSA) Pathogenesis, Treatment and Resistance. Int. J. Antimicrob. Agents 2012, 39, 96–104. [Google Scholar] [CrossRef]
- Baba, K.; Ishihara, K.; Ozawa, M.; Tamura, Y.; Asai, T. International Journal of Antimicrobial Agents Isolation of Meticillin-Resistant Staphylococcus aureus (MRSA) from Swine in Japan. Int. J. Antimicrob. Agents 2010, 36, 352–354. [Google Scholar] [CrossRef] [PubMed]
- Outbreak, H.W.; Toleman, M.S.; Reuter, S.; Coll, F.; Harrison, E.M.; Peacock, S.J. Local Persistence of Novel MRSA Lineage after Community-Acquired Clostridium Difficile Infection. Emerg. Infect. Dis. 2016, 22, 2011–2012. [Google Scholar]
- Huang, Y.; Chen, C. International Journal of Antimicrobial Agents Community-Associated Meticillin-Resistant Staphylococcus aureus in Children in Taiwan. Int. J. Antimicrob. Agents 2011, 38, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Peacock, S.J.; Paterson, G.K. Mechanisms of Methicillin Resistance in Staphylococcus aureus. Annu. Rev. Biochem. 2015, 84, 577–601. [Google Scholar] [CrossRef]
- Id, C.B.; Schwab, F.; Id, R.V. Infection Control Measures in Nosocomial MRSA Outbreaks—Results of a Systematic Analysis. PLoS ONE 2021, 16, e0249837. [Google Scholar] [CrossRef]
- Kumar, P. A Review on Quinoline Derivatives as Anti-Methicillin Resistant Staphylococcus aureus (MRSA) Agents. BMC Chem. 2020, 14, 1–14. [Google Scholar] [CrossRef]
- Jiang, W.; Li, B.; Zheng, X.; Liu, X.; Cen, Y.; Li, J.; Pan, X.; Cao, H.; Zheng, J.; Zhou, H. International Immunopharmacology Artesunate in Combination with Oxacillin Protect Sepsis Model Mice Challenged with Lethal Live Methicillin-Resistant Staphylococcus aureus (MRSA) via Its Inhibition on Proin Fl Ammatory Cytokines Release and Enhancement on Antibacterial Activity of Oxacillin. Int. Immunopharmacol. 2011, 11, 1065–1073. [Google Scholar] [CrossRef]
- Stryjewski, M.E.; Corey, G.R. Methicillin-Resistant Staphylococcus aureus: An Evolving Pathogen. Clin. Infect. Dis. 2014, 58 (Suppl. S1), 10–19. [Google Scholar] [CrossRef] [PubMed]
- Mrsa, C.S. Methicillin-Resistant Staphylococcus aureus Infections. Top HIV Med. 2008, 16, 151–155. [Google Scholar]
- O’Daniel, P.I.; Peng, Z.; Pi, H.; Testero, S.A.; Ding, D.; Spink, E.; Leemans, E.; Boudreau, M.A.; Yamaguchi, T.; Schroeder, V.A.; et al. Discovery of a New Class of Non-β-Lactam Inhibitors of Penicillin-Binding Proteins with Gram-Positive Antibacterial Activity. J. Am. Chem. Soc. 2014, 136, 3664–3672. [Google Scholar] [CrossRef] [PubMed]
- Sahare, P.; Moon, A. Penicillin Binding Proteins: An Insight Into Novel Antibacterial Drug Target. Int. J. Eng. Sci. Res. 2014, 5, 13–23. [Google Scholar]
- Goffin, C.; Ghuysen, J. Biochemistry and Comparative Genomics of SxxK Superfamily Acyltransferases Offer a Clue to the Mycobacterial Paradox: Presence of Penicillin-Susceptible Target Proteins versus Lack of Efficiency of Penicillin as Therapeutic Agent. Microbiol. Mol. Biol. Rev. 2002, 66, 702–738. [Google Scholar] [CrossRef] [PubMed]
- Sauvage, E.; Terrak, M.; Ayala, J.A.; Charlier, P. The Penicillin-Binding Proteins: Structure and Role in Peptidoglycan Biosynthesis. FEMS Microbiol. Rev. 2008, 32, 234–258. [Google Scholar] [CrossRef] [PubMed]
- EZervosen, A.; Sauvage, E.; Frère, J.; Charlier, P.; Luxen, A. Development of New Drugs for an Old Target—The Penicillin Binding Proteins. Molecules 2012, 17, 12478–12505. [Google Scholar] [CrossRef] [PubMed]
- Green, D.W. The Bacterial Cell Wall as a Source of Antibacterial Targets. Expert Opin. Ther. Targets 2002, 6, 1–20. [Google Scholar] [CrossRef]
- Resistance, B. MINIREVIEW Penicillin-Binding Proteins and Bacterial Resistance to Beta-Lactams. Antimicrob. Agents Chemother. 1993, 37, 2045–2053. [Google Scholar]
- Scheffers, D.; Pinho, M.G. Bacterial Cell Wall Synthesis: New Insights from Localization Studies. Microbiol. Mol. Biol. Rev. 2005, 69, 585–607. [Google Scholar] [CrossRef]
- Available online: https://orbi.uliege.be/bitstream/2268/73806/1/Frere_Joris_1985_CRC_299.pdf (accessed on 23 May 2023).
