The Molecular and Pathophysiological Functions of Members of the LNX/PDZRN E3 Ubiquitin Ligase Family
Abstract
1. Introduction
2. Structural Significance and Sequence Similarity of Members of the LNX/PDZRN E3 Ubiquitin Ligase Family
3. Molecular and Cellular Functions of LNX1 and LNX2
3.1. Recent Findings on the Roles of LNX1 and LNX2 in Synaptic Integrity
3.2. Role of LNX1 and LNX2 in Tissue and Organ Specification, and Vertebrate Development
3.3. Recent Findings on the Pathogenic Roles of LNX1 and LNX2
3.4. Role of LNX1 and LNX2 in Tumorigenesis
4. Molecular Functions of LNX3/PDZRN3/SEMCAP3
4.1. Cellular and Pathophysiological Functions of LNX3/PDZRN3/SEMCAP3
4.2. Other Pathological Roles of LNX3/PDZRN3/SEMCAP3, Particularly in Tumor Development
5. Possible Pathophysiological Roles of LNX4/PDZRN4
6. Conclusions
- (1)
- What are the molecular and cellular functions of LNX5?
- (2)
- What are the substrates of LNX4? Could LNX4 share substrates with LNX3?
- (3)
- Which are the post-translational modifiers of LNX proteins that may alter their molecular activities?
- (4)
- Why are only selected LNX1/2 substrates targeted by the UPS, whereas several miscellaneous proteins appear to bind to them? Are they all/are most of them LNX-binding proteins that can be modified through ubiquitylation?
- (5)
- Complete three-dimensional structures of individual LNX proteins must be validated in future studies for a better understanding of their molecular function.
- (6)
- Diverse types of genetically modified animals with altered LNX expression profiles should be established to evaluate the cellular functions of LNX proteins.
- (7)
- The developmental significance of LNX1 and LNX2 in teleosts and mammals appears to differ. The source of this difference should be identified.
- (8)
- Could LNX proteins serve as potential targets for drugs that interfere with their E3 ubiquitin ligase activity, inhibit their substrate-binding ability, or promote their function to facilitate substrate degradation?
- (9)
- Does the loss of N-terminal RING and Zn finger domains of LNX/PDZRN by alternative splicing event affect the function of E3 ubiquitin ligase activities of other variants retaining RING domain? It would be an interesting research topic by considering the discrete expression pattern of LNX1p70 and LNX1p80, and their opposite effects on the stability of EphB2 [56]. In addition, we also previously reported that N-terminal truncation of the endogenous lnx2a in zebrafish interfered with not only the E3 ubiquitin ligase activity of Lnx2a but also Lnx2b [21]. Thus it should be scrutinized in the future whether the dominant negative effects of N-terminal truncation of LNX/PDZRN by alternative splicing are critical yet generic mechanisms for fine-tuning of cellular proteostasis to be tightly controlled.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Li, W.; Bengtson, M.H.; Ulbrich, A.; Matsuda, A.; Reddy, V.A.; Orth, A.; Chanda, S.K.; Batalov, S.; Joazeiro, C.A.P. Genome-Wide and Functional Annotation of Human E3 Ubiquitin Ligases Identifies MULAN, a Mitochondrial E3 that Regulates the Organelle’s Dynamics and Signaling. PLoS ONE 2008, 3, e1487. [Google Scholar] [CrossRef]
- Clague, M.J.; Heride, C.; Urbé, S. The demographics of the ubiquitin system. Trends Cell Biol. 2015, 25, 417–426. [Google Scholar] [CrossRef]
- Lipkowitz, S.; Weissman, A.M. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer 2011, 11, 629–643. [Google Scholar] [CrossRef] [PubMed]
- Ro, H.; Hur, T.-L.; Rhee, M. Ubiquitin conjugation system for body axes specification in vertebrates. Anim. Cells Syst. 2015, 19, 87–95. [Google Scholar] [CrossRef]
- Bhat, K.P.; Greer, S.F. Proteolytic and non-proteolytic roles of ubiquitin and the ubiquitin proteasome system in transcriptional regulation. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2011, 1809, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Tsimokha, A.S.; Artamonova, T.O.; Diakonov, E.E.; Khodorkovskii, M.A.; Tomilin, A.N. Post-Translational Modifications of Extracellular Proteasome. Molecules 2020, 25, 3504. [Google Scholar] [CrossRef]
- Hanna, J.; Guerra-Moreno, A.; Ang, J.; Micoogullari, Y. Protein Degradation and the Pathologic Basis of Disease. Am. J. Pathol. 2019, 189, 94–103. [Google Scholar] [CrossRef]
- Thibaudeau, T.A.; Smith, D.M. A Practical Review of Proteasome Pharmacology. Pharmacol. Rev. 2019, 71, 170–197. [Google Scholar] [CrossRef]
- Rice, D.S.; Northcutt, G.M.; Kurschner, C. The Lnx Family Proteins Function as Molecular Scaffolds for Numb Family Proteins. Mol. Cell Neurosci. 2001, 18, 525–540. [Google Scholar] [CrossRef]
- Xie, Y.; Zhao, W.; Wang, W.; Zhao, S.; Tang, R.; Ying, K.; Zhou, Z.; Mao, Y. Identification of a human LNX protein containing multiple PDZ domains. Biochem. Genet. 2001, 39, 117–126. [Google Scholar] [CrossRef]
- Katoh, M.; Katoh, M. Identification and characterization of PDZRN3 and PDZRN4 genes in silico. Int. J. Mol. Med. 2004, 13, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M.; Katoh, M. Identification and characterization of human PDZRN4L gene and mouse Pdzrn4l gene in silico. Int. J. Mol. Med. 2004, 13, 923–927. [Google Scholar] [CrossRef] [PubMed]
- Romero, G.; Von Zastrow, M.; Friedman, P.A. Role of PDZ Proteins in Regulating Trafficking, Signaling, and Function of GPCRs: Means, Motif, and Opportunity. Adv. Pharm. 2011, 62, 279–314. [Google Scholar] [CrossRef]
- Wolting, C.D.; Griffiths, E.K.; Sarao, R.; Prevost, B.C.; Wybenga-Groot, L.E.; McGlade, C.J. Biochemical and Computational Analysis of LNX1 Interacting Proteins. PLoS ONE 2011, 6, e26248. [Google Scholar] [CrossRef]
- Guo, Z.; Song, E.; Ma, S.; Wang, X.; Gao, S.; Shao, C.; Hu, S.; Jia, L.; Tian, R.; Xu, T.; et al. Proteomics Strategy to Identify Substrates of LNX, a PDZ Domain-containing E3 Ubiquitin Ligase. J. Proteome Res. 2012, 11, 4847–4862. [Google Scholar] [CrossRef]
