The Functional Power of the Human Milk Proteome
Abstract
1. Introduction
2. Factors that Affect Milk Composition
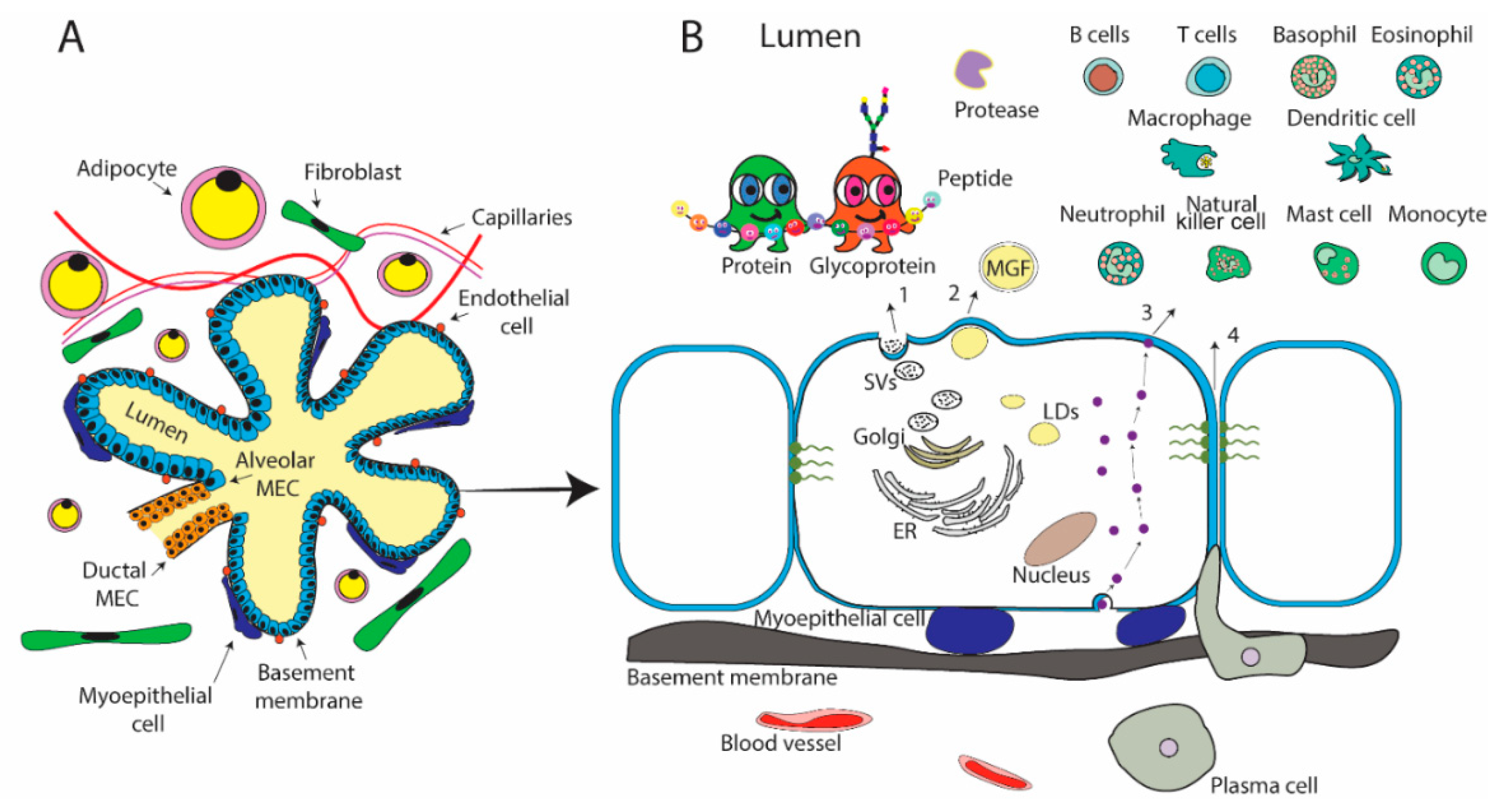
2.1. Mammary Gland Physiology
3. Proteins
4. Glycoproteins
5. Endogenous Peptides
6. Enzymes
6.1. Enzymes that Function in the Mammary Gland
6.2. Enzymes Present in Milk
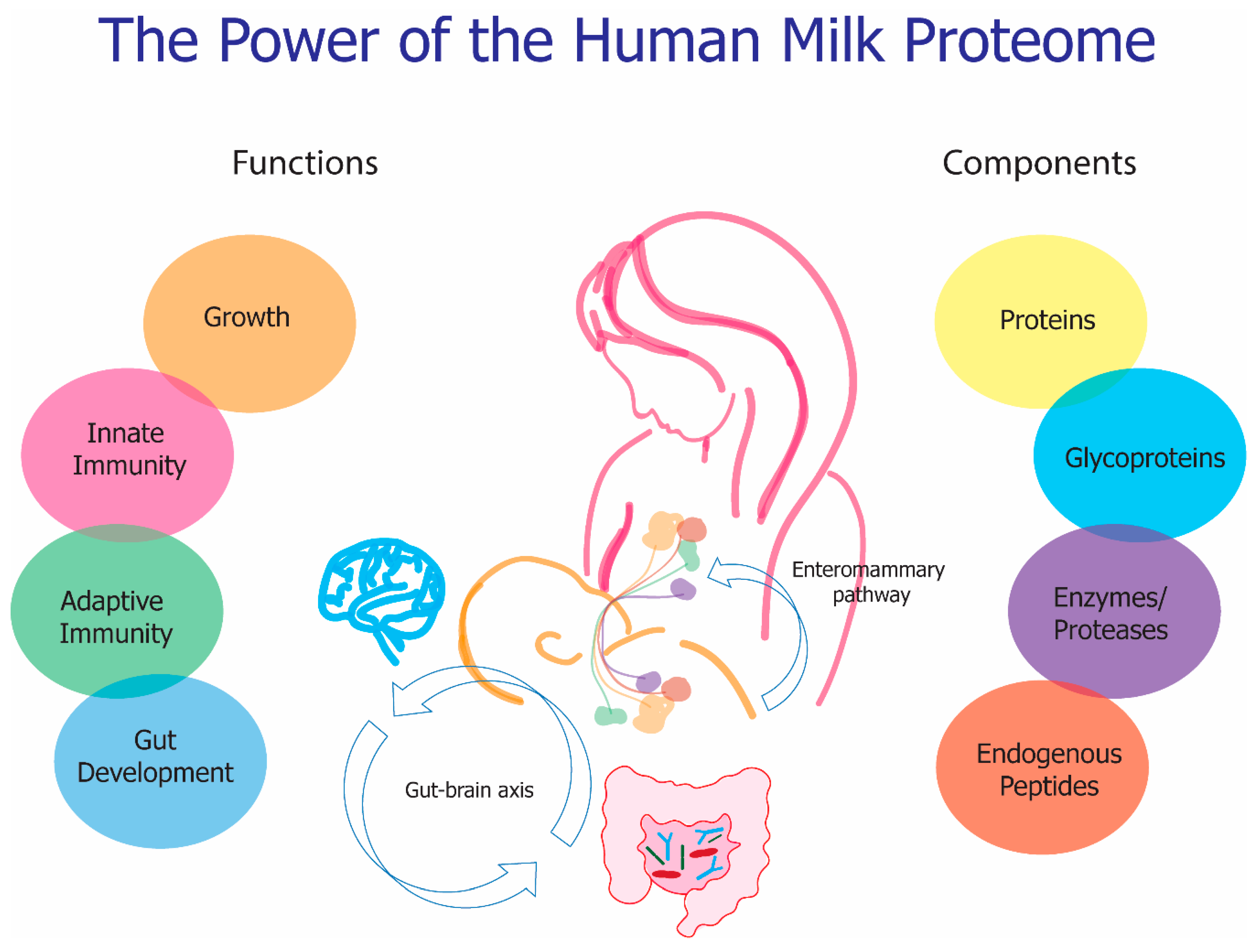
7. Functionality
7.1. Growth
7.2. Immune
7.2.1. Innate Immunity
7.2.2. Adaptive Immunity
7.2.3. Potential Allergens in Human Milk and Immunity
7.3. Gut Development
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Eidelman, A.I.; Schanler, R.J. Breastfeeding and the use of human milk. Pediatrics 2012, 129, 827–841. [Google Scholar] [CrossRef]
- Donovan, S.M. The Role of Lactoferrin in Gastrointestinal and Immune Development and Function: A Preclinical Perspective. J. Pediatr. 2016, 173, S16–S28. [Google Scholar] [CrossRef] [PubMed]
- Demmelmair, H.; Prell, C.; Timby, N.; Lonnerdal, B. Benefits of Lactoferrin, Osteopontin and Milk Fat Globule Membranes for Infants. Nutrients 2017, 9, 817. [Google Scholar] [CrossRef] [PubMed]
- Lonnerdal, B. Bioactive Proteins in Human Milk: Health, Nutrition, and Implications for Infant Formulas. J. Pediatr. 2016, 173, S4–S9. [Google Scholar] [CrossRef] [PubMed]
- Palmeira, P.; Carneiro-Sampaio, M. Immunology of breast milk. Revista da Associação Médica Brasileira 2016, 62, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Ballard, O.; Morrow, A.L. Human milk composition: Nutrients and bioactive factors. Pediatr. Clin. 2013, 60, 49–74. [Google Scholar] [CrossRef]
- Lemay, D.G.; Ballard, O.A.; Hughes, M.A.; Morrow, A.L.; Horseman, N.D.; Nommsen-Rivers, L.A. RNA sequencing of the human milk fat layer transcriptome reveals distinct gene expression profiles at three stages of lactation. PLoS ONE 2013, 8, e67531. [Google Scholar] [CrossRef]
- Gidrewicz, D.A.; Fenton, T.R. A systematic review and meta-analysis of the nutrient content of preterm and term breast milk. BMC Pediatr. 2014, 14, 216. [Google Scholar] [CrossRef]
- Donovan, S.M. Human Milk Proteins: Composition and Physiological Significance; Karger Publishers: Basel, Switzerland, 2019. [Google Scholar]
- Andreas, N.J.; Kampmann, B.; Mehring Le-Doare, K. Human breast milk: A review on its composition and bioactivity. Early Hum. Dev. 2015, 91, 629–635. [Google Scholar] [CrossRef]
- Xinliu, G.; Robert, J.M.; Jessica, G.W.; Barbara, S.D.; Ardythe, L.M.; Qiang, Z. Temporal changes in milk proteomes reveal developing milk functions. J. Proteome Res. 2012, 11, 3897–3907. [Google Scholar] [CrossRef]
- Zhang, Q.; Cundiff, J.; Maria, S.; McMahon, R.; Woo, J.; Davidson, B.; Morrow, A. Quantitative Analysis of the Human Milk Whey Proteome Reveals Developing Milk and Mammary-Gland Functions across the First Year of Lactation. Proteomes 2013, 1, 128–158. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; de Waard, M.; Verheijen, H.; Boeren, S.; Hageman, J.A.; van Hooijdonk, T.; Vervoort, J.; van Goudoever, J.B.; Hettinga, K. Changes over lactation in breast milk serum proteins involved in the maturation of immune and digestive system of the infant. J. Proteom. 2016, 147, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Garrigues, L.; Van den Toorn, H.; Stahl, B.; Heck, A.J.R. Discovery and quantification of non-human proteins in human milk. J. Proteom. Res. 2018, 18, 225–238. [Google Scholar] [CrossRef]
- Oftedal, O.T. The evolution of milk secretion and its ancient origins. Animal 2012, 6, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Weber, D.; Xu, W.; Durbin-Johnson, B.P.; Phinney, B.S.; Lonnerdal, B. Absolute Quantification of Human Milk Caseins and the Whey/Casein Ratio during the First Year of Lactation. J. Proteome Res. 2017, 16, 4113–4121. [Google Scholar] [CrossRef] [PubMed]
- Gan, J.; Robinson, R.C.; Wang, J.; Krishnakumar, N.; Manning, C.J.; Lor, Y.; Breck, M.; Barile, D.; German, J.B. Peptidomic profiling of human milk with LC-MS/MS reveals pH-specific proteolysis of milk proteins. Food Chem. 2019, 274, 766–774. [Google Scholar] [CrossRef] [PubMed]
- Lonnerdal, B.; Erdmann, P.; Thakkar, S.K.; Sauser, J.; Destaillats, F. Longitudinal evolution of true protein, amino acids and bioactive proteins in breast milk: A developmental perspective. J. Nutr. Biochem. 2017, 41, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ke, X.; Chen, Q.; Pan, X.; Zhang, J.; Mo, W.; Ren, Y. Quantification of lactoferrin in breast milk by ultra-high performance liquid chromatography-tandem mass spectrometry with isotopic dilution. RSC Adv. 2016, 6, 12280–12285. [Google Scholar] [CrossRef]
- Chatterton, D.E.; Nguyen, D.N.; Bering, S.B.; Sangild, P.T. Anti-inflammatory mechanisms of bioactive milk proteins in the intestine of newborns. Int. J. Biochem. Cell Biol. 2013, 45, 1730–1747. [Google Scholar] [CrossRef]
- Huang, J.; Kailemia, M.J.; Goonatilleke, E.; Parker, E.A.; Hong, Q.; Sabia, R.; Smilowitz, J.T.; German, J.B.; Lebrilla, C.B. Quantitation of human milk proteins and their glycoforms using multiple reaction monitoring (MRM). Anal. Bioanal. Chem. 2017, 409, 589–606. [Google Scholar] [CrossRef]
- Shin, K.; Hayasawa, H.; Lonnerdal, B. Purification and quantification of lactoperoxidase in human milk with use of immunoadsorbents with antibodies against recombinant human lactoperoxidase. Am. J. Clin. Nutr. 2001, 73, 984–989. [Google Scholar] [CrossRef] [PubMed]
- Cole, F.S.; Schneeberger, E.E.; Lichtenberg, N.A.; Colten, H.R. Complement biosynthesis in human breast-milk macrophages and blood monocytes. Immunology 1982, 46, 429–441. [Google Scholar] [PubMed]
