Olive Leaf Extract, from Olea europaea L., Reduces Palmitate-Induced Inflammation via Regulation of Murine Macrophages Polarization
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Sodium Palmitate (SP) Preparation
2.3. Olive Leaf Extract (OLE) Preparation
2.4. High-Performance Liquid Chromatography (HPLC)/Diode-Aray Detector (DAD) Analyses of OLE Polyphenols
2.5. Quantitative Real-Time PCR
2.6. Intracellular ROS Measurement
2.7. Cytokines Measurement
2.8. Nitrite Analysis
2.9. PGE2 Assay
2.10. Flow cytometry
2.11. Western Blot Analysis
2.12. MTT Assay
2.13. Bone Marrow Derived Macrophages (BMDMs) Generation and Treatment
2.14. Statistical Analysis
3. Results
3.1. Characterization of OLE Composition
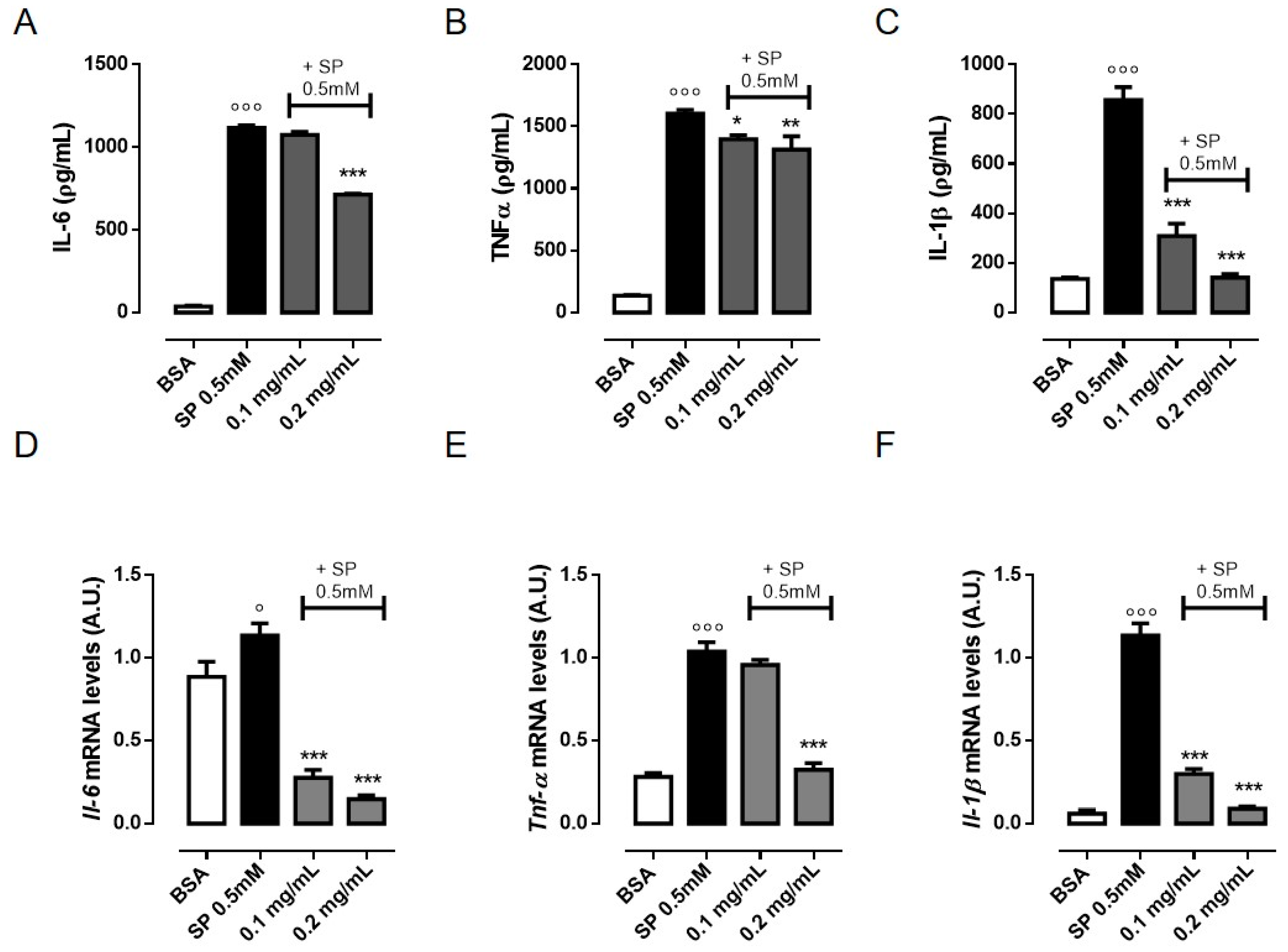
3.2. OLE Suppresses TNF-α, IL-6 and IL-1β Production in FFAs-Stimulated RAW 264.7 Macrophages
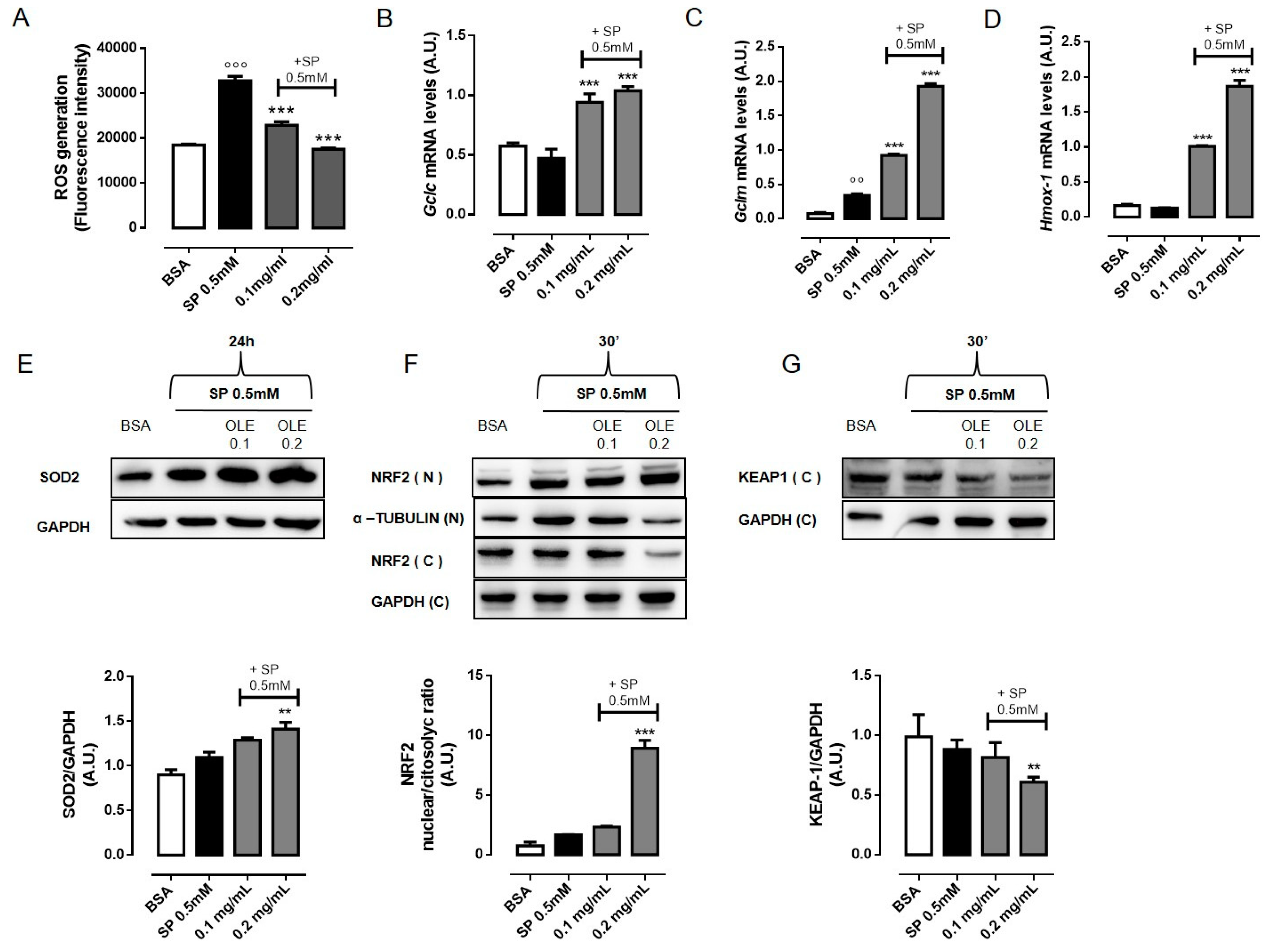
3.3. OLE Attenuates FFAs-Induced Oxidative Stress in RAW 264.7 Macrophages by Activating NRF2
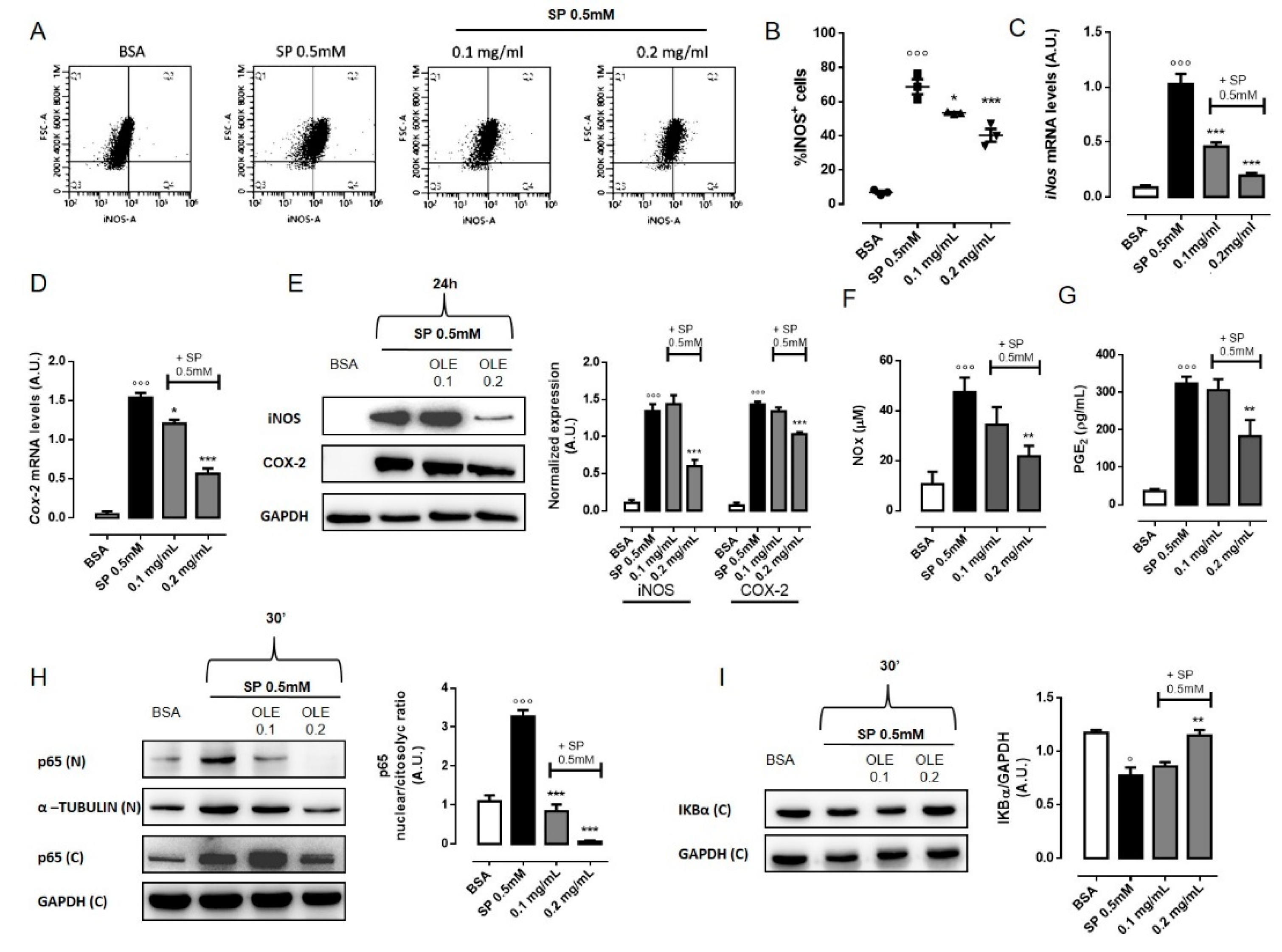
3.4. OLE Suppressed M1 Pro-Inflammatory Phenotype in FFAs-Stimulated RAW 264.7 Macrophages