- Lenihan, J.A.; Saha, O.; Young, P. Proteomic analysis reveals novel ligands and substrates for LNX1 E3 ubiquitin ligase. PLoS ONE 2017, 12, e0187352. [Google Scholar] [CrossRef]
- Young, P. LNX1/LNX2 proteins: Functions in neuronal signalling and beyond. Neuronal Signal. 2018, 2, NS20170191. [Google Scholar] [CrossRef] [PubMed]
- Pece, S.; Confalonieri, S.; Romano, P.R.; Di Fiore, P.P. NUMB-ing down cancer by more than just a NOTCH. Biochim. et Biophys. Acta (BBA)-Bioenerg. 2011, 1815, 26–43. [Google Scholar] [CrossRef]
- Nie, J.; McGill, M.A.; Dermer, M.; Dho, S.E.; Wolting, C.D.; McGlade, C.J. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 2002, 21, 93–102. [Google Scholar] [CrossRef]
- Nie, J.; Li, S.S.-C.; McGlade, C.J. A Novel PTB-PDZ Domain Interaction Mediates Isoform-specific Ubiquitylation of Mammalian Numb. J. Biol. Chem. 2004, 279, 20807–20815. [Google Scholar] [CrossRef]
- Won, M.; Ro, H.; Dawid, I.B. Lnx2 ubiquitin ligase is essential for exocrine cell differentiation in the early zebrafish pancreas. Proc. Nat. Acad. Sci. USA 2015, 112, 12426–12431. [Google Scholar] [CrossRef] [PubMed]
- Nayak, D.; Sivaraman, J. Structural basis for the indispensable role of a unique zinc finger motif in LNX2 ubiquitination. Oncotarget 2015, 6, 34342–34357. [Google Scholar] [CrossRef] [PubMed]
- Hekstra, D.R.; White, K.I.; Socolich, M.A.; Henning, R.W.; Šrajer, R.W.H.V.; Ranganathan, R. Electric-field-stimulated protein mechanics. Nat. Cell Biol. 2016, 540, 400–405. [Google Scholar] [CrossRef]
- Nayak, D.; Sivaraman, J. Structure of LNX1:Ubc13 ~ Ubiquitin Complex Reveals the Role of Additional Motifs for the E3 Ligase Activity of LNX1. J. Mol. Biol. 2018, 430, 1173–1188. [Google Scholar] [CrossRef] [PubMed]
- Metzger, M.B.; Pruneda, J.N.; Klevit, R.E.; Weissman, A.M. RING-type E3 ligases: Master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta (BBA)-Bioenerg. 2014, 1843, 47–60. [Google Scholar] [CrossRef]
- Weiss, A.; Baumgartner, M.; Radziwill, G.; Dennler, J.; Moelling, K. c-Src is a PDZ interaction partner and substrate of the E3 ubiquitin ligase Ligand-of-Numb protein X1. FEBS Lett. 2007, 581, 5131–5136. [Google Scholar] [CrossRef]
- Lazzari, E.; El-Halawany, M.; De March, M.; Valentino, F.; Cantatore, F.; Migliore, C.; Onesti, S.; Meroni, G. Analysis of the Zn-Binding Domains of TRIM32, the E3 Ubiquitin Ligase Mutated in Limb Girdle Muscular Dystrophy 2H. Cells 2019, 8, 254. [Google Scholar] [CrossRef]
- Maitland, M.E.R.; Onea, G.; Chiasson, C.A.; Wang, X.; Ma, J.; Moor, S.E.; Barber, K.R.; Lajoie, G.A.; Shaw, G.S.; Schild-Poulter, C. The mammalian CTLH complex is an E3 ubiquitin ligase that targets its subunit muskelin for degradation. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Lu, Z.; Je, H.-S.; Young, P.; Gross, J.; Lu, B.; Feng, G. Regulation of synaptic growth and maturation by a synapse-associated E3 ubiquitin ligase at the neuromuscular junction. J. Cell Biol. 2007, 177, 1077–1089. [Google Scholar] [CrossRef]
- Ro, H.; Dawid, I.B. Organizer restriction through modulation of Bozozok stability by the E3 ubiquitin ligase Lnx-like. Nat. Cell Biol. 2009, 11, 1121–1127. [Google Scholar] [CrossRef]
- Fu, B.; Xue, W.; Zhang, H.; Zhang, R.; Feldman, K.; Zhao, Q.; Zhang, S.; Shi, L.; Pavani, K.C.; Nian, W.; et al. MicroRNA-325-3p Facilitates Immune Escape of Mycobacterium tuberculosis through Targeting LNX1 via NEK6 Accumulation to Promote Anti-Apoptotic STAT3 Signaling. mBio 2020, 11. [Google Scholar] [CrossRef]
- Lee, C.-H.; Yang, J.-R.; Chen, C.-Y.; Tsai, M.-H.; Hung, P.-F.; Chen, S.-J.; Chiang, S.-L.; Chang, H.; Lin, P. Novel STAT3 Inhibitor LDOC1 Targets Phospho-JAK2 for Degradation by Interacting with LNX1 and Regulates the Aggressiveness of Lung Cancer. Cancers 2019, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.K.; Potts, P.R. A Comprehensive Guide to the MAGE Family of Ubiquitin Ligases. J. Mol. Biol. 2017, 429, 1114–1142. [Google Scholar] [CrossRef] [PubMed]
- Sang, M.; Wang, L.; Ding, C.; Zhou, X.; Wang, B.; Wang, L.; Lian, Y.; Shan, B. Melanoma-associated antigen genes-an update. Cancer Lett 2011, 302, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.M.; Gao, J.; Wang, J.; Yang, M.; Potts, P.R. MAGE-RING Protein Complexes Comprise a Family of E3 Ubiquitin Ligases. Mol. Cell 2010, 39, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Kozakova, L.; Vondrova, L.; Stejskal, K.; Charalabous, P.; Kolesar, P.; Lehmann, A.R.; Uldrijan, S.; Sanderson, C.M.; Zdrahal, Z.; Palecek, J.J. The melanoma-associated antigen 1 (MAGEA1) protein stimulates the E3 ubiquitin-ligase activity of TRIM31 within a TRIM31-MAGEA1-NSE4 complex. Cell Cycle 2015, 14, 920–930. [Google Scholar] [CrossRef] [PubMed]
- Park, R.; Kim, H.; Jang, M.; Jo, D.; Park, Y.-I.; Namkoong, S.; Lee, J.I.; Jang, I.-S.; Park, J. LNX1 contributes to tumor growth by down-regulating p53 stability. FASEB J. 2019, 33, 13216–13227. [Google Scholar] [CrossRef]
- Kansaku, A.; Hirabayashi, S.; Mori, H.; Fujiwara, N.; Kawata, A.; Ikeda, M.; Rokukawa, C.; Kurihara, H.; Hata, Y. Ligand-of-Numb protein X is an endocytic scaffold for junctional adhesion molecule 4. Oncogene 2006, 25, 5071–5084. [Google Scholar] [CrossRef]
- Takahashi, S.; Iwamoto, N.; Sasaki, H.; Ohashi, M.; Oda, Y.; Tsukita, S.; Furuse, M. The E3 ubiquitin ligase LNX1p80 promotes the removal of claudins from tight junctions in MDCK cells. J. Cell Sci. 2009, 122, 985–994. [Google Scholar] [CrossRef]
- Tsukita, S.; Tanaka, H.; Tamura, A. The Claudins: From Tight Junctions to Biological Systems. Trends Biochem. Sci. 2019, 44, 141–152. [Google Scholar] [CrossRef]