- Kassim, O.O.; Afolabi, O.; Ako-Nai, K.A.; Torimiro, S.E.; Littleton, G.K.; Mbogua, C.N.; Oke, O.; Turner, W.; Grissom, F. Immunoprotective factors in breast milk and sera of mother-infant pairs. Trop. Geogr. Med. 1986, 38, 362–366. [Google Scholar] [PubMed]
- Peterson, J.A.; Hamosh, M.; Scallan, C.D.; Ceriani, R.L.; Henderson, T.R.; Mehta, N.R.; Armand, M.; Hamosh, P. Milk Fat Globule Glycoproteins in Human Milk and in Gastric Aspirates of Mother’s Milk-Fed Preterm Infants. Pediatr. Res. 1998, 44, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Dallas, D.C.; Murray, N.M.; Gan, J. Proteolytic Systems in Milk: Perspectives on the Evolutionary Function within the Mammary Gland and the Infant. J. Mammary Gland Biol. Neoplasia 2015, 20, 133–147. [Google Scholar] [CrossRef]
- Demers-Mathieu, V.; Nielsen, S.D.; Underwood, M.A.; Borghese, R.; Dallas, D.C. Changes in Proteases, Antiproteases, and Bioactive Proteins From Mother’s Breast Milk to the Premature Infant Stomach. J. Pediatr. Gastroenterol. Nutr. 2018, 66, 318–324. [Google Scholar] [CrossRef]
- Khodabakhshi, A.; Ghayour-Mobarhan, M.; Rooki, H.; Vakili, R.; Hashemy, S.I.; Mirhafez, S.R.; Shakeri, M.T.; Kashanifar, R.; Pourbafarani, R.; Mirzaei, H.; et al. Comparative measurement of ghrelin, leptin, adiponectin, EGF and IGF-1 in breast milk of mothers with overweight/obese and normal-weight infants. Eur. J. Clin. Nutr. 2015, 69, 614–618. [Google Scholar] [CrossRef]
- Aydin, S.; Aydin, S.; Ozkan, Y.; Kumru, S. Ghrelin is present in human colostrum, transitional and mature milk. Peptides 2006, 27, 878–882. [Google Scholar] [CrossRef]
- Ilcol, Y.O.; Hizli, Z.B.; Ozkan, T. Leptin concentration in breast milk and its relationship to duration of lactation and hormonal status. Int. Breastfeed. J. 2006, 1, 21. [Google Scholar] [CrossRef]
- Gridneva, Z.; Kugananthan, S.; Rea, A.; Lai, C.T.; Ward, L.C.; Murray, K.; Hartmann, P.E.; Geddes, D.T. Human Milk Adiponectin and Leptin and Infant Body Composition over the First 12 Months of Lactation. Nutrients 2018, 10, 1125. [Google Scholar] [CrossRef]
- Martin, L.J.; Woo, J.G.; Geraghty, S.R.; Altaye, M.; Davidson, B.S.; Banach, W.; Dolan, L.M.; Ruiz-Palacios, G.M.; Morrow, A.L. Adiponectin is present in human milk and is associated with maternal factors. Am. J. Clin. Nutr. 2006, 83, 1106–1111. [Google Scholar] [CrossRef] [PubMed]
- Lubetzky, R.; Weisman, Y.; Dollberg, S.; Herman, L.; Mandel, D. Parathyroid hormone-related protein in preterm human milk. Breastfeed. Med. 2010, 5, 67–69. [Google Scholar] [CrossRef] [PubMed]
- Truchet, S.; Honvo-Houeto, E. Physiology of milk secretion. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 367–384. [Google Scholar] [CrossRef] [PubMed]
- McManaman, J.L.; Neville, M.C. Mammary physiology and milk secretion. Adv. Drug Deliv. Rev. 2003, 55, 629–641. [Google Scholar] [CrossRef]
- Truchet, S.; Chat, S.; Ollivier-Bousquet, M. Milk secretion: The role of SNARE proteins. J. Mammary Gland Biol. Neoplasia 2014, 19, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Linzell, J.L.; Peaker, M. Mechanism of milk secretion. Physiol. Rev. 1971, 51, 564–597. [Google Scholar] [CrossRef] [PubMed]
- Wooding, F.B. The mechanism of secretion of the milk fat globule. J. Cell Sci. 1971, 9, 805–821. [Google Scholar]
- Lee, H.; Padhi, E.; Hasegawa, Y.; Larke, J.; Parenti, M.; Wang, A.; Hernell, O.; Lonnerdal, B.; Slupsky, C. Compositional Dynamics of the Milk Fat Globule and Its Role in Infant Development. Front. Pediatr. 2018, 6, 313. [Google Scholar] [CrossRef]
- Hunziker, W.; Kraehenbuhl, J.P. Epithelial transcytosis of immunoglobulins. J. Mammary Gland Biol. Neoplasia 1998, 3, 287–302. [Google Scholar] [CrossRef]
- Monks, J.; Neville, M.C. Albumin transcytosis across the epithelium of the lactating mouse mammary gland. J. Physiol. 2004, 560, 267–280. [Google Scholar] [CrossRef]
- Ollivier-Bousquet, M. Transferrin and prolactin transcytosis in the lactating mammary epithelial cell. J. Mammary Gland Biol. Neoplasia 1998, 3, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Ninkina, N.; Kukharsky, M.S.; Hewitt, M.V.; Lysikova, E.A.; Skuratovska, L.N.; Deykin, A.V.; Buchman, V.L. Stem cells in human breast milk. Hum. Cell 2019, 32, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Verd, S.; Ginovart, G.; Calvo, J.; Ponce-Taylor, J.; Gaya, A. Variation in the Protein Composition of Human Milk during Extended Lactation: A Narrative Review. Nutrients 2018, 10, 1124. [Google Scholar] [CrossRef] [PubMed]
- Shennan, D.B.; Peaker, M. Transport of milk constituents by the mammary gland. Physiol. Rev. 2000, 80, 925–951. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Simala-Grant, J.L.; Taylor, D.E. Fucosylation in prokaryotes and eukaryotes. Glycobiology 2006, 16, 158R–184R. [Google Scholar] [CrossRef] [PubMed]
- Georgi, G.; Bartke, N.; Wiens, F.; Stahl, B. Functional glycans and glycoconjugates in human milk. Am. J. Clin. Nutr. 2013, 98, 578S–585S. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, S.; Rincon, G.; Islas-Trejo, A.; Medrano, J.F. Transcriptional profiling of bovine milk using RNA sequencing. BMC Genom. 2012, 13, 45. [Google Scholar] [CrossRef] [PubMed]
- Riskin, A.; Almog, M.; Peri, R.; Halasz, K.; Srugo, I.; Kessel, A. Changes in immunomodulatory constituents of human milk in response to active infection in the nursing infant. Pediatr. Res. 2012, 71, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Hassiotou, F.; Hepworth, A.R.; Metzger, P.; Tat Lai, C.; Trengove, N.; Hartmann, P.E.; Filgueira, L. Maternal and infant infections stimulate a rapid leukocyte response in breastmilk. Clin. Transl. Immunol. 2013, 2, e3. [Google Scholar] [CrossRef]