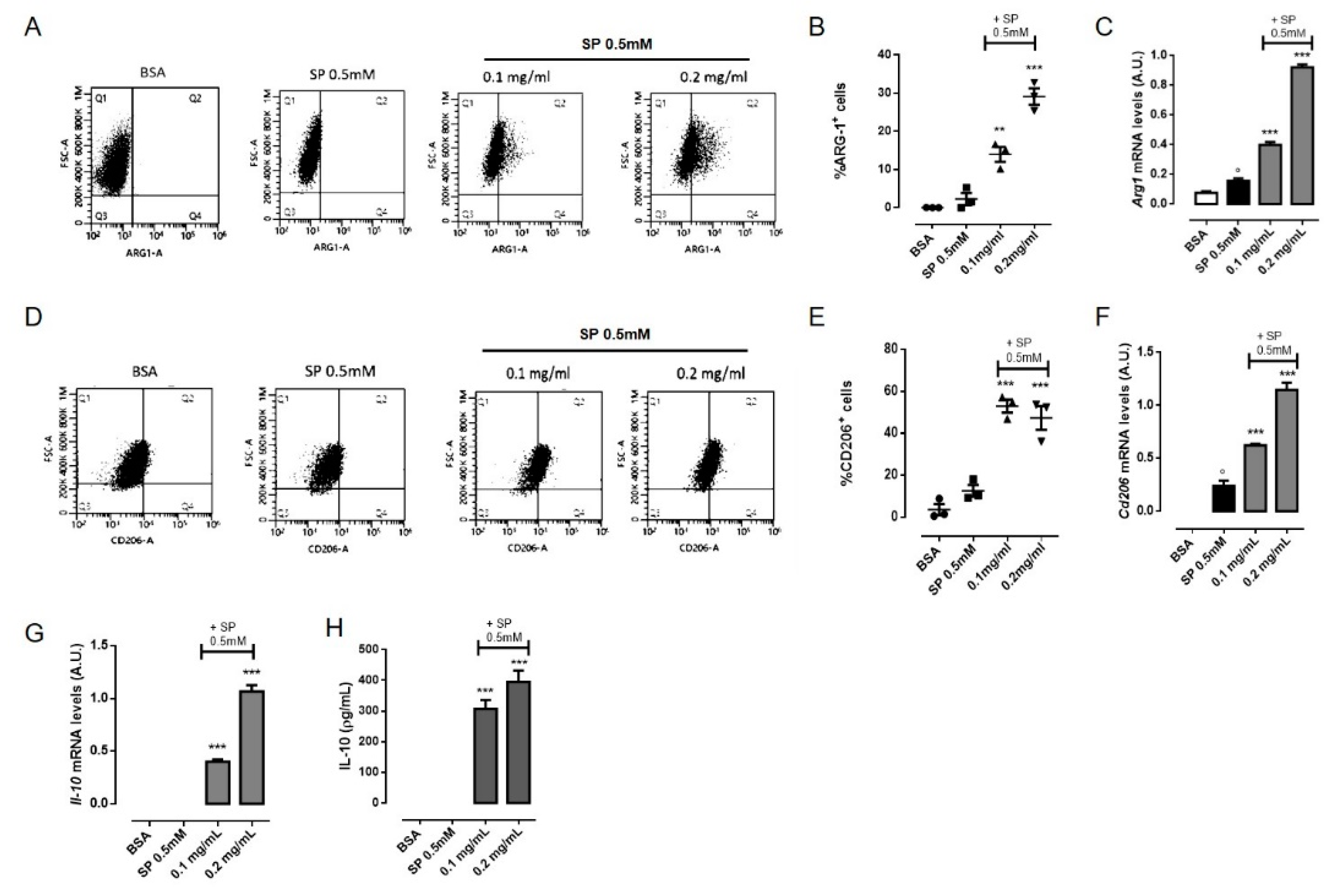
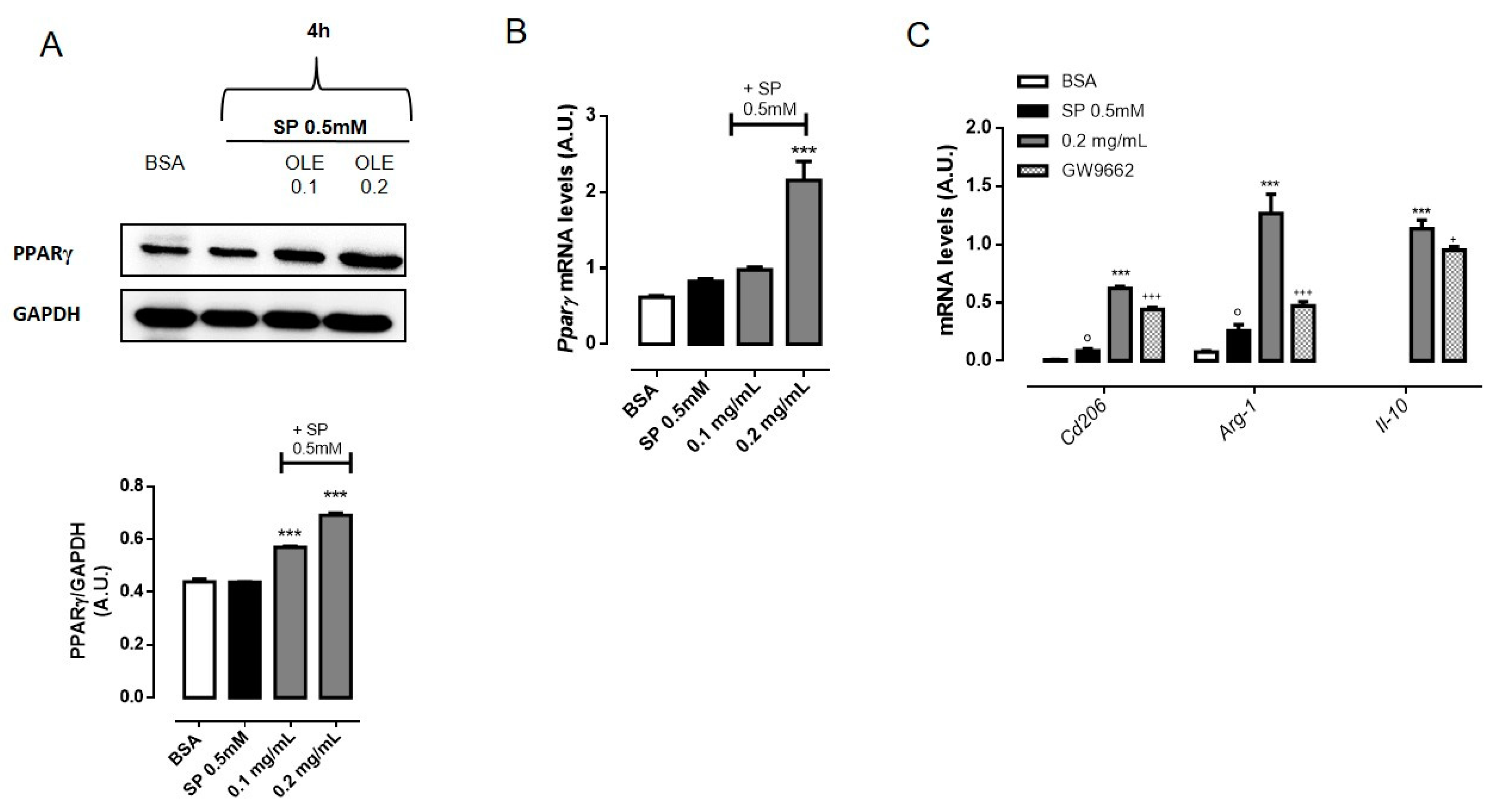
3.5. OLE Promoted M2 Polarization in FFAs-Stimulated RAW264.7 Macrophages via PPARγ Activation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Available online: http://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 28 May 2020).