- Sollerbrant, K.; Raschperger, E.; Mirza, M.; Engström, U.; Philipson, L.; Ljungdahl, P.O.; Pettersson, R.F. The Coxsackievirus and Adenovirus Receptor (CAR) Forms a Complex with the PDZ Domain-containing Protein Ligand-of-Numb Protein-X (LNX). J. Biol. Chem. 2002, 278, 7439–7444. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.; Raschperger, E.; Philipson, L.; Pettersson, R.F.; Sollerbrant, K. The cell surface protein coxsackie- and adenovirus receptor (CAR) directly associates with the Ligand-of-Numb Protein-X2 (LNX2). Exp. Cell Res. 2005, 309, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Mirza, M.; Hreinsson, J.; Strand, M.-L.; Hovatta, O.; Söder, O.; Philipson, L.; Pettersson, R.F.; Sollerbrant, K. Coxsackievirus and adenovirus receptor (CAR) is expressed in male germ cells and forms a complex with the differentiation factor JAM-C in mouse testis. Exp. Cell Res. 2006, 312, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Goodenough, D.A.; Paul, D.L. Gap Junctions. Cold Spring Harb. Perspect. Biol. 2009, 1, a002576. [Google Scholar] [CrossRef] [PubMed]
- Skerrett, I.M.; Williams, J.B. A structural and functional comparison of gap junction channels composed of connexins and innexins. Dev. Neurobiol. 2017, 77, 522–547. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, R. Connexins in health and disease. Clin. Exp. Neuroimmunol. 2018, 9, 30–36. [Google Scholar] [CrossRef]
- Nagy, J.I.; Pereda, A.E.; Rash, J.E. Electrical synapses in mammalian CNS: Past eras, present focus and future directions. Biochim. et Biophys. Acta (BBA)-Biomembr. 2018, 1860, 102–123. [Google Scholar] [CrossRef]
- Laing, J.G.; Tadros, P.N.; Westphale, E.M.; Beyer, E.C. Degradation of Connexin43 Gap Junctions Involves both the Proteasome and the Lysosome. Exp. Cell Res. 1997, 236, 482–492. [Google Scholar] [CrossRef]
- Falk, M.M.; Kells, R.M.; Berthoud, V.M. Degradation of connexins and gap junctions. FEBS Lett. 2014, 588, 1221–1229. [Google Scholar] [CrossRef]
- Lynn, B.D.; Li, X.; Hormuzdi, S.G.; Griffiths, E.K.; McGlade, C.J.; Nagy, J.I. E3 ubiquitin ligases LNX 1 and LNX 2 localize at neuronal gap junctions formed by connexin36 in rodent brain and molecularly interact with connexin36. Eur. J. Neurosci. 2018, 48, 3062–3081. [Google Scholar] [CrossRef]
- Fujita, Y.; Krause, G.; Scheffner, M.; Zechner, D.; Leddy, H.E.M.; Behrens, J.; Sommer, T.; Birchmeier, W. Hakai, a c-Cbl-like protein, ubiquitinates and induces endocytosis of the E-cadherin complex. Nat. Cell Biol. 2002, 4, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Traweger, A.; Fang, D.; Liu, Y.-C.; Stelzhammer, W.; Krizbai, I.A.; Fresser, F.; Bauer, H.-C.; Bauer, H. The Tight Junction-specific Protein Occludin Is a Functional Target of the E3 Ubiquitin-protein Ligase Itch. J. Biol. Chem. 2002, 277, 10201–10208. [Google Scholar] [CrossRef] [PubMed]
- Kaabeche, K.; Guenou, H.; Bouvard, D.; Didelot, N.; Listrat, A.; Marie, P.J. Cbl-mediated ubiquitination of 5 integrin subunit mediates fibronectin-dependent osteoblast detachment and apoptosis induced by FGFR2 activation. J. Cell Sci. 2005, 118, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- Knights, A.J.; Funnell, A.P.W.; Crossley, M.; Pearson, R.C. Holding Tight: Cell Junctions and Cancer Spread. Trends Cancer Res. 2012, 8, 61–69. [Google Scholar] [PubMed]
- D’Agostino, M.; Tornillo, G.; Caporaso, M.G.; Barone, M.V.; Ghigo, E.; Bonatti, S.; Mottola, G. Ligand of Numb proteins LNX1p80 and LNX2 interact with the human glycoprotein CD8α and promote its ubiquitylation and endocytosis. J. Cell Sci 2011, 124, 3545–3556. [Google Scholar] [CrossRef]
- Liu, X.-D.; Zhu, X.-N.; Halford, M.M.; Xu, T.-L.; Henkemeyer, M.; Xu, N.-J. Retrograde regulation of mossy fiber axon targeting and terminal maturation via postsynaptic Lnx1. J. Cell Biol. 2018, 217, 4007–4024. [Google Scholar] [CrossRef]
- Liu, X.-D.; Ai, P.-H.; Zhu, X.-N.; Pan, Y.-B.; Halford, M.M.; Henkemeyer, M.; Feng, D.-F.; Xu, T.-L.; Sun, S.; Xu, N.-J. Hippocampal Lnx1–NMDAR multiprotein complex mediates initial social memory. Mol. Psychiatry 2019, 1–14. [Google Scholar] [CrossRef]
- Rocha-Muñoz, A.; Núñez, E.; Arribas-González, E.; López-Corcuera, B.; Aragón, C.; de Juan-Sanz, J. E3 ubiquitin ligases LNX1 and LNX2 are major regulators of the presynaptic glycine transporter GlyT2. Sci Rep. 2019, 9, 14944. [Google Scholar] [CrossRef]
- Zafra, F.; Ibáñez, I.; Giménez, C. Glycinergic transmission: Glycine transporter GlyT2 in neuronal pathologies. Neuronal Signal. 2016, 1, NS20160009. [Google Scholar] [CrossRef]
- Robinson, P.J. Differential stimulation of protein kinase C activity by phorbol ester or calcium/phosphatidylserine in vitro and in intact synaptosomes. J. Biol. Chem. 1992, 267, 21637–21644. [Google Scholar]
- Vaughan, P.F.T.; Walker, J.H.; Peers, C. The regulation of neurotransmitter secretion by protein kinase C. Mol. Neurobiol. 1998, 18, 125–155. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.-J.; Henkemeyer, M. Ephrin-B3 reverse signaling through Grb4 and cytoskeletal regulators mediates axon pruning. Nat. Neurosci. 2009, 12, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Nolt, M.J.; Lin, Y.; Hruska, M.; Murphy, J.; Sheffler-Colins, S.I.; Kayser, M.S.; Passer, J.; Bennett, M.V.L.; Zukin, R.S.; Dalva, M.B. EphB controls NMDA receptor function and synaptic targeting in a subunit-specific manner. J. Neurosci. 2011, 31, 5353–5364. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Wei, P.; Jin, L.; Zheng, T.; Chen, W.-Y.; Liu, X.-Y.; Shi, X.-D.; Hao, J.-R.; Sun, N.; Gao, C. Overexpression of EphB2 in hippocampus rescues impaired NMDA receptors trafficking and cognitive dysfunction in Alzheimer model. Cell Death Dis. 2017, 8, e2717. [Google Scholar] [CrossRef] [PubMed]
- Fekany, K.; Yamanaka, Y.; Leung, T.; Sirotkin, H.I.; Topczewski, J.; Gates, M.A.; Hibi, M.; Renucci, A.; Stemple, D.; Radbill, A.; et al. The zebrafish bozozok locus encodes Dharma, a homeodomain protein essential for induction of gastrula organizer and dorsoanterior embryonic structures. Development 1999, 126, 1427–1438. [Google Scholar]
- Leung, T.; Bischof, J.; Söll, I.; Niessing, D.; Zhang, D.; Ma, J.; Jäckle, H.; Driever, W. bozozok directly represses bmp2b transcription and mediates the earliest dorsoventral asymmetry of bmp2b expression in zebrafish. Development 2003, 130, 3639–3649. [Google Scholar] [CrossRef]