- Anderson, J.S.; Matsuhashi, M.; Haskin, M.A.; Strominger, J.L. Biosythesis of the Peptidoglycan of Bacterial Cell Walls. II. Phospholipid Carriers in the Reaction Sequence. J. Biol. Chem. 1967, 242, 3180–3190. [Google Scholar] [CrossRef] [PubMed]
- Rohs, P.D.A.; Buss, J.; Sim, S.I.; Squyres, G.R.; Srisuknimit, V.; Smith, M.; Cho, H.; Sjodt, M.; Kruse, A.C.; Garner, E.C.; et al. A Central Role for PBP2 in the Activation of Peptidoglycan Polymerization by the Bacterial Cell Elongation Machinery. PLoS Genet. 2018, 14, e1007726. [Google Scholar] [CrossRef]
- Contreras-martel, C.; Vernet, T. Penicillin-Binding Proteins and b-Lactam Resistance. FEMS Microbiol. Rev. 2008, 32, 361–385. [Google Scholar] [CrossRef]
- Dave, K.; Palzkill, T.; Pratt, R.F. Neutral β-Lactams Inactivate High Molecular Mass Penicillin-Binding Proteins of Class B1, Including PBP2a of MRSA. ACS Med. Chem. Lett. 2014, 5, 154–157. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arbeloa, A.; Hugonnet, J.; Sentilhes, A.; Josseaume, N.; Dubost, L.; Monsempes, C.; Blanot, D.; Brouard, J.; Arthur, M. Synthesis of Mosaic Peptidoglycan Cross-Bridges by Hybrid Peptidoglycan Assembly Pathways in Gram-Positive Bacteria. J. Biol. Chem. 2004, 279, 41546–41556. [Google Scholar] [CrossRef] [PubMed]
- Lovering, A.L.; Safadi, S.S.; Strynadka, N.C.J. Structural Perspective of Peptidoglycan Biosynthesis and Assembly. Annu. Rev. Biochem. 2012, 81, 451–478. [Google Scholar] [CrossRef] [PubMed]
- Pinho, M.G.; Errington, J. Recruitment of Penicillin-Binding Protein PBP2 to the Division Site of Staphylococcus aureus is Dependent on Its Transpeptidation Substrates. Mol. Microbiol. 2005, 55, 799–807. [Google Scholar] [CrossRef]
- Wu, H.W.; De Lencastre, H.; Tomasz, S.A. Recruitment of the MecA Gene Homologue of Staphylococcus sciuri into a Resistance Determinant and Expression of the Resistant Phenotype in Staphylococcus aureus. J. Bacteriol. 2001, 183, 2417–2424. [Google Scholar] [CrossRef]
- De Jonge, B.L.M.; Chang, Y.S.; Gage, D.; Tomasz, A. Peptidoglycan Composition of a Highly Methicillin-Resistant Staphylococcus aureus Strain. The Role of Penicillin Binding Protein 2A. J. Biol. Chem. 1992, 267, 11248–11254. [Google Scholar] [CrossRef]
- Oliveira, D.C.; Wu, S.W. Genetic Organization of the Downstream Region of the MecA Element in Methicillin-Resistant Staphylococcus aureus Isolates Carrying Different Polymorphisms of This Region. Antimicrob. Agents Chemother. 2000, 44, 1906–1910. [Google Scholar] [CrossRef]
- De Lencastre, H.; De Jonge, B.L.M.; Matthews, P.R.; Tomasz, A. Review Molecular Aspects of Methicillin Resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 1994, 33, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K. Nucleotide Sequence of the Structural Gene for the Penicillin-Binding Protein 2 of Staphylococcus aureus and the Presence of a Homologous Gene in Other Staphylococci. FEMS Microbiol. Lett. 1994, 117, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Filipe, R.R.; Pinho, M.G.; Tomasz, A. Complementation of the Essential Peptidoglycan Transpeptidase Function of Penicillin-Binding Protein 2 (PBP2) by the Drug Resistance Protein PBP2A in Staphylococcus aureus. J. Bacteriol. 2001, 183, 6525–6531. [Google Scholar] [CrossRef]
- Fuda, C.; Hesek, D.; Lee, M.; Morio, K.I.; Nowak, T.; Mobashery, S. Activation for Catalysis of Penicillin-Binding Protein 2a from Methicillin-Resistant Staphylococcus aureus by Bacterial Cell Wall. J. Am. Chem. Soc. 2005, 127, 2056–2057. [Google Scholar] [CrossRef] [PubMed]
- Hakenbeck, R.; Coyette, J. Resistant Penicillin-Binding Proteins. Cell. Mol. Life Sci. 1998, 54, 332–340. [Google Scholar] [CrossRef] [PubMed]
- Fuda, C.; Suvorov, M.; Vakulenko, S.B.; Mobashery, S. The Basis for Resistance to β-Lactam Antibiotics by Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus. J. Biol. Chem. 2004, 279, 40802–40806. [Google Scholar] [CrossRef]
- Stapleton, P.D.; Taylor, P.W. Europe PMC Funders Group Methicillin Resistance in Staphylococcus aureus: Methicillin Resistance. Microbiology 2007, 85 Pt 1, 1–14. [Google Scholar]
- Srisuknimit, V.; Qiao, Y.; Schaefer, K.; Kahne, D.; Walker, S. Peptidoglycan Cross-Linking Preferences of Staphylococcus aureus Penicillin-Binding Proteins Have Implications for Treating MRSA Infections. J. Am. Chem. Soc. 2017, 139, 9791–9794. [Google Scholar] [CrossRef]
- Contreras-martel, C.; Martins, A.; Ecobichon, C.; Trindade, D.M.; Matteï, P.; Hicham, S.; Hardouin, P.; El Ghachi, M.; Boneca, I.G.; Dessen, A. Molecular Architecture of the PBP2–MreC Core Bacterial Cell Wall Synthesis Complex. Nat. Commun. 2017, 8, 776. [Google Scholar] [CrossRef]
- Schleifer, K.H.; Kandler, O. Peptidoglycan Types of Bacterial Cell Walls and Their Taxonomic Implications. Bacteriol. Rev. 1972, 36, 407–477. [Google Scholar] [CrossRef]
