Multifunctional Derivatives of Spiropyrrolidine Tethered Indeno-Quinoxaline Heterocyclic Hybrids as Potent Antimicrobial, Antioxidant and Antidiabetic Agents: Design, Synthesis, In Vitro and In Silico Approaches
Abstract
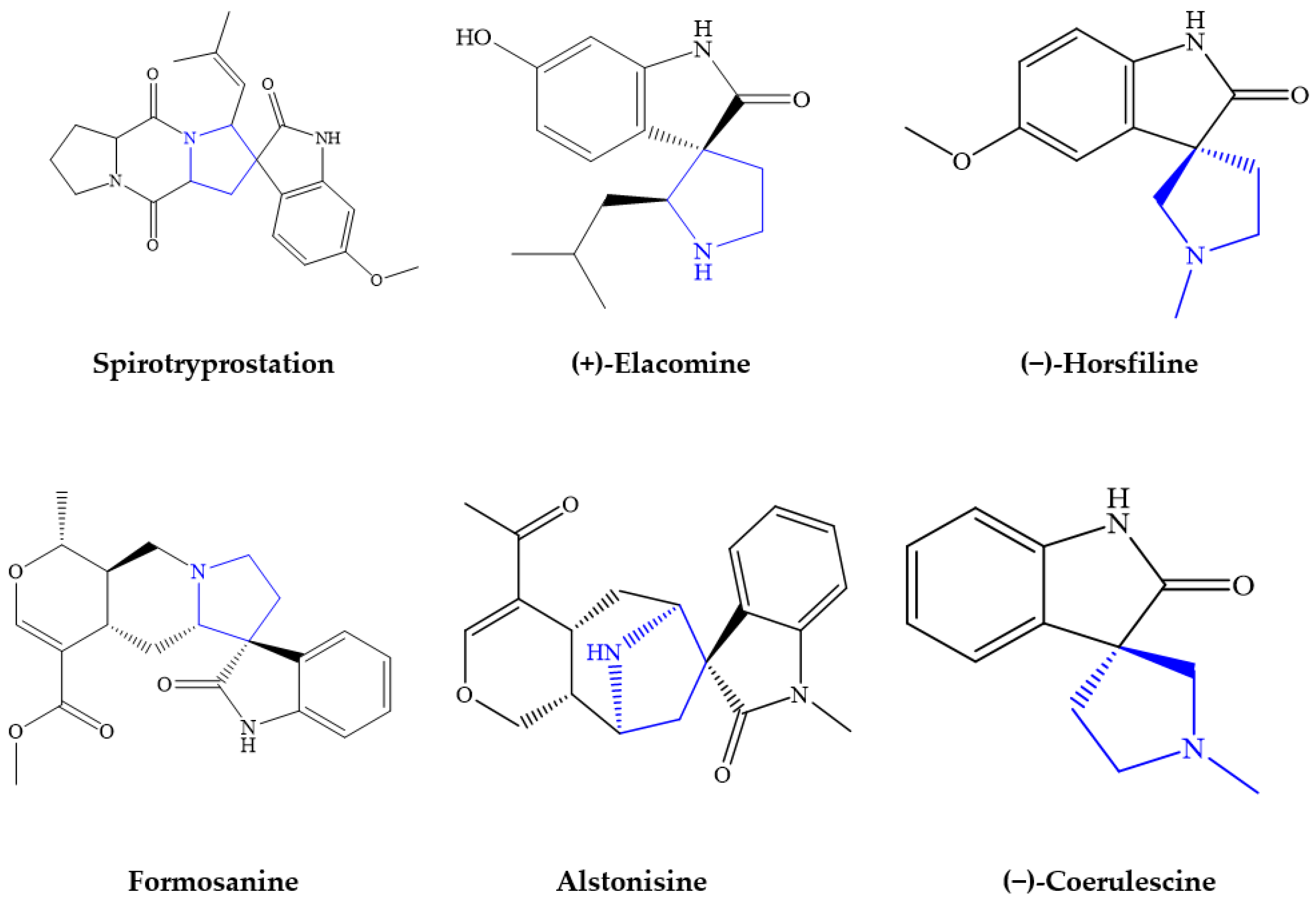
1. Introduction
2. Results and Discussion
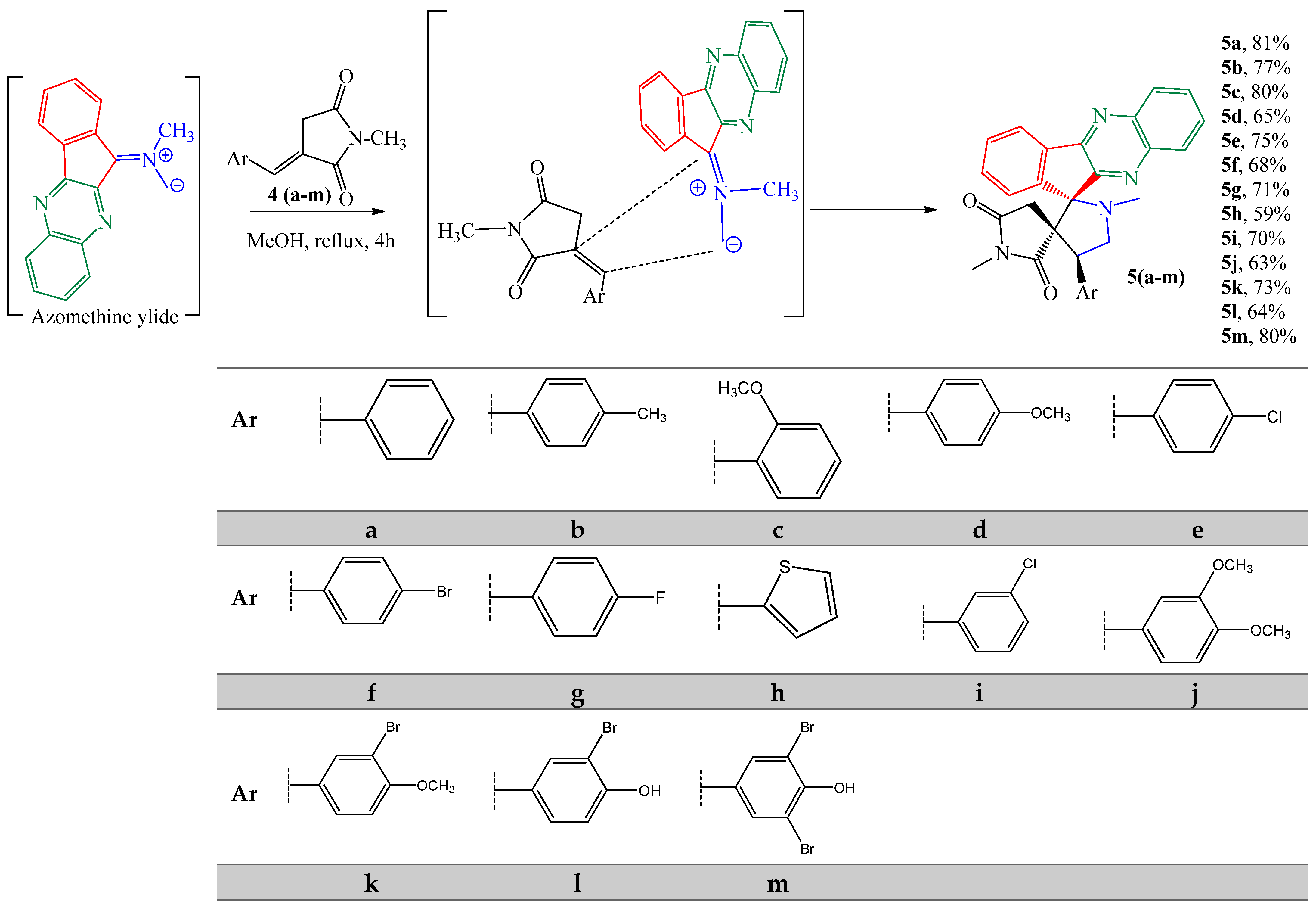
2.1. Chemistry
2.2. Spectroscopic of the Isomeric Cycloadducts
2.3. Biological Screening
2.3.1. Antimicrobial Activity vs. Structure Activity Relationship Studies
2.3.2. Antioxidant Activity vs. Structure Activity Relationship Studies
2.3.3. Antidiabetic Activity vs. Structure Activity Relationship Studies
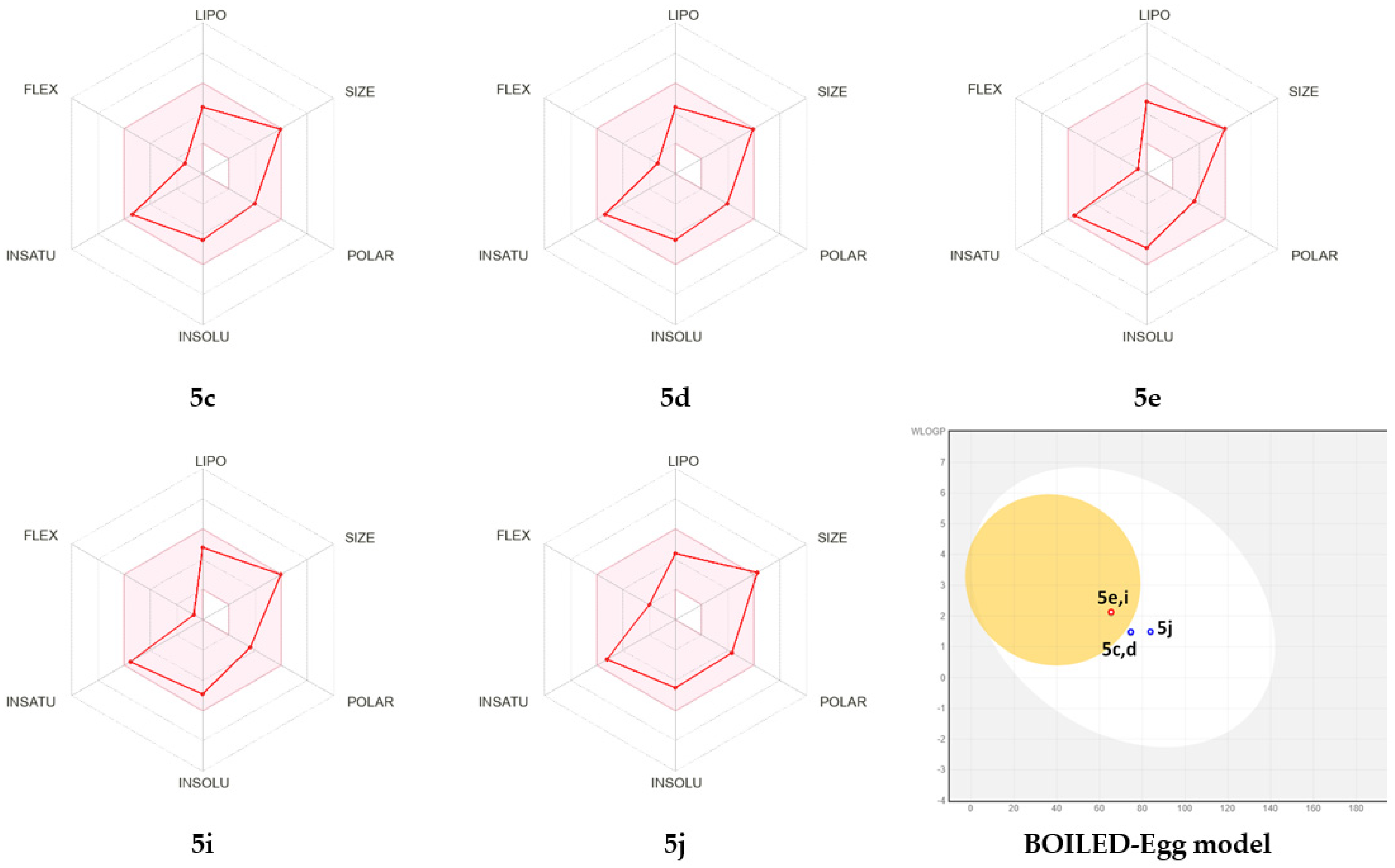
2.4. Computational Studies
Druglikeness and Pharmacokinetics
2.5. Molecular Docking and Dynamic Simulation
2.5.1. Molecular Docking
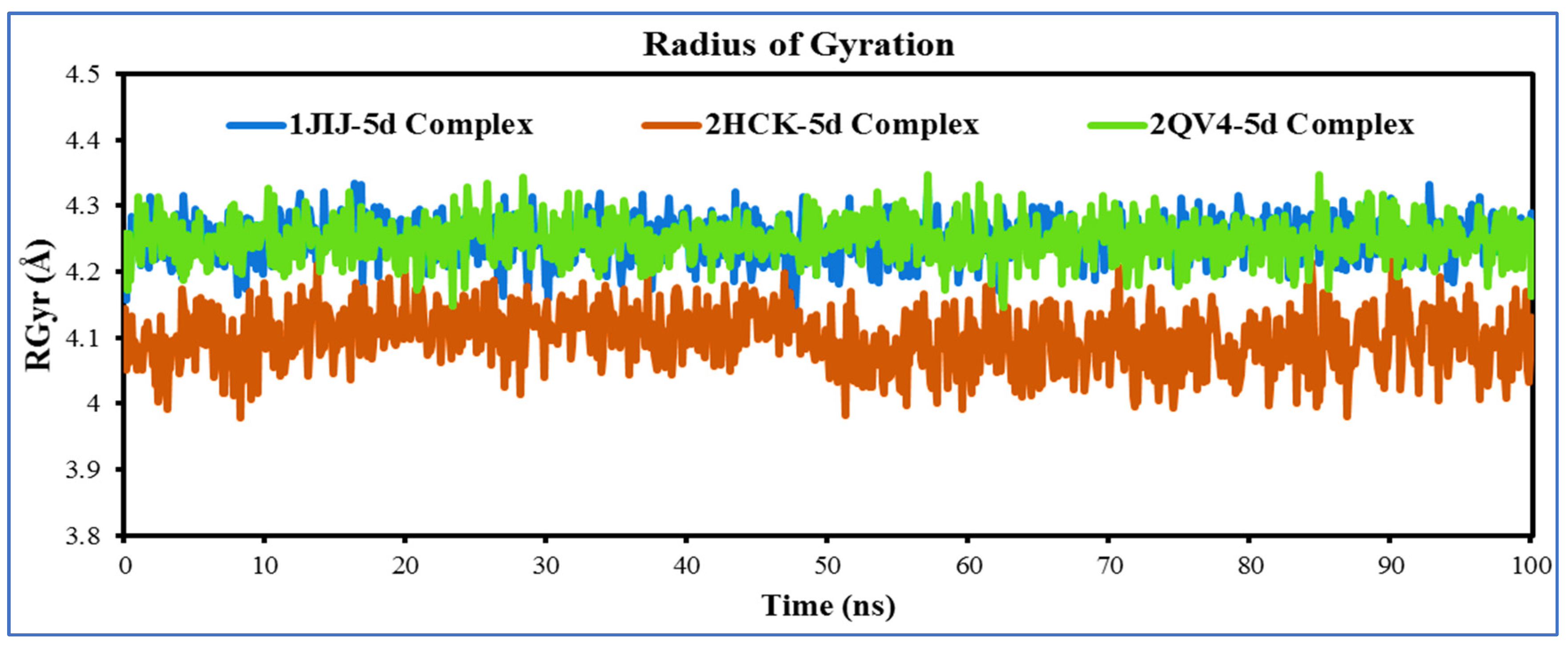
2.5.2. Molecular Dynamic Simulations
2.5.3. PCA Analysis
2.5.4. MMGBSA Binding Free Energy Analysis
3. Materials and Methods
3.1. General Experimental Methods
General Procedure for the Preparation of Spiro-Indenoquinoxaline Pyrrolizidines 5a–m
3.2. Pharmacological Study
3.2.1. Antimicrobial Activity
3.2.2. Antioxidant Activity
3.2.3. α-Amylase Inhibitory Assay
3.3. Computational Study
3.3.1. Molecular Docking
3.3.2. Molecular Dynamics (MD) Simulation
3.3.3. ADME Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Lammie, S.L.; Hughes, J.M. Antimicrobial resistance, food safety, and one health: The need for convergence. Annu. Rev. Food Sci. Technol. 2016, 7, 287–312. [Google Scholar] [CrossRef] [PubMed]
- Rana, M.S.; Usman, M.; Salman, M.; Alam, M.M.; Ikram, A.; Umair, M.; Qadir, M. Potential impact of COVID-19 pandemic on escalating antimicrobial resistance in Pakistan. J. Infect. 2021, 83, 12–13. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Álvarez, S.; Sanz, S.; Olarte, C.; Hidalgo-Sanz, R.; Carvalho, I.; Fernández-Fernández, R.; Torres, C. Antimicrobial Resistance in Escherichia coli from the Broiler Farm Environment, with Detection of SHV-12-Producing Isolates. Antibiotics 2022, 11, 444. [Google Scholar] [CrossRef] [PubMed]
- Larsson, D.G.; Flach, C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022, 20, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, S.; Rahman, N.A.A.; Peile, E.; Rahman, M.; Sartelli, M.; Hassali, M.A.; Haque, M. Microbial resistance movements: An overview of global public health threats posed by antimicrobial resistance, and how best to counter. Front. Public Health 2020, 8, 535668. [Google Scholar] [CrossRef]
- Hajlaoui, H.; Arraouadi, S.; Mighri, H.; Ghannay, S.; Aouadi, K.; Adnan, M.; Kadri, A. HPLC-MS profiling, antioxidant, antimicrobial, antidiabetic, and cytotoxicity activities of Arthrocnemum indicum (Willd.) Moq. extracts. Plants 2022, 11, 232. [Google Scholar] [CrossRef]
- Ghannay, S.; Snoussi, M.; Messaoudi, S.; Kadri, A.; Aouadi, K. Novel enantiopure isoxazolidine and C-alkyl imine oxide derivatives as potential hypoglycemic agents: Design, synthesis, dual inhibitors of α-amylase and α-glucosidase, ADMET and molecular docking study. Bioorg. Chem. 2020, 104, 104270. [Google Scholar] [CrossRef]
- Aouadi, K.; Hajlaoui, H.; Arraouadi, S.; Ghannay, S.; Snoussi, M.; Kadri, A. Phytochemical Profiling, Antimicrobial and α-Glucosidase Inhibitory Potential of Phenolic-Enriched Extracts of the Aerial Parts from Echium humile Desf.: In Vitro Combined with In Silico Approach. Plants 2022, 11, 1131. [Google Scholar] [CrossRef]
