Abstract
A series of 3-ethyl(methyl)-2-thioxo-2,3-dihydrobenzo[g]quinazolines (1–17) were synthesized, characterized, and evaluated in vitro for their antiangiogenesis VEGFR-2-targeting, antiproliferative, and antiapoptotic activities against breast MCF-7 and liver HepG2 cells. Flow cytometry was used to determine cancer-cell cycle distributions, and apoptosis was detected using annexin-V-FITC (V) and propidium iodide (PI) dyes. Fluorescence microscopy, in combination with Hoechst staining was used to detect DNA fragmentation. Most of the tested benzo[g]quinazolines demonstrated promising activity (IC50 = 8.8 ± 0.5–10.9 ± 0.9 μM) and (IC50 = 26.0 ± 2.5–40.4 ± 4.1 μM) against MCF-7 and HepG2, respectively. Doxorubicin was used as a reference drug. Compounds 13–15 showed the highest activity against both cancer cell lines. Differential effects were detected by cell-cycle analysis, indicating similarities in the actions of 13 and 14 against both MCF7 and HepG2, involving the targeting of G1 and S phases, respectively. Compound 15 showed similar indices against both cells, indicating that its cytotoxicity toward the examined cancer cells could be unselective. Interestingly, 14 and 15 showed the highest apoptosis (30.76% and 25.30%, respectively) against MCF-7. The DNA fragmentation results agreed well with the apoptosis detected by flow cytometry. In terms of antiangiogenesis activity, as derived from VEGFR-2 inhibition, 13 and 15 were comparable to sorafenib and effected 1.5- and 1.4-fold inhibition relative to the standard sorafenib. A docking study was conducted to investigate the interaction between the synthesized benzo[g]quinazolines and the ATP-binding site within the catalytic domain of VEGFR-2.
1. Introduction
Angiogenesis plays a substantial role in cancer development and relies on the appearance of specific mediators that start a cascade of events resulting in microvessel formation [1]. Vascular endothelial growth factor receptor-2 (VEGFR-2) is one of the main targets within the angiogenesis-related kinases and is the main endothelial species required for tumor neovascularization [2,3,4]. Such kinases exert their biological effects by protecting against apoptosis-inducing conditions, increasing vessel permeability, and promoting endothelial cell proliferation enhancement, migration, and differentiation. Fighting cancer angiogenesis is one of the main focuses of effective cancer remedy. Thus, obstruction of erratic angiogenesis by specific inhibitors that target receptor tyrosine kinases is of pronounced interest in medicinal chemistry research. By specifically preventing angiogenesis by VEGFR-2 inhibitors, Figure 1A represents an attractive strategy for the treatment of VEGFR-2-mediated tumor growth, as well as addressing drug resistance [2,3,4].
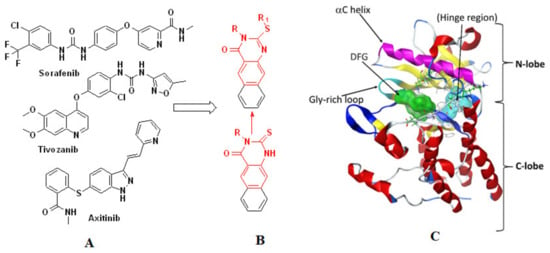
Figure 1.
The reported VEGFR-2 inhibitors (A) the designed VEGFR-2 inhibitors (B) and the site for binding of ATP for VEGFR-2 catalytic domain (C).
VEGFR-2 inhibitors are classified into three types (I, II, and III). Type-I are ATP-competitive inhibitors such as sunitinib, which bind to the region that accommodates ATP’s adenine ring [5]. Sorafenib belongs to the second type of inhibitors (II), which cannot bind at the adenine binding site but instead bind adjacent to the hydrophobic pocket [6]. Type-III inhibitors, such as vatalanib, are covalent inhibitors that bind to cysteine amino acid residues and prevent binding of ATP at the binding site [7,8]. Blocking the receptor dimerization and autophosphorylation processes provides a basis for discovering novel VEGFR-2 inhibitors. For VEGFR-2, the catalytic domain is the ATP site for binding. It is a bi-lobed structure with a small N-lobe and a large C-lobe. These lobes are linked via a linker that consists of a hinge region and a convex motif region. The binding of ATP is dependent on the confirmation of these lobes (Figure 1C) [9,10].
Molecules featuring the benzo[g]quinazoline scaffold are widely studied in medicinal chemistry because of their numerous biological and chemical properties [11,12,13,14,15,16,17,18,19,20]. In our studies on benzo[g]quinazoline chemistry, many derivatives have been identified as potent α-glucosidase inhibitors with promising antidiabetic effects [17] and antimicrobial agents that show excellent activities against both Gram-positive and -negative bacteria along with strong antifungal activities [11,14,15]. Furthermore, some derivatives of benzo[g]quinazolines show high antiviral activity against herpes simplex (HSV-1 and 2), coxsackie (CVB4), and HAV viruses [12,19]. Benzo[h]quinazolines have been reported as potent tyrosine kinase inhibitors that exhibit significant cytotoxicity against human carcinoma HT29, HCT116, and A549 cell lines [21], whereas benzo[f]quinazoline derivatives have been demonstrated to be potent thymidylate synthase inhibitors [22]. Moreover, benzo[g]quinazolines bearing sulfonamide, alkoxy, amino, and thioxo functional groups have shown high affinities toward the anaplastic lymphoma kinase receptor and demonstrate potent effects against EGFR and HER2 cells [16].
In our previous studies, we have evaluated some 2-thioxobenzo[g]quinazoline derivatives for their cytotoxicity activities against several carcinoma cell lines, i.e., A549, PC-3, HCT-116, Hep-G2, and MCF-7 [13]. These results encouraged us to consider the benzo[g]quinazoline core as a template for the design of further derivatives with potent cytotoxic activities. Moreover, chemical modification of 2-thioxobenzo[g]quinazoline structures provided us with valuable insight into the properties required for potent benzo[g]quinazoline-based anticancer agents. VEGF/VEGFR-2 binding is an early event in the angiogenic cascade, so targeting VEGFR-2 receptors and/or its ligand VEGF are considered good strategies for arresting cancer metastasis. We continued our attention in drug discovery, regarding the antiangiogenic therapy, besides our attention to exploring more anticancer agents.
Herein, 3-ethyl(methyl)-2-thioxobenzo[g]quinazolines (1–17) were synthesized and evaluated for their cytotoxicities against human breast MCF-7 and liver HepG2 carcinoma cells and their abilities to trigger apoptosis and restrict cancer-cell migration. Furthermore, the most active benzoquinazolines were evaluated as antiangiogenic chemotherapeutic agents targeting the VEGFR-2 tyrosine kinase receptor (Figure 1B). Finally, a molecular docking study was conducted to rationalize the structure-activity relationship (SAR) of the synthesized benzo[g]quinazolines and predict their interactions with the ATP-binding site in the catalytic domain of VEGFR-2. The QSAR was carried out to explore the relationships between the benzo[g]quinazolines molecular structures and their cytotoxicity findings by developing prediction models among the known anti HepG2 and MCF-7 agents then to predict the cytotoxicity of the benzo[g]quinazolines.
2. Results and Discussion
2.1. Chemistry
Using previously reported synthetic procedures for preparation of their analogues [13,17,23], the current 3-ethyl(methyl)-2-thioxobenzo[g]quinazolin-4(3H)-ones (1–17) were obtained in good yields (see Section 3.1.1). The structures of all products were established using HREI-MS and NMR analyses. The benzoquinazoline core was confirmed by observation of the three aromatic resonance pairs in the range of 8.8–7.5 ppm. The first is two singlets for H-5 and H-10; the second is for H-6 and H-9 in the form of two broad doublets, and the third is two identical broad triplets at around 7.60 and 7.50 ppm for H-7 and H-8, respectively [13,17,23,24]. The core structure was confirmed from the 12 typical 13C-resonances, including the most downfield ones at around 161.0 and 156.0 ppm, assignable for C-4 and C-2, respectively. In the case of the 2-thioxo derivatives (1, 2), C-2 was interpreted at 175.0–176.0 ppm. Further confirmation was achieved according to the intrinsic NMR data corresponding to the S-alkyl/benzyl or N-alkyl (Me or Et) substituents located at positions 2 and 3, respectively. Concerning the 2-S-benzyl derivatives (4–8, 10–13), the CH2-group presented singlets at 4.5–4.65 and 35.0–36.0 ppm in the 1H and 13C NMR spectra, respectively. The 3-N-ethyl products (3, 9) were unambiguously identified from the characteristic A2 × 3 quartet and triplet around 4.10 and 1.27 ppm with 13C-resonances around 41.0 and 13.7 ppm, respectively, while 2-S-ethyl resonances were observed at 3.3 and 1.4 in the 1H NMR and at 26.5 and 14.5 ppm in 13C NMR spectra. The 3-N-Me group was assigned to the singlets around 3.5 ppm and 30.5 ppm in the 1H and 13C NMR spectra, respectively (1–8, 16). The 2-hydrazinyl products (16, 17) were confirmed through two characteristic singlets at 9.55 and 6.22 ppm for –NH-NH2, whereas the aryl C-CH3 group was assigned at about 2.3 and 21.4 in the 1H and 13C NMR, respectively (12, 5). Similarly, the confirmation of 14 and 15 based on the characteristic resonances of the N-ethylpiperidine, propyl and isoindoline moieties (see experimental data).