- Contreras-martel, C.; Amoroso, A.; Woon, E.C.Y.; Zervosen, A.; Inglis, S.; Martins, A.; Verlaine, O.; Rydzik, A.M.; Job, V. Structure-Guided Design of Cell Wall Biosynthesis Inhibitors That Overcome β -Lactam Resistance in Staphylococcus aureus (MRSA). ACS Chem. Biol. 2011, 6, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Peck, T.E.; Hill, S.A. Pharmacology for Anaesthesia and Intensive Care, 4th ed.; Cambrdige University Press: Cambridge, UK, 2014. [Google Scholar]
- Hrast, M.; Rožman, K.; Jukič, M.; Patin, D.; Gobec, S.; Sova, M. Synthesis and Structure–Activity Relationship Study of Novel Quinazolinone-Based Inhibitors of MurA. Bioorg. Med. Chem. Lett. 2017, 27, 3529–3533. [Google Scholar] [CrossRef] [PubMed]
- Nikolaidis, I.; Dessen, A. Resistance to Antibiotics Targeted to the Bacterial Cell Wall. Protein Sci. 2014, 23, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.J. Structural Basis for the Coordination of Cell Division with the Synthesis of the Bacterial Cell Envelope. Protein Sci. 2019, 28, 2042–2054. [Google Scholar] [CrossRef]
- Caveney, N.A.; Li, F.K.; Strynadka, N.C. Enzyme Structures of the Bacterial Peptidoglycan and Wall Teichoic Acid Biogenesis Pathways. Curr. Opin. Struct. Biol. 2018, 53, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Macheboeuf, P.; Contreras-martel, C.; Job, V.; Dideberg, O. Penicillin Binding Proteins: Key Players in Bacterial Cell Cycle and Drug Resistance Processes. FEMS Microbiol. Rev. 2006, 30, 673–691. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Vyas, R.; Tewari, R. Peptidoglycan Biosynthesis Machinery: A Rich Source of Drug Targets. Crit. Rev. Biotechnol. 2011, 31, 295–336. [Google Scholar] [CrossRef] [PubMed]
- Naclerio, G.A.; Sintim, H.O. Multiple Ways to Kill Bacteria via Inhibiting Novel Cell Wall or Membrane Targets. Future Med. Chem. 2020, 12, 1253–1279. [Google Scholar] [CrossRef]
- Kovac, A. Cytoplasmic Steps of Peptidoglycan Biosynthesis. FEMS Microbiol. Rev. 2008, 32, 168–207. [Google Scholar] [CrossRef]
- Rogers, H.J.; Perkins, H.R.; Ward, J.B. Biosynthesis of Peptidoglycan. Microb. Cell Walls Membr. 1980, 239–297. [Google Scholar] [CrossRef]
- Monteiro, J.M.; Covas, G.; Rausch, D.; Filipe, S.R.; Schneider, T.; Sahl, H.G.; Pinho, M.G. The Pentaglycine Bridges of Staphylococcus aureus Peptidoglycan are Essential for Cell Integrity. Sci. Rep. 2019, 9, 5010. [Google Scholar] [CrossRef]
- Goldman, R.C.; Gange, D. Inhibition of Transglycosylation Peptidoglycan Synthesis Involved in Bacterial. Curr. Med. Chem. 2000, 7, 801–820. [Google Scholar] [CrossRef] [PubMed]
- Gondokesumo, M.E.; Kurniawan, I.M. Molecular Docking Study of Sappan Wood Extract to Inhibit PBP2A Enzyme on Methicillin-Resistant Staphylococcus aureus (MRSA). J. Basic Clin. Physiol. Pharmacol. 2019, 30, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.E.; Young, K.D. Contributions of PBP 5 and DD-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 2001, 183, 3055–3064. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.T. Enzymes in the D-Alanine Branch of Bacterial Cell Wall Peptidoglycan Assembly. J. Biol. Chem. 1989, 264, 2393–2396. [Google Scholar] [CrossRef] [PubMed]
- Berger-Bächi, B.; Tschierske, M. Role of Fem Factors in Methicillin Resistance. Drug Resist. Updates 1998, 1, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Kourtis, A.P.; Hatfield, K.; Baggs, J.; Mu, Y.; See, I.; Epson, E.; Nadle, J.; Kainer, M.A.; Dumyati, G. Vital Signs: Epidemiology and Recent Trends in Methicillin-Resistant and in Methicillin-Susceptible Staphylococcus aureus Bloodstream Infections—United States. Morb. Mortal. Wkly. Rep. 2020, 68, 214–219. [Google Scholar] [CrossRef]
- Cedraro, N.; Cannalire, R.; Astolfi, A.; Mangiaterra, G.; Felicetti, T.; Vaiasicca, S.; Cernicchi, G.; Massari, S.; Biavasco, F.; Sabatini, S. From Quinoline to Quinazoline-Based S. aureus NorA Efflux Pump Inhibitors by Coupling a Focused Scaffold Hopping Approach and a Pharmacophore Search. ChemMedChem 2021, 16, 3044–3059. [Google Scholar] [CrossRef]
- Lade, H.; Kim, J.S. Bacterial Targets of Antibiotics in Methicillin-Resistant Staphylococcus aureus. Antibiotics 2021, 10, 398. [Google Scholar] [CrossRef]
- Kaczor, A.; Witek, K.; Podlewska, S.; Czekajewska, J.; Lubelska, A.; Zesławska, E.; Nitek, W.; Latacz, G.; Alibert, S.; Pagès, J.M.; et al. 5-Arylideneimidazolones with Amine at Position 3 as Potential Antibiotic Adjuvants against Multidrug Resistant Bacteria. Molecules 2019, 24, 438. [Google Scholar] [CrossRef]
- Jousselin, A.; Manzano, C.; Biette, A.; Reed, P.; Pinho, M.G.; Rosato, A.E.; Kelley, W.L.; Renzoni, A. The Staphylococcus aureus Chaperone PrsA Is a New Auxiliary Factor of Oxacillin Resistance Affecting Penicillin-Binding Protein 2A. Antimicrob. Agents Chemother. 2016, 60, 1656–1666. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.T.; Walshe, R.; Devocelle, M. A Computational Model of Antibiotic-Resistance Mechanisms in Methicillin-Resistant Staphylococcus aureus (MRSA). J. Theor. Biol. 2008, 254, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, S.B.; Adlan, T.A.; Khalafalla, N.A.; Abdalla, N.I.; Ali, Z.S.A.; Munir, K.A.A.; Hassan, M.M.; Elnour, M.A.B. Proteomics and Docking Study Targeting Penicillin-Binding Protein and Penicillin-Binding Protein2a of Methicillin-Resistant Staphylococcus aureus Strain SO-1977 Isolated from Sudan. Evol. Bioinform. 2019, 15. [Google Scholar] [CrossRef] [PubMed]