- Hajlaoui, H.; Arraouadi, S.; Noumi, E.; Aouadi, K.; Adnan, M.; Khan, M.A.; Kadri, A.; Snoussi, M. Antimicrobial, antioxidant, anti-acetylcholinesterase, antidiabetic, and pharmacokinetic properties of Carum carvi L. and Coriandrum sativum L. essential oils alone and in combination. Molecules 2021, 26, 3625. [Google Scholar] [CrossRef]
- Kadri, A.; Aouadi, K. In vitro antimicrobial and α-glucosidase inhibitory potential of enantiopure cycloalkylglycine derivatives: Insights into their in silico pharmacokinetic, druglikeness, and medicinal chemistry properties. J. Appl. Pharm. Sci. 2020, 10, 107–115. [Google Scholar]
- Kim, E.J.; Ha, K.H.; Kim, D.J.; Choi, Y.H. Diabetes and the risk of infection: A national cohort study. Diabetes Metab. J. 2019, 43, 804. [Google Scholar] [CrossRef] [PubMed]
- Sundarram, A.; Murthy, T.P.K. α-amylase production and applications: A review. J. Appl. Environ. Microbiol. 2014, 2, 166–175. [Google Scholar]
- Ghabi, A.; Brahmi, J.; Alminderej, F.; Messaoudi, S.; Vidald, S.; Kadri, A.; Aouadi, K. Multifunctional isoxazolidine derivatives as α-amylas e and α-glucosidase inhibitors. Bioorg. Chem. 2020, 98, 103713. [Google Scholar] [CrossRef] [PubMed]
- Dirir, A.M.; Daou, M.; Yousef, A.F.; Yousef, L.F. A review of alpha-glucosidase inhibitors from plants as potential candidates for the treatment of type-2 diabetes. Phytochem. Rev. 2022, 21, 1049–1079. [Google Scholar] [CrossRef]
- Tseng, C.H.; Chen, Y.R.; Tzeng, C.C.; Liu, W.; Chou, C.K.; Chiu, C.C.; Chen, Y.L. Discovery of indeno[1,2-b]quinoxaline derivatives as potential anticancer agents. Eur. J. Med. Chem. 2016, 27, 258–273. [Google Scholar] [CrossRef] [PubMed]
- Tseng, C.-H.; Tzeng, C.-C.; Yang, C.-L.; Lu, P.-J.; Chen, H.-L.; Li, H.-Y.; Chuang, Y.-C.; Yang, C.-N.; Chen, Y.-L. Synthesis and antiproliferative evaluation of certain indeno [1, 2-c] quinoline derivatives. Part 2. J. Med. Chem. 2010, 53, 6164–6179. [Google Scholar] [CrossRef]
- Schepetkin, I.A.; Khlebnikov, A.I.; Potapov, A.S.; Kovrizhina, A.R.; Matveevskaya, V.V.; Belyanin, M.L.; Atochin, D.N.; Zanoza, S.O.; Gaidarzhy, N.M.; Lyakhov, S.A.; et al. Synthesis, biological evaluation, and molecular modeling of 11H-indeno[1,2-b]quinoxalin-11-one derivatives and tryptanthrin-6-oxime as c-Jun N-terminal kinase inhibitors. Eur. J. Med. Chem. 2019, 161, 179–191. [Google Scholar] [CrossRef]
- Almansour, A.I.; Arumugam, N.; Soliman, S.M.; Krishnamoorthy, B.S.; Halet, J.-F.; Priya, R.V.; Suresh, J.; Al-thamili, D.M.; Al-aizari, F.A.; Kumar, R.S. Stereoselective synthesis, structure and DFT studies on fluoro- and nitro- substituted spirooxindole-pyrrolidine heterocyclic hybrids. J. Mol. Struct. 2021, 1237, 130396. [Google Scholar] [CrossRef]
- Bhaskar, G.; Arun, Y.; Balachandran, C.; Saikumar, C.; Perumal, P.T. Synthesis of novel spirooxindole derivatives by one pot multicomponent reaction and their antimicrobial activity. Eur. J. Med. Chem. 2012, 51, 79–91. [Google Scholar] [CrossRef]
- Haddad, S.; Boudriga, S.; Akhaja, T.N.; Raval, J.P.; Porzio, F.; Soldera, A.; Askri, M.; Knorr, M.; Rousselin, Y.; Kubicki, M.M.; et al. A Strategic Approach to the Synthesis of Functionalized Spirooxindole Pyrrolidine Derivatives: In vitro Antibacterial, Antifungal, Antimalarial and Antitubercular studies. New J. Chem. 2015, 39, 520–528. [Google Scholar] [CrossRef]
- Rajanarendar, E.; Ramakrishna, S.; Reddy, K.G.; Nagaraju, D.; Reddy, Y.N. A facile synthesis, anti-inflammatory and analgesic activity of isoxazolyl-2,3-dihydrospiro[benzo[f] isoindole-1,30-indoline]-20,4,9-triones. Bioorg. Med. Chem. Lett. 2013, 23, 3954–3958. [Google Scholar] [CrossRef] [PubMed]