- Minihane, A.M.; Vinoy, S.; Russell, W.R.; Baka, A.; Roche, H.M.; Tuohy, K.M.; Teeling, J.L.; Blaak, E.E.; Fenech, M.; Vauzour, D.; et al. Low-grade inflammation, diet composition and health: Current research evidence and its translation. Br. J. Nutr. 2015, 114, 999–1012. [Google Scholar] [CrossRef] [PubMed]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W., Jr. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Investig. 2003, 112, 1796–1808. [Google Scholar] [CrossRef] [PubMed]
- Lauterbach, M.A.; Wunderlich, F.T. Macrophage function in obesity-induced inflammation and insulin resistance. Pflug. Arch. Eur. J. Physiol. 2017, 469, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Chawla, A.; Nguyen, K.D.; Goh, Y.P. Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol. 2011, 11, 738–749. [Google Scholar] [CrossRef]
- Castoldi, A.; Naffah de Souza, C.; Camara, N.O.; Moraes-Vieira, P.M. The Macrophage Switch in Obesity Development. Front. Immunol. 2015, 6, 637. [Google Scholar] [CrossRef]
- Thomas, D.; Apovian, C. Macrophage functions in lean and obese adipose tissue. Metab. Clin. Exp. 2017, 72, 120–143. [Google Scholar] [CrossRef]
- Clement, K.; Viguerie, N.; Poitou, C.; Carette, C.; Pelloux, V.; Curat, C.A.; Sicard, A.; Rome, S.; Benis, A.; Zucker, J.D.; et al. Weight loss regulates inflammation-related genes in white adipose tissue of obese subjects. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2004, 18, 1657–1669. [Google Scholar] [CrossRef]
- Fujisaka, S.; Usui, I.; Bukhari, A.; Ikutani, M.; Oya, T.; Kanatani, Y.; Tsuneyama, K.; Nagai, Y.; Takatsu, K.; Urakaze, M.; et al. Regulatory mechanisms for adipose tissue M1 and M2 macrophages in diet-induced obese mice. Diabetes 2009, 58, 2574–2582. [Google Scholar] [CrossRef]
- van Dijk, S.J.; Feskens, E.J.; Bos, M.B.; Hoelen, D.W.; Heijligenberg, R.; Bromhaar, M.G.; de Groot, L.C.; de Vries, J.H.; Muller, M.; Afman, L.A. A saturated fatty acid-rich diet induces an obesity-linked proinflammatory gene expression profile in adipose tissue of subjects at risk of metabolic syndrome. Am. J. Clin. Nutr. 2009, 90, 1656–1664. [Google Scholar] [CrossRef]
- Ghanim, H.; Abuaysheh, S.; Sia, C.L.; Korzeniewski, K.; Chaudhuri, A.; Fernandez-Real, J.M.; Dandona, P. Increase in plasma endotoxin concentrations and the expression of Toll-like receptors and suppressor of cytokine signaling-3 in mononuclear cells after a high-fat, high-carbohydrate meal: Implications for insulin resistance. Diabetes Care 2009, 32, 2281–2287. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, J.Y.; Kim, O.Y.; Ham, B.M.; Kim, H.J.; Kwon, D.Y.; Jang, Y.; Lee, J.H. Metabolic profiling of plasma in overweight/obese and lean men using ultra performance liquid chromatography and Q-TOF mass spectrometry (UPLC-Q-TOF MS). J. Proteome Res. 2010, 9, 4368–4375. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Kokoeva, M.V.; Inouye, K.; Tzameli, I.; Yin, H.; Flier, J.S. TLR4 links innate immunity and fatty acid-induced insulin resistance. J. Clin. Investig. 2006, 116, 3015–3025. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.T.; Favelyukis, S.; Nguyen, A.K.; Reichart, D.; Scott, P.A.; Jenn, A.; Liu-Bryan, R.; Glass, C.K.; Neels, J.G.; Olefsky, J.M. A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via Toll-like receptors 2 and 4 and JNK-dependent pathways. J. Biol. Chem. 2007, 282, 35279–35292. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.H.; Ho, K.T.; Lu, S.H.; Huang, C.N.; Shiau, M.Y. Regulation of glucose/lipid metabolism and insulin sensitivity by interleukin-4. Int. J. Obes. 2012, 36, 993–998. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Hao, H.; Xie, Z.; Cheng, Y.; Yin, Y.; Xie, M.; Huang, H.; Gao, J.; Liu, H.; Tong, C.; et al. M2 macrophages infusion ameliorates obesity and insulin resistance by remodeling inflammatory/macrophages’ homeostasis in obese mice. Mol. Cell. Endocrinol. 2017, 443, 63–71. [Google Scholar] [CrossRef]
- Das, L.; Bhaumik, E.; Raychaudhuri, U.; Chakraborty, R. Role of nutraceuticals in human health. J. Food Sci. Technol. 2012, 49, 173–183. [Google Scholar] [CrossRef]