- Ro, H.; Dawid, I.B. Lnx-2b restricts gsc expression to the dorsal mesoderm by limiting Nodal and Bozozok activity. Biochem. Biophys. Res. Commun. 2010, 402, 626–630. [Google Scholar] [CrossRef] [PubMed]
- Ro, H.; Dawid, I.B. Modulation of Tcf3 repressor complex composition regulates cdx4 expression in zebrafish. EMBO J 2011, 30, 2894–2907. [Google Scholar] [CrossRef]
- Le Cam, L.; Linares, L.K.; Paul, C.; Julien, E.; Lacroix, M.; Hatchi, E.; Triboulet, R.; Bossis, G.; Shmueli, A.; Rodriguez, M.S.; et al. E4F1 Is an Atypical Ubiquitin Ligase that Modulates p53 Effector Functions Independently of Degradation. Cell 2006, 127, 775–788. [Google Scholar] [CrossRef]
- Davidson, A.J.; Ernst, P.; Wang, Y.; Dekens, M.P.S.; Kingsley, P.D.; Palis, J.; Korsmeyer, S.J.; Daley, G.Q.; Zon, L.I. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nat. Cell Biol. 2003, 425, 300–306. [Google Scholar] [CrossRef]
- Young, T.; Deschamps, J. Chapter 8 Hox, Cdx, and Anteroposterior Patterning in the Mouse Embryo. Cur. Top. Dev. BioL. 2009, 88, 235–255. [Google Scholar] [CrossRef]
- Hayward, A.G., 2nd; Joshi, P.; Skromne, I. Spatiotemporal analysis of zebrafish hox gene regulation by Cdx4. Dev. Dyn. 2015, 244, 1564–1573. [Google Scholar] [CrossRef] [PubMed]
- Amack, J.D.; Yost, H. The T Box Transcription Factor No Tail in Ciliated Cells Controls Zebrafish Left-Right Asymmetry. Curr. Biol. 2004, 14, 685–690. [Google Scholar] [CrossRef]
- Essner, J.J.; Amack, J.D.; Nyholm, M.K.; Harris, E.B.; Yost, H.J. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development 2005, 132, 1247–1260. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Bessho, Y. Left-right asymmetry in zebrafish. Cell Mol. Life Sci 2012, 69, 3069–3077. [Google Scholar] [CrossRef] [PubMed]
- Long, S.; Ahmad, N.; Rebagliati, M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development 2003, 130, 2303–2316. [Google Scholar] [CrossRef]
- Kim, M.; Rhee, M.; Ro, H. Lnx2b, an E3 ubiquitin ligase, in dorsal forerunner cells and Kupffer’s vesicle is required for specification of zebrafish left-right laterality. Ani. Cell. Syst. 2014, 18, 333–339. [Google Scholar] [CrossRef]
- Bekri, A.; Liao, M.; Drapeau, P. Glycine Regulates Neural Stem Cell Proliferation During Development via Lnx1-Dependent Notch Signaling. Front. Mol. Neurosci. 2019, 12, 44. [Google Scholar] [CrossRef]
- Samarut, E.; Bekri, A.; Edrapeau, P. Transcriptomic Analysis of Purified Embryonic Neural Stem Cells from Zebrafish Embryos Reveals Signaling Pathways Involved in Glycine-Dependent Neurogenesis. Front. Mol. Neurosci. 2016, 9, 22. [Google Scholar] [CrossRef]
- Lenihan, J.A.; Saha, O.; Mansfield, L.M.; Young, P. Tight, cell type-specific control of LNX expression in the nervous system, at the level of transcription, translation and protein stability. Gene 2014, 552, 39–50. [Google Scholar] [CrossRef]
- Lenihan, J.A.; Saha, O.; Heimer-McGinn, V.; Cryan, J.F.; Feng, G.; Young, P.W. Decreased Anxiety-Related Behaviour but Apparently Unperturbed NUMB Function in Ligand of NUMB Protein-X (LNX) 1/2 Double Knockout Mice. Mol. Neurobiol. 2017, 54, 8090–8109. [Google Scholar] [CrossRef] [PubMed]
- Burgner, D.P.; Davila, S.; Breunis, W.B.; Shingadia, D.; Filippini, L.; Bonnard, C.; Ling, L.; Wright, V.J.; Thalamuthu, A.; Odam, M.; et al. A Genome-Wide Association Study Identifies Novel and Functionally Related Susceptibility Loci for Kawasaki Disease. PLoS Genet. 2009, 5, e1000319. [Google Scholar] [CrossRef] [PubMed]
- Mehraj, V.; Boucherit, N.; Ben Amara, A.; Capo, C.; Bonatti, S.; Mege, J.-L.; Mottola, G.; Ghigo, E. The ligands of Numb proteins X1 and X2 are specific markers for chronic Q fever. FEMS Immunol. Med. Microbiol. 2012, 64, 98–100. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.J.; Lee, K.Y.; Cho, Y.-Y.; Pugliese, A.; Kim, H.G.; Jeong, C.-H.; Bode, A.M.; Dong, Z. Role of NEK6 in Tumor Promoter-induced Transformation in JB6 C141 Mouse Skin Epidermal Cells. J. Biol. Chem. 2010, 285, 28126–28133. [Google Scholar] [CrossRef] [PubMed]
- Jee, H.J.; Kim, H.-J.; Kim, A.J.; Song, N.; Kim, M.; Lee, H.-J.; Yun, J. The inhibition of Nek6 function sensitizes human cancer cells to premature senescence upon serum reduction or anticancer drug treatment. Cancer Lett. 2013, 335, 175–182. [Google Scholar] [CrossRef] [PubMed]
- Sampson, J.; O’Regan, L.; Dyer, M.J.S.; Bayliss, R.; Fry, A.M. Hsp72 and Nek6 Cooperate to Cluster Amplified Centrosomes in Cancer Cells. Cancer Res. 2017, 77, 4785–4796. [Google Scholar] [CrossRef]
- Camps, J.; Pitt, J.J.; Emons, G.; Hummon, A.B.; Case, C.M.; Grade, M.; Jones, T.L.; Nguyen, Q.T.; Ghadimi, B.M.; Beissbarth, T.; et al. Genetic amplification of the NOTCH modulator LNX2 upregulates the WNT/β-catenin pathway in colorectal cancer. Cancer Res. 2013, 73, 2003–2013. [Google Scholar] [CrossRef]
- Day, C.M.; Hickey, S.M.; Song, Y.; Plush, S.E.; Garg, S. Novel Tamoxifen Nanoformulations for Improving Breast Cancer Treatment: Old Wine in New Bottles. Molecules 2020, 25, 1182. [Google Scholar] [CrossRef]
- Ma, L.; Wang, L.; Shan, Y.; Nafees, M.; Ihab, E.; Zhang, R.; Wang, F.; Yin, W. Suppression of cancer stemness by upregulating Ligand-of-Numb protein X1 in colorectal carcinoma. PLoS ONE 2017, 12, e0188665. [Google Scholar] [CrossRef][Green Version]
- Chen, J.; Xu, J.; Zhao, W.; Hu, G.; Cheng, H.; Kang, Y.; Xie, Y.; Lu, Y. Characterization of human LNX, a novel ligand of Numb protein X that is downregulated in human gliomas. Int. J. Biochem. Cell Biol. 2005, 37, 2273–2283. [Google Scholar] [CrossRef]
- Blom, T.; Roselli, A.; Tanner, M.; Nupponen, N.N. Mutation and copy number analysis of LNX1 and Numbl in nervous system tumors. Cancer Genet. Cytogenet. 2008, 186, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Kohn, K.W.; Zeeberg, B.M.; Reinhold, W.C.; Pommier, Y. Gene Expression Correlations in Human Cancer Cell Lines Define Molecular Interaction Networks for Epithelial Phenotype. PLoS ONE 2014, 9, e99269. [Google Scholar] [CrossRef]