- Babic, M.; Hujer, A.M.; Bonomo, R.A.; Stokes, L.; Medical, A. What’s New in Antibiotic Resistance ? Focus on Beta-Lactamases. Drug Resist. Updates 2006, 9, 142–156. [Google Scholar] [CrossRef] [PubMed]
- Wielders, C.L.C.; Fluit, A.C.; Brisse, S.; Verhoef, J.; Schmitz, F.J. MecA Gene Is Widely Disseminated in Staphylococcus aureus Population. J. Clin. Microbiol. 2002, 40, 3970–3975. [Google Scholar] [CrossRef] [PubMed]
- Tsubakishita, S.; Kuwahara-arai, K.; Sasaki, T.; Hiramatsu, K. Origin and Molecular Evolution of the Determinant of Methicillin Resistance in Staphylococci. Antimicrob. Agents Chemother. 2010, 54, 4352–4359. [Google Scholar] [CrossRef] [PubMed]
- Webster, J.A.; Bannerman, T.L.; Hubner, R.J.; Ballard, D.N.; Cole, E.M.; Bruce, J.L.; Fiedler, F.; Schubert, K.; Kloos, W.E. Identification of the Staphylococcus sciuri Species Group with EcoRI Fragments Containing RRNA Sequences and Description of Staphylococcus vitulus sp. Nov. Int. J. Syst. Evol. Microbiol. 1994, 460, 454–460. [Google Scholar]
- Schleifer, K.H.; Geyer, U.; Kilpper-balz, R.; Devriese, L.A. Elevation of Staphylococcus sciuri subsp. Lentus (Kloos et al.) to Species Status: Staphylococcus lentus (Kloos et al.) Comb. Nov. Syst. Appl. Microbiol. 1983, 387, 382–387. [Google Scholar] [CrossRef]
- Ouchenane, Z.; Agabou, A.; Smati, F.; Rolain, J.M.; Raoult, D. Staphylococcal Cassette Chromosome Mec Characterization of Methicillin-Resistant Staphylococcus aureus Strains Isolated at the Military Hospital of Constantine/Algeria. Pathol. Biol. 2013, 61, 280–281. [Google Scholar] [CrossRef]
- Chromosome, S.C. Crossm Evolutionary Origin of the Staphylococcal Cassette Chromosome. Antimicrob. Agents Chemother. 2017, 61, 1–16. [Google Scholar]
- Katayama, Y.; Ito, T.; Hiramatsu, K. A New Class of Genetic Element, Staphylococcus Cassette Chromosome Mec, Encodes Methicillin Resistance in Staphylococcus aureus. Nippon Saikingaku Zasshi 2000, 44, 1549–1555. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, D.; Peters, B.M.; Li, L.; Li, B.; Xu, Z.; Shirliff, M.E. Staphylococcal Chromosomal Cassettes Mec (SCCmec): A Mobile Genetic Element in Methicillin-Resistant Staphylococcus aureus. Microb. Pathog. 2016, 101, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Ma, X.X.; Takeuchi, F.; Okuma, K.; Yuzawa, H.; Hiramatsu, K. Novel Type V Staphylococcal Cassette Chromosome Mec Driven by a Novel Cassette Chromosome Recombinase, CcrC. Antimicrob. Agents Chemother. 2004, 48, 2637–2651. [Google Scholar] [CrossRef]
- Ender, M.; Berger-Bächi, B.; McCallum, N. Variability in SCCmecN1 Spreading among Injection Drug Users in Zurich, Switzerland. BMC Microbiol. 2007, 7, 62. [Google Scholar] [CrossRef]
- Chongtrakool, P.; Ito, T.; Ma, X.X.; Trakulsomboon, S.; Tiensasitorn, C.; Jamklang, M.; Chavalit, T.; Song, J.; Chongtrakool, P.; Ito, T.; et al. Staphylococcal Cassette Chromosome Mec (SCC Mec) Typing of Methicillin-Resistant Staphylococcus aureus Strains Isolated in 11 Asian Countries: A Proposal for a New Nomenclature for SCC Mec Elements Staphylococcal Cassette Chromosome Mec (SCC Mec) Typing of Methicillin-Resistant Staphylococcus aureus Strains Isolated in 11 Asian Countries: A Proposal for a New Nomenclature for SCC Mec Elements. Antimicrob. Agents Chemother. 2006, 50, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Tsubakishita, S.; Kuwahara-arai, K.; Baba, T.; Hiramatsu, K. Staphylococcal Cassette Chromosome Mec -Like Element in Macrococcus caseolyticus. Antimicrob. Agents Chemother. 2010, 54, 1469–1475. [Google Scholar] [CrossRef] [PubMed]
- Uehara, Y. Current Status of Staphylococcal Cassette. Antibiotics 2022, 11, 86. [Google Scholar] [CrossRef] [PubMed]
- Berger-Bächi, B.; Rohrer, S. Factors Influencing Methicillin Resistance in Staphylococci. Arch. Microbiol. 2002, 178, 165–171. [Google Scholar] [CrossRef]
- Fishovitz, J.; Hermoso, J.A.; Chang, M.; Mobashery, S. Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus. IUBMB Life 2014, 66, 572–577. [Google Scholar] [CrossRef]
- Lim, D.; Strynadka, N.C.J. Structural Basis for the β-Lactam Resistance of PBP2a from Methicillin-Resistant Staphylococcus aureus. Nat. Struct. Biol. 2002, 9, 870–876. [Google Scholar] [CrossRef]
- Kumar, K.M.; Anitha, P.; Sivasakthi, V.; Bag, S.; Lavanya, P.; Anbarasu, A.; Ramaiah, S. In Silico Study on Penicillin Derivatives and Cephalosporins for Upper Respiratory Tract Bacterial Pathogens. 3 Biotech 2014, 4, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.C.; Wong, M.T.Y.; Essex, J.W. Molecular Dynamics Simulations of Antibiotic Ceftaroline at the Allosteric Site of Penicillin-Binding Protein 2a (PBP2a). Isr. J. Chem. 2020, 60, 754–763. [Google Scholar] [CrossRef]