- Romani, A.; Ieri, F.; Urciuoli, S.; Noce, A.; Marrone, G.; Nediani, C.; Bernini, R. Health Effects of Phenolic Compounds Found in Extra-Virgin Olive Oil, By-Products, and Leaf of Olea europaea L. Nutrients 2019, 11, 1776. [Google Scholar] [CrossRef]
- Japon-Lujan, R.; Luque-Rodriguez, J.M.; Luque de Castro, M.D. Dynamic ultrasound-assisted extraction of oleuropein and related biophenols from olive leaves. J. Chromatogr. A 2006, 1108, 76–82. [Google Scholar] [CrossRef]
- Zuntar, I.; Putnik, P.; Bursac Kovacevic, D.; Nutrizio, M.; Supljika, F.; Poljanec, A.; Dubrovic, I.; Barba, F.J.; Rezek Jambrak, A. Phenolic and Antioxidant Analysis of Olive Leaves Extracts (Olea europaea L.) Obtained by High Voltage Electrical Discharges (HVED). Foods 2019, 8. [Google Scholar] [CrossRef]
- Santiago-Mora, R.; Casado-Diaz, A.; De Castro, M.D.; Quesada-Gomez, J.M. Oleuropein enhances osteoblastogenesis and inhibits adipogenesis: The effect on differentiation in stem cells derived from bone marrow. Osteoporos. Int. 2011, 22, 675–684. [Google Scholar] [CrossRef]
- Drira, R.; Chen, S.; Sakamoto, K. Oleuropein and hydroxytyrosol inhibit adipocyte differentiation in 3 T3-L1 cells. Life Sci. 2011, 89, 708–716. [Google Scholar] [CrossRef] [PubMed]
- De Cicco, P.; Panza, E.; Armogida, C.; Ercolano, G.; Taglialatela-Scafati, O.; Shokoohinia, Y.; Camerlingo, R.; Pirozzi, G.; Calderone, V.; Cirino, G.; et al. The Hydrogen Sulfide Releasing Molecule Acetyl Deacylasadisulfide Inhibits Metastatic Melanoma. Front. Pharmacol. 2017, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.-J. Phenolic compositions, and antioxidant performance of olive leaf and fruit (Olea europaea L.) extracts and their structure–activity relationships. J. Funct. Foods 2015, 16, 460–471. [Google Scholar] [CrossRef]
- Febbraio, M.A. Role of interleukins in obesity: Implications for metabolic disease. Trends Endocrinol. Metab. 2014, 25, 312–319. [Google Scholar] [CrossRef]
- Korbecki, J.; Bajdak-Rusinek, K. The effect of palmitic acid on inflammatory response in macrophages: An overview of molecular mechanisms. Inflamm. Res. Off. J. Eur. Histamine Res. Soc. 2019, 68, 915–932. [Google Scholar] [CrossRef]
- Karpe, F.; Dickmann, J.R.; Frayn, K.N. Fatty acids, obesity, and insulin resistance: Time for a reevaluation. Diabetes 2011, 60, 2441–2449. [Google Scholar] [CrossRef]
- Masschelin, P.M.; Cox, A.R.; Chernis, N.; Hartig, S.M. The Impact of Oxidative Stress on Adipose Tissue Energy Balance. Front. Physiol. 2019, 10, 1638. [Google Scholar] [CrossRef]
- Houstis, N.; Rosen, E.D.; Lander, E.S. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature 2006, 440, 944–948. [Google Scholar] [CrossRef]
- Manna, P.; Jain, S.K. Obesity, Oxidative Stress, Adipose Tissue Dysfunction, and the Associated Health Risks: Causes and Therapeutic Strategies. Metab. Syndr. Relat. Disord. 2015, 13, 423–444. [Google Scholar] [CrossRef]
- Liu, Y.; Qi, W.; Richardson, A.; Van Remmen, H.; Ikeno, Y.; Salmon, A.B. Oxidative damage associated with obesity is prevented by overexpression of CuZn- or Mn-superoxide dismutase. Biochem. Biophys. Res. Commun. 2013, 438, 78–83. [Google Scholar] [CrossRef]
- Kobayashi, A.; Kang, M.I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef] [PubMed]
- Lumeng, C.N.; DelProposto, J.B.; Westcott, D.J.; Saltiel, A.R. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes 2008, 57, 3239–3246. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-kappaB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tugal, D.; Liao, X.; Jain, M.K. Transcriptional control of macrophage polarization. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1135–1144. [Google Scholar] [CrossRef]
- Martinez, F.O.; Helming, L.; Gordon, S. Alternative activation of macrophages: An immunologic functional perspective. Annu. Rev. Immunol. 2009, 27, 451–483. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Ricardo-Gonzalez, R.R.; Goforth, M.H.; Morel, C.R.; Subramanian, V.; Mukundan, L.; Red Eagle, A.; Vats, D.; Brombacher, F.; Ferrante, A.W.; et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 2007, 447, 1116–1120. [Google Scholar] [CrossRef]