- Lonnerdal, B. Nutritional and physiologic significance of human milk proteins. Am. J. Clin. Nutr. 2003, 77, 1537s–1543s. [Google Scholar] [CrossRef]
- Kramer, M.S. Breastfeeding and allergy: The evidence. Ann. Nutr. Metab. 2011, 59 (Suppl. 1), 20–26. [Google Scholar] [CrossRef] [PubMed]
- Iyengar, S.R.; Walker, W.A. Immune factors in breast milk and the development of atopic disease. J. Pediatr. Gastroenterol. Nutr. 2012, 55, 641–647. [Google Scholar] [CrossRef] [PubMed]
- Matheson, M.C.; Allen, K.J.; Tang, M.L. Understanding the evidence for and against the role of breastfeeding in allergy prevention. Clin. Exp. Allergy 2012, 42, 827–851. [Google Scholar] [CrossRef] [PubMed]
- Matheson, M.C.; Erbas, B.; Balasuriya, A.; Jenkins, M.A.; Wharton, C.L.; Tang, M.L.; Abramson, M.J.; Walters, E.H.; Hopper, J.L.; Dharmage, S.C. Breast-feeding and atopic disease: A cohort study from childhood to middle age. J. Allergy Clin. Immunol. 2007, 120, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- LÖNnerdal, B.O.; Atkinson, S. CHAPTER 5—Nitrogenous Components of Milk: A. Human Milk Proteins. In Handbook of Milk Composition; Jensen, R.G., Ed.; Academic Press: San Diego, CA, USA, 1995. [Google Scholar]
- Carlson, S.E. Human milk nonprotein nitrogen: Occurrence and possible functions. Adv. Pediatr. 1985, 32, 43–70. [Google Scholar] [PubMed]
- Atkinson, S.A.; LÖNnerdal, B.O. B-Nonprotein Nitrogen Fractions of Human Milk. In Handbook of Milk Composition; Jensen, R.G., Ed.; Academic Press: San Diego, CA, USA, 1995. [Google Scholar]
- Froehlich, J.W.; Dodds, E.D.; Barboza, M.; McJimpsey, E.L.; Seipert, R.R.; Francis, J.; An, H.J.; Freeman, S.; German, J.B.; Lebrilla, C.B. Glycoprotein expression in human milk during lactation. J. Agric. Food Chem. 2010, 58, 6440–6448. [Google Scholar] [CrossRef] [PubMed]
- Lairson, L.L.; Henrissat, B.; Davies, G.J.; Withers, S.G. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef]
- Henrissat, B.; Surolia, A.; Stanley, P. A Genomic View of Glycobiology. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Eds.; Cold Spring Harbor Press: Cold Spring Harbor, NY, USA, 2015; pp. 89–97. [Google Scholar]
- Chung, C.Y.; Majewska, N.I.; Wang, Q.; Paul, J.T.; Betenbaugh, M.J. SnapShot: N-Glycosylation Processing Pathways across Kingdoms. Cell 2017, 171, 258. [Google Scholar] [CrossRef]
- Joshi, H.J.; Narimatsu, Y.; Schjoldager, K.T.; Tytgat, H.L.P.; Aebi, M.; Clausen, H.; Halim, A. SnapShot: O-Glycosylation Pathways across Kingdoms. Cell 2018, 172, 632. [Google Scholar] [CrossRef]
- Schwarz, F.; Aebi, M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 2011, 21, 576–582. [Google Scholar] [CrossRef]
- Aebi, M. N-linked protein glycosylation in the ER. Biochim. Biophys. Acta 2013, 1833, 2430–2437. [Google Scholar] [CrossRef] [PubMed]
- Giuffrida, M.G.; Cavaletto, M.; Giunta, C.; Neuteboom, B.; Cantisani, A.; Napolitano, L.; Calderone, V.; Godovac-Zimmermann, J.; Conti, A. The unusual amino acid triplet Asn-Ile-Cys is a glycosylation consensus site in human alpha-lactalbumin. J. Protein Chem. 1997, 16, 747–753. [Google Scholar] [CrossRef] [PubMed]
- Levery, S.B.; Steentoft, C.; Halim, A.; Narimatsu, Y.; Clausen, H.; Vakhrushev, S.Y. Advances in mass spectrometry driven O-glycoproteomics. Biochim. Biophys. Acta 2015, 1850, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Orczyk-Pawilowicz, M.; Hirnle, L.; Berghausen-Mazur, M.; Katnik-Prastowska, I.M. Lactation stage-related expression of sialylated and fucosylated glycotopes of human milk alpha-1-acid glycoprotein. Breastfeed. Med. 2014, 9, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Goonatilleke, E.; Huang, J.; Xu, G.; Wu, L.; Smilowitz, J.T.; German, J.B.; Lebrilla, C.B. Human Milk Proteins and Their Glycosylation Exhibit Quantitative Dynamic Variations during Lactation. J. Nutr. 2019. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Furukawa, J.I.; Naito, S.; Higashino, K.; Numata, Y.; Shinohara, Y. Quantitative analysis of total serum glycome in human and mouse. Proteomics 2016, 16, 2747–2758. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Al-Shareffi, E.; Haltiwanger, R.S. Biological functions of fucose in mammals. Glycobiology 2017, 27, 601–618. [Google Scholar] [CrossRef] [PubMed]
- Sumer-Bayraktar, Z.; Grant, O.C.; Venkatakrishnan, V.; Woods, R.J.; Packer, N.H.; Thaysen-Andersen, M. Asn347 glycosylation of corticosteroid-binding globulin fine-tunes the host Immune response by modulating proteolysis by pseudomonas aeruginosa and neutrophil elastase. J. Biol. Chem. 2016, 291, 17727–17742. [Google Scholar] [CrossRef]
- Schrader, M. Origins, Technological Development, and Applications of Peptidomics. Methods Mol. Biol 2018, 1719, 3–39. [Google Scholar] [CrossRef]
- Dallas, D.C.; Smink, C.J.; Robinson, R.C.; Tian, T.; Guerrero, A.; Parker, E.A.; Smilowitz, J.T.; Hettinga, K.A.; Underwood, M.A.; Lebrilla, C.B.; et al. Endogenous human milk peptide release is greater after preterm birth than term birth. J. Nutr. 2015, 145, 425–433. [Google Scholar] [CrossRef]
- Wan, J.; Cui, X.W.; Zhang, J.; Fu, Z.Y.; Guo, X.R.; Sun, L.Z.; Ji, C.B. Peptidome analysis of human skim milk in term and preterm milk. Biochem. Biophys. Res. Commun. 2013, 438, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, A.; Dallas, D.C.; Contreras, S.; Chee, S.; Parker, E.A.; Sun, X.; Dimapasoc, L.; Barile, D.; German, J.B.; Lebrilla, C.B. Mechanistic peptidomics: Factors that dictate specificity in the formation of endogenous peptides in human milk. Mol. Cell. Proteom. 2014, 13, 3343–3351. [Google Scholar] [CrossRef] [PubMed]