- Wu, Z.-H.; Zhou, T.; Sun, H.-Y. DNA methylation-based diagnostic and prognostic biomarkers of nasopharyngeal carcinoma patients. Medicine 2020, 99, e20682. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Prives, C. Relevance of the p53-MDM2 axis to aging. Cell Death Differ. 2018, 25, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Shadfan, M.; Lopez-Pajares, V.; Yuan, Z.-M. MDM2 and MDMX: Alone and together in regulation of p53. Transl. Cancer Res. 2012, 1, 88–89. [Google Scholar]
- Karni-Schmidt, O.; Lokshin, M.; Prives, C. The Roles of MDM2 and MDMX in Cancer. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 617–644. [Google Scholar] [CrossRef] [PubMed]
- Colaluca, I.N.; Tosoni, D.; Nuciforo, P.; Senic-Matuglia, F.; Galimberti, V.; Viale, G.; Pece, S.; Di Fiore, P.P. NUMB controls p53 tumour suppressor activity. Nat. Cell Biol. 2008, 451, 76–80. [Google Scholar] [CrossRef]
- Juven-Gershon, T.; Shifman, O.; Unger, T.; Elkeles, A.; Haupt, Y.; Oren, M. The Mdm2 Oncoprotein Interacts with the Cell Fate Regulator Numb. Mol. Cell. Biol. 1998, 18, 3974–3982. [Google Scholar] [CrossRef]
- Carter, S.; Vousden, K.H. A role for Numb in p53 stabilization. Genome Biol. 2008, 9, 221. [Google Scholar] [CrossRef]
- Ko, J.-A.; Kimura, Y.; Matsuura, K.; Yamamoto, H.; Gondo, T.; Inui, M. PDZRN3 (LNX3, SEMCAP3) is required for the differentiation of C2C12 myoblasts into myotubes. J. Cell Sci. 2006, 119, 5106–5113. [Google Scholar] [CrossRef]
- Sewduth, R.N.; Jaspard-Vinassa, B.; Peghaire, C.; Guillabert, A.; Franzl, N.; Larrieu-Lahargue, F.; Moreau, C.; Fruttiger, M.; Dufourcq, P.; Couffinhal, T.; et al. The ubiquitin ligase PDZRN3 is required for vascular morphogenesis through Wnt/planar cell polarity signalling. Nat. Commun. 2014, 5, 4832. [Google Scholar] [CrossRef] [PubMed]
- Linares, L.K.; Hengstermann, A.; Ciechanover, A.; Müller, S.; Scheffner, M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc. Natl. Acad. Sci. USA 2003, 100, 12009–12014. [Google Scholar] [CrossRef] [PubMed]
- Diller, D.J.; Swanson, J.; Bayden, A.S.; Brown, C.J.; Thean, D.; Lane, D.P.; Partridge, A.W.; Sawyer, T.K.; Audie, J. Rigorous Computational and Experimental Investigations on MDM2/MDMX-Targeted Linear and Macrocyclic Peptides. Molecules 2019, 24, 4586. [Google Scholar] [CrossRef] [PubMed]
- Dente, L.; Gestri, G.; Tsang, M.; Kudoh, T.; Wilson, S.W.; Dawid, I.B.; Andreazzoli, M. Cloning and developmental expression of zebrafish pdzrn3. Int. J. Dev. Biol. 2011, 55, 989–993. [Google Scholar] [CrossRef] [PubMed]
- Marracci, S.; Vangelisti, A.; Raffa, V.; Andreazzoli, M.; Dente, L. pdzrn3 is required for pronephros morphogenesis in Xenopus laevis. Int. J. Dev. Biol. 2016, 60, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, Y.; Furihata, T.; Nakagawa, K.; Ohno, Y.; Reien, Y.; Ouchi, M.; Wakashin, H.; Tsuruoka, S.; Anzai, N. Sodium-coupled monocarboxylate transporter 1 interacts with the RING finger- and PDZ domain-containing protein PDZRN3. J. Physiol. Sci. 2019, 69, 635–642. [Google Scholar] [CrossRef]
- Yang, Y.; Mlodzik, M. Wnt-Frizzled/Planar Cell Polarity Signaling: Cellular Orientation by Facing the Wind (Wnt). Annu. Rev. Cell Dev. Biol. 2015, 31, 623–646. [Google Scholar] [CrossRef]
- Schnell, U.; Carroll, T.J. Planar cell polarity of the kidney. Exp. Cell Res. 2016, 343, 258–266. [Google Scholar] [CrossRef]
- Yusenko, M.V.; Nagy, A.; Kovacs, G. Molecular analysis of germline t(3;6) and t(3;12) associated with conventional renal cell carcinomas indicates their rate-limiting role and supports the three-hit model of carcinogenesis. Cancer Genet. Cytogenet. 2010, 201, 15–23. [Google Scholar] [CrossRef]
- Marunaka, K.; Furukawa, C.; Fujii, N.; Kimura, T.; Furuta, T.; Matsunaga, T.; Endo, S.; Hasegawa, H.; Anzai, N.; Yamazaki, Y.; et al. The RING finger- and PDZ domain-containing protein PDZRN3 controls localization of the Mg2+regulator claudin-16 in renal tube epithelial cells. J. Biol. Chem. 2017, 292, 13034–13044. [Google Scholar] [CrossRef]
- Hou, J.; Goodenough, D.A. Claudin-16 and claudin-19 function in the thick ascending limb. Curr. Opin. Nephrol. Hypertens. 2010, 19, 483–488. [Google Scholar] [CrossRef]
- Murea, M.; Lu, L.; Ma, L.; Hicks, P.J.; Divers, J.; McDonough, C.W.; Langefeld, C.D.; Bowden, N.W.; Freedman, B.I. Genome-wide association scan for survival on dialysis in African-Americans with type 2 diabetes. Am. J. Nephrol. 2011, 33, 502–509. [Google Scholar] [CrossRef]
- Sewduth, R.N.; Kovacic, H.; Jaspard-Vinassa, B.; Jecko, V.; Wavasseur, T.; Fritsch, N.; Pernot, M.; Jeaningros, S.; Roux, E.; Dufourcq, P.; et al. PDZRN3 destabilizes endothelial cell-cell junctions through a PKCζ-containing polarity complex to increase vascular permeability. Sci. Signal. 2017, 10, eaag3209. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, M. The Par3/Par6/aPKC complex and epithelial cell polarity. Exp. Cell Res. 2013, 319, 1357–1364. [Google Scholar] [CrossRef] [PubMed]
- Hakanen, J.; Ruiz-Reig, N.; Tissir, F. Linking Cell Polarity to Cortical Development and Malformations. Front. Cell. Neurosci. 2019, 13, 244. [Google Scholar] [CrossRef] [PubMed]
- Nagai-Tamai, Y.; Mizuno, K.; Hirose, T.; Suzuki, A.; Ohno, S. Regulated protein-protein interaction between aPKC and PAR-3 plays an essential role in the polarization of epithelial cells. Genes Cells 2002, 7, 1161–1171. [Google Scholar] [CrossRef]
- Lampugnani, M.G.; Orsenigo, F.; Rudini, N.; Maddaluno, L.; Boulday, G.; Chapon, F.; Dejana, E. CCM1 regulates vascular-lumen organization by inducing endothelial polarity. J. Cell Sci. 2010, 123, 1073–1080. [Google Scholar] [CrossRef]
- Willis, C.L.; Meske, D.S.; Davis, T.P. Protein Kinase C Activation Modulates Reversible Increase in Cortical Blood–Brain Barrier Permeability and Tight Junction Protein Expression during Hypoxia and Posthypoxic Reoxygenation. Br. J. Pharmacol. 2010, 30, 1847–1859. [Google Scholar] [CrossRef]