2.2. Biology
2.2.1. Antiproliferative Activity
Selected compounds were evaluated in vitro for their antiproliferative activities against HepG2 and MCF-7 human cancer-cell lines using MTT assays. Cell viabilities and IC50 values were obtained and compared with those of the control, doxorubicin (Table 1).

Table 1.
The antiproliferative IC50 values of the compounds 1–17 on the two human cell types.
All the compounds presented dose-dependent antiproliferative activities against both cell varieties (Figures S1 and S2). The IC50 values for compounds 1–17 are summarized in Table 1. For HepG2 cancer cells, benzoquinazolines 13, 14, and 15 exhibited potent activities (IC50 = 27.5 ± 2.1, 27.7 ± 2.5 and 26.0 ± 2.5 μM, respectively); compounds 6 and 16 had comparable activities (IC50 = 28.8 ± 2.6 and 28.9 ± 2.3 μM); 3–5, 8, 9, 11, 12, and 17 exhibited slightly lower activities; and compounds 1, 2, 7, and 10, showed moderate activities relative to that of the positive control (IC50 = 28.5 ± 1.9 μM). Regarding the breast cancer cells (MCF-7); benzoquinazolines 3, 4, 7, 9, 10, 13–15, and 17 were more potent (IC50 = 9.6 ± 0.5–10.2 ± 1.1 μM) than doxorubibcin (IC50 = 10.3 ± 0.8). The rest of the compounds showed slightly lower activities relative to that of the positive control (Table 1). Thus, our results demonstrated that compounds 13, 14, and 15 were the most active against the two examined cell lines.
The results in Table 1 indicate that the chemical changes to the lead structures 1 and 2 have considerable impacts on their cytotoxicity profiles for all the derivatives (3–17). Particularly, the S-alkylated products (3–5, 7–15) showed increasing cytotoxicities against MCF-7, and improved activity profiles against HepG2. Furthermore, hydrazinolysis of 1 and 2 into 16 and 17 resulted in remarkable changes in activity. Variations in the position of substitution on the benzyl ring (5–8, 11–13) also affected cytotoxicity, where the presence of the chlorine atom in 6 resulted in a significant increasing in cytotoxicity against HepG2 cells in comparison with the corresponding cyano and methoxy derivatives (7 and 8). Furthermore, the heteroalkyl moiety provided a significant enhancement in cytotoxicity, as indicated by compounds 14 and 15. Thus, the two cell lines studied appeared to be sensitive toward the antiproliferative properties of most of the investigated benzoquinazolines, with 13, 14, and 15 exhibiting the most noticeable effects
2.2.2. Flow Cytometry Cell-Cycle Analysis
Based on cytotoxicity screening (Table 1), we select compounds 13–15 for further investigation of anticancer activity against MCF-7 and HepG2 cancer cells. In addition, 10 was tested against MCF-7 cells. Analyzing cells by flow cytometry allows the detection of their different effects on cell-cycle distribution. Table 2 showed the percentage cell accumulation for the G0/G1, S, and G2/M phases at their IC50 concentrations, as well as apoptosis percentages for the pre-G1 phase (Figure 2A,B and Figure 3). The results revealed the capability of compound 10 to arrest MCF-7 cells at the G2/M phase (34.6% compared to 9.5% for the vehicle control). Compounds 13 and 14 exerted their effects similarly, with MCF-7 cell accumulation at the G1 phase of approximately 62.4% and 66.2%, respectively (Figure 4). Both were remarkably capable of elevating the percentages of HepG2 cells at the S phase (55.3% and 52.3%, respectively). The selectivities of 13 and 14 for the G1 phase in MCF-7 cells were highlighted in the present study, while both trigger some cell-cycle arrest at the S phase. Conversely, the cell-cycle-arrest of compound 15 did not show phase selectivity. Treating MCF-7 and HepG2 cells with 15 resulted in accumulations of 28.0% and 39.2%, respectively, indicating non-selectivity.

Table 2.
Flow cytometry analysis of HepG2 and MCF-7cell cycle and induction of apoptosis after treatment with either DMSO (0.0 μM) or displayed concentration the IC50 of compounds 10, 13–15.
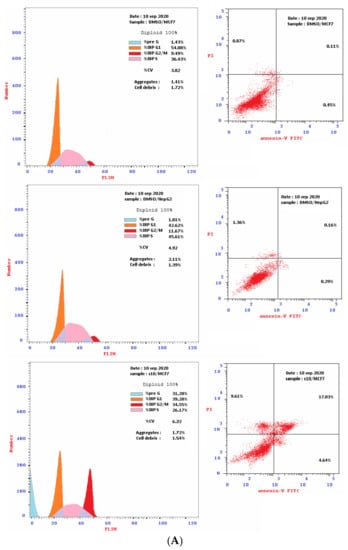
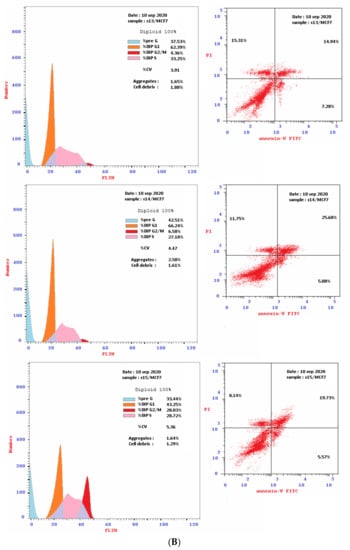
Figure 2.
(A): The flow cytometry cancer cell cycle distribution (Left) and apoptotic status (Right) for MCF-7 and HepG2 cancer cells. Monolayer of cells was treated separately, for 48 h with DMSO or the IC50 of compound 10. (B): The flow cytometry cancer cell cycle distribution (Left) and apoptotic status (Right) for MCF-7 breast cancer cells. Monolayer cells of MCF-7 were treated separately, for 48 h with the IC50 of compounds 13, 14 and 15.
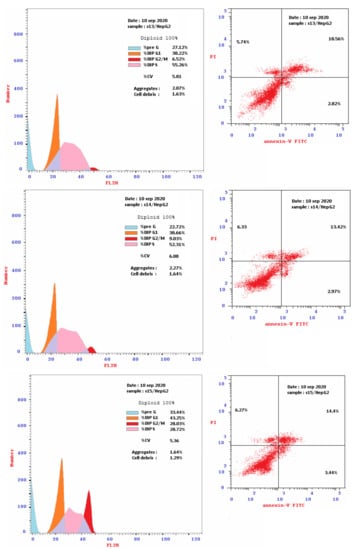
Figure 3.
Flow cytometry cancer cell cycle distribution (Left) and apoptotic status (Right) for HepG2 liver cancer cells. Monolayer cells of HepG2 were treated separately for 48 h with the IC50 of compounds 13, 14 and 15.
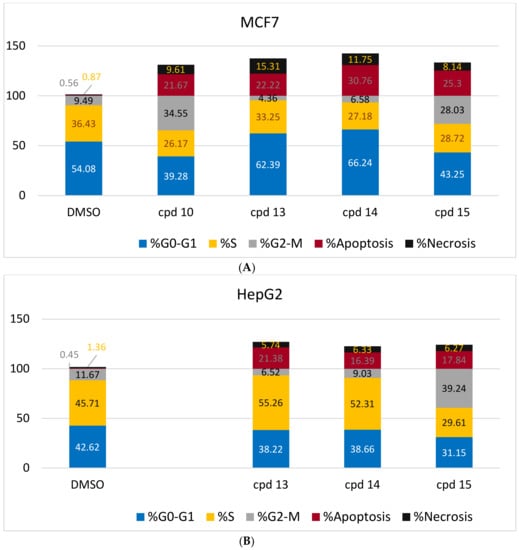
Figure 4.
Graphical representations of the cell cycle distributions of MCF-7 (A) and HepG2 (B) under untreated control conditions (DMSO) and in response to the IC50 treatment of compounds 10 or 13–15 for 48 h. The detailed analysis is given in Table 2.
2.2.3. Detection of Apoptosis
Double Staining with V and PI Dyes
To investigate the type of cell death induced, the treated MCF-7 and HepG2 cells were stained with V and PI. The apoptotic cells are indicated by V+/PI− and V+/PI+ in Figure 2 and Figure 3. Interestingly, apoptosis was evidenced by all the tested compounds (10, 13–15) against both MCF-7 and HepG2 cells. Table 2 reveals 21.67–30.76% apoptosis for MCF-7 cells, with the best induction being observed for 14 (Figure 4A). Induction of apoptosis in the HepG2 cells ranges from 16.39 to 21.38%, with 13 being the best inducer (Figure 4B). However, it should be denoted that, the percentages of the necrotic MCF-7 cells recorded increment upon the separate treatment, with the IC50 of the selected compounds (about 8–15%), while 13 was the worst. Conversely, their equivalents from the treated HepG2 showed about 6%.
Hoechst 33258 Nuclear Staining-DNA Fragmentation
The nuclear morphological changes were detected by fluorescence microscopy upon staining the treated cells with Hoechst 33258 dye. DMSO, as a vehicle, introduces a uniform light-blue staining and intact cell membranes (Figure 5). Conversely, cells treated for 48 h at the IC50 of each compound were reduced in size. Bright-blue fluorescence was observed due to chromatin condensation, except in the case of MCF-7 treated with compound 13, where elevation in the necrotic cells by 15% was observed (as shown by flow cytometry). Their morphology, size, and blue color did not differ more than those of the MCF-7 vehicle. Interestingly, 15 caused similar changes for both cells.