- Dallas, D.C.; Guerrero, A.; Khaldi, N.; Castillo, P.A.; Martin, W.F.; Smilowitz, J.T.; Bevins, C.L.; Barile, D.; German, J.B.; Lebrilla, C.B. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. J. Proteome Res. 2013, 12, 2295–2304. [Google Scholar] [CrossRef] [PubMed]
- Dingess, K.A.; van den Toorn, H.W.P.; Mank, M.; Stahl, B.; Heck, A.J.R. Toward an efficient workflow for the analysis of the human milk peptidome. Anal. Bioanal. Chem. 2019, 411, 1351–1363. [Google Scholar] [CrossRef] [PubMed]
- Dingess, K.A.; de Waard, M.; Boeren, S.; Vervoort, J.; Lambers, T.T.; van Goudoever, J.B.; Hettinga, K. Human milk peptides differentiate between the preterm and term infant and across varying lactational stages. Food Funct. 2017, 8, 3769–3782. [Google Scholar] [CrossRef] [PubMed]
- Wada, Y.; Lonnerdal, B. Bioactive peptides derived from human milk proteins-mechanisms of action. J. Nutr. Biochem. 2014, 25, 503–514. [Google Scholar] [CrossRef]
- Dallas, D.C.; Weinborn, V.; de Moura Bell, J.M.; Wang, M.; Parker, E.A.; Guerrero, A.; Hettinga, K.A.; Lebrilla, C.B.; German, J.B.; Barile, D. Comprehensive peptidomic and glycomic evaluation reveals that sweet whey permeate from colostrum is a source of milk protein-derived peptides and oligosaccharides. Food Res. Int. 2014, 63, 203–209. [Google Scholar] [CrossRef]
- Nielsen, S.D.; Beverly, R.L.; Dallas, D.C. Milk Proteins Are Predigested Within the Human Mammary Gland. J. Mammary Gland Biol. Neoplasia 2017, 22, 251–261. [Google Scholar] [CrossRef]
- Nielsen, S.D.; Beverly, R.L.; Qu, Y.; Dallas, D.C. Milk bioactive peptide database: A comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017, 232, 673–682. [Google Scholar] [CrossRef]
- Dallas, D.C.; Guerrero, A.; Parker, E.A.; Robinson, R.C.; Gan, J.; German, J.B.; Barile, D.; Lebrilla, C.B. Current peptidomics: Applications, purification, identification, quantification, and functional analysis. Proteomics 2015, 15, 1026–1038. [Google Scholar] [CrossRef]
- Hamosh, M. Bioactive factors in human milk. Pediatr. Clin. 2001, 48, 69–86. [Google Scholar] [CrossRef]
- Hamosh, M. C—Enzymes in Human Milk. In Handbook of Milk Composition; Jensen, R.G., Ed.; Academic Press: San Diego, CA, USA, 1995; pp. 388–427. [Google Scholar]
- Shahani, K.M.; Kwan, A.J.; Friend, B.A. Role and significance of enzymes in human milk. Am. J. Clin. Nutr. 1980, 33, 1861–1868. [Google Scholar] [CrossRef] [PubMed]
- Dallas, D.C.; German, J.B. Enzymes in Human Milk; Karger Publishers: Basel, Switzerland, 2017. [Google Scholar]
- Shahani, K.M. Milk enzymes: Their role and significance. J. Dairy Sci. 1966, 49, 907–920. [Google Scholar] [CrossRef]
- Jenness, R. The composition of human milk. Semin. Perinatol. 1979, 3, 225–239. [Google Scholar] [PubMed]
- Dallas, D.C.; Underwood, M.A.; Zivkovic, A.M.; German, J.B. Digestion of Protein in Premature and Term Infants. J. Nutr. Disord. 2012, 2, 112. [Google Scholar] [CrossRef]
- Timmer, J.C.; Zhu, W.; Pop, C.; Regan, T.; Snipas, S.J.; Eroshkin, A.M.; Riedl, S.J.; Salvesen, G.S. Structural and kinetic determinants of protease substrates. Nat. Struct. Mol. Biol. 2009, 16, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef]
- Heegaard, C.W.; Larsen, L.B.; Rasmussen, L.K.; Hojberg, K.E.; Petersen, T.E.; Andreasen, P.A. Plasminogen activation system in human milk. J. Pediatr. Gastroenterol. Nutr. 1997, 25, 159–166. [Google Scholar] [CrossRef]
- Beverly, R.L.; Underwood, M.A.; Dallas, D.C. Peptidomics Analysis of Milk Protein-Derived Peptides Released over Time in the Preterm Infant Stomach. J. Proteome Res. 2019, 18, 912–922. [Google Scholar] [CrossRef]
- Dallas, D.C.; Guerrero, A.; Khaldi, N.; Borghese, R.; Bhandari, A.; Underwood, M.A.; Lebrilla, C.B.; German, J.B.; Barile, D. A peptidomic analysis of human milk digestion in the infant stomach reveals protein-specific degradation patterns. J. Nutr. 2014, 144, 815–820. [Google Scholar] [CrossRef]
- Holton, T.A.; Vijayakumar, V.; Dallas, D.C.; Guerrero, A.; Borghese, R.A.; Lebrilla, C.B.; German, J.B.; Barile, D.; Underwood, M.A.; Shields, D.C.; et al. Following the digestion of milk proteins from mother to baby. J. Proteome Res. 2014, 13, 5777–5783. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Baerlocher, K.; Bauer, J.M.; Elmadfa, I.; Heseker, H.; Leschik-Bonnet, E.; Stangl, G.; Volkert, D.; Stehle, P. Revised Reference Values for the Intake of Protein. Ann. Nutr. Metab. 2019, 74, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Kibangou, I.; Bouhallab, S.; Bureau, F.; Allouche, S.; Thouvenin, G.; Bougle, D. Caseinophosphopeptide-bound iron: Protective effect against gut peroxidation. Ann. Nutr. Metab. 2008, 52, 177–180. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Nebot, M.J.; Barbera, R.; Alegria, A. Iron and zinc bioavailability in Caco-2 cells: Influence of caseinophosphopeptides. Food Chem. 2013, 138, 1298–1303. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Sandstrom, B.; Lonnerdal, B. The effect of casein phosphopeptides on zinc and calcium absorption from high phytate infant diets assessed in rat pups and Caco-2 cells. Pediatr. Res. 1996, 40, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Lonnerdal, B.; Glazier, C. Calcium binding by alpha-lactalbumin in human milk and bovine milk. J. Nutr. 1985, 115, 1209–1216. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Stuart, D.I.; Acharya, K.R. Alpha-lactalbumin possesses a distinct zinc binding site. J. Biol. Chem. 1993, 268, 19292–19298. [Google Scholar] [PubMed]