- Toumi, A.; Boudriga, S.; Hamden, K.; Sobeh, M.; Cheurfa, M.; Askri, M.; Knorr, M.; Strohmann, B.C.L. Synthesis, antidiabetic activity and molecular docking study of rhodanine-substitued spirooxindole pyrrolidine derivatives as novel α-amylase inhibitors. Bioorg. Chem. 2021, 106, 104507. [Google Scholar] [CrossRef] [PubMed]
- Hammouda, M.B.; Boudriga, S.; Hamden, K.; Askri, M.; Knorr, M.; Strohmann, C.; Brieger, L.; Krupp, A.; Anouar, E.H.; Snoussi, M.; et al. New Spiropyrrolothiazole Derivatives Bearing an Oxazolone Moiety as Potential Antidiabetic Agent: Design, Synthesis, Crystal Structure, Hirshfeld Surface Analysis, ADME and Molecular Docking Studies. J. Mol. Struct. 2022, 1254, 132398. [Google Scholar] [CrossRef]
- Kathirvelan, D.; Haribabu, J.; Reddy, B.S.R.; Balachandran, C.; Duraipandiyan, V. Facile and diastereoselective synthesis of 3,20-spiropyrrolidine-oxindoles derivatives, their molecular docking and antiproliferative activities. Bioorg. Med. Chem. Lett. 2015, 25, 389–399. [Google Scholar] [CrossRef]
- Arumugam, N.; Almansour, A.I.; Kumar, R.S.; Kotresha, D.; Saiswaroop, R.; Venketesh, S. Dispiropyrrolidinylpiperidone embedded indeno[1,2-b]quinoxaline heterocyclic hybrids: Synthesis, cholinesterase inhibitory activity and their molecular docking simulation. Bioorg. Med. Chem. 2019, 27, 2621–2628. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Yu, D.Q.; Liu, H.M. Spirooxindoles: Promising scaffolds for anticancer agents. Eur. J. Med. Chem. 2015, 97, 673–698. [Google Scholar] [CrossRef]
- Rajesh, S.M.; Perumal, S.; Menendez, J.C.; Yogeeswari, P.; Sriram, D. Antimycobacterial activity of spirooxindolo-pyrrolidine, pyrrolizine and pyrrolothiazole hybrids obtained by a three-component regio- and stereoselective 1,3-dipolar cycloaddition. Med. Chem. Commun. 2011, 2, 626–630. [Google Scholar] [CrossRef]
- Asad, M.; Arshad, M.N.; Asiri, A.M.; Khan, S.A.; Rehan, M.; Oves, M. Synthesis, Characterization, Molecular Docking and Antimicrobial Activity of Novel Spiropyrrolidine Derivatives. Polycycl. Aromat. Compd. 2021, 42, 5385–5397. [Google Scholar] [CrossRef]
- Almansour, A.I.; Kumar, R.S.; Beevi, F.; Nasrolahi Shirazi, A.; Osman, H.; Ismail, R.; Ali, M.A. Facile, regio-and diastereoselective synthesis of spiro-pyrrolidine and pyrrolizine derivatives and evaluation of their antiproliferative activities. Molecules 2014, 19, 10033–10055. [Google Scholar] [CrossRef]
- Arumugam, N.; Suresh babu, P.; Angamuthu, G.; Kotresha, D.; Manohar, T.S.; Venketesh, S. Spiropyrrolidine/spiroindolizino [6, 7-b] indole heterocyclic hybrids: Stereoselective synthesis, cholinesterase inhibitory activity and their molecular docking study. Bioorg. Chem. 2018, 79, 64–71. [Google Scholar] [CrossRef]
- Haddaji, F.; Papetti, A.; Noumi, E.; Colombo, R.; Deshpande, S.; Aouadi, K.; Adnan, M.; Kadri, A.; Selmi, B.; Snoussi, M. Bioactivities and in silico study of Pergularia tomentosa L. phytochemicals as potent antimicrobial agents targeting type IIA topoisomerase, TyrRS, and Sap1 virulence proteins. Environ. Sci. Poll. Res. 2021, 28, 25349–25367. [Google Scholar] [CrossRef] [PubMed]
- Othman, I.M.M.; Gad-Elkareem, M.A.M.; Anouar, E.H.; Snoussi, M.; Aouadi, K.; Kadri, A. Novel fused pyridine derivatives containing pyrimidine moiety as prospective tyrosyl-tRNA synthetase inhibitors: Design, synthesis, pharmacokinetics and molecular docking studies. J. Mol. Struct. 2020, 1219, 128651. [Google Scholar] [CrossRef]