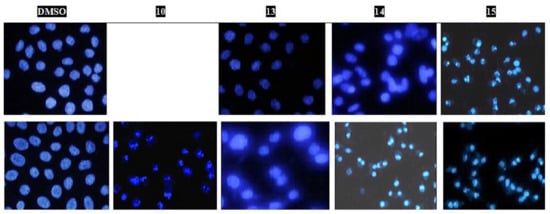
Figure 5.
Fluorescent microphotograph section of the treated HepG2 (A) and MCF-7 (B) cells with DMSO (0.0 μM) or the IC50 of the indicated compounds 10, 13–15 for 48 h. Cells were all stained by the Hoechst 33258.
In Vitro Inhibition of VEGFR-2
Compounds 10 and 13–15 were further evaluated for their antiangiogenic activities. Figure 6 and Table 3 show the determined IC50 values (46.6 ± 2.8 and 44.4 ± 2.6 nM, respectively)) for 13 and 15, which are comparable to that of sorafenib (31.1 ± 1.8 nM).
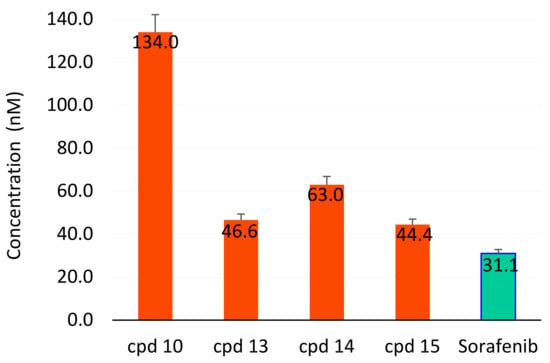
Figure 6.
IC50 of compounds 10, 13–15 (compared with sorafenib) as inhibitors to the angiogenic VEGFR2 enzyme compared with sorafenib. Green color shows the lowest concentration.

Table 3.
Effect of compounds 10, 13–15 on VEGFR-2 enzyme and their fold inactivation.
Our results confirmed the widely accepted fact that VEGF prevents apoptosis [25]. In the present study, inhibition of the VEGFR-2 enzyme and induction of apoptosis for both tested cell lines were observed for compounds 10 and 13–15. The VEGFR-2 inhibition activity relative to sorafenib increased in the order of 15 (1.4-fold) > 13 (1.5-fold) > 14 (2.0-fold) > 10 (4.5-fold). The induction of apoptosis for MCF-7 (Table 2) follows the order 14 (30.76%) > 15 (25.3%) > 13 (22.22%) > 10 (21.67%). Conversely, HepG2 apoptosis followed the order 13 (21.38%) > 15 (17.84%) > 14 (16.39%).
2.3. Molecular Docking
The molecular design of the benzo[g]quinazolines as VEGFR-2 inhibitors was based on the binding mode of sorafenib with VEGFR-2, which was obtained from the crystallographic complex available in the Protein Data Bank (4ASE.pdb, https://www.rcsb.org/structure/4ASE). The docking study was performed to investigate the interaction between the synthesized benzo[g]quinazolines and the ATP-binding site, which is located between the N-terminal and the C-terminal lobes within the catalytic domain of VEGFR-2 (Figure 7A,B). Many kinase inhibitors act as ATP mimetics and compete with cellular ATP for binding sites, thus suppressing autophosphorylation [9,10]. The VEGFR-2 ATP-binding pocket residues, as shown in Figure 7B, were HYD I (encapsulated by Leu-840, Phe-918, and Gly-922), the HYD II (encapsulated by Leu-889, Ile-892, Val-8 98, and Ile-1044), and the linker (encapsulated by Ala-866, Val-914, Leu-1035, and Cys-1045). Thus, ATP-binding site of VEGFR-2 was mainly established by hydrophobic residues in HYD I and HYD II (Figure 7 and Figure 8A).
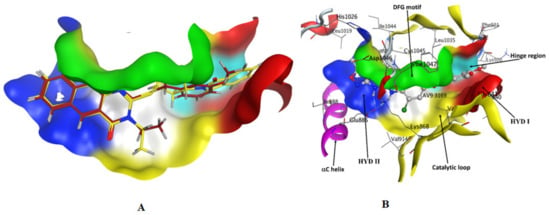
Figure 7.
(A) 3D visualization of 13 (red) and 15 (yellow) in fully active VEGFR2 ATP binding pocket residues; (B) 3D visualization of the fully active VEGFR2 ATP binding pocket residues. The residues are colored according to the regions.
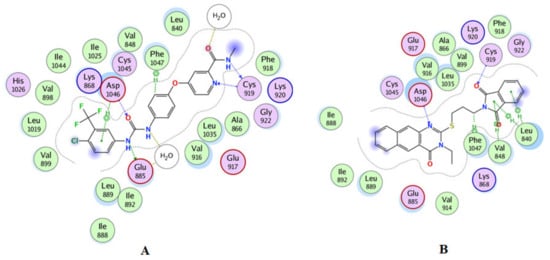
Figure 8.
(A) 2D interactions of the co-crystallized ligand inside the active site of 4ASE (sorafenib); (B) 2D interaction of compound 15 in the active site of VEGFR-2.
The docking protocol was confirmed by re-docking of the co-crystallized ligand in the vicinity of the active site of the enzyme with binding affinity = −11.0317 kcal/mol and RMSD = 0.34016 (Table 4, Figure 7B). The docking results revealed that 15 and 13 were the most potent inhibitors (Figure 7A). Compound 15 demonstrated a good Z-score in QSAR (Table S4) and a high binding affinity of −9.669 kcal/mol. Moreover, 15 tightly bound to the key amino acids Asp1046, Phe1047, and Cys919 through the formation of H bonding and Leu840, Val848, and Phe1047 via the formation of π-π interactions, as shown in Figure 8B.

Table 4.
Docking energy and interactions of compounds 1–17 and reference compounds with target protein 4ASE.
Compound 13 interacted via H bonds with Asp1046 and CYS1045 and with VAL848 via π-π interactions in the active site of VEGFR-2 (Figure 9A) with high binding affinity −8.133 kcal/mol (Table 4). Compound 14 bound with Phe918 and Cys919 via H bonds and with Leu 840 and Val 848 through π-π interactions with the high binding affinity −8.307 kcal/mol (Figure 9B). Moreover, the configuration results for compounds 13 and 15 revealed that both compounds were type-II inhibitors due to their binding in the DFG region adjacent to the HYD II region and called the allosteric site. The other compounds were identified as type-I inhibitors, as their interactions were mainly with the hinge-region residues Cys919 and Leu840 adjacent to HYD I, as shown in Table 4.
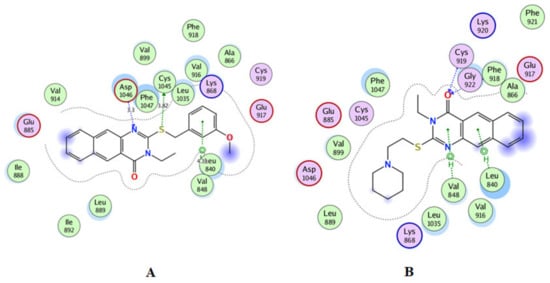
Figure 9.
2D interactions in the active site of VEGFR2 for compound 13 (A) and for compound 14 (B).
2.4. QSAR Study
Partial least squares (PLS) were employed to analyze the chemical space of the training set. After PLS with the 364 Molecular operating environment (MOE) descriptors for 71 training set modeling compounds, the first three most important components (PEOE_VSA+0, PEOE_VSA-1, and SMR_VSA1) and (PEOE_VSA+0, PEOE_VSA+1, and SMR_VSA-0) for HepG2 and MCF-7, respectively, were chosen to generate a 3D plot (Figure 10A,B) for these compounds. The compound activities were distinguished well, and were labeled in different colors according to their biological response. Therefore, we retained these three descriptors and combined them with two other descriptors to build QSAR models (Tables S1–S5).
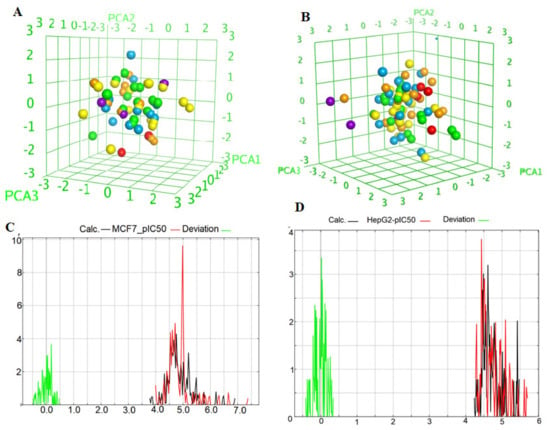
Figure 10.
(A,B) 3D chemical space plot of the selected training compounds of HepG2 and MCF-7 divided by the important descriptors; (C,D) Curve plot of the training set for MCF-7 and HepG2 (n = 10), pIC50 exp: red curve; pIC50 cal: black curve; Deviation: green curve.