- Bouhlel, M.A.; Derudas, B.; Rigamonti, E.; Dievart, R.; Brozek, J.; Haulon, S.; Zawadzki, C.; Jude, B.; Torpier, G.; Marx, N.; et al. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007, 6, 137–143. [Google Scholar] [CrossRef]
- Rigamonti, E.; Chinetti-Gbaguidi, G.; Staels, B. Regulation of macrophage functions by PPAR-alpha, PPAR-gamma, and LXRs in mice and men. Arterioscler. Thromb. Vasc. Biol. 2008, 28, 1050–1059. [Google Scholar] [CrossRef]
- Ryu, S.J.; Choi, H.S.; Yoon, K.Y.; Lee, O.H.; Kim, K.J.; Lee, B.Y. Oleuropein suppresses LPS-induced inflammatory responses in RAW 264.7 cell and zebrafish. J. Agric. Food Chem. 2015, 63, 2098–2105. [Google Scholar] [CrossRef]
- Mao, X.; Xia, B.; Zheng, M.; Zhou, Z. Assessment of the anti-inflammatory, analgesic and sedative effects of oleuropein from Olea europaea L. Cell. Mol. Biol. 2019, 65, 52–55. [Google Scholar] [CrossRef]
- Jung, Y.C.; Kim, H.W.; Min, B.K.; Cho, J.Y.; Son, H.J.; Lee, J.Y.; Kim, J.Y.; Kwon, S.B.; Li, Q.; Lee, H.W. Inhibitory Effect of Olive Leaf Extract on Obesity in High-fat Diet-induced Mice. In Vivo 2019, 33, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Kim, S.H.; Kim, Y.S.; Ryu, S.Y.; Hwang, J.T.; Yang, H.J.; Kim, G.H.; Kwon, D.Y.; Kim, M.S. Luteolin inhibits adipogenic differentiation by regulating PPARgamma activation. BioFactors 2009, 35, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Cho, A.S.; Jeon, S.M.; Kim, M.J.; Yeo, J.; Seo, K.I.; Choi, M.S.; Lee, M.K. Chlorogenic acid exhibits anti-obesity property and improves lipid metabolism in high-fat diet-induced-obese mice. Food Chem. Toxicol. 2010, 48, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Song, S.J.; Keum, N.; Park, T. Olive leaf extract attenuates obesity in high-fat diet-fed mice by modulating the expression of molecules involved in adipogenesis and thermogenesis. Evid. Based Complementary Altern. Med. Ecam 2014, 2014, 971890. [Google Scholar] [CrossRef]
- Lumeng, C.N.; Bodzin, J.L.; Saltiel, A.R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Investig. 2007, 117, 175–184. [Google Scholar] [CrossRef]
- Riera-Borrull, M.; Cuevas, V.D.; Alonso, B.; Vega, M.A.; Joven, J.; Izquierdo, E.; Corbi, A.L. Correction: Palmitate Conditions Macrophages for Enhanced Responses toward Inflammatory Stimuli via JNK Activation. J. Immunol. 2019, 203, 580. [Google Scholar] [CrossRef]
- Ko, H.-J.; Lo, C.-Y.; Wang, B.-J.; Chiou, R.Y.-Y.; Lin, S.-M. Theaflavin-3, 3′-digallate, a black tea polyphenol, attenuates adipocyte-activated inflammatory response of macrophage associated with the switch of M1/M2-like phenotype. J. Funct. Foods 2014, 11, 36–48. [Google Scholar] [CrossRef]
- Feng, X.; Weng, D.; Zhou, F.; Owen, Y.D.; Qin, H.; Zhao, J.; Wen, Y.; Huang, Y.; Chen, J.; Fu, H.; et al. Activation of PPARgamma by a Natural Flavonoid Modulator, Apigenin Ameliorates Obesity-Related Inflammation Via Regulation of Macrophage Polarization. EBioMedicine 2016, 9, 61–76. [Google Scholar] [CrossRef]
- Feng, X.; Qin, H.; Shi, Q.; Zhang, Y.; Zhou, F.; Wu, H.; Ding, S.; Niu, Z.; Lu, Y.; Shen, P. Chrysin attenuates inflammation by regulating M1/M2 status via activating PPARgamma. Biochem. Pharmacol. 2014, 89, 503–514. [Google Scholar] [CrossRef]
- Pardo, V.; Gonzalez-Rodriguez, A.; Guijas, C.; Balsinde, J.; Valverde, A.M. Opposite cross-talk by oleate and palmitate on insulin signaling in hepatocytes through macrophage activation. J. Biol. Chem. 2015, 290, 11663–11677. [Google Scholar] [CrossRef]
- Xiu, F.; Diao, L.; Qi, P.; Catapano, M.; Jeschke, M.G. Palmitate differentially regulates the polarization of differentiating and differentiated macrophages. Immunology 2016, 147, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Fain, J.N. Release of interleukins and other inflammatory cytokines by human adipose tissue is enhanced in obesity and primarily due to the nonfat cells. Vitam. Horm. 2006, 74, 443–477. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Gris, D.; Lei, Y.; Jha, S.; Zhang, L.; Huang, M.T.; Brickey, W.J.; Ting, J.P. Fatty acid-induced NLRP3-ASC inflammasome activation interferes with insulin signaling. Nat. Immunol. 2011, 12, 408–415. [Google Scholar] [CrossRef] [PubMed]