- Ben Mefteh, F.; Daoud, A.; Bouket, A.C.; Thissera, B.; Kadri, Y.; Cherif-Silini, H.; Eshelli, M.; Alenezi, F.N.; Vallat, A.; Oszako, T.; et al. Date Palm Tree’s Root-Derived Endophytes as Fungal Cell Factories for Diverse Bioactive Metabolites. Int. J. Mol. Sci. 2018, 19, 1986. [Google Scholar] [CrossRef] [PubMed]
- Ghannay, S.; Kadri, A.; Aouadi, K. Synthesis, in vitro antimicrobial assessment, and computational investigation of pharmacokinetic and bioactivity properties of novel trifluoromethylated compounds using in silico ADME and toxicity prediction tools. Monatsh. Chem. 2020, 151, 267–280. [Google Scholar] [CrossRef]
- Othman, I.M.M.; Gad-Elkareem, M.A.M.; Anouar, E.H.; Aouadi, K.; Kadri, A.; Snoussi, M. Design, synthesis ADMET and molecular docking of new imidazo[4,5-b]pyridine-5-thione derivatives as potential tyrosyl-tRNA synthetase inhibitors. Bioorg. Chem. 2020, 102, 104105. [Google Scholar] [CrossRef]
- Othman, I.M.M.; Gad-Elkareem, M.A.M.; Radwan, H.A.; Badraoui, R.; Aouadi, K.; Snoussi, M.; Kadri, A. Synthesis, Structure-Activity Relationship and in silico Studies of Novel Pyrazolothiazole and Thiazolopyridine Derivatives as Prospective Antimicrobial and Anticancer Agents. ChemistrySelect 2021, 6, 7860–7872. [Google Scholar] [CrossRef]
- Alminderej, F.; Bakari, S.; Almundarij, T.I.; Snoussi, M.; Aouadi, K.; Kadri, A. Antimicrobial and Wound Healing Potential of a New Chemotype from Piper cubeba L. Essential Oil and In Silico Study on S. aureus tyrosyl-tRNA Synthetase Protein. Plants 2021, 10, 205–224. [Google Scholar] [CrossRef]
- Othman, I.M.M.; Gad-Elkareem, M.A.M.; Anouar, E.H.; Aouadi, K.; Snoussi, M.; Kadri, A. New substituted pyrazolones and dipyrazolotriazines as promising tyrosyl-tRNA synthetase and peroxiredoxin-5 inhibitors: Design, synthesis, molecular docking and structure-activity relationship (SAR) analysis. Bioorg. Chem. 2021, 109, 104704. [Google Scholar] [CrossRef]
- Mseddi, K.; Alimi, F.; Noumi, E.; Veettil, V.N.; Deshpande, S.; Adnan, M.; Hamdi, A.; Elkahoui, S.; Alghamdi, A.; Kadri, A.; et al. Thymus musilii Velen. as a promising source of potent bioactive compounds with its pharmacological properties: In vitro and in silico analysis. Arab. J. Chem. 2020, 13, 6782–6801. [Google Scholar] [CrossRef]
- Felhi, S.; Hajlaoui, H.; Ncir, M.; Bakari, S.; Ktari, N.; Saoudi, M.; Gharsallah, N.; Kadri, A. Nutritional, phytochemical and antioxidant evaluation and FT-IR analysis of freeze-dried extracts of Ecballium elaterium fruit juice from three localities. Food Sci. Technol. 2016, 36, 646–655. [Google Scholar] [CrossRef]
- Felhi, S.; Saoudi, M.; Daoud, A.; Hajlaoui, H.; Ncir, M.; Chaabane, R.; El Feki, A.; Gharsallah, N.; Kadri, A. Investigation of phytochemical contents, in vitro antioxidant and antibacterial behavior and in vivo anti-inflammatory potential of Ecballium elaterium methanol fruits extract. Food Sci. Technol. 2017, 37, 558–563. [Google Scholar] [CrossRef]
- Bakari, S.; Hajlaoui, H.; Daoud, A.; Mighri, H.; Ross-Garcia, J.M.; Gharsallah, N.; Kadri, A. Phytochemicals, antioxidant and antimicrobial potentials and LC-MS analysis of hydroalcoholic extracts of leaves and flowers of Erodium glaucophyllum collected from Tunisian Sahara. Food Sci. Biotechnol. 2018, 38, 310–317. [Google Scholar] [CrossRef]
