Artificial Intelligence Compared to Radiologists for the Initial Diagnosis of Prostate Cancer on Magnetic Resonance Imaging: A Systematic Review and Recommendations for Future Studies
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Search
2.2. Selection Criteria
2.3. Data Extraction
2.4. Risk of Bias Assessment
2.5. Data Synthesis
3. Results
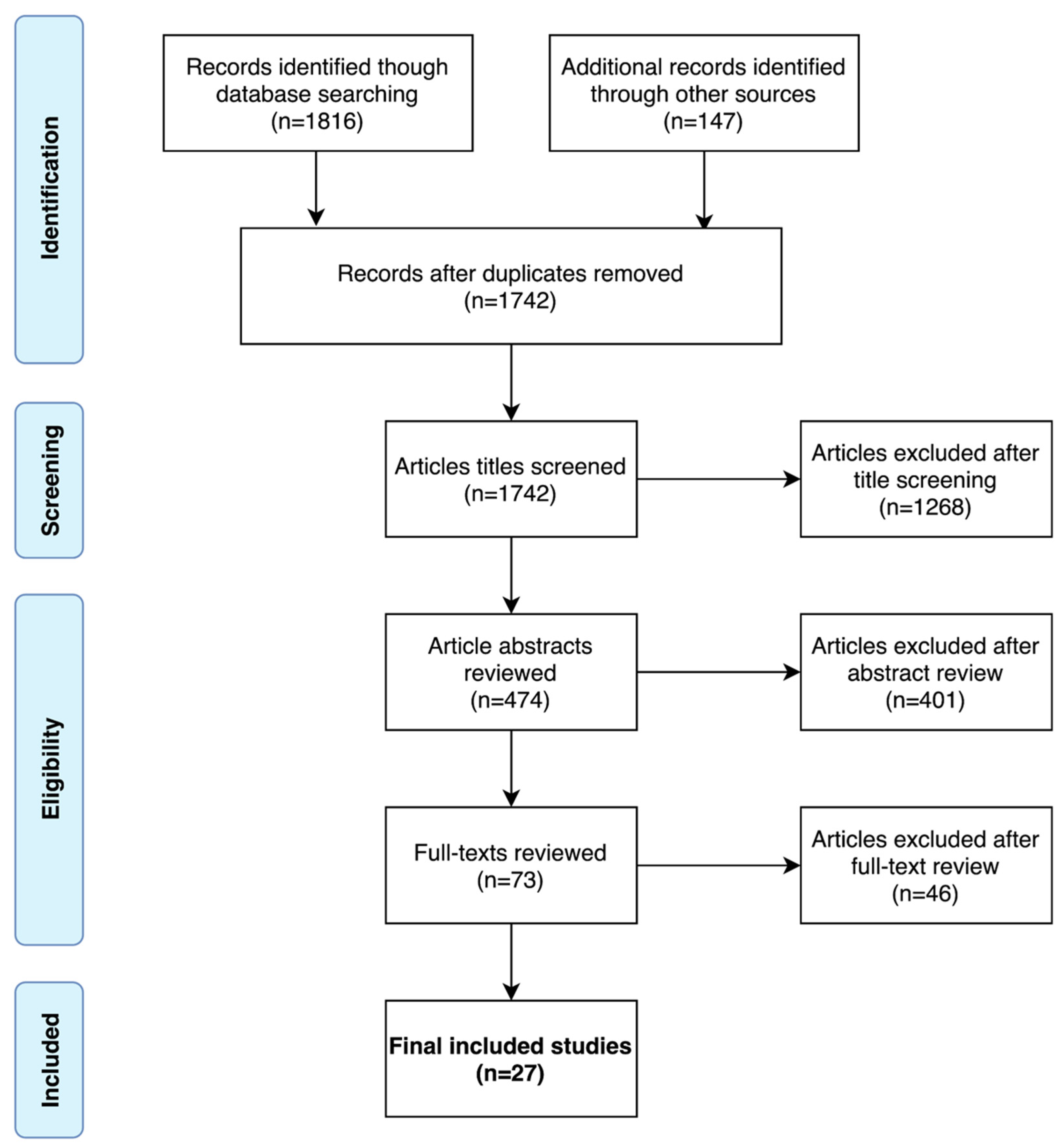
3.1. Literature Search
3.2. Patient and Study Characteristics
3.3. Reader and CAD System Characteristics
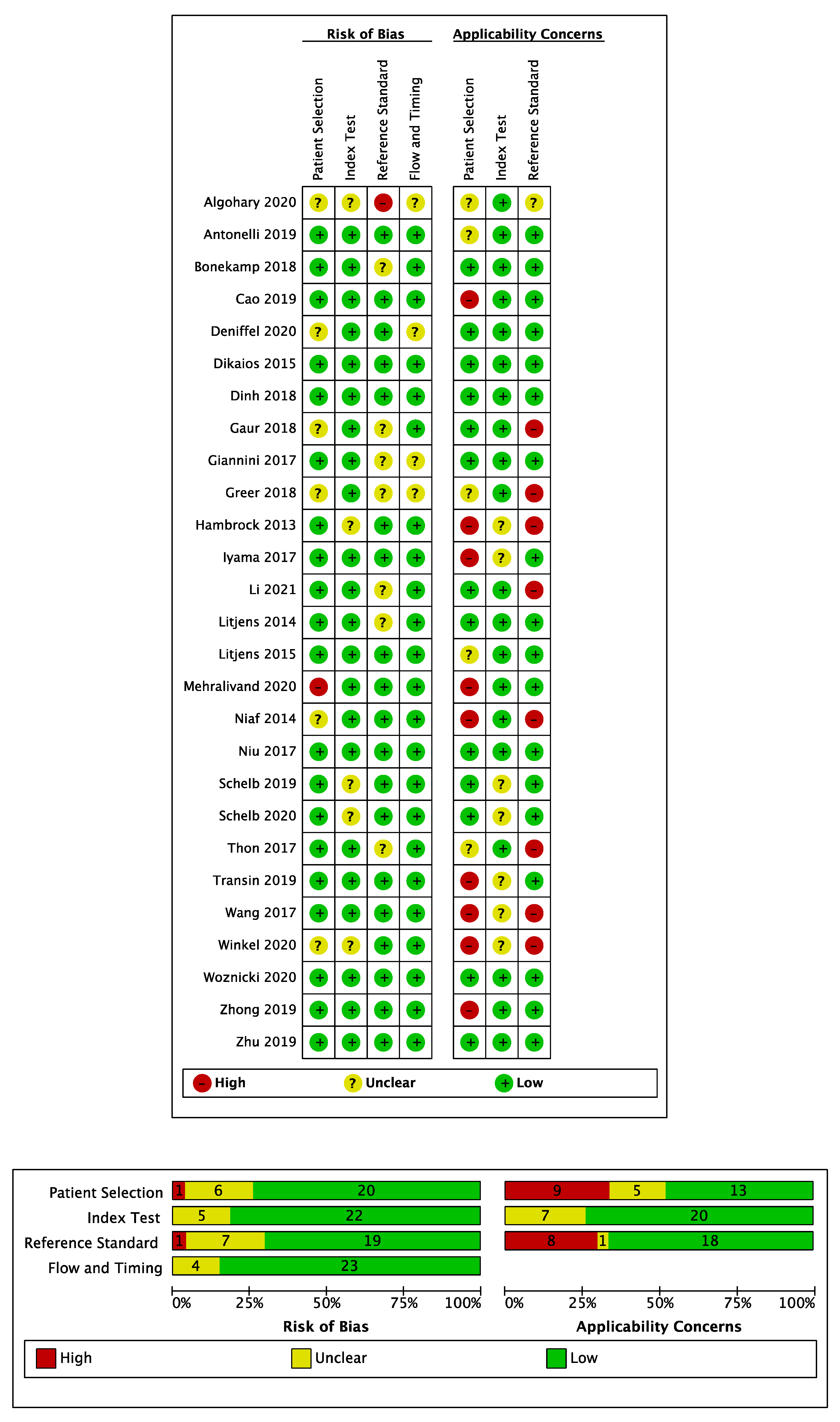
3.4. Risk of Bias Assessment
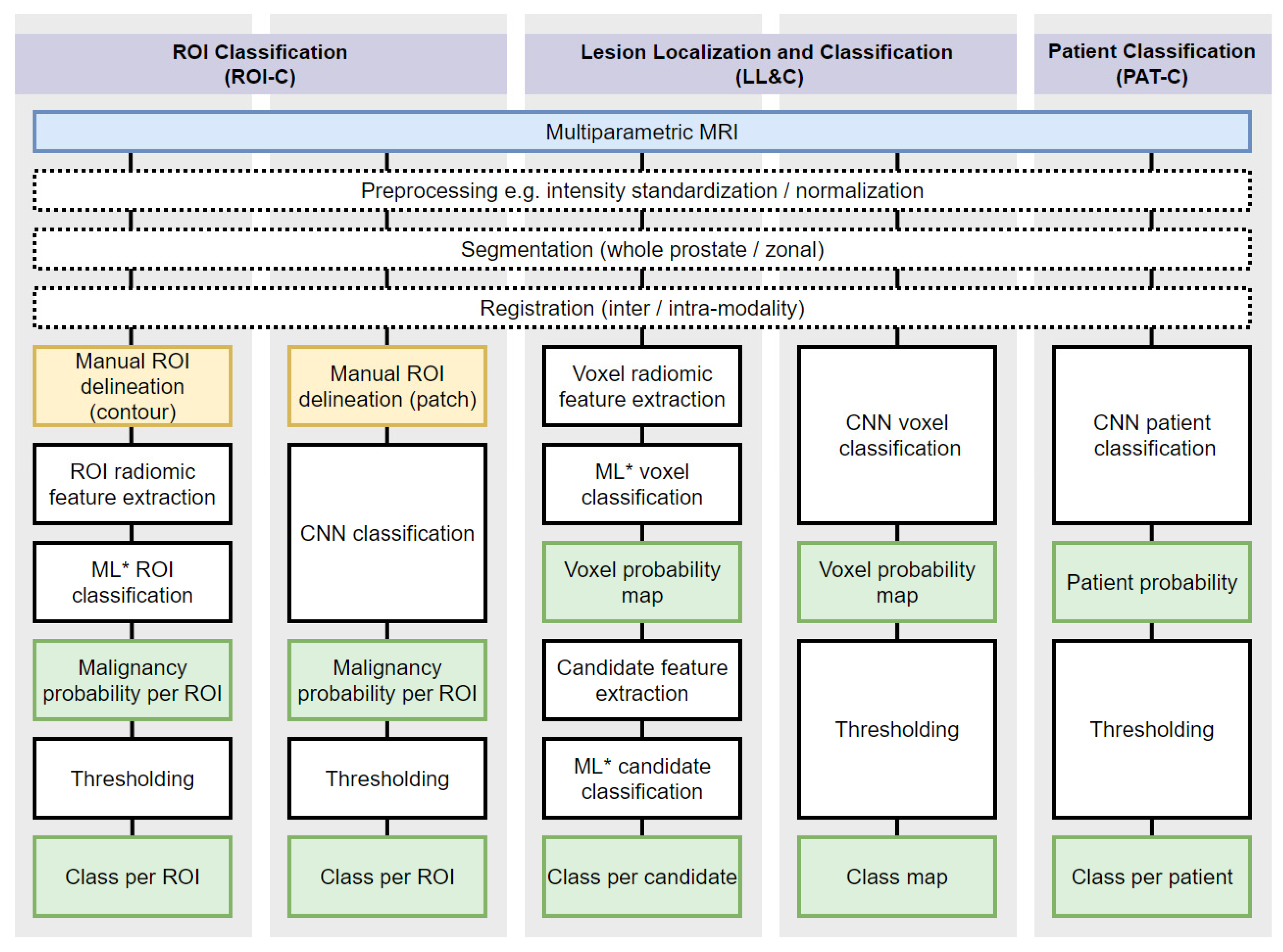
3.5. ROI Classification Summary of Results
3.6. Lesion Localization and Classification Summary of Results
3.7. Patient Classification Summary of Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Appendix A
| # | Search Term |
|---|---|
| 1 | exp Prostatic Neoplasms/ |
| 2 | Prostat * Cancer *.mp. |
| 3 | Prostat * Neoplasm *.mp. |
| 4 | Prostat * Malignanc *.mp. |
| 5 | Prostat * Tumo ?r.mp. |