- Adkins, Y.; Lonnerdal, B. Mechanisms of vitamin B(12) absorption in breast-fed infants. J. Pediatr. Gastroenterol. Nutr. 2002, 35, 192–198. [Google Scholar] [CrossRef]
- Suzuki, Y.A.; Shin, K.; Lonnerdal, B. Molecular cloning and functional expression of a human intestinal lactoferrin receptor. Biochemistry 2001, 40, 15771–15779. [Google Scholar] [CrossRef]
- Lopez, V.; Suzuki, Y.A.; Lonnerdal, B. Ontogenic changes in lactoferrin receptor and DMT1 in mouse small intestine: Implications for iron absorption during early life. Biochem. Cell Biol. 2006, 84, 337–344. [Google Scholar] [CrossRef]
- Lonnerdal, B.; Bryant, A. Absorption of iron from recombinant human lactoferrin in young US women. Am. J. Clin. Nutr. 2006, 83, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Blackberg, L.; Hernell, O. Further characterization of the bile salt-stimulated lipase in human milk. FEBS Lett. 1983, 157, 337–341. [Google Scholar] [CrossRef]
- Li, X.; Lindquist, S.; Lowe, M.; Noppa, L.; Hernell, O. Bile salt-stimulated lipase and pancreatic lipase-related protein 2 are the dominating lipases in neonatal fat digestion in mice and rats. Pediatr. Res. 2007, 62, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Hamosh, M. Lipid metabolism in premature infants. Neonatology 1987, 52 (Suppl. 1), 50–64. [Google Scholar] [CrossRef]
- Segurel, L.; Bon, C. On the Evolution of Lactase Persistence in Humans. Annu. Rev. Genom. Hum. Genet. 2017, 18, 297–319. [Google Scholar] [CrossRef]
- Sevenhuysen, G.P.; Holodinsky, C.; Dawes, C. Development of salivary alpha-amylase in infants from birth to 5 months. Am. J. Clin. Nutr. 1984, 39, 584–588. [Google Scholar] [CrossRef]
- Hamosh, M. Enzymes in milk: Their function in the mammary gland, in milk, and in the infant. Biol. Hum. Milk 1988, 15, 45–61. [Google Scholar]
- Heitlinger, L.A.; Lee, P.C.; Dillon, W.P.; Lebenthal, E. Mammary amylase: A possible alternate pathway of carbohydrate digestion in infancy. Pediatr. Res. 1983, 17, 15–18. [Google Scholar] [CrossRef]
- Chen, Y.; Zheng, Z.; Zhu, X.; Shi, Y.; Tian, D.; Zhao, F.; Liu, N.; Hüppi, P.S.; Troy, F.A.; Wang, B. Lactoferrin Promotes Early Neurodevelopment and Cognition in Postnatal Piglets by Upregulating the BDNF Signaling Pathway and Polysialylation. Mol. Neurobiol. 2015, 52, 256–269. [Google Scholar] [CrossRef]
- Yang, C.; Zhu, X.; Liu, N.; Chen, Y.; Gan, H.; Troy, F.A.; Wang, B. Lactoferrin up-regulates intestinal gene expression of brain-derived neurotrophic factors BDNF, UCHL1 and alkaline phosphatase activity to alleviate early weaning diarrhea in postnatal piglets. J. Nutr. Biochem. 2014, 25, 834–842. [Google Scholar] [CrossRef]
- Timby, N.; Domellof, E.; Hernell, O.; Lonnerdal, B.; Domellof, M. Neurodevelopment, nutrition, and growth until 12 mo of age in infants fed a low-energy, low-protein formula supplemented with bovine milk fat globule membranes: A randomized controlled trial. Am. J. Clin. Nutr. 2014, 99, 860–868. [Google Scholar] [CrossRef] [PubMed]
- Friedman, N.J.; Zeiger, R.S. The role of breast-feeding in the development of allergies and asthma. J. Allergy Clin. Immunol. 2005, 115, 1238–1248. [Google Scholar] [CrossRef] [PubMed]
- Brandtzaeg, P. The mucosal immune system and its integration with the mammary glands. J. Pediatr. 2010, 156, S8–S15. [Google Scholar] [CrossRef] [PubMed]
- Field, C.J. The immunological components of human milk and their effect on immune development in infants. J. Nutr. 2005, 135, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed]
- Morkbak, A.L.; Poulsen, S.S.; Nexo, E. Haptocorrin in humans. Clin. Chem. Lab. Med. 2007, 45, 1751–1759. [Google Scholar] [CrossRef]
- Adkins, Y.; Lonnerdal, B. Potential host-defense role of a human milk vitamin B-12-binding protein, haptocorrin, in the gastrointestinal tract of breastfed infants, as assessed with porcine haptocorrin in vitro. Am. J. Clin. Nutr. 2003, 77, 1234–1240. [Google Scholar] [CrossRef]
- Jensen, H.R.; Laursen, M.F.; Lildballe, D.L.; Andersen, J.B.; Nexo, E.; Licht, T.R. Effect of the vitamin B12-binding protein haptocorrin present in human milk on a panel of commensal and pathogenic bacteria. BMC Res. Notes 2011, 4, 208. [Google Scholar] [CrossRef]
- Callewaert, L.; Michiels, C.W. Lysozymes in the animal kingdom. J. Biosci. 2010, 35, 127–160. [Google Scholar] [CrossRef]
- Ellison, R.T.; Giehl, T.J. Killing of gram-negative bacteria by lactoferrin and lysozyme. J. Clin. Investig. 1991, 88, 1080–1091. [Google Scholar] [CrossRef]
- Grapov, D.; Lemay, D.G.; Weber, D.; Phinney, B.S.; Azulay Chertok, I.R.; Gho, D.S.; German, J.B.; Smilowitz, J.T. The human colostrum whey proteome is altered in gestational diabetes mellitus. J. Proteome Res. 2015, 14, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Niyonsaba, F.; Ushio, H.; Okuda, D.; Nagaoka, I.; Ikeda, S.; Okumura, K.; Ogawa, H. Synergistic effect of antibacterial agents human beta-defensins, cathelicidin LL-37 and lysozyme against Staphylococcus aureus and Escherichia coli. J. Derm. Sci. 2005, 40, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Nash, J.A.; Ballard, T.N.; Weaver, T.E.; Akinbi, H.T. The peptidoglycan-degrading property of lysozyme is not required for bactericidal activity in vivo. J. Immunol. 2006, 177, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jiang, A.; Yu, H.; Xiong, Y.; Zhou, G.; Qin, M.; Dou, J.; Wang, J. Human Lysozyme Synergistically Enhances Bactericidal Dynamics and Lowers the Resistant Mutant Prevention Concentration for Metronidazole to Helicobacter pylori by Increasing Cell Permeability. Molecules 2016, 21, 1435. [Google Scholar] [CrossRef] [PubMed]