- Lovering, A.L.; Gretes, M.C.; Safadi, S.S.; Danel, F.; De Castro, L.; Page, M.G.P.; Strynadka, N.C.J. Structural Insights into the Anti-Methicillin-Resistant Staphylococcus aureus (MRSA) Activity of Ceftobiprole. J. Biol. Chem. 2012, 287, 32096–32102. [Google Scholar] [CrossRef] [PubMed]
- Hermoso, J.A.; Chang, M.; Mobashery, S. Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis. J. Am. Chem. Soc. 2018, 139, 2102–2110. [Google Scholar] [CrossRef]
- My, N.H.; Hirao, H.; Van, D.U.; Morokuma, K. Computational Studies of Bacterial Resistance to β-Lactam Antibiotics: Mechanism of Covalent Inhibition of the Penicillin-Binding Protein 2a (PBP2a). J. Chem. Inf. Model. 2011, 51, 3226–3234. [Google Scholar] [CrossRef] [PubMed]
- Janardhanan, J.; Bouley, R.; Peng, Z.; Batuecas-mordillo, M.; Meisel, J.E.; Ding, D.; Schroeder, V.A.; Wolter, W.R.; Mahasenan, K.V.; Hermoso, J.A.; et al. Crossm The Quinazolinone Allosteric Inhibitor of PBP 2a Synergizes. Antimicrob. Agents Chemother. 2019, 63, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bouley, R.; Kumarasiri, M.; Peng, Z.; Otero, L.H.; Song, W.; Mark, A.; Schroeder, V.A.; Wolter, W.R.; Lastochkin, E.; Antunes, N.T.; et al. Discovery of Antibiotic (E)-3-(3-Carboxyphenyl)-2-(4-cyanostyryl)quinazolin-4(3H)-One. J. Am. Chem. Soc. 2015, 137, 1738–1741. [Google Scholar] [CrossRef]
- Otero, L.H.; Rojas-Altuve, A.; Llarrull, L.I.; Carrasco-López, C.; Kumarasiri, M.; Lastochkin, E.; Fishovitz, J.; Dawley, M.; Hesek, D.; Lee, M.; et al. How Allosteric Control of Staphylococcus aureus Penicillin Binding Protein 2a Enables Methicillin Resistance and Physiological Function. Proc. Natl. Acad. Sci. USA 2013, 110, 16808–16813. [Google Scholar] [CrossRef]
- Lv, N.; Kong, Q.; Zhang, H.; Li, J. Discovery of Novel Staphylococcus aureus Penicillin Binding Protein 2a Inhibitors by Multistep Virtual Screening and Biological Evaluation. Bioorg. Med. Chem. Lett. 2021, 41, 2–9. [Google Scholar] [CrossRef]
- Lavanya, P.; Ramaiah, S.; Anbarasu, A. A Molecular Docking and Dynamics Study to Screen Potent Anti-Staphylococcal Compounds Against Ceftaroline Resistant MRSA. J. Cell Biochem. 2016, 117, 542–548. [Google Scholar] [CrossRef]
- Matys, A.; Podlewska, S.; Witek, K.; Witek, J.; Bojarski, A.J.; Schabikowski, J.; Otrębska-Machaj, E.; Latacz, G.; Szymańska, E.; Kieć-Kononowicz, K.; et al. Imidazolidine-4-One Derivatives in the Search for Novel Chemosensitizers of Staphylococcus aureus MRSA: Synthesis, Biological Evaluation and Molecular Modeling Studies. Eur. J. Med. Chem. 2015, 101, 313–325. [Google Scholar] [CrossRef] [PubMed]
- Eom, S.; Kang, S.; Lee, D.; Myeong, J.; Lee, J.; Kim, H.; Kim, K.; Je, J.; Jung, W.; Kim, Y. Synergistic Antibacterial Effect and Antibacterial Action Mode of Chitosan—Ferulic Acid Conjugate against Methicillin-Resistant Staphylococcus aureus. J. Microbiol. Biotechnol. 2016, 26, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Kumar, D.; Raj, V. Studies on Imidazole and Its Derivatives with Particular Emphasis on Their Chemical/Biological Applications as Bioactive Molecules/Intermediated to Bioactive Molecule. Curr. Synth. Syst. Biol. 2017, 5, 1–10. [Google Scholar] [CrossRef]
- Bush, K. Synergistic MRSA Combinations. Nat. Publ. Group 2015, 11, 832–833. [Google Scholar] [CrossRef] [PubMed]
- Stryjewski, M.E.; Jones, R.N.; Corey, G.R. Ceftaroline: Clinical and microbiology experience with focus on methicillin-resistant Staphylococcus aureus after regulatory approval in the USA. Diagn. Microbiol. Infect. Dis. 2014, 81, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Soler-Gómez, A.; Arca-Suárez, J.; González-Gómez, M.; Galán-Sánchez, F.; Rodríguez-Iglesias, M. Antimicrobial activity of ceftaroline against methicillin-resistant Staphylococcus aureus (MRSA) isolates from Spain and molecular characterisation of borderline susceptible and resistant strains. J. Glob. Antimicrob. Resist. 2018, 15, 171–172. [Google Scholar] [CrossRef] [PubMed]
- Saravolatz, L.D.; Stein, G.E.; Johnson, L.B. Ceftaroline: A Novel Cephalosporin with Activity against Methicillin-Resistant Staphylococcus aureus. Clin. Infect. Dis. 2011, 52, 1156–1163. [Google Scholar] [CrossRef]
- Soriano, A. Ceftaroline. Rev. Esp. Quimioter. 2021, 34, 29–31. [Google Scholar] [CrossRef]
- Hernandez, P.O.; Lema, S.; Tyring, S.K.; Mendoza, N. Ceftaroline in Complicated Skin and Skin-Structure Infections. Infect. Drug Resist. 2012, 5, 23–35. [Google Scholar] [CrossRef]
- Schaumburg, F.; Peters, G.; Alabi, A.; Becker, K.; Idelevich, E.A. Missense Mutations of PBP2a are Associated with Reduced Susceptibility to Ceftaroline and Ceftobiprole in African MRSA. J. Antimicrob. Chemother. 2016, 71, 41–44. [Google Scholar] [CrossRef]
- Livermore, D.M. Can β-Lactams Be Re-Engineered to Beat MRSA? Clin. Microbiol. Infect. 2006, 12 (Suppl. S2), 11–16. [Google Scholar] [CrossRef] [PubMed]