Before building the QSAR models, all descriptor’s correlations must be considered. If one descriptor is collinear with one or more descriptors, i.e., the correlation result is higher than 0.5, these descriptors should not be considered together. The selected descriptors had little mutual correlation and can be used in the same equation (Tables S3 and S5). PEOE_VSA+5 and PEOE_VSA-1 exemplified the sum of van der Waals surface areas, where the partial charges are in the range (0.25, 0.30) and (−0.10, −0.05), respectively. SMR_VSA1, SMR_VSA4, and SMR_VSA7 were molar refractivity descriptors and had a high correlation with molecular polarizability [26]. They were calculated in the ranges (0.11–0.26), (0.39–0.44), and >0.56, respectively. The correlation coefficients for HepG2 and MCF-7 and their data had acceptable R2, XR2, RMSE, and the cross-validated RMSE values for all compounds, as shown by Equations (5) and (6) (Table S2). A high value of q2 (for instance, q2 > 0.5), as an indicator or even as the ultimate proof that model, is considered highly predictive by many authors. The Equations (3) and (4) models (Table S2) have the following statistical results: = 0.62709, XRMSE = 0.21027, = 0.7277, and XRMSE = 0.2822, which suggests that these models are highly predictive and reliable for the prediction of biological activity. The pIC50 data indicate that the range is wide, from 4.2 to 0.56 and 4.97 to 5.02 for HepG2 and MCF-7, respectively (Table S6). The structures of the target benzoquinazolines were diverse, with different substitutions. Thus, the width of the data and the variety of the structures can help to increase the prediction ability and universality of the QSAR model. Figure 10C,D shows that the curve for the experimental pIC50 is similar to that of the calculated pIC50, and the deviations mainly fluctuate from −0.5 to 0.5 for both HepG2 and MCF-7. Approximately 80% of the deviations are around 0 for both, indicating that the QSAR model had high quality and stability from the training set.
The predicted pIC50 values for the training and test sets were plotted against the experimental pIC50 values (Figure 11 and Figure 12). The predicted pIC50 results obtained for both the modeling set were in good agreement with the experimental results obtained for biological activity against HepG2 and MCF-7. The residual values obtained between predicted and experimental pIC50 values were very low. The coefficient of determination (R2) was close to 1.0, indicating that the model represents a very high percentage of the response variable (descriptor) variation, certainly high enough for a robust QSAR model. The obtained values (0.71947 and 0.85989) indicated that 71.947 and 85.989% of the variation for HepG2 and MCF-7, respectively, resided in the residual, indicating that the model is very good.
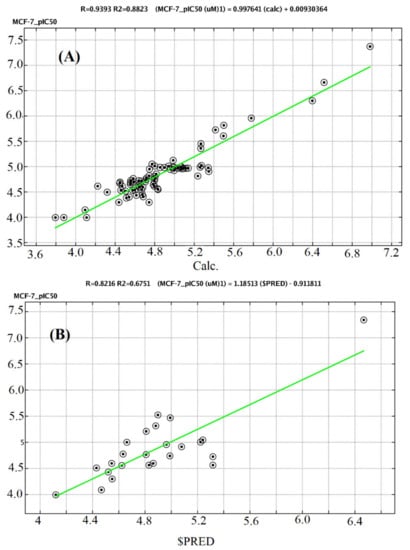
Figure 11.
A plot of observed versus predicted pIC50 (MCF-7) of the training set (A) and testing set (B).
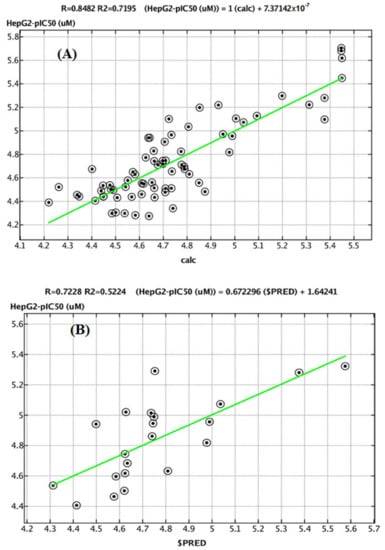
Figure 12.
A plot of observed versus predicted pIC50 (HepG2) of the training set (A) and testing set (B).
3. Materials and Methods
3.1. Chemistry
3.1.1. Synthesis of 1–17
The general procedures for the preparation of compounds 1–17 were previously reported for the preparation of their analogues [13,17]. Briefly, in the basic medium, the reaction of ethyl(methyl)isothiocyanate with 3-amino-2-naphthaoic acid under a refluxing condition in DMF for 3–5 h afforded the parent intermediates 1 and 2 in good yields (Scheme 1). When compound 1 or 2 was treated with the appropriate alkyl/heteroalkyl halide in the presence of a base at 80 °C for 20 h, benzo[g]quinazolines 3–15 were obtained in good to high yields. Hydrazinolysis of 1 and 2 in boiling DMF for 15–18 h afforded 16 and 17, respectively. Their characterization data are reported herein as follows:
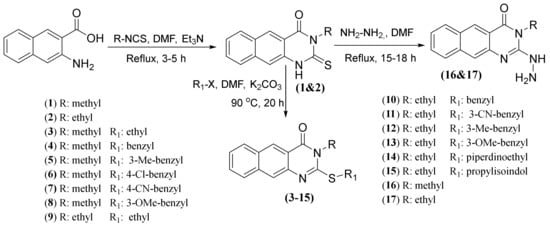
Scheme 1.
The synthetic routes for 2-thioxobenzo[g]quinazolines 1–17.
3-Methyl-2-thioxo-2,3-dihydrobenzo[g]quinazolin-4(1H)-one (1)
White amorphous powder (86%); mp > 300 °C; 1H NMR (500 MHz, DMSO-d6): δ 13.02 (br s, 1H, -NH), 8.71 (s, 1H, H-5), 8.14 (br s, 1H, H-6), 7.95 (br s, 1H, H-9), 7.77 (s, 1H, H-10), 7.64 (br s, 1H, H-7), 7.51 (br s, 1H, H-8), 3.72 (s, 3H, N-CH3); 13C NMR (125 MHz, DMSO-d6): δ 175.9 (C-2), 160.2 (C-4), 136.6 (C-4b,9a), 135.3 (C-5a), 130.5 (C-5), 129.9 (C-6), 129.5 (C-8), 127.5 (C-4a), 125.9 (C-9), 115.8 (C-7), 111.3 (C-10), 33.5 (N-CH3); HRMS-MS: m/z calcd. for C13H10N2OS (M)•+ 242.0514, found 242.0531 [23,24].
3-Ethyl-2-thioxo-2,3-dihydrobenzo[g]quinazolin-4(1H)-one (2)
White amorphous powder (82%); mp 296 °C; 1H NMR (700 MHz, DMSO-d6): δ 12.96 (s, 1H, -NH-), 8.69 (s, 1H, H-5), 8.12 (br d, J = 8 Hz, 1H, H-6), 7.93 (br d, J = 8 Hz, 1H, H-9), 7.74 (s, 1H, H-10), 7.62 (br t, J = 7.4 Hz, 1H, H-7), 7.50 (br t, J = 7.4 Hz, 1H, H-8), 4.49 (t, q = 7 Hz, 2H, H-1′), 1.27 (t, J = 7 Hz, 3H, H-2′); 13C NMR (175 MHz, DMSO-d6): δ 175.4 (C-2), 159.5 (C-4), 136.6 (C-4b,9a), 135.3 (C-5a), 130.0 (C-5), 129.9 (C-6), 129.7 (C-8), 127.7 (C-4a), 126.1 (C-9), 116.0 (C-7), 111.5 (C-10), 41.4 (C-1′), 12.7 (C-2′); HRMS-MS: m/z calcd. for C14H12N2OS (M)•+ 256.0670, found 256.0693 [23,24].
2-(Ethylthio)-3-methylbenzo[g]quinazolin-4(3H)-one (3)
White amorphous powder (75%); mp 115–117 °C; 1H NMR (500 MHz, DMSO-d6): δ 8.76 (s, 1H, H-5), 8.17 (br d, J = 8.3 Hz, 1H, H-6), 8.05 (s, 1H, H-10), 8.04 (br d, J = 8.3 Hz, 1H, H-9), 7.63 (br t, J = 7.3 Hz, 1H, H-7), 7.53 (br t, J = 7.3 Hz, 1H, H-8), 3.45 (s, 3H, N-CH3), 3.28 (q, J = 7.3 Hz, 2H, H-1′), 1.41 (t, J = 7.3 Hz, 3H, H-2′); 13C NMR (125 MHz, DMSO-d6): δ 161.5 (C-4), 156.5 (C-2), 142.8 (C-4b), 136.8 (C-5a), 130.8 (C-9a), 129.7 (C-5), 128.9 (C-6), 128.3 (C-8), 128.1 (C-4a), 126.3 (C-9), 123.1 (C-7), 118.8 (C-10), 30.4 (N-CH3), 26.4 (C-1′), 14.5 (C-2′); HRMS-MS: m/z calcd. for C15H14N2OS (M)•+ 270.0827, found 270.0872 [24].
2-(Benzylthio)-3-methylbenzo[g]quinazolin-4(3H)-one (4)
Pale yellow amorphous powder (71%); mp 150–152 °C; 1H NMR (700 MHz, DMSO-d6): δ 8.81 (s, 1H, H-5), 8.19 (br s, 2H, H-6, 10), 8.11 (br s, 1H, H-9), 7.67 (br s, 1H, H-7), 7.58 (br s, 3H, H-8, 2′/6′), 7.36 (br s, 2H, H-3′/5′), 7.28 (br s, 1H, H-4′), 4.61 (s, 2H, -CH2-7′), 3.52 (s, 3H, N-CH3); 13C NMR (175 MHz, DMSO-d6): δ 161.5 (C-4), 156.2 (C-2), 142.7 (C-4b), 137.3 (C-1′), 136.8 (C-9a), 130.9 (C-5a), 130.1 (C-5), 129.9 (C-3′/5′), 129.8 (C-6), 129.1 (C-8), 128.9 (C-2′/6′), 128.4 (C-4a), 128.1 (C-4′), 126.4 (C-9), 123.2 (C-7), 118.8 (C-10), 35.9 (-CH2-7′), 30.4 (N-CH3); HRMS-MS: m/z calcd. for C20H16N2OS (M)•+ 332.0983, found 332.0991.