- Ragland, S.A.; Criss, A.K. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017, 13, e1006512. [Google Scholar] [CrossRef] [PubMed]
- Serna, M.; Giles, J.L.; Morgan, B.P.; Bubeck, D. Structural basis of complement membrane attack complex formation. Nat. Commun. 2016, 7, 10587. [Google Scholar] [CrossRef] [PubMed]
- Dunkelberger, J.R.; Song, W.C. Complement and its role in innate and adaptive immune responses. Cell Res. 2010, 20, 34–50. [Google Scholar] [CrossRef]
- Ogundele, M. Role and significance of the complement system in mucosal immunity: Particular reference to the human breast milk complement. Immunol. Cell Biol. 2001, 79, 1–10. [Google Scholar] [CrossRef]
- Sharma, S.; Singh, A.K.; Kaushik, S.; Sinha, M.; Singh, R.P.; Sharma, P.; Sirohi, H.; Kaur, P.; Singh, T.P. Lactoperoxidase: Structural insights into the function, ligand binding and inhibition. Int. J. Biochem. Mol. Biol. 2013, 4, 108–128. [Google Scholar]
- Sarr, D.; Toth, E.; Gingerich, A.; Rada, B. Antimicrobial actions of dual oxidases and lactoperoxidase. J. Microbiol. 2018, 56, 373–386. [Google Scholar] [CrossRef]
- Kussendrager, K.D.; van Hooijdonk, A.C. Lactoperoxidase: Physico-chemical properties, occurrence, mechanism of action and applications. Br. J. Nutr 2000, 84 (Suppl. 1), S19–S25. [Google Scholar] [CrossRef] [PubMed]
- Newburg, D.S. Neonatal protection by an innate immune system of human milk consisting of oligosaccharides and glycans. J. Anim. Sci. 2009, 87, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Lis-Kuberka, J.; Orczyk-Pawilowicz, M. Sialylated Oligosaccharides and Glycoconjugates of Human Milk. The Impact on Infant and Newborn Protection, Development and Well-Being. Nutrients 2019, 11, 306. [Google Scholar] [CrossRef] [PubMed]
- Fouda, G.G.; Jaeger, F.H.; Amos, J.D.; Ho, C.; Kunz, E.L.; Anasti, K.; Stamper, L.W.; Liebl, B.E.; Barbas, K.H.; Ohashi, T.; et al. Tenascin-C is an innate broad-spectrum, HIV-1-neutralizing protein in breast milk. Proc. Natl. Acad. Sci. USA 2013, 110, 18220–18225. [Google Scholar] [CrossRef] [PubMed]
- Barboza, M.; Pinzon, J.; Wickramasinghe, S.; Froehlich, J.W.; Moeller, I.; Smilowitz, J.T.; Ruhaak, L.R.; Huang, J.; Lonnerdal, B.; German, J.B.; et al. Glycosylation of human milk lactoferrin exhibits dynamic changes during early lactation enhancing its role in pathogenic bacteria-host interactions. Mol. Cell. Proteom. 2012, 11. [Google Scholar] [CrossRef] [PubMed]
- Ruvoen-Clouet, N.; Mas, E.; Marionneau, S.; Guillon, P.; Lombardo, D.; Le Pendu, J. Bile-salt-stimulated lipase and mucins from milk of ‘secretor’ mothers inhibit the binding of Norwalk virus capsids to their carbohydrate ligands. Biochem. J. 2006, 393, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Yolken, R.H.; Peterson, J.A.; Vonderfecht, S.L.; Fouts, E.T.; Midthun, K.; Newburg, D.S. Human milk mucin inhibits rotavirus replication and prevents experimental gastroenteritis. J. Clin. Investig. 1992, 90, 1984–1991. [Google Scholar] [CrossRef]
- Liu, B.; Yu, Z.; Chen, C.; Kling, D.E.; Newburg, D.S. Human milk mucin 1 and mucin 4 inhibit Salmonella enterica serovar Typhimurium invasion of human intestinal epithelial cells in vitro. J. Nutr. 2012, 142, 1504–1509. [Google Scholar] [CrossRef]
- Schroten, H.; Stapper, C.; Plogmann, R.; Kohler, H.; Hacker, J.; Hanisch, F.G. Fab-independent antiadhesion effects of secretory immunoglobulin A on S-fimbriated Escherichia coli are mediated by sialyloligosaccharides. Infect. Immun. 1998, 66, 3971–3973. [Google Scholar]
- Bode, L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 2012, 22, 1147–1162. [Google Scholar] [CrossRef]
- Nanthakumar, N.; Meng, D.; Goldstein, A.M.; Zhu, W.; Lu, L.; Uauy, R.; Llanos, A.; Claud, E.C.; Walker, W.A. The mechanism of excessive intestinal inflammation in necrotizing enterocolitis: An immature innate immune response. PLoS ONE 2011, 6, e17776. [Google Scholar] [CrossRef] [PubMed]
- Buescher, E.S. Anti-inflammatory characteristics of human milk: How, where, why. Adv. Exp. Med. Biol. 2001, 501, 207–222. [Google Scholar] [CrossRef] [PubMed]
- Langjahr, P.; Diaz-Jimenez, D.; De la Fuente, M.; Rubio, E.; Golenbock, D.; Bronfman, F.C.; Quera, R.; Gonzalez, M.J.; Hermoso, M.A. Metalloproteinase-dependent TLR2 ectodomain shedding is involved in soluble toll-like receptor 2 (sTLR2) production. PLoS ONE 2014, 9, e104624. [Google Scholar] [CrossRef] [PubMed]
- Kitchens, R.L.; Thompson, P.A. Modulatory effects of sCD14 and LBP on LPS-host cell interactions. J. Endotoxin Res. 2005, 11, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Ando, K.; Hasegawa, K.; Shindo, K.; Furusawa, T.; Fujino, T.; Kikugawa, K.; Nakano, H.; Takeuchi, O.; Akira, S.; Akiyama, T.; et al. Human lactoferrin activates NF-kappaB through the Toll-like receptor 4 pathway while it interferes with the lipopolysaccharide-stimulated TLR4 signaling. FEBS J. 2010, 277, 2051–2066. [Google Scholar] [CrossRef] [PubMed]
- Aziz, M.; Jacob, A.; Matsuda, A.; Wang, P. Review: Milk fat globule-EGF factor 8 expression, function and plausible signal transduction in resolving inflammation. Apoptosis 2011, 16, 1077–1086. [Google Scholar] [CrossRef] [PubMed]
- Stroinigg, N.; Srivastava, M.D. Modulation of toll-like receptor 7 and LL-37 expression in colon and breast epithelial cells by human beta-defensin-2. Allergy Asthma Proc. 2005, 26, 299–309. [Google Scholar] [PubMed]
- Lonnerdal, B.; Kvistgaard, A.S.; Peerson, J.M.; Donovan, S.M.; Peng, Y.M. Growth, Nutrition, and Cytokine Response of Breast-fed Infants and Infants Fed Formula With Added Bovine Osteopontin. J. Pediatr. Gastroenterol. Nutr. 2016, 62, 650–657. [Google Scholar] [CrossRef]