- Ghannay, S.; Bakari, S.; Ghabi, A.; Kadri, A.; Msaddek, M.; Aouadi, K. Stereoselective synthesis of enantiopure N-substituted pyrrolidin-2,5-dione derivatives by 1,3-dipolar cycloaddition and assessment of their in vitro antioxidant and antibacterial activities. Bioorg. Med. Chem. Lett. 2017, 27, 2302–2307. [Google Scholar] [CrossRef] [PubMed]
- Ghannay, S.; Bakari, S.; Msaddek, M.; Vidal, S.; Kadri, A.; Aouadi, K. Design, synthesis, molecular properties and in vitro antioxidant and antibacterial potential of novel enantiopure isoxazolidine derivatives. Arab. J. Chem. 2020, 13, 2121–2131. [Google Scholar] [CrossRef]
- Noumi, E.; Snoussi, M.; Anouar, E.H.; Alreshidi, M.; Veettil, V.N.; Elkahoui, S.; Adnan, M.; Patel, M.; Kadri, A.; Aouadi, K.; et al. HR-LCMS-Based Metabolite Profiling, Antioxidant, and Anticancer Properties of Teucrium polium L. Methanolic Extract: Computational and In Vitro Study. Antioxidants 2020, 9, 1089. [Google Scholar] [CrossRef]
- Alminderej, F.; Bakari, S.; Almundarij, T.I.; Snoussi, M.; Aouadi, K.; Kadri, A. Antioxidant Activities of a New Chemotype of Piper cubeba L. Fruit Essential Oil (Methyleugenol/Eugenol): In Silico Molecular Docking and ADMET Studies. Plants 2020, 9, 1534. [Google Scholar] [CrossRef] [PubMed]
- Brahmi, J.; Ghannay, S.; Bakari, S.; Kadri, A.; Aouadi, K.; Msaddek, M.; Vidal, S. Unprecedented stereoselective synthesis of 3-methylisoxazolidine-5-aryl-1,2,4-oxadiazoles via 1,3-dipolar cycloaddition and study of their in vitro antioxidant activity. Synth. Commun. 2016, 46, 2037–2044. [Google Scholar] [CrossRef]
- Badraoui, R.; Rebai, T.; Elkahoui, S.; Alreshidi, M.; Veettil, V.N.; Noumi, E.; Al-Motair, K.A.; Aouadi, K.; Kadri, A.; De Feo, V.; et al. Allium subhirsutum L. as a Potential Source of Antioxidant and Anticancer Bioactive Molecules: HR-LCMS Phytochemical Profiling, In Vitro and In Vivo Pharmacological Study. Antioxidants 2020, 9, 1003. [Google Scholar] [CrossRef] [PubMed]
- Kutyashev, I.B.; Barkov, A.Y.; Zimnitskiy, N.S.; Korotaev, V.Y.; Sosnovskikh, V.Y. Different behavior of azomethine ylides derived from 11 H-indeno [1, 2-b] quinoxalin-11-one and proline/sarcosine in reactions with 3-nitro-2 H-chromenes. Chem. Heterocycl. 2019, 55, 861–874. [Google Scholar] [CrossRef]
- Zimnitskiy, N.S.; Barkov, A.Y.; Ulitko, M.V.; Kutyashev, I.B.; Korotaev, V.Y.; Sosnovskikh, V.Y. An expedient synthesis of novel spiro [indenoquinoxaline-pyrrolizidine]-pyrazole conjugates with anticancer activity from 1, 5-diarylpent-4-ene-1, 3-diones through the 1, 3-dipolar cycloaddition/ cyclocondensation sequence. New J. Chem. 2020, 44, 16185–16199. [Google Scholar] [CrossRef]
- Rajesh, S.M.; Bala, B.D.; Perumal, S. Multi-component, 1, 3-dipolar cycloaddition reactions for the chemo-, regio-and stereoselective synthesis of novel hybrid spiroheterocycles in ionic liquid. Tetrahedron. Lett. 2012, 53, 5367–5371. [Google Scholar] [CrossRef]
- Ayipo, Y.O.; Ahmad, I.; Najib, Y.S.; Sheu, S.K.; Patel, H.; Mordi, M.N. Molecular modelling and structure-activity relationship of a natural derivative of o-hydroxybenzoate as a potent inhibitor of dual NSP3 and NSP12 of SARS-CoV-2: In silico study. J. Biomol. Struct. Dyn. 2022. ahead of print. [Google Scholar] [CrossRef]
- Abdelgawad, M.A.; Oh, J.M.; Parambi, D.G.T.; Kumar, S.; Musa, A.; Ghoneim, M.M.; Nayl, A.A.; El-Ghorab, A.H.; Ahmad, I.; Patel, H.; et al. Development of bromo- and fluoro-based α, β-unsaturated ketones as highly potent MAO-B inhibitors for the treatment of Parkinson’s disease. J. Mol. Struct. 2022, 1266, 133545. [Google Scholar] [CrossRef]