- Oubaha, M.; Lin, M.I.; Margaron, Y.; Filion, D.; Price, E.N.; Zon, L.I.; Côté, J.-F.; Gratton, J.-P. Formation of a PKCζ/β-catenin complex in endothelial cells promotes angiopoietin-1–induced collective directional migration and angiogenic sprouting. Blood 2012, 120, 3371–3381. [Google Scholar] [CrossRef]
- Riddell, M.; Nakayama, A.; Hikita, T.; Mirzapourshafiyi, F.; Kawamura, T.; Pasha, A.; Li, M.; Masuzawa, M.; Looso, M.; Steinbacher, T.; et al. aPKC controls endothelial growth by modulating c-Myc via FoxO1 DNA-binding ability. Nat. Commun. 2018, 9, 5357. [Google Scholar] [CrossRef]
- Flynn, M.; Saha, O.; Young, P. Molecular evolution of the LNX gene family. BMC Evol. Biol. 2011, 11, 235. [Google Scholar] [CrossRef]
- Honda, T.; Yamamoto, H.; Ishii, A.; Inui, M. PDZRN3 Negatively Regulates BMP-2–induced Osteoblast Differentiation through Inhibition of Wnt Signaling. Mol. Biol. Cell 2010, 21, 3269–3277. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Hernández, J.M.; García-González, E.G.; Brun, C.E.; Rudnicki, M.A. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin. Cell Dev. Biol. 2017, 72, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Pownall, M.E.; Gustafsson, M.K.; Emerson, C.P. Myogenic Regulatory Factors and the Specification of Muscle Progenitors in Vertebrate Embryos. Annu. Rev. Cell Dev. Biol. 2002, 18, 747–783. [Google Scholar] [CrossRef] [PubMed]
- Francetic, T.; Li, Q. Skeletal myogenesis and Myf5 activation. Transcription 2011, 2, 109–114. [Google Scholar] [CrossRef]
- Honda, T.; Inui, M. PDZRN3 regulates differentiation of myoblasts into myotubes through transcriptional and posttranslational control of Id2. J. Cell. Physiol. 2018, 234, 2963–2972. [Google Scholar] [CrossRef]
- Venuti, J.M.; Morris, J.H.; Vivian, J.L.; Olson, E.N.; Klein, W.H. Myogenin is required for late but not early aspects of myogenesis during mouse development. J. Cell Biol. 1995, 128, 563–576. [Google Scholar] [CrossRef]
- Langlands, K.; Yin, X.; Anand, G.; Prochownik, E.V. Differential Interactions of Id Proteins with Basic-Helix-Loop-Helix Transcription Factors. J. Biol. Chem. 1997, 272, 19785–19793. [Google Scholar] [CrossRef]
- Yokoyama, S.; Ito, Y.; Ueno-Kudoh, H.; Shimizu, H.; Uchibe, K.; Albini, S.; Mitsuoka, K.; Miyaki, S.; Kiso, M.; Nagai, A.; et al. A Systems Approach Reveals that the Myogenesis Genome Network Is Regulated by the Transcriptional Repressor RP58. Dev. Cell 2009, 17, 836–848. [Google Scholar] [CrossRef]
- Kalaszczynska, I.; Geng, Y.; Iino, T.; Mizuno, S.-I.; Choi, Y.; Kondratiuk, I.; Silver, D.P.; Wolgemuth, D.J.; Akashi, K.; Sicinski, P. Cyclin A Is Redundant in Fibroblasts but Essential in Hematopoietic and Embryonic Stem Cells. Cell 2009, 138, 352–365. [Google Scholar] [CrossRef]
- Loukil, A.; Cheung, C.T.; Bendris, N.; Lemmers, B.; Peter, M.; Blanchard, J.M. Cyclin A2: At the crossroads of cell cycle and cell invasion. World J. Biol. Chem. 2015, 6, 346–350. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Inui, M. PDZRN3 protects against apoptosis in myoblasts by maintaining cyclin A2 expression. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Popescu, N.; DiPaolo, J.; Amsbaugh, S. Integration sites of human papillomavirus 18 DNA sequences on HeLa cell chromosomes. Cytogenet. Genome Res. 1987, 44, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Banks, L. PDZRN3/LNX3 Is a Novel Target of Human Papillomavirus Type 16 (HPV-16) and HPV-18 E6. J. Virol. 2015, 89, 1439–1444. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shipley, J.M.; Waxman, D.J. Down-regulation of STAT5b transcriptional activity by ligand-activated peroxisome proliferator-activated receptor (PPAR) alpha and PPARgamma. Mol. Pharmacol. 2003, 64, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Honda, T.; Ishii, A.; Inui, M. Regulation of adipocyte differentiation of 3T3-L1 cells by PDZRN3. Am. J. Physiol. Physiol. 2013, 304, C1091–C1097. [Google Scholar] [CrossRef] [PubMed]
- Lefterova, M.I.; Haakonsson, A.K.; Lazar, M.A.; Mandrup, S. PPARγ and the global map of adipogenesis and beyond. Trends Endocrinol. Metab. 2014, 25, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Burden, S.J.; Yumoto, N.; Zhang, W. The Role of MuSK in Synapse Formation and Neuromuscular Disease. Cold Spring Harb. Perspect. Biol. 2013, 5, a009167. [Google Scholar] [CrossRef] [PubMed]
- Han, J.-Y.; Shin, E.S.; Lee, Y.-S.; Ghang, H.Y.; Kim, S.-Y.; Hwang, J.-A.; Kim, J.Y.; Lee, J.S. A genome-wide association study for irinotecan-related severe toxicities in patients with advanced non-small-cell lung cancer. Pharmacogenom. J. 2012, 13, 417–422. [Google Scholar] [CrossRef]
- De Man, F.M.; Goey, A.K.L.; Van Schaik, R.H.N.; Mathijssen, R.H.J.; Bins, S. Individualization of Irinotecan Treatment: A Review of Pharmacokinetics, Pharmacodynamics, and Pharmacogenetics. Clin. Pharmacokinet. 2018, 57, 1229–1254. [Google Scholar] [CrossRef]
- Gajek, G.; Marciniak, B.; Lewkowski, J.; Kontek, R. Antagonistic Effects of CAPE (a Component of Propolis) on the Cytotoxicity and Genotoxicity of Irinotecan and SN38 in Human Gastrointestinal Cancer Cells In Vitro. Molecules 2020, 25, 658. [Google Scholar] [CrossRef] [PubMed]
- Zoetemelk, M.; Ramzy, G.M.; Rausch, M.; Nowak-Sliwinska, P. Drug-Drug Interactions of Irinotecan, 5-Fluorouracil, Folinic Acid and Oxaliplatin and Its Activity in Colorectal Carcinoma Treatment. Molecules 2020, 25, 2614. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.; Socci, N.D.; Dhall, D.; D’Angelica, M.; DeMatteo, R.P.; Allen, P.J.; Singh, B.; Fong, Y.; Blumgart, L.H.; Klimstra, D.S.; et al. Genome wide analysis and clinical correlation of chromosomal and transcriptional mutations in cancers of the biliary tract. J. Exp. Clin. Cancer Res. 2009, 28, 62. [Google Scholar] [CrossRef] [PubMed]