- Famulener, L. On the transmission of immunity from mother to offspring: A study upon serum hemolysins in goats [with discussion]. J. Infect. Dis. 1912, 10, 332–368. [Google Scholar] [CrossRef]
- Hurley, W.L.; Theil, P.K. Perspectives on immunoglobulins in colostrum and milk. Nutrients 2011, 3, 442–474. [Google Scholar] [CrossRef]
- Mix, E.; Goertsches, R.; Zett, U.K. Immunoglobulins-basic considerations. J. Neurol. 2006, 253 (Suppl. 5), V9–V17. [Google Scholar] [CrossRef] [PubMed]
- Van de Perre, P. Transfer of antibody via mother’s milk. Vaccine 2003, 21, 3374–3376. [Google Scholar] [CrossRef]
- Johansen, F.E.; Braathen, R.; Brandtzaeg, P. Role of J chain in secretory immunoglobulin formation. Scand. J. Immunol. 2000, 52, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Newburg, D.S. Innate immunity and human milk. J. Nutr. 2005, 135, 1308–1312. [Google Scholar] [CrossRef] [PubMed]
- Asano, M.; Komiyama, K. Polymeric immunoglobulin receptor. J. Oral Sci. 2011, 53, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Hanson, L.; Silfverdal, S.A.; Stromback, L.; Erling, V.; Zaman, S.; Olcen, P.; Telemo, E. The immunological role of breast feeding. Pediatr. Allergy Immunol. 2001, 12 (Suppl. 14), 15–19. [Google Scholar] [CrossRef]
- Goldman, A.S. Evolution of the mammary gland defense system and the ontogeny of the immune system. J. Mammary Gland Biol. Neoplasia 2002, 7, 277–289. [Google Scholar] [CrossRef]
- Mantis, N.J.; Rol, N.; Corthesy, B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011, 4, 603–611. [Google Scholar] [CrossRef]
- Michaelsen, T.E.; Emilsen, S.; Sandin, R.H.; Granerud, B.K.; Bratlie, D.; Ihle, O.; Sandlie, I. Human Secretory IgM Antibodies Activate Human Complement and Offer Protection at Mucosal Surface. Scand. J. Immunol. 2017, 85, 43–50. [Google Scholar] [CrossRef]
- Wilcox, C.R.; Holder, B.; Jones, C.E. Factors Affecting the FcRn-Mediated Transplacental Transfer of Antibodies and Implications for Vaccination in Pregnancy. Front. Immunol. 2017, 8, 1294. [Google Scholar] [CrossRef]
- Jiang, X.; Hu, J.; Thirumalai, D.; Zhang, X. Immunoglobulin Transporting Receptors Are Potential Targets for the Immunity Enhancement and Generation of Mammary Gland Bioreactor. Front. Immunol. 2016, 7, 214. [Google Scholar] [CrossRef] [PubMed]
- Gasparoni, A.; Avanzini, A.; Ravagni Probizer, F.; Chirico, G.; Rondini, G.; Severi, F. IgG subclasses compared in maternal and cord serum and breast milk. Arch. Dis. Child. 1992, 67, 41–43. [Google Scholar] [CrossRef] [PubMed]
- Rojas, R.; Apodaca, G. Immunoglobulin transport across polarized epithelial cells. Nat. Rev. Mol. Cell Biol. 2002, 3, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Siegrist, C.A. Mechanisms by which maternal antibodies influence infant vaccine responses: Review of hypotheses and definition of main determinants. Vaccine 2003, 21, 3406–3412. [Google Scholar] [CrossRef]
- Munoz, F.M. Current Challenges and Achievements in Maternal Immunization Research. Front. Immunol. 2018, 9, 436. [Google Scholar] [CrossRef] [PubMed]
- Blanchard-Rohner, G.; Eberhardt, C. Review of maternal immunisation during pregnancy: Focus on pertussis and influenza. Swiss Med. Wkly. 2017, 147, w14526. [Google Scholar] [CrossRef] [PubMed]
- Donnally, H.H. The question of the elimination of foreign protein (egg-white) in woman’s milk. J. Immunol. 1930, 19, 15–40. [Google Scholar]
- Kilshaw, P.J.; Cant, A.J. The passage of maternal dietary proteins into human breast milk. Int. Arch. Allergy Appl. Immunol. 1984, 75, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Chirdo, F.G.; Rumbo, M.; Anon, M.C.; Fossati, C.A. Presence of high levels of non-degraded gliadin in breast milk from healthy mothers. Scand. J. Gastroenterol. 1998, 33, 1186–1192. [Google Scholar] [PubMed]
- Palmer, D.J.; Gold, M.S.; Makrides, M. Effect of maternal egg consumption on breast milk ovalbumin concentration. Clin. Exp. Allergy 2008, 38, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Coscia, A.; Orru, S.; Di Nicola, P.; Giuliani, F.; Varalda, A.; Peila, C.; Fabris, C.; Conti, A.; Bertino, E. Detection of cow’s milk proteins and minor components in human milk using proteomics techniques. J. Matern. Fetal Neonatal Med. 2012, 25 (Suppl. 4), 54–56. [Google Scholar] [CrossRef] [PubMed]
- Bernard, H.; Ah-Leung, S.; Drumare, M.F.; Feraudet-Tarisse, C.; Verhasselt, V.; Wal, J.M.; Creminon, C.; Adel-Patient, K. Peanut allergens are rapidly transferred in human breast milk and can prevent sensitization in mice. Allergy 2014, 69, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Metcalfe, J.R.; Marsh, J.A.; D’Vaz, N.; Geddes, D.T.; Lai, C.T.; Prescott, S.L.; Palmer, D.J. Effects of maternal dietary egg intake during early lactation on human milk ovalbumin concentration: A randomized controlled trial. Clin. Exp. Allergy 2016, 46, 1605–1613. [Google Scholar] [CrossRef] [PubMed]
- Baiz, N.; Macchiaverni, P.; Tulic, M.K.; Rekima, A.; Annesi-Maesano, I.; Verhasselt, V. Early oral exposure to house dust mite allergen through breast milk: A potential risk factor for allergic sensitization and respiratory allergies in children. J. Allergy Clin. Immunol. 2017, 139, 369–372. [Google Scholar] [CrossRef] [PubMed]
- van Odijk, J.; Kull, I.; Borres, M.P.; Brandtzaeg, P.; Edberg, U.; Hanson, L.A.; Host, A.; Kuitunen, M.; Olsen, S.F.; Skerfving, S.; et al. Breastfeeding and allergic disease: A multidisciplinary review of the literature (1966–2001) on the mode of early feeding in infancy and its impact on later atopic manifestations. Allergy 2003, 58, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Dogaru, C.M.; Nyffenegger, D.; Pescatore, A.M.; Spycher, B.D.; Kuehni, C.E. Breastfeeding and childhood asthma: Systematic review and meta-analysis. Am. J. Epidemiol. 2014, 179, 1153–1167. [Google Scholar] [CrossRef] [PubMed]