- Vidaillac, C.; Rybak, M.J. Ceftobiprole: First Cephalosporin with Activity against Methicillin-Resistant Staphylococcus aureus. Pharmacotherapy 2009, 29, 511–525. [Google Scholar] [CrossRef] [PubMed]
- Nigo, M.; Lorena Diaz, L.P.; Carvajal, T.T.; Tran, R.R.; Panesso, D.; Garavito, J.D.; Miller, W.R.; Wanger, A.; Weinstock, G.; Munita, J.M.; et al. Ceftaroline-Resistant, Daptomycin-Tolerant, and Heterogeneous Vancomycin-Intermediate Methicillin- Resistant Staphylococcus aureus Causing Infective Endocarditis. Antimicrob. Agents Chemother. 2017, 61, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.; Storey, C.; Stubbings, W.J.; Hoyle, A.M.; Hobbs, J.K.; Chopra, I. Antistaphylococcal Activity of the Novel Cephalosporin. J. Antimicrob. Chemother. 2005, 55, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, P.R.; Pesesky, M.W.; Bouley, R.; Ballard, A.; Biddy, B.A.; Suckow, M.A.; Wolter, W.R.; Schroeder, V.A.; Burnham, C.D.; Mobashery, S.; et al. Combinations Suppress Resistance in MRSA. Nat. Chem. Biol. 2015, 11, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, M.-A.; Dokla, E.; Serya, R.; Abouzid, K. Identification of Novel Pyrazole and Benzimidazole Based Derivatives as PBP2a Inhibitors: Design, Synthesis, and Biological Evaluation. Arch. Pharm. Sci. Ain Shams Univ. 2019, 3, 228–245. [Google Scholar] [CrossRef]
- Yadav, G.; Ganguly, S. Structure Activity Relationship (SAR) Study of Benzimidazole Scaffold for Different Biological Activities: A Mini-Review. Eur. J. Med. Chem. 2015, 97, 419–443. [Google Scholar] [CrossRef]
- Malasala, S.; Ahmad, N.; Akunuri, R.; Shukla, M.; Kaul, G.; Dasgupta, A.; Madhavi, Y.; Chopra, S.; Nanduri, S. Synthesis and evaluation of new quinazoline-benzimidazole hybrids as potent anti-microbial agents against multidrug resistant Staphylococcus aureus and Mycobacterium tuberculosis. Eur. J. Med. Chem. 2020, 212, 112996. [Google Scholar] [CrossRef]
- Tunçbilek, M.; Kiper, T.; Altanlar, N. Synthesis and in Vitro Antimicrobial Activity of Some Novel Substituted Benzimidazole Derivatives Having Potent Activity against MRSA. Eur. J. Med. Chem. 2009, 44, 1024–1033. [Google Scholar] [CrossRef]
- Buommino, E.; De Marino, S.; Sciarretta, M.; Auria, M.V.D.; Festa, C. With Oxacillin against Methicillin-Resistant Staphylococcus aureus. Antibiotics 2021, 16, 1258. [Google Scholar] [CrossRef]
- Verma, S.K.; Verma, R.; Kumar, K.S.S.; Banjare, L.; Shaik, A.B.; Bhandare, R.R.; Rakesh, K.P.; Rangappa, K.S. A Key Review on Oxadiazole Analogs as Potential Methicillin-Resistant Staphylococcus aureus (MRSA) Activity: Structure-Activity Relationship Studies. Eur. J. Med. Chem. 2021, 219, 113442. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, M.A.W.; Dokla, E.M.E.; Serya, R.A.T.; Abouzid, K.A.M. Penicillin Binding Protein 2a: An Overview and a Medicinal Chemistry Perspective. Eur. J. Med. Chem. 2020, 199, 112312. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, M.A.A.; Abdeljawaad, K.A.A.; Abdelrahman, A.H.M.; Alzahrani, O.R.; Alshabrmi, F.M.; Khalaf, E.; Moustafa, M.F.; Alrumaihi, F.; Allemailem, K.S.; Soliman, M.E.S.; et al. Non-β-Lactam Allosteric Inhibitors Target Methicillin-Resistant Staphylococcus aureus: An in Silico Drug Discovery Study. Antibiotics 2021, 10, 934. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, S.I.; Chaudhari, H.K. Design, Synthesis, in-Silico Studies and Biological Screening of Quinazolinone Analogues as Potential Antibacterial Agents against MRSA. Bioorg. Med. Chem. 2019, 27, 2676–2688. [Google Scholar] [CrossRef]
- Bouley, R.; Ding, D.; Peng, Z.; Bastian, M.; Lastochkin, E.; Song, W.; Suckow, M.A.; Schroeder, V.A.; Wolter, W.R.; Mobashery, S.; et al. Structure-Activity Relationship for the 4(3H)-Quinazolinone Antibacterials. J. Med. Chem. 2016, 59, 5011–5021. [Google Scholar] [CrossRef] [PubMed]
- Gunasekharan, M.; Choi, T.I.; Rukayadi, Y.; Latif, M.A.M.; Karunakaran, T.; Faudzi, S.M.M.; Kim, C.H. Preliminary Insight of Pyrrolylated-Chalcones as New Anti-Methicillin-Resistant Staphylococcus aureus (Anti-Mrsa) Agents. Molecules 2021, 26, 5314. [Google Scholar] [CrossRef] [PubMed]
- Chem, M.; Marchand-brynaert, J. Unprecedented inhibition of resistant penicillin bindingproteins by bis-2-oxoazetidinylmacrocycles. MedChemComm 2012, 3, 344–351. [Google Scholar] [CrossRef]
- Nanjundaswamy, S.; Bindhu, S.; Renganathan, R.R.A.; Nagashree, S.; Mallu, P.; Rai, V.R.; Bindhu, S.; Renganathan, R.R.A.; Nagashree, S. Design, Synthesis of Pyridine Coupled Pyrimidinone/Pyrimidinthione as Anti-MRSA Agent: Validation by Molecular Docking and Dynamics Simulation. J. Biomol. Struct. Dyn. 2021, 40, 12106–12117. [Google Scholar] [CrossRef]
- Zhang, P.; Gopala, L.; Zhang, S.; Cai, G. European Journal of Medicinal Chemistry An Unanticipated Discovery towards Novel Naphthalimide Corbelled Aminothiazoximes as Potential Anti-MRSA Agents and Allosteric Modulators for PBP2a. Eur. J. Med. Chem. 2022, 229, 114050. [Google Scholar] [CrossRef]