- Hartmaier, R.; Albacker, L.A.; Chmielecki, J.; Bailey, M.; He, J.; Goldberg, M.E.; Ramkissoon, S.; Suh, J.; Elvin, J.A.; Chiacchia, S.; et al. High-Throughput Genomic Profiling of Adult Solid Tumors Reveals Novel Insights into Cancer Pathogenesis. Cancer Res. 2017, 77, 2464–2475. [Google Scholar] [CrossRef]
- Suurmeijer, A.J.H.; Dickson, B.C.; Swanson, D.; Zhang, L.; Sung, Y.-S.; Cotzia, P.; Fletcher, C.D.; Antonescu, C.R. A novel group of spindle cell tumors defined by S100 and CD34 co-expression shows recurrent fusions involving RAF1, BRAF, and NTRK1/2 genes. Gen. Chr. Can. 2018, 57, 611–621. [Google Scholar] [CrossRef]
- Mok, Y.; Kimpo, M.S.; Chen, H.; Kuick, C.H.; Chang, K.T.; Lee, V.K.M. Spindle cell tumour with S100 and CD34 co-expression showing PDZRN3-RAF1 rearrangement-a recently described entity. Histopathology 2019, 74, 1109–1111. [Google Scholar] [CrossRef]
- Picco, G.; Chen, E.; Garcia-Alonso, L.; Behan, F.M.; Gonçalves, E.; Bignell, G.; Matchan, A.; Fu, B.; Banerjee, R.; Anderson, E.; et al. Functional linkage of gene fusions to cancer cell fitness assessed by pharmacological and CRISPR-Cas9 screening. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef]
- Prall, O.W.J.; Nastevski, V.; Xu, H.; McEvoy, C.R.E.; Vissers, J.H.A.; Byrne, D.J.; Takano, E.; Yerneni, S.; Ellis, S.; Green, T.; et al. RAF1 rearrangements are common in pancreatic acinar cell carcinomas. Mod. Pathol. 2020, 33, 1811–1821. [Google Scholar] [CrossRef]
- Pim, D.; Bergant, M.; Boon, S.S.; Ganti, K.; Kranjec, C.; Massimi, P.; Subbaiah, V.K.; Thomas, M.; Tomaić, V.; Banks, L. Human papillomaviruses and the specificity of PDZ domain targeting. FEBS J. 2012, 279, 3530–3537. [Google Scholar] [CrossRef]
- Nagasaka, K.; Kawana, K.; Osuga, Y.; Fujii, T. PDZ Domains and Viral Infection: Versatile Potentials of HPV-PDZ Interactions in relation to Malignancy. BioMed Res. Int. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Thatte, J.; Massimi, P.; Thomas, M.; Boon, S.S.; Banks, L. The Human Papillomavirus E6 PDZ Binding Motif Links DNA Damage Response Signaling to E6 Inhibition of p53 Transcriptional Activity. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Rozenblatt-Rosen, O.; Deo, R.C.; Padi, M.; Adelmant, G.; Calderwood, M.A.; Rolland, T.; Grace, M.; Dricot, A.; Askenazi, M.; Tavares, M.; et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nat. Cell Biol. 2012, 487, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Belotti, E.; Polanowska, J.; Daulat, A.M.; Audebert, S.; Thomé, V.; Lissitzky, J.-C.; Lembo, F.; Blibek, K.; Omi, S.; Lenfant, N.; et al. The Human PDZome: A Gateway to PSD95-Disc Large-Zonula Occludens (PDZ)-mediated Functions. Mol. Cell. Proteom. 2013, 12, 2587–2603. [Google Scholar] [CrossRef] [PubMed]
- Huibregtse, J.M.; Scheffner, M.; Howley, P.M. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 1993, 13, 775–784. [Google Scholar] [CrossRef]
- Nakagawa, S.; Huibregtse, J.M. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell Biol 2000, 20, 8244–8253. [Google Scholar] [CrossRef]
- Mantovani, F.; Banks, L. The Human Papillomavirus E6 protein and its contribution to malignant progression. Oncogene 2001, 20, 7874–7887. [Google Scholar] [CrossRef] [PubMed]
- Yim, E.-K.; Park, J.S. The Role of HPV E6 and E7 Oncoproteins in HPV-associated Cervical Carcinogenesis. Cancer Res. Treat. 2005, 37, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Pol, S.B.V.; Klingelhutz, A.J. Papillomavirus E6 oncoproteins. Virology 2013, 445, 115–137. [Google Scholar] [CrossRef]
- Jemal, A.; Bray, F.; Center, M.M.; Ferlay, J.; Ward, E.; Forman, D. Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [Google Scholar] [CrossRef]
- Hu, T.; Yang, H.; Han, Z. PDZRN4 acts as a suppressor of cell proliferation in human liver cancer cell lines. Cell Biochem. Funct. 2015, 33, 443–449. [Google Scholar] [CrossRef]
- Lu, Y.-L.; Yang, X.; Liu, Y.-K. Reduced PDZRN4 promotes breast cancer progression and predicts poor prognosis. Int. J. Clin. Exp. Pathol 2019, 12, 142–153. [Google Scholar]
- Hua, Y.; Ma, X.; Liu, X.; Yuan, X.; Qin, H.; Zhang, X. Abnormal expression of mRNA, microRNA alteration and aberrant DNA methylation patterns in rectal adenocarcinoma. PLoS ONE 2017, 12, e0174461. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, C.; Zhang, Y.; Xu, L.; Fang, W.; Zhu, Y.; Zheng, Y.; Chen, X.; Xie, X.; Hu, X.; et al. Genomic signatures reveal DNA damage response deficiency in colorectal cancer brain metastases. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Stein, L.; Rothschild, J.; Luce, J.; Cowell, J.K.; Thomas, G.; Bogdanova, T.I.; Tronko, M.D.; Hawthorn, L. Copy Number and Gene Expression Alterations in Radiation-Induced Papillary Thyroid Carcinoma from Chernobyl Pediatric Patients. Thyroid 2010, 20, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Baranzini, S.E.; Wang, J.; Gibson, R.A.; Galwey, N.; Naegelin, Y.; Barkhof, F.; Radue, E.-W.; Lindberg, R.L.; Uitdehaag, B.M.; Johnson, M.R.; et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum. Mol. Genet. 2008, 18, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Cavanillas, M.L.; Fernandez, O.; Comabella, M.; Alcina, A.; Fedetz, M.; Izquierdo, G.; Lucas, M.; Cenit, M.C.; Arroyo, R.; Vandenbroeck, K.; et al. Replication of top markers of a genome-wide association study in multiple sclerosis in Spain. Genes Immun. 2010, 12, 110–115. [Google Scholar] [CrossRef] [PubMed]
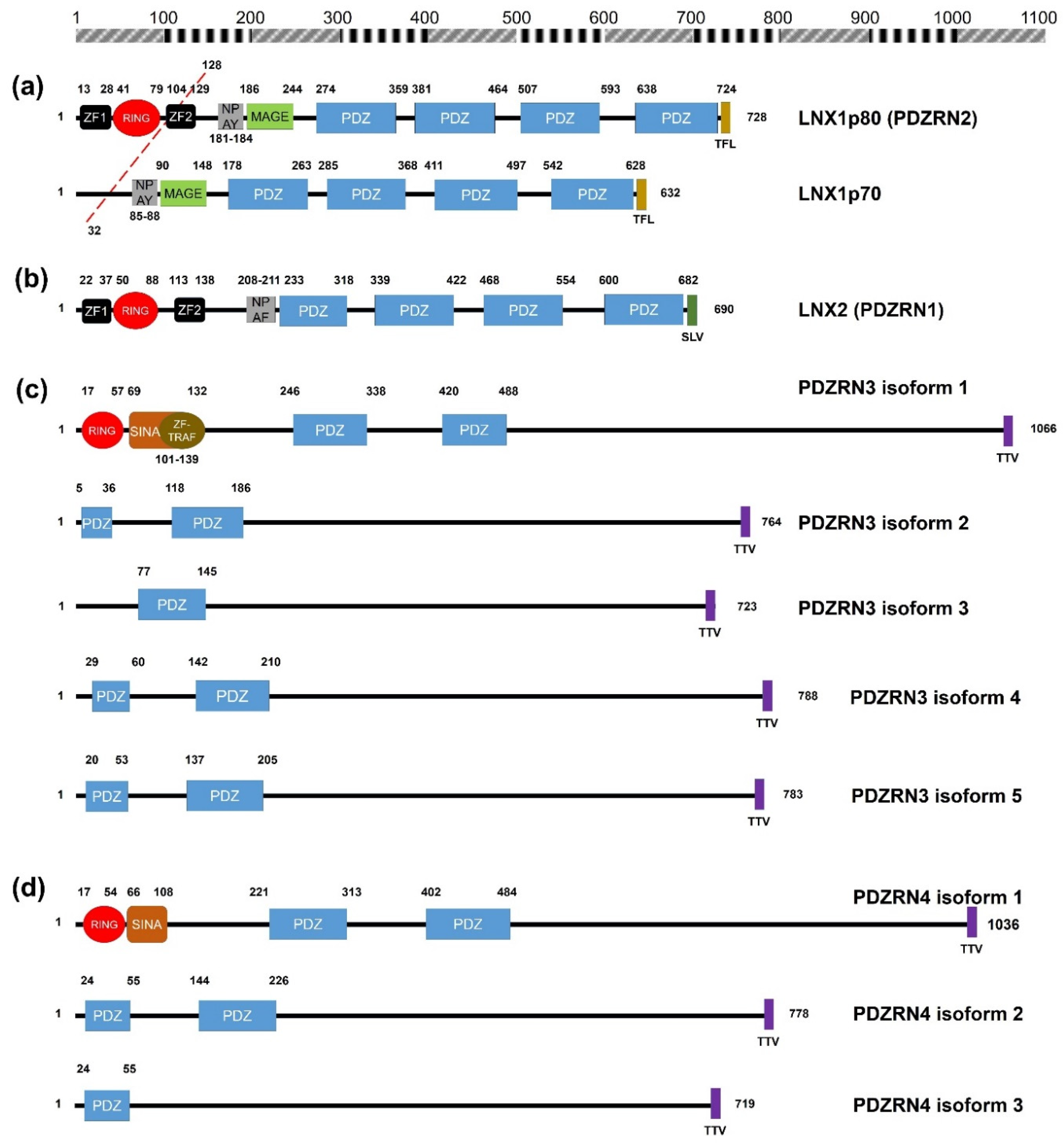
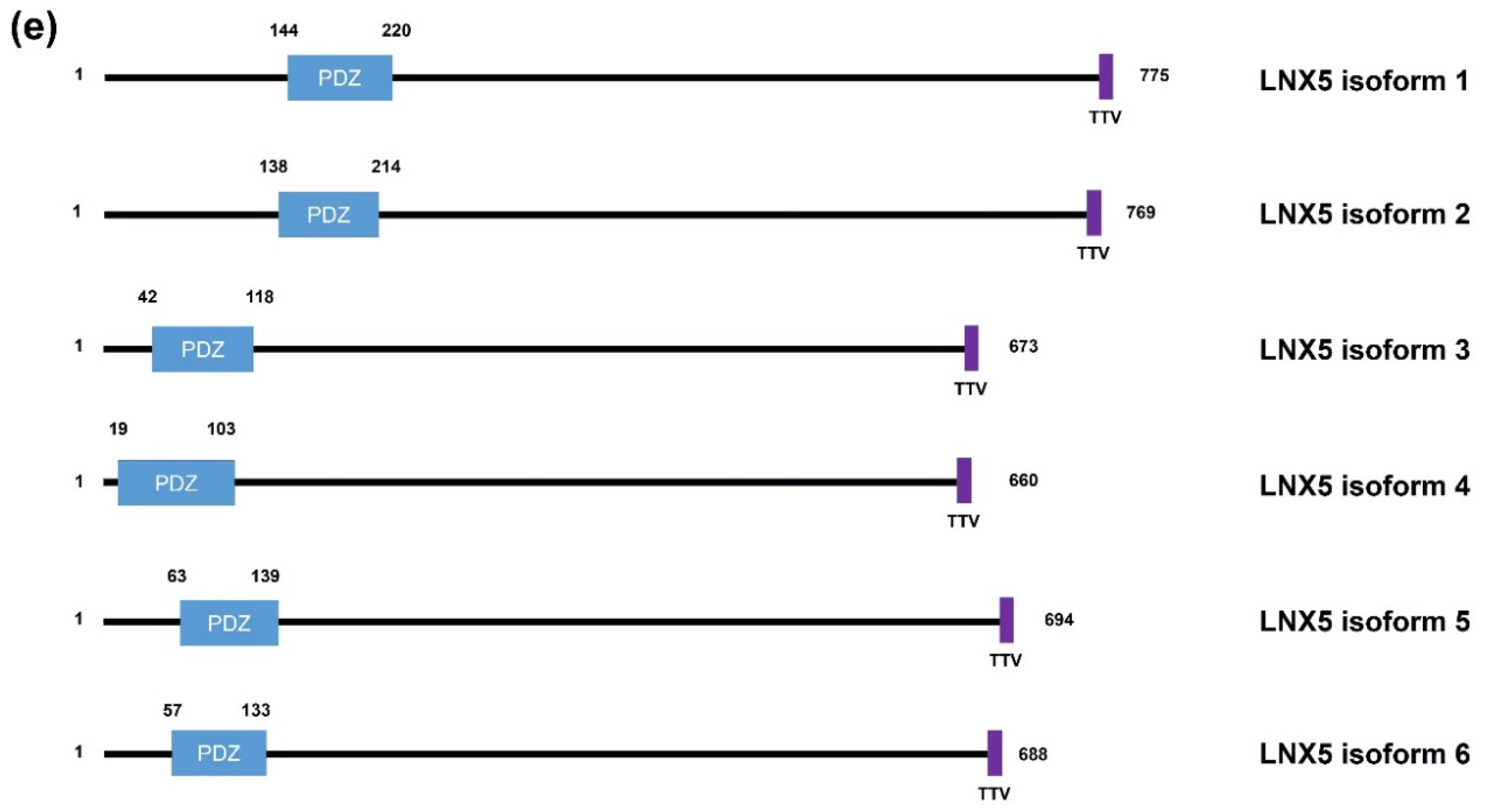
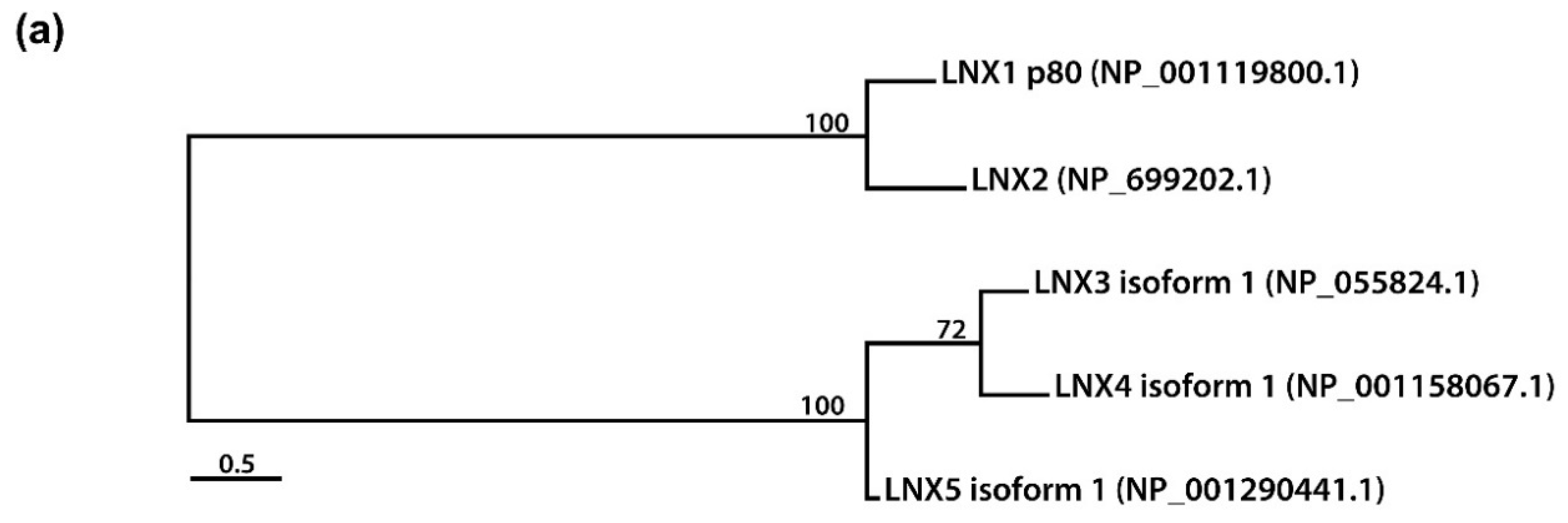
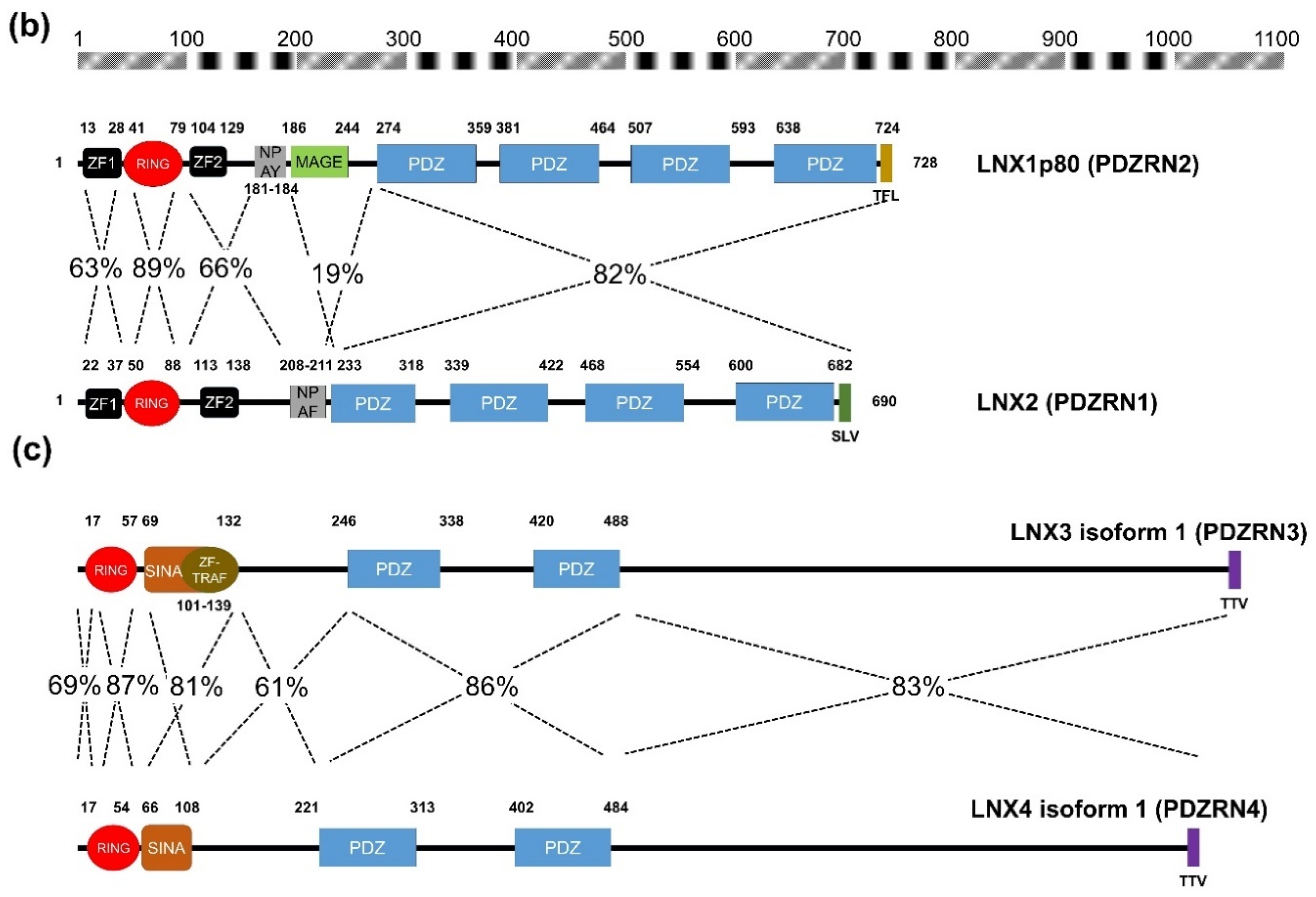

| Acronym | Explanation |
|---|---|
| BBB | Blood-brain barrier |
| bHLH | Basic helix-loop-helix |
| BMP | Bone morphogenetic protein |
| CA3 | Cornu ammonis 3 |
| CAR | Coxsackievirus and adenovirus receptor |
| Cdx4 | Caudal related homeobox transcription factor 4 |
| CLDN16 | Claudin-16 |
| CNS | Central nervous system |
| CNTN3 | Contactin-3 |