- Mavroudi, A.; Xinias, I. Dietary interventions for primary allergy prevention in infants. Hippokratia 2011, 15, 216–222. [Google Scholar] [PubMed]
- Ierodiakonou, D.; Garcia-Larsen, V.; Logan, A.; Groome, A.; Cunha, S.; Chivinge, J.; Robinson, Z.; Geoghegan, N.; Jarrold, K.; Reeves, T.; et al. Timing of Allergenic Food Introduction to the Infant Diet and Risk of Allergic or Autoimmune Disease: A Systematic Review and Meta-analysis. JAMA 2016, 316, 1181–1192. [Google Scholar] [CrossRef] [PubMed]
- Jeurink, P.V.; Knipping, K.; Wiens, F.; Baranska, K.; Stahl, B.; Garssen, J.; Krolak-Olejnik, B. Importance of maternal diet in the training of the infant’s immune system during gestation and lactation. Crit. Rev. Food Sci. Nutr. 2019, 59, 1311–1319. [Google Scholar] [CrossRef] [PubMed]
- Pabst, O.; Mowat, A.M. Oral tolerance to food protein. Mucosal Immunol. 2012, 5, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Neu, J. Gastrointestinal maturation and implications for infant feeding. Early Hum. Dev. 2007, 83, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Kaetzel, C.S. Cooperativity among secretory IgA, the polymeric immunoglobulin receptor, and the gut microbiota promotes host-microbial mutualism. Immunol. Lett. 2014, 162, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Karav, S.; Le Parc, A.; Leite Nobrega de Moura Bell, J.M.; Frese, S.A.; Kirmiz, N.; Block, D.E.; Barile, D.; Mills, D.A. Oligosaccharides Released from Milk Glycoproteins Are Selective Growth Substrates for Infant-Associated Bifidobacteria. Appl. Environ. Microbiol. 2016, 82, 3622–3630. [Google Scholar] [CrossRef] [PubMed]
| Proteins | Protein Name | Total | Colostrum | Early | Transitional | Mature | References | Function |
| Total protein | 203–1752 | 360–1690 | 606–1675 | 203–1752 | 362–1632 | [16] | ||
| Total caseins | 19–591 | 42–507 | 103–355 | 87–591 | 19–743 | [16] | ||
| Ratio whey/casein | 90:10 | 78:22 | 72:28 | 60:40 | [9,16] | |||
| Whey Proteins | ||||||||
| α-Lactalbumin | 275–372 | 300–560 | NA | 420 | 275–372 | [17,18] | Lactose synthesis | |
| Lactoferrin | 97–291 | 291 | NA | 180 | 97 | [17,19] | Antimicrobial; Gut development | |
| Osteopontin | 6–149 | 149 | NA | NA | 6–22 | [3,20] | Cell adhesion | |
| sIgA | 22–545 | 545 | NA | 150 | 22–130 | [17,18] | Adaptive immunity | |
| IgG | 2–7 | NA | NA | 5 | 2–7 | [18,21] | Adaptive immunity | |
| sIgM | 1–3 | NA | NA | 12 | 1–3 | [18,21] | Adaptive immunity | |
| Lysozyme | 3–110 | 32 | NA | 30 | 3–110 | [17,18] | Antimicrobial | |
| α1-Antitrypsin | 2–5 | NA | NA | NA | 2–5 | [21] | Protease inhibitor | |
| Serum albumin | 35–69 | 35 | NA | 62 | 37–69 | [18] | Transport | |
| Lactoperoxidase | 70 * | NA | NA | NA | 70 *,# | [22] | Antimicrobial | |
| Haptocorrin | 70–700 * | NA | NA | NA | 70–700 * | [3] | Vitamin B12 transport | |
| Complement C3 | 11–12 | NA | 11 | NA | 12 | [23,24] | Innate immunity | |
| Complement C4 | 5 | NA | 5 | NA | 5 | [23,24] | Innate immunity | |
| Complement factor B | 2 | NA | 2 | NA | NA | [23] | Innate immunity | |
| Casein Proteins | ||||||||
| β-casein | 4–442 | 4–364 | 18–204 | 6–414 | 5–442 | [17] | Calcium transport | |
| α-S1-casein | 4–168 | 12–58 | 15–85 | 9–110 | 4–168 | [16] | Calcium transport | |
| κ-casein | 10–172 | 25–150 | 47–134 | 10–172 | 10–134 | [16] | Calcium transport | |
| MFGM Proteins | ||||||||
| Mucin 1 | 13–294 § | NA | NA | 13–250 § | 35–294 § | [25] | Growth promoter | |
| Lactadherin | 3–33 § | NA | NA | 4–33 § | 3–13 § | [25] | Cell adhesion | |
| Butyrophilin subfamily 1 | 500–10,000 *,§ | NA | NA | 800–8200 *,§ | 500–10,000 *,§ | [25] | Regulation of immune response | |
| Bile salt-activated lipase | 10–20 | NA | NA | NA | NA | [3] | Lipid digestion | |
| Enzymes | Total protease activity | 0.76–1.38 † | 1.38 † | NA | NA | 0.76 † | [26] | |
| Thrombin | 7100 **,§ | NA | NA | NA | 7100 **,§,# | [27] | Coagulation | |
| Plasmin | 14600 **,§ | NA | NA | NA | 14,600 **,§,# | [27] | Proteolysis | |
| Elastase | 200 **,§ | NA | NA | NA | 200 **,§,# | [27] | Proteolysis | |
| Hormone peptides | Total endogenous peptides | 1–2 | NA | NA | NA | NA | [3] | |
| Ghrelin | 7–16 ** | 6–9 ** | NA | 7–10 ** | 13–16 ** | [28,29] | Appetite stimulator | |
| Leptin | 16–194 ** | 16–700 ** | NA | 20–84 ** | 165–194 ** | [28,30,31] | Energy regulator | |
| Epidermal growth factor | 4–5 ** | NA | NA | NA | 4–5 ** | [28] | Stimulates magnesium reabsorption | |
| Insulin-like growth factor-1 | 6–12 * | NA | NA | NA | 6–12 * | [28] | Insulin regulator and growth-promoting | |
| Adiponectin | 420–8790 ** | NA | NA | 661–2156 ** | 420–8790 ** | [31,32] | Glucose and fat regulator | |
| Parathyroid | 1029–5840 ‡ | 1029 ‡ | 4584 ‡ | 5840 ‡ | NA | [33] | Epidermis development | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, J.; Dingess, K.A. The Functional Power of the Human Milk Proteome. Nutrients 2019, 11, 1834. https://doi.org/10.3390/nu11081834
Zhu J, Dingess KA. The Functional Power of the Human Milk Proteome. Nutrients. 2019; 11(8):1834. https://doi.org/10.3390/nu11081834
Chicago/Turabian StyleZhu, Jing, and Kelly A. Dingess. 2019. "The Functional Power of the Human Milk Proteome" Nutrients 11, no. 8: 1834. https://doi.org/10.3390/nu11081834
APA StyleZhu, J., & Dingess, K. A. (2019). The Functional Power of the Human Milk Proteome. Nutrients, 11(8), 1834. https://doi.org/10.3390/nu11081834