- Toney, J.H.; Hammond, G.G.; Leiting, B.; Pryor, K.A.D.; Wu, J.K.; Cuca, G.C.; Pompliano, D.L. Soluble Penicillin-Binding Protein 2a: β-Lactam Binding and Inhibition by Non-β-Lactams Using a 96-Well Format. Anal. Biochem. 1998, 255, 113–119. [Google Scholar] [CrossRef]
- Kurosu, M.; Siricilla, S.; Mitachi, K. Advances in MRSA drug discovery: Where are we and where do we need to be? Expert Opin. Drug Discov. 2013, 8, 1095–1116. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Cheng, G.; Dai, M.; Wu, Q.; Yuan, Z. Inhibitors Targeting on Cell Wall Biosynthesis Pathway of MRSA. Mol. Biosyst. 2012, 8, 2828–2838. [Google Scholar] [CrossRef] [PubMed]
- Shiota, S.; Shimizu, M.; Sugiyama, J.; Morita, Y.; Mizushima, T.; Tsuchiya, T. Mechanisms of Action of Corilagin and Tellimagrandin I That Remarkably Potentiate the Activity of β-Lactams against Methicillin-Resistant Staphylococcus aureus. Microbiol. Immunol. 2004, 48, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Kurazono, M.; Ida, T.; Yamada, K.; Hirai, Y.; Maruyama, T.; Shitara, E.; Yonezawa, M. In Vitro Activities of ME1036 (CP5609), a Novel Parenteral Carbapenem, against Methicillin-Resistant Staphylococci. Antimicrob. Agents Chemother. 2004, 48, 2831–2837. [Google Scholar] [CrossRef] [PubMed]
- Erol, M.; Celik, I.; Temiz-Arpaci, O.; Kaynak-Onurdag, F.; Okten, S. Design, Synthesis, Molecular Docking, Density Functional Theory and Antimicrobial Studies of Some Novel Benzoxazole Derivatives as Structural Bioisosteres of Nucleotides. J. Biomol. Struct. Dyn. 2021, 39, 3080–3091. [Google Scholar] [CrossRef] [PubMed]
- Karthik, C.S.; Ananda, A.P.; Nagashree, S.; Mallu, P.; Rai, V.R. Design, Synthesis, and in-Silico Studies of Pyrazolylpyridine Analogues: A Futuristic Antibacterial Contender against Coagulase Positive. J. Mol. Struct. 2022, 1255, 132400. [Google Scholar] [CrossRef]
- Sangeetha, M.; Saranya, T.S.; Sathianarayanan, S.; Hima Vyshnavi, A.M.; Krishnan Namboori, P.K. Design and Development of Potential Flavonoid Moiety for Pbp2a Inhibition for Mrsa Therapy-A Computational Technique. Biomed. Pharmacol. J. 2020, 13, 687–692. [Google Scholar] [CrossRef]
- Li, Q.Q.; Kang, O.H.; Kwon, D.Y. Study on Demethoxycurcumin as a Promising Approach to Reverse Methicillin-Resistance of Staphylococcus aureus. Int. J. Mol. Sci. 2021, 22, 3778. [Google Scholar] [CrossRef]
- Neto, D.A.; Ramos, A.; Max, W.; Silva, J.; Silva, E.; Coelho, B.; Odorico, M.; Moraes, D. Microbial Pathogenesis Anti-MRSA Activity of Curcumin in Planktonic Cells and Biofilms and Determination of Possible Action Mechanisms. Microb. Pathog. 2021, 155, 104892. [Google Scholar] [CrossRef]
- Rani, N.; Saravanan, V.; Lakshmi, P.T.V. Allosteric Site Mediated Active Site Inhibition of PBP2a Using Quercetin 3-O-Rutinoside and Its Combination. J. Biomol. Struct. Dyn. 2015, 34, 1778–1796. [Google Scholar] [CrossRef]
- Rani, N.; Vijayakumar, S.; Thanga Velan, L.P.; Arunachalam, A. Quercetin 3-O-Rutinoside Mediated Inhibition of PBP2a: Computational and Experimental Evidence to Its Anti-MRSA Activity. Mol. Biosyst. 2014, 10, 3229–3237. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Li, Z.; Kang, O.-H.; Mun, S.-H.; Seo, Y.-S.; Kong, R.; Shin, D.-W.; Liu, X.-Q.; Kwon, D.-Y. Antimicrobial Activity and Synergism of Ursolic Acid 3-O-α-l-Arabinopyranoside with Oxacillin against Methicillin-Resistant Staphylococcus aureus. Int. J. Mol. Med. 2017, 40, 1285–1293. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tozar, T.; Costa, S.S.; Udrea, A.M.; Nastasa, V.; Couto, I.; Viveiros, M.; Pascu, M.L.; Romanitan, M.O. Anti-Staphylococcal Activity and Mode of Action of Thioridazine Photoproducts. Sci. Rep. 2020, 10, 18043. [Google Scholar] [CrossRef] [PubMed]
- Negi, B.; Kumar, D.; Kumbukgolla, W.; Jayaweera, S.; Ponnan, P.; Singh, R.; Agarwal, S.; Rawat, D.S. Anti-Methicillin Resistant Staphylococcus aureus Activity, Synergism with Oxacillin and Molecular Docking Studies of Metronidazole-Triazole Hybrids. Eur. J. Med. Chem. 2016, 115, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Zhang, X.; He, Y.; Sun, W.; Feng, W.; Liu, J.; Hu, Z.; Xu, Q.; Zhu, H.; Zhang, J.; et al. Aspermerodione, a Novel Fungal Metabolite with an Unusual 2,6-Dioxabicyclo[2.2.1]Heptane Skeleton, as an Inhibitor of Penicillin-Binding Protein 2a. Sci. Rep. 2018, 8, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Alhadrami, H.A.; Hamed, A.A.; Hassan, H.M.; Belbahri, L.; Rateb, M.E.; Sayed, A.M. Flavonoids as Potential Anti-MRSA Agents through Modulation of PBP2A: A Computational and Experimental Study. Antibiotics 2020, 9, 562. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Z.; Tangadanchu, V.K.R.; Battini, N.; Bheemanaboina, R.R.Y.; Zang, Z.L.; Zhang, S.L.; Zhou, C.H. Indole-Nitroimidazole Conjugates as Efficient Manipulators to Decrease the Genes Expression of Methicillin-Resistant Staphylococcus aureus. Eur. J. Med. Chem. 2019, 179, 723–735. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.W.; Huang, W.C.; Lin, C.Y.; Wang, W.H.; Hung, L.C.; Chen, Y.H. Tellimagrandin Ii, a Type of Plant Polyphenol Extracted from Trapa Bispinosa Inhibits Antibiotic Resistance of Drug-Resistant Staphylococcus aureus. Int. J. Mol. Sci. 2019, 20, 5790. [Google Scholar] [CrossRef]