| 6 | Prostat * carcinoma *.mp. |
| 7 | 1 or 2 or 3 or 4 or 5 or 6 |
| 8 | exp Magnetic Resonance Imaging/ |
| 9 | Magnetic Resonance Imaging.mp. |
| 10 | Magnetic Resonance.mp. |
| 11 | MRI.mp. |
| 12 | MR.mp. |
| 13 | 8 or 9 or 10 or 11 or 12 |
| 14 | exp Artificial Intelligence/ |
| 15 | exp Diagnosis, Computer-assisted/ |
| 16 | Artificial Intelligence.mp. |
| 17 | AI.mp. |
| 18 | Computer Assisted.mp. |
| 19 | Computer Diagnosis.mp. |
| 20 | Computer Aided Diagnosis.mp. |
| 21 | Computer Aided Detection.mp. |
| 22 | CAD *.mp. |
| 23 | Machine Learning.mp. |
| 24 | Deep Learning.mp. |
| 25 | Neural Network *.mp. |
| 26 | Convolutional Neural Network *.mp. |
| 27 | CNN.mp. |
| 28 | Support Vector Machine *.mp. |
| 29 | SVM.mp. |
| 30 | Automatic Classif *.mp. |
| 31 | 14 o 15 or 16 or 17 or 18 or 19 or 20 or 21 or 22 or 23 or 24 or 25 or 26 or 27 or 28 or 29 or 30 |
| 32 | 7 and 13 and 31 |
| 33 | 32 Limit to Human Studies |
| 34 | 33 Limit to English Language |