- Everett, B.M.; Donath, M.Y.; Pradhan, A.D.; Thuren, T.; Pais, P.; Nicolau, J.C.; Glynn, R.J.; Libby, P.; Ridker, P.M. Anti-Inflammatory Therapy With Canakinumab for the Prevention and Management of Diabetes. J. Am. Coll. Cardiol. 2018, 71, 2392–2401. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Yamamoto, M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox Signal. 2005, 7, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.K.; Yeager, R.L.; Tanaka, Y.; Klaassen, C.D. Enhanced expression of Nrf2 in mice attenuates the fatty liver produced by a methionine-and choline-deficient diet. Toxicol. Appl. Pharmacol. 2010, 245, 326–334. [Google Scholar] [CrossRef]
- Yu, Z.; Shao, W.; Chiang, Y.; Foltz, W.; Zhang, Z.; Ling, W.; Fantus, I.G.; Jin, T. Oltipraz upregulates the nuclear factor (erythroid-derived 2)-like 2 [corrected](NRF2) antioxidant system and prevents insulin resistance and obesity induced by a high-fat diet in C57BL/6J mice. Diabetologia 2011, 54, 922–934. [Google Scholar] [CrossRef]
- Bronte, V.; Zanovello, P. Regulation of immune responses by L-arginine metabolism. Nat. Rev. Immunol. 2005, 5, 641–654. [Google Scholar] [CrossRef]
- Wahli, W.; Michalik, L. PPARs at the crossroads of lipid signaling and inflammation. Trends Endocrinol. Metab. 2012, 23, 351–363. [Google Scholar] [CrossRef]
- Martin, H. Role of PPAR-gamma in inflammation. Prospects for therapeutic intervention by food components. Mutat. Res. 2009, 669, 57–63. [Google Scholar] [CrossRef]
- Marciano, D.P.; Chang, M.R.; Corzo, C.A.; Goswami, D.; Lam, V.Q.; Pascal, B.D.; Griffin, P.R. The therapeutic potential of nuclear receptor modulators for treatment of metabolic disorders: PPARgamma, RORs, and Rev-erbs. Cell Metab. 2014, 19, 193–208. [Google Scholar] [CrossRef] [PubMed]
- Aparicio-Soto, M.; Montserrat-de la Paz, S.; Sanchez-Hidalgo, M.; Cardeno, A.; Bermudez, B.; Muriana, F.J.G.; Alarcon-de-la-Lastra, C. Virgin olive oil and its phenol fraction modulate monocyte/macrophage functionality: A potential therapeutic strategy in the treatment of systemic lupus erythematosus. Br. J. Nutr. 2018, 120, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Svobodova, M.; Andreadou, I.; Skaltsounis, A.L.; Kopecky, J.; Flachs, P. Oleuropein as an inhibitor of peroxisome proliferator-activated receptor gamma. Genes Nutr. 2014, 9, 376. [Google Scholar] [CrossRef] [PubMed]
| Phenolic Compound | Calibration Curve | Content in OLE (mg/g) * |
|---|---|---|
| Apigenin | Y = 4 × 107x + 118566 | 0.01 ± 0.01 |
| Apigenin-7-O-glucoside | Y = 5 × 107x − 11131 | 0.43 ± 0.02 |
| Hydroxy-tyrosol | Y = 650564x − 932.11 | 26.65 ± 0.08 |
| Luteolin | Y = 6 × 107x − 2632.8 | 0.36 ± 0.01 |
| Oleuropein | Y = 3 × 106x − 11515 | 174.64 ± 2.32 |
| Rutin | Y = 2 × 107x + 396.8 | 0.24 ± 0.02 |
| Tyrosol | Y = 8 × 106x − 4951.2 | 0.64 ± 0.08 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Cicco, P.; Maisto, M.; Tenore, G.C.; Ianaro, A. Olive Leaf Extract, from Olea europaea L., Reduces Palmitate-Induced Inflammation via Regulation of Murine Macrophages Polarization. Nutrients 2020, 12, 3663. https://doi.org/10.3390/nu12123663
De Cicco P, Maisto M, Tenore GC, Ianaro A. Olive Leaf Extract, from Olea europaea L., Reduces Palmitate-Induced Inflammation via Regulation of Murine Macrophages Polarization. Nutrients. 2020; 12(12):3663. https://doi.org/10.3390/nu12123663
Chicago/Turabian StyleDe Cicco, Paola, Maria Maisto, Gian Carlo Tenore, and Angela Ianaro. 2020. "Olive Leaf Extract, from Olea europaea L., Reduces Palmitate-Induced Inflammation via Regulation of Murine Macrophages Polarization" Nutrients 12, no. 12: 3663. https://doi.org/10.3390/nu12123663
APA StyleDe Cicco, P., Maisto, M., Tenore, G. C., & Ianaro, A. (2020). Olive Leaf Extract, from Olea europaea L., Reduces Palmitate-Induced Inflammation via Regulation of Murine Macrophages Polarization. Nutrients, 12(12), 3663. https://doi.org/10.3390/nu12123663