3-Methyl-2-((3-methylbenzyl)thio)benzo[g]quinazolin-4(3H)-one (5)
White amorphous powder (70%); mp 155–157 °C; 1H NMR (500 MHz, DMSO-d6): δ 8.81 (s, 1H, H-5), 8.20 (br d, J = 8.4 Hz, 1H, H-6), 8.18 (s, 1H, H-10), 8.10 (br d, J = 8.4 Hz, 1H, H-9), 7.66 (br t, J = 7.6 Hz, 1H, H-7), 7.56 (br t, J = 7.6 Hz, 1H, H-8), 7.38 (br s, 1H, H-2′), 7.36 (br d, J = 7.5 Hz, 1H, H-6′), 7.24 (br t, J = 7.5 Hz, 1H, H-5′), 7.08 (br d, J = 7.5 Hz, 1H, H-4′), 4.57 (s, 2H, -CH2-7′), 3.52 (s, 3H, N-CH3), 2.30 (s, 3H, Ar-Me); 13C NMR (125 MHz, DMSO-d6): δ 161.5 (C-4), 156.2 (C-2), 142.7 (C-4b), 138.1 (C-1′), 136.9 (C-5a), 136.8 (C-3′), 130.9 (C-9a), 130.5 (C-5), 129.8 (C-6), 129.0 (C-2′), 128.9 (C-8), 128.6 (C-5′), 128.4 (C-4a), 128.1 (C-4′), 126.9 (C-9), 126.4 (C-6′), 123.2 (C-7), 118.9 (C-10), 36.0 (-CH2-7′), 30.4 (N-CH3), 21.4 (Ar-Me); HRMS-MS: m/z calcd for C21H18N2OS (M)•+ 346.1140, found 346.1155.
2-((4-Chlorobenzyl)thio)-3-methylbenzo[g]quinazolin-4(3H)-one (6)
Pale yellow amorphous powder (78%); mp 190–192 °C; 1H NMR (500 MHz, DMSO-d6): δ 8.81 (s, 1H, H-5), 8.20 (br d, J = 8.3 Hz, 1H, H-6), 8.19 (s, 1H, H-10), 8.10 (br d, J = 8.3 Hz, 1H, H-9), 7.65 (br t, J = 7.3 Hz, 1H, H-7), 7.59 (d, J = 8.4 Hz, 2H, H-3′/5′), 7.56 (br t, J = 7.3 Hz, 1H, H-8), 7.41 (d, J = 8.4 Hz, 2H, H-2′/6′), 4.60 (s, 2H, -CH2-7′), 3.52 (s, 3H, N-CH3); 13C NMR (125 MHz, DMSO-d6): δ 161.5 (C-4), 155.9 (C-2), 142.6 (C-4b), 136.8 (C-5a), 132.4 (C-1′), 131.7 (C-3′/5′), 131.2 (C-4′), 130.9 (C-9a), 129.8 (C-5), 129.1 (C-6), 128.8 (C-2′/6′), 128.4 (C-8), 128.1 (C-4a), 126.4 (C-9), 123.2 (C-7), 118.7 (C-10), 35.0 (-CH2-7′), 30.4 (N-CH3); HRMS-MS: m/z calcd. for C20H15ClN2OS (M)•+ 366.0594, found 366.0607.
4-(((3-Methyl-4-oxo-3,4-dihydrobenzo[g]quinazolin-2-yl)thio)methyl)benzonitrile (7)
White amorphous powder (75%); mp 201–203 °C; 1H NMR (500 MHz, DMSO-d6): δ 8.79 (s, 1H, H-5), 8.19 (br d, J = 7.5 Hz, 1H, H-6), 8.18 (s, 1H, H-10), 8.07 (br d, J = 7.5 Hz, 1H, H-9), 7.94 (d, J = 8.5 Hz, 2H, H-3′/5′), 7.73 (d, J = 8.5 Hz, 2H, H-2′/6′), 7.65 (br t, J = 7.5 Hz, 1H, H-7), 7.55 (br t, J = 7.5 Hz, 1H, H-8), 4.64 (s, 2H, -CH2-7′), 3.51 (s, 3H, N-CH3); 13C NMR (125 MHz, DMSO-d6): δ 161.5 (C-4), 155.6 (C-2), 143.8 (C-4b), 141.4 (C-1′), 136.8 (C-9a), 132.7 (C-3′/5′), 131.0 (C-5a), 130.7 (C-2′/6′), 129.7 (C-5), 128.8 (C-6), 128.4 (C-8), 128.1 (C-4a), 126.4 (C-9), 123.2 (C-7), 119.0 (C≡N), 118.8 (C-10), 110.9 (C-4′), 35.5 (-CH2-7′), 30.4 (N-CH3),; HRMS-MS: m/z calcd. for C21H15N3OS (M)•+ 357.0936, found 357.0950.
2-((3-Methoxybenzyl)thio)-3-methylbenzo[g]quinazolin-4(3H)-one (8)
White amorphous powder (75%); mp 188–190 °C; 1H NMR (500 MHz, DMSO-d6): δ 8.82 (s, 1H, H-5), 8.21 (br d, J = 8.7 Hz, 1H, H-6), 8.20 (s, 1H, H-10), 8.11 (br d, J = 8.7 Hz, 1H, H-9), 7.67 (br t, J = 7.3 Hz, 1H, H-7), 7.57 (br t, J = 7.3 Hz, 1H, H-8), 7.27 (br t, J = 8.1 Hz, 1H, H-5′), 7.17 (br s, 1H, H-2′), 7.14 (br d, J = 8.1 Hz, 1H, H-6′), 6.86 (br d, J = 8.3 Hz, 1H, H-4′), 4.59 (s, 2H, -CH2-7′), 3.75 (s, 3H, -OCH3), 3.53 (s, 3H, N-CH3); 13C NMR (125 MHz, DMSO-d6): δ 161.5 (C-4), 159.7 (C-3′), 156.2 (C-2), 142.7 (C-4b), 138.8 (C-1′), 136.8 (C-5a), 130.9 (C-9a), 130.1 (C-5′), 129.8 (C-5), 129.1 (C-6), 128.4 (C-8), 128.1 (C-4a), 126.4 (C-9), 123.2 (C-7), 122.1 (C-6′), 118.8 (C-10), 115.5 (C-4′), 113.4 (C-2′), 55.5 (-OCH3), 35.9 (-CH2-7′), 30.4 (N-CH3); HRMS-MS: m/z calcd. for C21H18N2O2S (M)•+ 362.1089, found 362.1104.
3-Ethyl-2-(ethylthio)benzo[g]quinazolin-4(3H)-one (9)
White amorphous powder (73%); mp 107–109 °C; 1H NMR (500 MHz, DMSO-d6): δ 8.79 (s, 1H, H-5), 8.18 (br d, J = 8.3 Hz, 1H, H-6), 8.10 (s, 1H, H-10), 8.08 (br d, J = 8.3 Hz, 1H, H-9), 7.65 (br t, J = 7.8 Hz, 1H, H-7), 7.55 (br t, J = 7.8 Hz, 1H, H-8), 4.10 (q, J = 7.1 Hz, 2H, H-1″), 3.31 (q, J = 7.3 Hz, 2H, H-1′), 1.43 (t, J = 7.3 Hz, 3H, H-2′), 1.31 (t, J = 7.1 Hz, 3H, H-2″); 13C NMR (125 MHz, DMSO-d6): δ 161.1 (C-4), 155.7 (C-2), 142.8 (C-4b), 136.8 (C-5a), 130.9 (C-9a), 129.7 (C-5), 129.0 (C-6), 128.3 (C-8), 128.1 (C-4a), 126.4 (C-9), 123.2 (C-7), 118.9 (C-10), 40.5 (C-1″ hidden by DMSOd6 signal), 26.4 (C-1′), 14.5 (C-2′), 13.7 (C-2″); HRMS-MS: m/z calcd. for C16H16N2OS (M)•+ 284.0983, found 284.1003 [24].