- Ghosh, S.; Das, S.; Ahmad, I.; Patel, H. In silico validation of anti-viral drugs obtained from marine sources as a potential target against SARS-CoV-2 Mpro. J. Indian Chem. Soc. 2021, 98, 100272. [Google Scholar] [CrossRef]
- Farhan, M.M.; Guma, M.A.; Rabeea, M.A.; Ahmad, I.; Patel, H. Synthesizes, Characterization, Molecular docking and in vitro Bioactivity study of new compounds containing Triple Beta Lactam Rings. J. Mol. Struct. 2022, 1269, 133781. [Google Scholar] [CrossRef]
- Ahmad, I.; Akand, S.R.; Shaikh, M. Synthesis, molecular modelling study of the methaqualone analogues as anti-convulsant agent with improved cognition activity and minimized neurotoxicity. J. Mol. Struct. 2022, 1251, 131972. [Google Scholar] [CrossRef]
- Osmaniye, D.; Karaca, Ş.; Kurban, B.; Baysal, M.; Ahmad, I.; Patel, H.; Özkay, Y.; Asım Kaplancıklı, Z. Design, synthesis, molecular docking and molecular dynamics studies of novel triazolothiadiazine derivatives containing furan or thiophene rings as anticancer agents. Bioorg. Chem. 2022, 122, 105709. [Google Scholar] [CrossRef]
- Girase, R.; Ahmad, I.; Pawara, R.; Patel, H. Optimizing cardio, hepato and phospholipidosis toxicity of the Bedaquiline by chemoinformatics and molecular modelling approach. SAR and QSAR in environmental research, 1–21. Advance 2022. online publication. [Google Scholar] [CrossRef]
- Ahmad, I.; Pawara, R.H.; Girase, R.T.; Pathan, A.Y.; Jagatap, V.R.; Desai, N.; Ayipo, Y.O.; Surana, S.J.; Patel, H. Synthesis, Molecular Modeling Study, and Quantum-Chemical-Based Investigations of Isoindoline-1, 3-diones as Antimycobacterial Agents. ACS Omega 2022, 7, 21820–21844. [Google Scholar] [CrossRef]
- Piang-Siong, W.; de Caro, P.; Marvilliers, A.; Chasseray, X.; Payet, B.; Shum Cheong Sing, A.; Illien, B. Contribution of trans-aconitic acid to DPPH scavenging ability in different media. Food Chem. 2017, 214, 447–452. [Google Scholar] [CrossRef]
- Tuberoso, C.I.G.; Boban, M.; Bifulco, E.; Budimir, D.; Pirisi, F.M. Antioxidant capacity and vasodilatory properties of Mediterranean food: The case of Cannonau wine, myrtle berries liqueur and strawberry-tree honey. Food Chem. 2013, 140, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Bonilla, P.; Gandia-Herrero, F.; Matencio, A.; Garcia-Carmona, F.; Lopez-Nicolas, J.M. Comparative study of the antioxidant capacity of four stilbenes using ORAC, ABTS+, and FRAP techniques. Food Anal. Method 2017, 10, 2994–3000. [Google Scholar] [CrossRef]
- Amamou, S.; Lazreg, H.; Hafsa, J.; Majdoub, H.; Rihouey, C.; Le Cerf, D.; Achour, L. Effect of extraction condition on the antioxidant, antiglycation and α-amylase inhibitory activities of Opuntia macrorhiza fruit peels polysaccharides. LWT 2020, 127, 109411. [Google Scholar] [CrossRef]
- Patel, H.; Ansari, A.; Pawara, R.; Ansari, I.; Jadhav, H.; Surana, S. Design and synthesis of novel 2,4-disubstituted aminopyrimidines: Reversible non-covalent T790M EGFR inhibitors. J. Recept. Signal Transduct. Res. 2018, 38, 393–412. [Google Scholar] [CrossRef]
- Patel, H.; Ahmad, I.; Jadhav, H.; Pawara, R.; Lokwani, D.; Surana, S. Investigating the Impact of Different Acrylamide (Electrophilic Warhead) on Osimertinib’s Pharmacological Spectrum by Molecular Mechanic and Quantum Mechanic Approach. Comb. Chem. High Throughput Screen. 2021, 25, 149–166. [Google Scholar] [CrossRef]
- Shaw, D.E. Research, Schrödinger Release (2021-1). Desmond Molecular Dynamics System. Maestro-Desmond Interoperability Tools. Available online: https://www.schrodinger.com/products/desmond (accessed on 7 September 2022).