- Chowdhury, A.; Paul, P.; Bhattacharjee, A.; Das Talukdar, A.; Choudhury, M.D. Virtual High Throughput Screening of Carbapenem Derivatives as New Generation Carbapenemase and Penicillin Binding Protein Inhibitors: A Hunt to Save Drug of Last Resort. Comb. Chem. High Throughput Screen. 2015, 18, 18–23. [Google Scholar] [CrossRef]
- Lian, X.; Xia, Z.; Li, X.; Karpov, P.; Jin, H.; Tetko, I.V.; Xia, J.; Wu, S. Bioorganic Chemistry Anti-MRSA Drug Discovery by Ligand-Based Virtual Screening and Biological Evaluation. Bioorg. Chem. 2021, 114, 105042. [Google Scholar] [CrossRef]
- Skariyachan, S.; Krishnan, R.S.; Siddapa, S.B.; Salian, C.; Bora, P.; Sebastian, D. Computer Aided Screening and Evaluation of Herbal Therapeutics against MRSA Infections. Bioinformation 2011, 7, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Abedi, F.; Balmeh, N.; Sanjari, S.; Plantago, A. Informatics in Medicine Unlocked In-Silico Investigation of Antibacterial Herbal Compounds in Order to Find New Antibiotic against Staphylococcus aureus and Its Resistant Subtypes. Inform. Med. Unlocked 2022, 28, 100843. [Google Scholar] [CrossRef]
- Chang, M.; Mahasenan, K.V.; Hermoso, J.A.; Mobashery, S. Unconventional Antibacterials and Adjuvants. Acc. Chem. Res. 2021, 54, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.R. Chapter 23 Screening Ligands by X-Ray Crystallography. Struct. Genom. Drug Discov. 2014, 1140, 315–323. [Google Scholar] [CrossRef]
- Fishovitz, J.; Rojas-altuve, A.; Otero, L.H.; Dawley, M.; Carrasco-lo, C.; Chang, M.; Hermoso, J.A.; Mobashery, S. Disruption of allosteric response as an unprecedented mechanism of resistance to antibiotics. J. Am. Chem. Soc. 2014, 16, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Zhang, A.; Xu, Y. Synthesis and Antibacterial Activity of Novel Icariin Derivatives. Pharm. Int. J. Pharm. Sci. 2019, 74, 73–78. [Google Scholar] [CrossRef]
- Mart, A.; Pascual, M.; Sheth, C.C.; Veses, V. Icariin in Combination with Amoxycillin-Clavulanate and Ampicillin, but Not Vancomycin, Increases Antibiotic Sensitivity and Growth Inhibition against Methicillin-Resistant Staphylococcus aureus. Antibiotics 2022, 11, 233. [Google Scholar]
- Marchand-brynaert, J.; Urbach, A.; Dive, G.; Tinant, B. European Journal of Medicinal Chemistry Large Ring 1, 3-Bridged 2-Azetidinones: Experimental and Theoretical Studies. Eur. J. Med. Chem. 2009, 44, 2071–2080. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Bae, J.S.; Otto, M. Pathogenicity and Virulence of Staphylococcus aureus ABSTRACT. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]

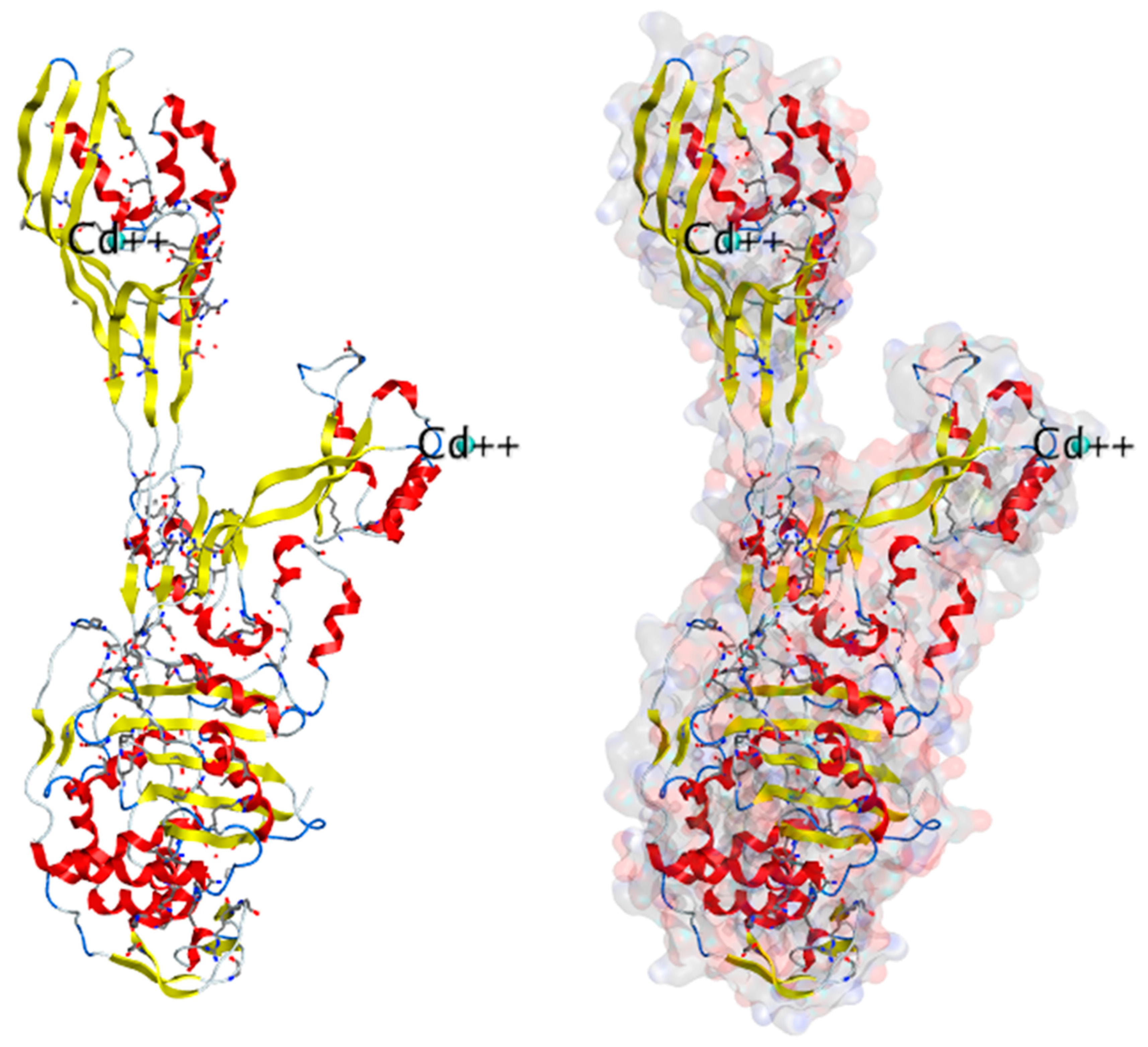



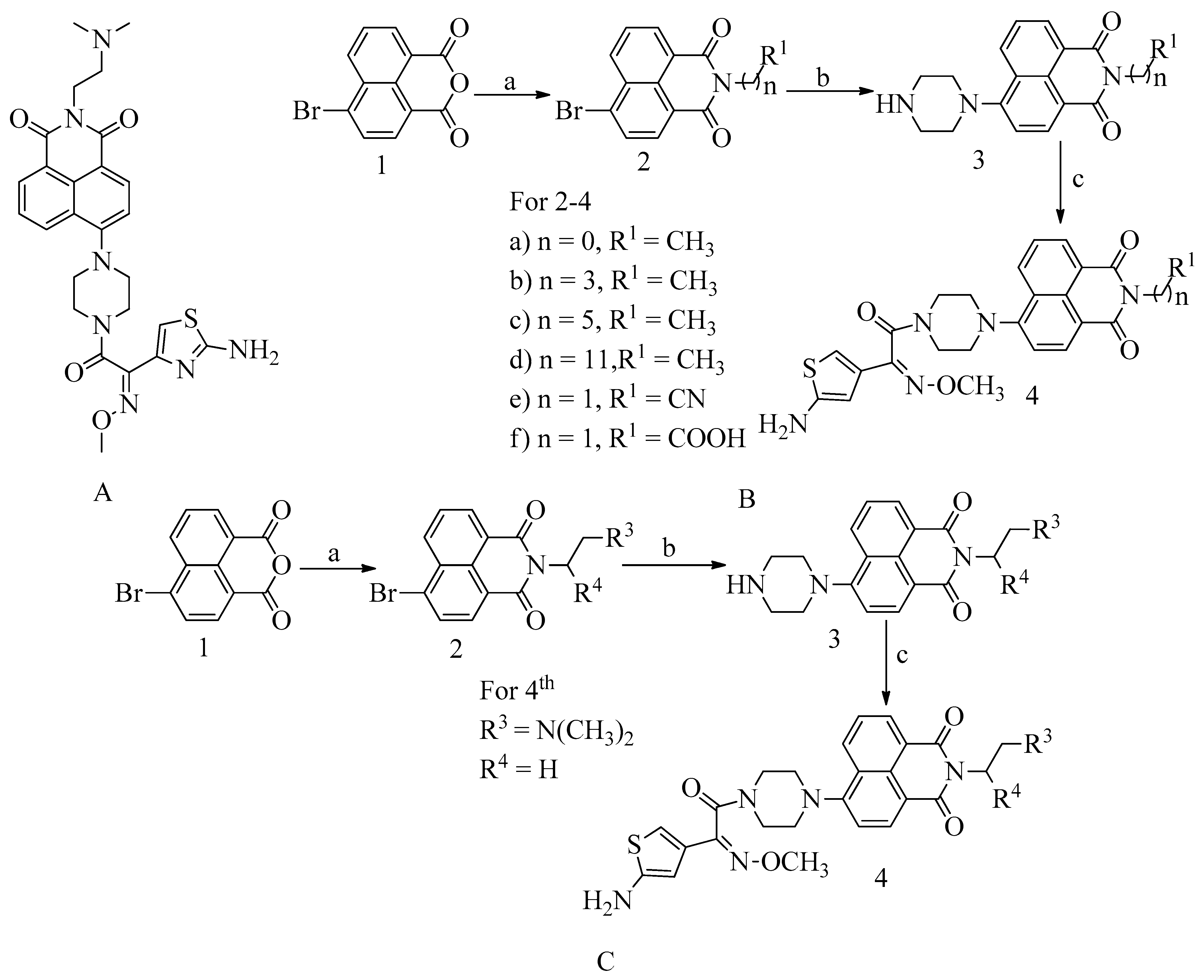


| Sr. No. | Release Year | PDB ID | Resolution | Bound Ligand | Species | Ref. |
|---|---|---|---|---|---|---|
| 1 | 2019 | 6Q9N | 2.5 | 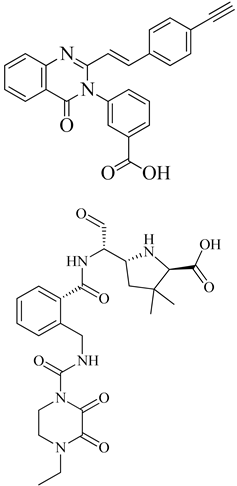 | S. aureus subsp. Mu50 | [121] |
| 2 | 2019 | 6H5O | 2.82 | 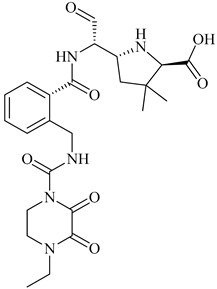 | S. aureus Subsp. Mu50 | [122] |
| 3 | 2015 | 4CJN | 1.947 |  | S. aureus | [123] |
| 4 | 2013 | 3ZG0 | 2.6 | 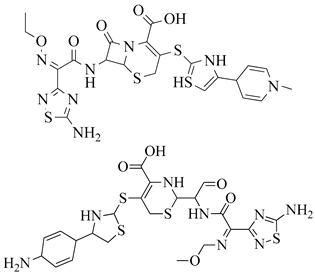 | S. aureus Subsp. Mu50 | [124] |
| 5 | 2013 | 3ZFZ | 2.25 |  | S. aureus subsp. Mu50 | [124] |
| 6 | 2002 | 1MWT | 2.45 | 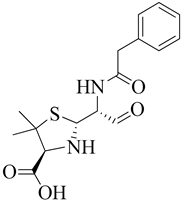 | S. aureus | [115] |
| 7 | 2002 | 1MWU | 2.60 | 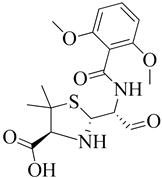 | S. aureus | [115] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ambade, S.S.; Gupta, V.K.; Bhole, R.P.; Khedekar, P.B.; Chikhale, R.V. A Review on Five and Six-Membered Heterocyclic Compounds Targeting the Penicillin-Binding Protein 2 (PBP2A) of Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules 2023, 28, 7008. https://doi.org/10.3390/molecules28207008
Ambade SS, Gupta VK, Bhole RP, Khedekar PB, Chikhale RV. A Review on Five and Six-Membered Heterocyclic Compounds Targeting the Penicillin-Binding Protein 2 (PBP2A) of Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules. 2023; 28(20):7008. https://doi.org/10.3390/molecules28207008
Chicago/Turabian StyleAmbade, Shraddha S., Vivek Kumar Gupta, Ritesh P. Bhole, Pramod B. Khedekar, and Rupesh V. Chikhale. 2023. "A Review on Five and Six-Membered Heterocyclic Compounds Targeting the Penicillin-Binding Protein 2 (PBP2A) of Methicillin-Resistant Staphylococcus aureus (MRSA)" Molecules 28, no. 20: 7008. https://doi.org/10.3390/molecules28207008
APA StyleAmbade, S. S., Gupta, V. K., Bhole, R. P., Khedekar, P. B., & Chikhale, R. V. (2023). A Review on Five and Six-Membered Heterocyclic Compounds Targeting the Penicillin-Binding Protein 2 (PBP2A) of Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules, 28(20), 7008. https://doi.org/10.3390/molecules28207008