2-(Benzylthio)-3-ethylbenzo[g]quinazolin-4(3H)-one (10)
White amorphous powder (74%); mp 132–134 °C; 1H NMR (700 MHz, DMSO-d6): δ 8.77 (s, 1H, H-5), 8.16 (br d, J = 8.4 Hz, 1H, H-6), 8.15 (s, 1H, H-10), 8.07 (br d, J = 8.4 Hz, 1H, H-9), 7.64 (br t, J = 8 Hz, 1H, H-7), 7.56 (br d, 2H, J = 7.4 Hz, H-2′/6′), 7.54 (br t, J = 8 Hz, 1H, H-8), 7.35 (t-like, J = 7.4 Hz, 2H, H-3′,5′), 7.28 (br d, J = 7.4 Hz, 1H, H-4′), 4.59 (s, 2H, -CH2-7′), 4.07 (q, J = 7 Hz, 2H, H-1″), 1.27 (t, J = 7 Hz, 3H, H-2″); 13C NMR (175 MHz, DMSO-d6): δ 161.1 (C-4), 155.2 (C-2), 142.6 (C-4b), 137.2 (C-1′), 136.8 (C-9a), 130.9 (C-5a), 129.9 (C-5), 129.8 (C-3′,5′), 129.7 (C-6), 128.9 (C-8), 128.7 (C-2′,6′), 128.3 (C-4a), 127.9 (C-4′), 126.4 (C-9), 123.3 (C-7), 118.9 (C-10), 40.5 (C-1″), 35.9 (-CH2-7′), 13.8 (C-2″); HRMS-MS: m/z Calcd for C21H18N2OS (M)•+ 346.1140, found 346.1154
3-(((3-Ethyl-4-oxo-3,4-dihydrobenzo[g]quinazolin-2-yl)thio)methyl)benzonitrile (11)
Pale yellow amorphous powder (81%); mp 140–142 °C; 1H NMR (700 MHz, DMSO-d6): δ 8.80 (s, 1H, H-5), 8.19 (br s, 2H, H-6, 10), 8.08 (br d, J = 8.4 Hz, 1H, H-9), 8.07 (br s, 1H, H-2′), 7.95 (br d, J = 7.3 Hz, 1H, H-6′), 7.74 (br d, J = 7.1 Hz, 1H, H-4′), 7.67 (br t, J = 7.3 Hz, 1H, H-7), 7.57 (br s, 2H, H-8, 5′), 4.66 (s, 2H, H-7′) 4.10 (q, J = 6.9 Hz, 2H, H-1″), 1.29 (t, J = 6.9 Hz, 3H, H-2″); 13C NMR (175 MHz, DMSO-d6; δ 161.1 (C-4), 155.3 (C-2), 142.6 (C-4b), 139.8 (C-1′), 136.8 (C-5a), 134.8 (C-6′), 133.4 (C-2′), 131.5 (C-4′), 130.9 (C-9a), 130.4 (C-5), 130.1 (C-5′), 129.1 (C-6), 128.4 (C-8), 128.1 (C-4a), 126.5 (C-9), 123.2 (C-7), 118.9 (C-10), 119.1 (C≡N), 111.7 (C-3′), 41.0 (C-1″), 35.6 (-CH2-7′), 13.7 (C-2″); HRMS-MS: m/z calcd. for C22H17N3OS (M)•+ 371.1092, found 371.1108.
3-Ethyl-2-((3-methylbenzyl)thio)benzo[g]quinazolin-4(3H)-one (12)
White amorphous powder (70%); mp 186–188 °C; 1H NMR (700 MHz, DMSO-d6): δ 8.77 (s, 1H, H-5), 8.16 (br d, J = 8.3 Hz, 1H, H-6), 8.14 (s, 1H, H-10), 8.07 (br d, J = 8.3 Hz, 1H, H-9), 7.64 (br t, J = 7.6 Hz, 1H, H-7), 7.54 (br t, J = 7.6 Hz, 1H, H-8), 7.36 (br s, 1H, H-2′), 7.34 (br d, J = 7.4 Hz, 1H, H-6′), 7.23 (br t, J = 7.4 Hz, 1H, H-5′), 7.08 (br d, J = 7.4 Hz, 1H, H-4′), 4.54 (s, 2H, -CH2-7′), 4.06 (q, J = 7 Hz, 2H, H-1″), 2.29 (s, 3H, Ar-Me), 1.27 (t, J = 7 Hz, 3H, H-2″); 13C NMR (175 MHz, DMSO-d6): δ 161.1 (C-4), 155.3 (C-2), 142.6 (C-4b), 138.1 (C-1′), 136.9 (C-5a), 136.8 (C-3′), 130.9 (C-9a), 130.5 (C-5), 129.7 (C-6), 129.0 (C-2′), 128.9 (C-8), 128.6 (C-5′), 128.4 (C-4a), 128.1 (C-4′), 126.9 (C-9), 126.4 (C-6′), 123.2 (C-7), 118.9 (C-10), 41.0 (C-1″), 36.0 (-CH2-7′), 21.4 (Ar-Me), 13.8 (C-2″); HRMS-MS: m/z calcd. for C22H20N2OS (M)•+ 360.1296, found 360.1312.
3-Ethyl-2-((3-methoxybenzyl)thio)benzo[g]quinazolin-4(3H)-one (13)
White amorphous powder (80%); mp 110–112 °C; 1H NMR (700 MHz, DMSO-d6): δ 8.80 (s, 1H, H-5), 8.19 (br d, J = 8.4 Hz, 1H, H-6), 8.17 (s, 1H, H-10), 8.09 (br d, J = 8.4 Hz, 1H, H-9), 7.66 (br t, J = 7.5 Hz, 1H, H-7), 7.56 (br t, J = 7.5 Hz, 1H, H-8), 7.27 (br t, J = 7.6 Hz, 1H, H-5′), 7.16 (br s, 1H, H-2′), 7.13 (br d, J = 7.6 Hz, 1H, H-6′), 6.85 (br d, J = 7.6 Hz, 1H, H-4′), 4.58 (s, 2H, -CH2-7′), 4.09 (q, J = 7 Hz, 2H, H-1″), 3.74 (s, 3H, -OCH3), 1.28 (t, J = 7 Hz, 3H, H-2″); 13C NMR (175 MHz, DMSO-d6): δ 161.1 (C-4), 159.7 (C-3′), 155.3 (C-2), 142.6 (C-4b), 138.7 (C-1′), 136.8 (C-5a), 130.9 (C-9a), 130.1 (C-5′), 129.8 (C-5), 129.1 (C-6), 128.4 (C-8), 128.1 (C-4a), 126.5 (C-9), 123.2 (C-7), 122.0 (C-6′), 118.9 (C-10), 115.5 (C-4′), 113.4 (C-2′), 55.5 (-OCH3), 41.0 (C-1″), 35.9 (-CH2-7′), 13.8 (C-2″); HRMS-MS: m/z calcd. for C22H20N2O2S (M)•+ 376.1245, found 376.1250.
3-Ethyl-2-((2-(piperidin-1-yl)ethyl)thio)benzo[g]quinazolin-4(3H)-one (14)
Pale yellow amorphous powder (50%); mp190–192 °C; 1H NMR (500 MHz, DMSO-d6): δ 8.80 (s, 1H, H-5), 8.16 (m, 2H, H-6, 10), 8.07 (br d, J = 7.5 Hz, 1H, H-9), 7.65 (br t, J = 7.5 Hz, 1H, H-7), 7.53 (br t, J = 7.5 Hz, 1H, H-8), 4.13 (br s, 2H, H-1″), 3.47 (br s, 2H, CH2-7′), 2.72 (br s, 2H, CH2-8′), 2.50 (m hidden by DMSO-6 signal, 4H, H-2′/6′), 1.54 (m, 4H, H-3′/5′), 1.42 (m, 2H, H-4′), 1.28 (br s, 3H, H-2″); 13C NMR (125 MHz, DMSO-d6): δ 161.1 (C-4), 155.7 (C-2), 142.7 (C-4b), 136.8 (C-5a), 130.0 (C-9a), 129.9 (C-5), 129.0 (C-6), 128.4 (C-8), 128.1 (C-4a), 126.4 (C-9), 123.1 (C-7), 118.9 (C-10), 56.5 (C-2′/6′), 54.1 (CH2-7′), 41.4 (C-1″), 31.2 (CH2-8′), 25.7 (C-3′/5′), 23.5 (C-4′), 13.7 (C-2″); HRMS-MS: m/z calcd. for C21H25N3OS (M)•+ 367.1718, found 367.1733.
2-(3-((3-Ethyl-4-oxo-3,4-dihydrobenzo[g]quinazolin-2-yl)thio)propyl)isoindoline -1,3-dione (15)
White amorphous powder (76%); mp 102–1704 °C; 1H NMR (700 MHz, DMSO-d6): δ 8.78 (s, 1H, H-5), 8.18 (br s, 1H, H-6), 7.96–7.80 (m, 5H, H-9, 5′/6′, 4′/7′), 7.73 (s, 1H, H-10), 7.65 (br s, 1H, H-7), 7.55 (br s, 1H, H-8), 4.10 (br s, 2H, H-1″), 3.79 (br s,2H, CH2-8′), 3.35 (br s hidden by H2O-signal, 2H, CH2-10′), 2.14 (br s, 2H, CH2-9′), 1.28 (br s, 3H, H-2″); 13C NMR (175 MHz, DMSO-d6): δ 168.6 (C-1′/3′), 161.1 (C-4), 155.4 (C-2), 142.6 (C-4b), 136.7 (C-5a), 134.9 (C-3a′/7a′), 132.2 (C-5′/6′), 130.9 (C-9a), 129.8 (C-5), 129.1 (C-6), 128.4 (C-8), 127.9 (C-4a), 126.4 (C-9), 123.6 (C-4′/7′), 122.9 (C-7), 118.9 (C-10), 40.5 (CH2-1″, hidden by DMSO-d6 signals), 37.1 (CH2-8′), 29.1 (CH2-10′), 28.4 (CH2-9′), 13.7 (CH3-2″); HRMS-MS: m/z calcd. for C25H21N3O3S (M)•+ 443.1304, found 443.1321.
Compounds 16 and 17 were reported [23].
3.2. Biology
3.2.1. MTT Assay
The antiproliferative activity of the test compounds against HepG2, and MCF-7 human cancer-cell lines were evaluated by MTT assay. The detailed methodology procedure was reported [27,28,29].