- Chaudhari, B.; Patel, H.; Thakar, S.; Ahmad, I.; Bansode, D. Optimizing the Sunitinib for cardio-toxicity and thyro-toxicity by scaffold hopping approach. Silico Pharmacol. 2022, 10, 10. [Google Scholar] [CrossRef]
- Ayipo, Y.O.; Alananzeh, W.A.; Ahmad, I.; Patel, H.; Mordi, M.N. Structural modelling and in silico pharmacology of β-carboline alkaloids as potent 5-HT1A receptor antagonists and reuptake inhibitors. J. Biomol. Struct. Dyn. 2022, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, K.K.; Ahmad, I.; Pati, S.; Ghosh, A.; Sarkar, T.; Rabha, B.; Patel, H.; Baishya, D.; Edinur, H.A.; Abdul Kari, Z.; et al. Potent Bioactive Compounds from Seaweed Waste to Combat Cancer Through Bioinformatics Investigation. Front. Nutr. 2022, 9, 889276. [Google Scholar] [CrossRef]
- Snoussi, M.; Redissi, A.; Mosbah, A.; De Feo, V.; Adnan, M.; Aouadi, K.; Alreshidi, M.; Patel, M.; Kadri, A.; Noumi, E. Emetine, a potent alkaloid for the treatment of SARS-CoV-2 targeting papain-like protease and non-structural proteins: Pharmacokinetics, molecular docking and dynamic studies. J. Biomol. Struct. Dyn 2021. ahead of print. [Google Scholar] [CrossRef]
- Aouadi, K.; Hajlaoui, H.; Arraouadi, S.; Ghannay, S.; Snoussi, M.; Kadri, A. HPLC/MS Phytochemical Profiling with Antioxidant Activities of Echium humile Desf. Extracts: ADMET Prediction and Computational Study Targeting Human Peroxiredoxin 5Receptor. Agronomy 2021, 11, 2165. [Google Scholar] [CrossRef]






| Entry | Solvent | T (°C) | Time (h) | Yield b (%) |
|---|---|---|---|---|
| 1 | CH3CN | 25 | 8 | - |
| 2 | CH3CN | 80 | 4 | 35 |
| 3 | CH3CN | 80 | 12 | 43 |
| 4 | MeOH | 25 | 8 | - |
| 5 | MeOH | 64 | 4 | 81 |
| 6 | MeOH | 64 | 24 | 81 |
| Entry | MBC (mM), MFC (mM), MBC/MIC and MFC/MIC | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram-Positive Bacteria | Gram-Negative Bacteria | Fungi | ||||||||||||||||
| S. aureus ATCC 25923 | M. luteus NCIMB 8166 | E. coli ATCC 25922 | P. aeruginosa ATCC 27853 | C. albicans ATCC 90028 | C. krusei ATCC 6258 | |||||||||||||
| MIC | MBC | MBC/ MIC | MIC | MBC | MBC/MIC | MIC | MBC | MBC/MIC | MIC | MBC | MBC/MIC | MIC | MFC | MFC/MIC | MIC | MFC | MFC/MIC | |
| 5a | 135.12 | 270.24 | 2 | 135.12 | - | - | 270.24 | 540.49 | 2 | 135.12 | 540.49 | 4 | 135.12 | 135.12 | 1 | 67.56 | 67.56 | 1 |
| 5b | 65.57 | 524.58 | 8 | 524.58 | - | - | 262.29 | 524.58 | 2 | 131.14 | 524.58 | 4 | 131.14 | 262.29 | 2 | 65.57 | 65.57 | 1 |
| 5c | 31.71 | 507.54 | 16 | 126.88 | - | - | 253.77 | 507.54 | 4 | 253.77 | 507.54 | 2 | 63.44 | 63.44 | 1 | 63.44 | 63.44 | 1 |
| 5d | 3.95 | 126.88 | 32 | 63.44 | 253.77 | 4 | 126.88 | 507.54 | 4 | 126.88 | 507.54 | 4 | 31.71 | 126.88 | 4 | 15.85 | 15.85 | 1 |
| 5e | 15.71 | 251.51 | 16 | 251.51 | 251.51 | 1 | 251.51 | 503.02 | 2 | 125.75 | 503.02 | 4 | 62.87 | 62.87 | 1 | 31.42 | 31.42 | 1 |
| 5f | 115.43 | 230.86 | 2 | 230.86 | - | - | 230.86 | 461.73 | 2 | 115.43 | 230.86 | 2 | 57.71 | 57.71 | 1 | 57.71 | 57.71 | 1 |
| 5g | 16.25 | 260.12 | 16 | 130.06 | - | - | 260.12 | 520.25 | 2 | 130.06 | 260.12 | 2 | 65.03 | 65.03 | 1 | 32.50 | 32.50 | 1 |
| 5h | 33.33 | 533.53 | 16 | 133.38 | 133.38 | 1 | 266.76 | 533.53 | 2 | 133.38 | 266.76 | 2 | 66.69 | 266.76 | 4 | 66.69 | 66.69 | 1 |
| 5i | 125.75 | 503.02 | 4 | 251.51 | 251.51 | 1 | 251.51 | 503.02 | 2 | 125.75 | 503.02 | 4 | 125.75 | 503.02 | - | 31.42 | 31.42 | 1 |
| 5j | 14.94 | 478.38 | 32 | 239.19 | 478.38 | 2 | 239.19 | 478.38 | 2 | 239.19 | 478.38 | 2 | 119.59 | 239.19 | 2 | 59.79 | 59.79 | 1 |
| 5k | 54.68 | 437.47 | 8 | 218.73 | - | - | 218.73 | 437.47 | 2 | 218.73 | 437.47 | 2 | 218.73 | 218.73 | 1 | 54.68 | 54.68 | 1 |
| 5l | 28.02 | 448.47 | 16 | 112.11 | 224.23 | 2 | 224.23 | 448.47 | 2 | 112.11 | 448.47 | 4 | 56.05 | 448.47 | 8 | 28.02 | 28.02 | 1 |
| 5m | 24.54 | 196.43 | 8 | 98.21 | - | - | 196.43 | 196.43 | 1 | 98.21 | 392.87 | 4 | 98.21 | 98.21 | 1 | 49.10 | 49.10 | 1 |
| Tetracycline | 576.01 | 288.00 | 2 | 576.01 | 1152.02 | 2 | 281.25 | 288.00 | 1 | 576.01 | 1152.02 | 2 | - | - | - | - | - | - |
| Amphotericin B | - | - | - | - | - | - | - | 5.41 | 5.41 | 1 | 5.41 | 5.41 | 1 | |||||
| Entry | IC50 (mM) | |||
|---|---|---|---|---|
| DPPH | ABTS | FRAP | α-Amylase | |
| 5a | 15.36 ± 0.65 | 49.19 ± 0.46 | 17.02 ± 0.52 | 1.19 ± 0.02 |
| 5b | 33.74 ± 0.004 | 57.51 ± 0.55 | 63.26 ± 0.21 | 2.00 ± 0.30 |
| 5c | 3.26 ± 0.32 | 7.03 ± 0.07 | 3.69 ± 0.72 | 0.92 ± 0.10 |
| 5d | 7.44 ± 0.15 | 9.78 ± 0.30 | 8.09 ± 0.82 | 0.55 ± 0.38 |
| 5e | 6.12 ± 0.01 | 12.16 ± 0.18 | 6.54 ± 0.18 | 1.91 ± 0.37 |
| 5f | 16.13 ± 0.39 | 18.12 ± 0.53 | 7.93 ± 0.44 | 1.69 ± 0.28 |
| 5g | 26.03 ± 0.50 | 7.19 ± 0.11 | 20.54 ± 0.60 | 1.94 ± 0.37 |
| 5h | 19.13 ± 0.23 | 18.04 ± 0.13 | 6.32 ± 0.68 | 1.90 ± 0.054 |
| 5i | 7.80 ± 0.32 | 11.39 ± 0.36 | 5.98 ± 0.52 | 1.33 ± 0.72 |