| DFC | Dorsal forerunner cell |
| Dvl | Disheveled |
| E6AP | E6-associated protein |
| EphB | Ephrin type-B receptor |
| GlyT2 | Sodium- and chloride-dependent glycine transporter 2 |
| Gro/TLE | Groucho/ transducin-like enhancer of split |
| GST | Glutathione S-transferase |
| GWAS | Genome-wide association study |
| HCC | hepatocellular carcinoma |
| Hdac1 | Histone deacetylase 1 |
| HECT | Homologous to the E6-AP Carboxyl Terminus |
| HOX | Homeobox |
| HPV | Human papillomavirus |
| Id2 | Inhibitor of DNA binding 2 |
| IL | Interleukin |
| JAM4 | Junctional adhesion molecule 4 |
| KV | Kupffer’s vesicle |
| LDOC1 | Leucine zipper downregulated in cancer 1 |
| LNX | Ligand of numb protein-X |
| MAGE | Melanoma-associated antigen |
| MC | Monocarboxylate |
| MDM2 | Murine double minute 2 homolog |
| MDMX | Murine double minute X |
| MF | Mossy fiber |
| MHC | Myogenin heavy chain |
| MRF | Myogenic regulatory factor |
| Mrf4 | Myogenic regulatory factor 4 |
| MUPP1 | Multiple PDZ domain protein 1 |
| Myf5 | Myogenic factor 5 |
| MyoD | Myoblast determination protein 1 |
| NEK6 | NIMA-related kinases 6, Serine/threonine-protein kinase NEK6 |
| NMDAR | N-methyl-D-aspartate receptor |
| PCP | Planar cell polarity |
| PDZ | Post-synaptic density protein-95, Disc large tumour suppressor, Zonula occludens-1 (PSD95, DLGA, ZO-1) |
| PDZK1 | PDZ-containing kidney protein 1 (Na(+)/H(+) exchange regulatory cofactor NHE-RF3) |
| PDZRN | PDZ and RING |
| pJAK2 | Phospho-Janus kinase 2 |
| PKC | Protein kinase C |
| PPAR | Peroxisome proliferator-activated receptors |
| RAF-1 | V-Raf-1 Murine Leukemia Viral Oncogene Homolog 1 |
| RING | Really interesting new gene |
| SEMCAP | Semaphoring cytoplasmic domain-associated protein |
| SINA | Seven in absentia |
| SMCT1 (SLC5A8) | Sodium-coupled monocarboxylate transporter 1 |
| SNP | Single nucleotide polymorphism |
| STAT | Signal Transducer and Transcription |
| STXBP5 | Syntaxin binding protein 5 |
| TCF3 | T-cell factor 3 |
| TRAF | Tumor necrosis factor receptor (TNF-R)-associated factor |
| UPS | Ubiquitin proteasome system |
| Wnt | Wingless and Int-1 |
| Zn | Zinc finger |
| Associated Proteins | LNX | Domain Involved | Methods Used | Major Functions | Consequences | References |
|---|---|---|---|---|---|---|
| NEK6 | LNX1 | RING and 3rd PDZ | Co-IP, GST pull down, Yeast two hybrid, | Serine/threonine kinase | K48-linked polyubiquitylated NEK6 undergoes UPS-dependent degradation | [31] (2020) |
| LDOC 1 | LNX1 | Co-IP | Tumor suppressor | LNX1 uses LDOC1 as a scaffold protein to indirectly target phospho-JAK2 destruction | [32] (2019) | |
| MDM2 | LNX1 | Co-IP | E3 ubiquitin ligase | LNX1 may indirectly interact with MDM2 | [37] (2019) | |
| p53 | LNX1 | Co-IP | Tumor suppressor | LNX1 indirectly mediates p53 destruction | [37] (2019) | |
| Connexin 36 | LNX1 | 2nd PDZ | Co-IP, GST pull down, Ni-NTA pull down | Gap junction protein | Ubiquitylated connexin36 undergoes lysosomal-dependent degradation. | [50] (2018) |
| Connexin 36 | LNX2 | 2nd PDZ | Co-IP, Ni-NTA pull down | Gap junction protein | Ubiquitylated connexin36 undergoes lysosomal-dependent degradation. | [50] (2018) |
| EphB1 | LNX1 p70 | N-terminal region ahead of 1st PDZ domain | Co-IP | Receptor tyrosine kinase | Stabilization | [56] (2018) |
| EphB2 | LNX1 p80 | Receptor tyrosine kinase | Degradation | [56] (2018) | ||
| EphB2 | LNX1 p70 | 2nd PDZ | Co-IP, GST-pull down | Receptor tyrosine kinase | Stabilization | [56] (2018), [57] (2019) |
| GluN1 | LNX1 | Co-IP | Glutamate receptor subunit | LNX1 helps form a NMDAR complex by recruiting GluN1 and GluN2B | [57] (2019) | |
| GluN2B | LNX1 | 1st PDZ | Co-IP, GST pull down | Glutamate receptor subunit | LNX1 helps form a NMDAR complex by recruiting GluN1 and GluN2B | [57] (2019) |
| GlyT2 | LNX1, LNX2 | 2nd PDZ | Co-IP | Glycine transporter | Polyubiquitylated and degraded | [58] (2019) |
| Associated Proteins | LNX | Domain Involved | Methods Used | Major Functions | Consequences | References |
|---|---|---|---|---|---|---|
| Dvl3 | LNX3 | C-terminal PDZ binding motif | Co-IP, Yeast two hybrid | Component of Wnt signaling | K63-linked polyubiquitylated Dvl3 undergoes endocytosis with Frizzled to activate non-canonical Wnt signaling | [101] (2014) |
| SMCT1(SLC5A8) | LNX3 | 1st PDZ | Co-IP, Yeast two hybrid | Sodium-coupled monocarboxylate (MC) transporter | The potentiation of SMCT1 transporter activity is inhibited by LNX3 | [106] (2019) |
| CLDN16 | LNX3 | PDZ domain | Co-IP, GST pull down, Yeast two hybrid | Tight junction protein | Monoubiquitylated CLDN16 become subjected to endocytosis | [110] (2017) |
| MUPP1, PKCζ, Par3 | LNX3 | Co-IP | Scaffold protein Serine/threonine kinase Polarity protein | Destruction of MUPP1 by LNX3 through UPS inhibits polarity complex formation composed of PAR3, PKCζ, and MUPP1 | [113] (2017) | |
| E6 | LNX3 | GST pull down, Peptide screen of human PDZome | Oncoprotein | E6 targets LNX3 for UPS-dependent destruction | [134] (2015), [152] (2012), [153] (2013) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, J.; Won, M.; Ro, H. The Molecular and Pathophysiological Functions of Members of the LNX/PDZRN E3 Ubiquitin Ligase Family. Molecules 2020, 25, 5938. https://doi.org/10.3390/molecules25245938
Hong J, Won M, Ro H. The Molecular and Pathophysiological Functions of Members of the LNX/PDZRN E3 Ubiquitin Ligase Family. Molecules. 2020; 25(24):5938. https://doi.org/10.3390/molecules25245938
Chicago/Turabian StyleHong, Jeongkwan, Minho Won, and Hyunju Ro. 2020. "The Molecular and Pathophysiological Functions of Members of the LNX/PDZRN E3 Ubiquitin Ligase Family" Molecules 25, no. 24: 5938. https://doi.org/10.3390/molecules25245938
APA StyleHong, J., Won, M., & Ro, H. (2020). The Molecular and Pathophysiological Functions of Members of the LNX/PDZRN E3 Ubiquitin Ligase Family. Molecules, 25(24), 5938. https://doi.org/10.3390/molecules25245938