3.2.2. Cell-Cycle Analysis by Flow Cytometry
To investigate whether the tested compounds could arrest HepG2 and MCF-7 mitotic cell division, cell distributions at cell-cycle phases were analyzed by flow cytometry following their treatment with the vehicle or the indicated compound at its IC50 concentration. Cells and samples were prepared as previously reported [30], and a PI (ab139418) Flow Cytometry Kit (Abcam, Cambridge, UK) was used in accordance with the manufacturer’s protocol. In brief, both HepG2 and MCF-7 cells were separate, plated into six-well plates at a density of 0.6 × 106 cells/well in DMEM growth medium supplemented with 10% FBS. Cells were incubated for 24 h at 37 °C and 5% CO2 prior to exposure to either the vehicle (DMSO) as the control or 13, 14, or 15, at their IC50 concentrations. In addition, MCF-7 cells were subjected to compound 10 at 9.61 μM (its recorded IC50) rather than HepG2 due to its higher IC50 (36.2 μM) against the latter cells. All cells were incubated for 48 h, washed with cold PBS after being harvested to single-cell suspensions by trypsinization, and fixed with 70% ethanol. The fixed cells were rehydrated in PBS prior to being lysed in a prepared solution of PI, Triton X-100, and RNase. Finally, the cells were kept at 37 °C for 15 min and analyzed using a FACS Caliber flow cytometer (BD Biosciences and Co., Franklin Lakes, NJ, USA).
3.2.3. Measurements of Apoptosis Using Double Staining with Annexin-V FITC/PI Dyes
Apoptosis was detected for both HepG2 and MCF-7cells using the same preparation and sample treatment conditions as those used for cell-cycle analysis. The manufacturer’s instructions for the annexin-V FITC apoptosis detection kit (K101, Biovision, Milpitas, CA, USA) were followed. In brief, treated cells were detached after 48 h by trypsinization, washed twice with cold PBS, and stained with 5 μL of annexin-V fluorescein isothiocyanate (annexin-V FITC) and 5 μL of PI in the binding buffer for 15 min at 37 °C. Finally, the processed cells were analyzed using a FACS Caliber flow cytometer (BD Biosciences and Co., Franklin Lakes, NJ, USA).
3.2.4. Hoechst 33258 Nuclear Staining-DNA Fragmentation
In the case of detecting the induction of apoptosis in HepG2 and MCF7 cells by the fluorescent microscopy, also same conditions of preparations and sample treatment as in the section of cell-cycle analysis were followed, while the instructions of Hoechst 33258 Staining Dye Solution, Cat. No. #ab228550 (Abcam, Cambridge, UK) were performed. Fluorescent micrographs were taken by a Fluorescence Microscope (LF-302) Cat. No. #2917 at 40× objective lens.
3.2.5. In Vitro Inhibition of VEGFR-2
Angiogenesis inhibition was investigated using an enzymatic VEGFR-2 (KDR) Kinase Assay Kit Cat. #40325 assay following the manufacturer’s instructions [31]. A series of 10-fold dilutions were prepared from compounds 10, 13–15 to provide the concentration range 0.0–10.0 μM. Sorafenib (Cat No. 284461-73-0, Santa Cruz, Dallas, TE, USA) was used as a standard inhibitor to the VEGFR2 enzyme. At the indicated end of the experiment, the luminescence was measured by a Tecan spark microplate reader. A dose-response curve was constructed for every tested compound, and the IC50 value, i.e., the concentration that inhibits VEGFR-2 enzyme activity to 50%, was also determined using Prism v.6 curve fitting software (GraphPad Software, Inc., San Diego, CA, USA).
3.3. Docking Study
MOE software was used in this study [5]. The 4ASE [8] (sorafenib) crystal structures of VEGFR-2 were used to study the binding modes between the docked-selected compounds and amino acid residues in the active site of VEGFR-2. The protein structure of VEGFR-2 was obtained from the Protein Data Bank [PDB Code 4ASE].). The structures of the synthesized and reference compounds were constructed using MOE-Builder. The related 3D structures were also obtained, and the energies of the identified molecules were minimized using the default parameters of MOE energy minimization algorithm.
A maximum of 10 conformations were allowed for each ligand using the default parameters of MOE (placement, triangle matcher; rescoring 1, London dG; refinement, force field; rescoring 2, GBVI/WSA dG). The 300 top-ranked conformations of the docked compounds were saved in a separate database.
3.4. QSAR Methodology
From several published reports and the chembl-database website, 110 and 90 known MCF-7 and HepG2, respectively, compounds were collected and their activities described in terms of inhibitory activity (pIC50 nM) [32,33,34,35]. The 17 benzoquinazoluine compounds and the reference compound were imported as .mdb files into a molecular database in MOE Version 2015.1001) including molecular structures and activity values.
PLS regression was employed as a statistical method to evaluate structure-activity relationships in QSAR. The QSAR models were built and cross-validation was conducted to check the reasonable QSAR models. Various statistical parameters (Tables S1 and S2) were utilized for validation of model suitability for the prediction of the anti-proliferative activities of the target compounds. This includes the correlation coefficient (R2), which describes the fraction of the total variation attributed to the model, and the cross-validation coefficient parameter ().
4. Conclusions
In conclusion, new benzo[g]quinazolines (1–17) were synthesized and evaluated for their in vitro antiproliferative activity against two selected human cancer cell lines, namely HepG2 and MCF-7. Most of the benzoquinazolines demonstrated promising activity. Compounds 13–15 showed the highest activity against HepG2 and MCF-7, and their effects were further investigated by cell-cycle analysis, revealing a similarity in the activity characteristics of 13 and 14 against both MCF-7 and HepG2, involving the targeting of the G1 and S phases, respectively. The best induction of apoptosis in MCF-7 cells was observed for compound 14. Molecular docking confirmed that 13 and 15 are type-II VEGFR-2 inhibitors and experimentally, gave 1.5- and 1.4-fold inhibition relative to the standard sorafenib. In QSAR analysis, the predicted pIC50 values of the active compounds showed a significant correlation with the experimental values from principal component analysis plots. These results strongly demonstrate the anticancer activity of benzoquinazolines 10, 13–15 in vitro.
Supplementary Materials
The following are available online. Table S1: Equations 1–6. Table S2: Minimum recommended values of validated parameters for generally acceptable QSAR. Table S3. Correlation matrix of from PLS analysis for HepG2 variables. Table S4. Correlation matrix of from PLS analysis for MCF7 variables. Table S5. Chemical structure of 2-thioxobenzo[g]quinazoline derivatives and their cytotoxicity activities on HepG2 and MCF7. Table S6: Statistical Validation Results of QSAR Model for HepG2 & MCF7 cell line. Figure S1: Dose-dependent antiproliferative data of the compounds on the HepG-2 human cancer type, according to the MTT assay after 48 h of exposure. Figure S2: Dose dependent antiproliferative data of the compounds on the MCF-7 human cancer type according to the MTT assay after 48 h of exposure.
Author Contributions
R.A.-S. and H.A.A. designed the work, data analyzed, synthesized the targets compounds, and wrote the manuscript; H.M.A. designed and performed the biological experiments (MTT assay); M.M.S. designed, analyzed, and wrote the experimental cell cycle analysis, apoptosis, DNA fragmentation, and the antiangiogenic parts; M.M. analyzed the NMR data and revised the manuscript; A.H.B. performed the docking and QSAR studies, and A.M.N. wrote and revised the manuscript. All authors revised and approved the final draft and data analysis. All authors have read and agreed to the published version of the manuscript.