| 5j | 19.20 ± 0.19 | 11.56 ± 0.05 | 5.98 ± 0.17 | 0.95 ± 0.14 |
| 5k | 123.44 ± 0.45 | 7.69 ± 0.36 | 6.03 ± 0.12 | 2.19 ± 0.23 |
| 5l | 15.28 ± 0.37 | 15.19 ± 0.31 | 3.26 ± 0.45 | 2.16 ± 0.35 |
| 5m | 47.29 ± 0.06 | 14.56 ± 0.26 | 7.38 ± 0.14 | 1.40 ± 0.11 |
| Trolox | 31.24 ± 3.67 | 99.88 ± 0.31 | 41.87 ± 2.07 | - |
| Acarbose | - | - | - | 1.19 ± 0.02 |
| Entry | 5c | 5d | 5e | 5i | 5j |
|---|---|---|---|---|---|
| Physicochemical Properties/Lipophilicity/Druglikeness | |||||
| Molecular weight | 492.57 | 492.57 | 496.99 | 496.99 | 522.59 |
| Num. heavy atoms | 37 | 37 | 36 | 36 | 39 |
| Num. arom. heavy atoms | 12 | 12 | 12 | 12 | 12 |
| Num. rotatable bonds | 0.33 | 0.33 | 0.31 | 0.31 | 0.35 |
| Num. H-bond acceptors | 2 | 2 | 1 | 1 | 3 |
| Num. H-bond donors | 6 | 6 | 5 | 5 | 7 |
| Molar Refractivity | 0 | 0 | 0 | 0 | 0 |
| TPSA | 155.61 | 155.61 | 154.12 | 154.12 | 162.10 |
| Consensus Log Po/w | 2.77 | 2.76 | 3.21 | 3.29 | 2.73 |
| Lipinski’s Rule | Yes | Yes | Yes | Yes | Yes |
| Bioavailability Score | 0.55 | 0.55 | 0.55 | 0.55 | 0.55 |
| Pharmacokinetics | |||||
| GI absorption | High | High | High | High | High |
| BBB permeant | No | No | Yes | Yes | No |
| P-gp substrate | Yes | Yes | No | No | Yes |
| CYP1A2 inhibitor | No | No | Yes | Yes | No |
| CYP2C19 inhibitor | Yes | Yes | Yes | Yes | Yes |
| CYP2C9 inhibitor | No | No | No | No | No |
| CYP2D6 inhibitor | No | No | No | No | No |
| CYP3A4 inhibitor | Yes | Yes | Yes | Yes | Yes |
| Log Kp (cm/s) | −7.74 | −7.74 | −7.30 | −7.30 | −7.95 |
| Compound | Docking Score | Glide Emodel | Glide Energy | Prime Energy | MMGBSA ΔG Bind |
|---|---|---|---|---|---|
| S. aureus Tyrosyl-tRNA Synthetase (1JIJ) | |||||
| 5d | −6.843 | −57.042 | −46.196 | −12980.2 | −43.02 |
| 5e | −4.834 | −30.317 | −27.625 | −12967.4 | −20.76 |
| 5j | −5.529 | −56.637 | −44.992 | −12980.4 | −47.93 |
| Reference a | −7.973 | −98.597 | −68.426 | 13103.87 | −63.92 |
| Tyrosine Kinase (2HCK) | |||||
| 5e | −5.804 | −41.828 | −39.753 | −17858.1 | −58.03 |
| 5d | −5.732 | −45.891 | −37.914 | −17853 | −53.11 |
| 5i | −5.288 | −41.24 | −36.51 | −17847.7 | −54.74 |
| 5c | −5.022 | −32.357 | −30.204 | −17827.1 | −34.18 |
| Reference a | −8.551 | −67.224 | −45.767 | −17945.56 | −59.30 |
| Human Pancreatic α-Amylase (2QV4) | |||||
| 5d | −6.182 | −41.759 | −35.02 | −22748.8 | −59.16 |
| 5c | −6.172 | −36.006 | −33.572 | −22749.8 | −51.25 |
| 5j | −6.146 | −46.534 | −38.443 | −22751.3 | −56.64 |
| Reference a | −8.141 | −99.527 | −74.699 | −22821.47 | −76.71 |
| MMGBSA Components (kcal/mol) | 1JIJ-5d Complex | 2HCK-5d Complex | 2QV4-5d Complex |
|---|---|---|---|
| ΔG Bind | −53.25 | −34.15 | −45.92 |
| ΔG Bind Coulomb | −9.72 | −6.28 | −3.55 |
| ΔG Bind H bond | −0.54 | −0.19 | −0.33 |
| ΔG Bind Lipo | −18.56 | −12.93 | −20.37 |
| ΔG Bind Solv GB | 34.23 | 19.24 | 16.57 |
| ΔG Bind VDW | −58.70 | −34.61 | −37.90 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouali, N.; Hammouda, M.B.; Ahmad, I.; Ghannay, S.; Thouri, A.; Dbeibia, A.; Patel, H.; Hamadou, W.S.; Hosni, K.; Snoussi, M.; et al. Multifunctional Derivatives of Spiropyrrolidine Tethered Indeno-Quinoxaline Heterocyclic Hybrids as Potent Antimicrobial, Antioxidant and Antidiabetic Agents: Design, Synthesis, In Vitro and In Silico Approaches. Molecules 2022, 27, 7248. https://doi.org/10.3390/molecules27217248
Bouali N, Hammouda MB, Ahmad I, Ghannay S, Thouri A, Dbeibia A, Patel H, Hamadou WS, Hosni K, Snoussi M, et al. Multifunctional Derivatives of Spiropyrrolidine Tethered Indeno-Quinoxaline Heterocyclic Hybrids as Potent Antimicrobial, Antioxidant and Antidiabetic Agents: Design, Synthesis, In Vitro and In Silico Approaches. Molecules. 2022; 27(21):7248. https://doi.org/10.3390/molecules27217248
Chicago/Turabian StyleBouali, Nouha, Manel Ben Hammouda, Iqrar Ahmad, Siwar Ghannay, Amira Thouri, Amal Dbeibia, Harun Patel, Walid Sabri Hamadou, Karim Hosni, Mejdi Snoussi, and et al. 2022. "Multifunctional Derivatives of Spiropyrrolidine Tethered Indeno-Quinoxaline Heterocyclic Hybrids as Potent Antimicrobial, Antioxidant and Antidiabetic Agents: Design, Synthesis, In Vitro and In Silico Approaches" Molecules 27, no. 21: 7248. https://doi.org/10.3390/molecules27217248
APA StyleBouali, N., Hammouda, M. B., Ahmad, I., Ghannay, S., Thouri, A., Dbeibia, A., Patel, H., Hamadou, W. S., Hosni, K., Snoussi, M., Adnan, M., Hassan, M. I., Noumi, E., Aouadi, K., & Kadri, A. (2022). Multifunctional Derivatives of Spiropyrrolidine Tethered Indeno-Quinoxaline Heterocyclic Hybrids as Potent Antimicrobial, Antioxidant and Antidiabetic Agents: Design, Synthesis, In Vitro and In Silico Approaches. Molecules, 27(21), 7248. https://doi.org/10.3390/molecules27217248