Funding
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through the research group project no. RG-1435-068.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Viallard, C.; Larrivée, B. Tumor angiogenesis and vascular normalization: Alternative therapeutic targets. Angiogenesis 2017, 20, 409–426. [Google Scholar] [CrossRef] [PubMed]
- El-GazzarRania, M.G.; El-HazekNashwa, R.M.; Zaher, N.H.; El-Ghazaly, M.A. Design and synthesis of novel pyridazinoquinazoline derivatives as potent VEGFR-2 inhibitors: In vitro and in vivo study. Bioorg. Chem. 2019, 92, 103251. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.A.; Serya, R.A.; Lasheen, D.S.; Abdel-Aziz, A.K.; Esmat, A.; Mansour, A.M.; Singab, A.N.; Abouzid, A.M. Discovery of potent VEGFR-2 inhibitors based on furopyrimidine and thienopyrimidne scaffolds as cancer targeting agents. Sci. Rep. 2016, 15, 24460. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhou, J.; Wu, J.; Shen, Y.; Li, X. Anti-angiogenic therapy: Strategies to develop potent VEGFR-2 tyrosine kinase inhibitors and future prospect. Curr. Med. Chem. 2016, 23, 1000–1040. [Google Scholar] [CrossRef] [PubMed]
- Modi, S.J.; Kulkarni, V.M. Vascular Endothelial Growth Factor Receptor (VEGFR-2)/KDR inhibitors. Med. Chem. Perspect. Med. Drug Discov. 2019, 2, 100009. [Google Scholar] [CrossRef]
- Liu, Y.; Gray, N.S. Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2006, 2, 358–364. [Google Scholar] [CrossRef]
- Kwak, E.L.; Sordella, R.; Bell, D.W.; Godin-Heymann, N.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Driscoll, D.R.; Fidias, P.; Lynch, T.J. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc. Natl. Acad. Sci. USA 2005, 102, 7665–7670. [Google Scholar] [CrossRef]
- Ghorab, M.M.; Alsaid, M.S.; Soliman, A.M.; Ragab, F.A. VEGFR-2 inhibitors and apoptosis inducers: Synthesis and molecular design of new benzo[g]quinazolin bearing benzenesulfonamide moiety. J. Enzyme Inhib. Med. Chem. 2017, 32, 893–907. [Google Scholar] [CrossRef]
- Huang, L.; Huang, Z.; Bai, Z.; Xie, R.; Sun, L.; Lin, K. Development and strategies of VEGFR-2/KDR inhibitors. Future Med. Chem. 2012, 4, 1839–1852. [Google Scholar] [CrossRef]
- Kornev, A.P.; Haste, N.M.; Taylor, S.S.; Ten Eyck, L.F. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. USA 2006, 103, 17783–17788. [Google Scholar] [CrossRef]
- Al-Salahi, R.; Abuelizz, H.; Eldib, R.; Wadi, M.; Alotaibi, M.; Marzouk, M. Antimicrobial activity of newly synthesized 2-methylthio-benzo[g][1,2,4]-triazolo[1,5-a]quinazolines. Med. Chem. 2016, 12, 760–766. [Google Scholar] [CrossRef] [PubMed]
- Al-Salahi, R.; Abuelizz, H.; Ghabbour, H.; Eldib, R.; Marzouk, M. Molecular Docking and Antiviral Evaluation of 2-Thioxo-benzo[g]quinazolin-4(3H)-one derivatives. Chem. Cent. J. 2016, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Al-Salahi, R.; El-dib, R.; Marzouk, M. Synthesis and in vitro cytotoxicity evaluation of new 2-thioxo-benzo[g]quinazolin-4(3H)–one derivatives. Heterocycles 2015, 91, 1735–1751. [Google Scholar] [CrossRef]
- Al-salahi, R.; Abuelizz, H.; Eldib, R.; Marzouk, M. Antimicrobial of 2-Thioxo-benzo[g]quinazolin-4(3H)-one derivatives. Med. Chem. 2017, 13, 85–92. [Google Scholar] [CrossRef]
- Abuelizz, H.A.; El-Dib, R.A.; Marzouk, M.; Al-Salahi, R. In vitro evaluation of new 2-phenoxy-benzo[g][1,2,4]triazolo[1,5-a]quinazoline derivatives as antimicrobial agents. Microbial Pathog. 2018, 117, 60–67. [Google Scholar] [CrossRef]
- Al-Salahi, R.; Moustapha, M.E.; Abuelizz, H.A.; Alharthi, A.I.; Alburikan, K.A.; Ibrahim, I.T.; Marzouk, M.; Motaleb, M.A. Radioiodination and biodistribution of newly synthesized 3-benzyl-2-([3-methoxybenzyl]thio) benzo[g]quinazolin-4-(3H)-one in tumor bearing mice. Saudi Pharm. J. 2018, 26, 1120–1126. [Google Scholar] [CrossRef]
- Al-Salahi, R.; Ahmad, R.; ElHassane, A.; Marzouk, M.; Abuelizz, H.A. 3-Benzyl(phenethyl)-2-thioxobenzo[g]quinazolines as a new class of potent α-glucosidase inhibitors: Synthesis and molecular docking study. Future Med. Chem. 2018, 10, 1889–1905. [Google Scholar] [CrossRef]
- Almehizia, A.A.; Abuelizz, H.A.; Taie, H.A.A.; ElHassane, A.; Marzouk, M.; Al-Salahi, R. Investigation the antioxidant activity of benzo[g]triazolo quinazolines correlated with a DFT study. Saudi Pharm. J. 2019, 27, 133–137. [Google Scholar] [CrossRef]
- Al-Salahi, R.; ElHassane, A.; Marzouk, M.; Abuelizz, H.A. Anti-HAV evaluation and molecular docking of newly synthesized 3-benzyl(phenethyl)benzo[g]quinazolines. Bioorg. Med. Chem. Lett. 2019, 29, 1614–1619. [Google Scholar] [CrossRef]
- Al-Salahi, R.; Taie, H.A.A.; Bakheit, A.H.; Marzouk, M.; Almehizia, A.A.; Herqash, R.; Abuelizz, H.A. Antioxidant activities and molecular docking of 2-thioxobenzo[g]quinazoline derivatives. Pharmacol. Rep. 2019, 71, 695–700. [Google Scholar] [CrossRef]
- Nowak, M.; Malinowski, Z.; Jóźwiak, A.J.; Fornal, E.; Błaszczyk, A.; Kontek, R. Substituted benzoquinazolinones. Part 1: Synthesis of 6-aminobenzo[h]quinazolinones via Buchwalde Hartwig amination from 6-bromobenzo[h]quinzolinones. Tetrahedron 2014, 70, 5153–5160. [Google Scholar] [CrossRef]
- Pendergast, W.; Dickerson, S.H.; Dev, I.K.; Ferone, R.; Duch, D.S.; Smith, G.K. Benzo[flquinazoline inhibitors of thymidylate synthase: Methyleneamino-linked aroylglutamate derivatives. J. Med. Chem. 1994, 37, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Abuelizz, H.A.; Marzouk, M.; Bakheit, A.H.; Al-Salahi, R. Investigation of some benzoquinazoline and quinazoline derivatives as novel inhibitors of HCV-NS3/4A protease: Biological, molecular docking and QSAR studies. RSC Adv. 2020, 10, 35820–35830. [Google Scholar] [CrossRef]
- Khodair, A.I.; Elsafi, M.A.; Al-Issa, S.A. Simple and efficient synthesis of novel 3-substituted 2-thioxo-2,3-dihydro-1H-benzo[g]quinazolin-4-ones and their reactions with alkyl halides and α-glycolpy ranosyl bromides. J. Heterocycl. Chem. 2019, 56, 2358. [Google Scholar] [CrossRef]
- Fallah, A.; Sadeghinia, A.; Kahroba, H.; Samadi, A.; Heidari, H.R.; Bradaran, B.; Zeinali, S.; Molavi, O. Therapeutic targeting of angiogenesis molecular pathways in angiogenesis-dependent diseases. Biomed. Pharmacother. 2019, 110, 775–785. [Google Scholar] [CrossRef]
- Wan, J.; Zhang, L.; Yang, G.; Zhan, C.-G. Quantitative structure− activity relationship for cyclic imide derivatives of protoporphyrinogen oxidase inhibitors: A study of quantum chemical descriptors from density functional theory. J. Chem. Inf. Comput. Sci. 2004, 44, 2099–2105. [Google Scholar] [CrossRef]
- Kassem, A.F.; Nassar, I.F.; Abdel-Aal, M.T.; Awad, H.M.; El-Sayed, W.A. Synthesis and anticancer activity of new ((Furan-2-yl)-1,3,4-thiadiazolyl)-1,3,4-oxadiazole acyclic sugar derivatives. Chem. Pharm. Bull. 2019, 67, 888–895. [Google Scholar] [CrossRef]
- Abuelizz, H.A.; Awad, A.M.; Marzouk, M.; Nasr, F.A.; Bakheit, A.H.; Naglah, A.M.; Al-Salahi, R. Exploiting the 4-Hydrazinobenzoic acid moiety for the development of anticancer agents: Synthesis and biological profile. Bioorg. Chem. 2020, 102, 104098. [Google Scholar] [CrossRef]
- Alminderej, F.M.; Elganzory, H.H.; El-Bayaa, M.N.; Awad, H.M.; El-Sayed, W.A. Synthesis and cytotoxic activity of new 1,3,4-thiadiazole thiogl ycosides and 1,2,3-triazolyl-1,3,4-thiadiazole N-glycosides. Molecules 2019, 24, 3738. [Google Scholar] [CrossRef]
- Alaaeldin, G.M.F.; Soltan, M.M.; Esam, A.R.; Hanna, A.G. Synthesis and biological evaluation of novel 5-chloro-N-(4-sulfamoylbenzyl)salicylamide derivatives as tubulin polymerization inhibitors. MedChemComm 2018, 9, 1511–1528. [Google Scholar]
- Barua, A.; Yellapa, A.; Bahr, J.M.; Machado, S.A.; Bitterman, P.; Basu, S.; Sharma, S.; Abramowicz, J.S. VEGFR2-targeted ultrasound imaging agent anhances the detection of ovarian tumors at early stage in laying hens, a preclinical model of spontaneous ovarian cancer. Ultrasonic Imaging 2015, 37, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, K.; Deushi, T.; Yoshida, H.; Miyashita, Y.; Sasaki, S.I. Chemometric QSAR studies of antifungal azoxy compounds. J. Comput. Aided Mol. Des. 1994, 8, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Asadollahi, T.; Dadfarnia, S.; Shabani, A.M.H.; Ghasemi, J.B.; Sarkhosh, M. QSAR models for CXCR2 receptor antagonists based on the genetic algorithm for data preprocessing prior to application of the PLS linear regression method and design of the new compounds using in silico virtual screening. Molecules 2011, 16, 1928–1955. [Google Scholar] [CrossRef]
- Umar, B.A.; Uzairu, A.; Shallangwa, G.A.; Sani, U. QSAR modeling for the prediction of pGI50 activity of compounds on LOX IMVI cell line and ligand-based design of potent compounds using in silico virtual screening. Netw. Modeling Anal. Health Inform. Bioinform. 2019, 8, 22. [Google Scholar] [CrossRef]
- Veerasamy, R.; Rajak, H.; Jain, A.; Sivadasan, S.; Varghese, C.P.; Agrawal, R.K. Validation of QSAR models-strategies and importance. Int. J. Drug Des. Discov. 2011, 3, 511–519. [Google Scholar]
Sample Availability: Samples of the benzo[g]quinazoline compounds 1–17 are available from the authors at College of pharmacy- king Saud University. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).