References
- Bjurlin, M.A.; Carroll, P.R.; Eggener, S.; Fulgham, P.F.; Margolis, D.J.; Pinto, P.A.; Rosenkrantz, A.; Rubenstein, J.N.; Rukstalis, D.B.; Taneja, S.; et al. Update of the Standard Operating Procedure on the Use of Multiparametric Magnetic Resonance Imaging for the Diagnosis, Staging and Management of Prostate Cancer. J. Urol. 2020, 203, 706–712. [Google Scholar] [CrossRef]
- Mottet, N.; Bergh, R.C.V.D.; Briers, E.; Broeck, T.V.D.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer—2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Turkbey, B.; Rosenkrantz, A.B.; Haider, M.A.; Padhani, A.R.; Villeirs, G.; Macura, K.J.; Tempany, C.M.; Choyke, P.L.; Cornud, F.; Margolis, D.J.; et al. Prostate Imaging Reporting and Data System Version 2.1: 2019 Update of Prostate Imaging Reporting and Data System Version 2. Eur. Urol. 2019, 76, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Harada, T.; Abe, T.; Kato, F.; Matsumoto, R.; Fujita, H.; Murai, S.; Miyajima, N.; Tsuchiya, K.; Maruyama, S.; Kudo, K.; et al. Five-point Likert scaling on MRI predicts clinically significant prostate carcinoma. BMC Urol. 2015, 15, 91. [Google Scholar] [CrossRef] [Green Version]
- Alabousi, M.; Salameh, J.-P.; Gusenbauer, K.; Samoilov, L.; Jafri, A.; Yu, H.; Alabousi, A. Biparametric vs multiparametric prostate magnetic resonance imaging for the detection of prostate cancer in treatment-naïve patients: A diagnostic test accuracy systematic review and meta-analysis. BJU Int. 2019, 124, 209–220. [Google Scholar] [CrossRef]
- Ahmed, H.U.; Bosaily, A.E.-S.; Brown, L.C.; Gabe, R.; Kaplan, R.S.; Parmar, M.K.; Collaco-Moraes, Y.; Ward, K.; Hindley, R.G.; Freeman, A.; et al. Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): A paired validating confirmatory study. Lancet 2017, 389, 815–822. [Google Scholar] [CrossRef] [Green Version]
- Park, K.J.; Choi, S.H.; Lee, J.S.; Kim, J.K.; Kim, M.-H.; Jeong, I.G. Risk Stratification of Prostate Cancer According to PI-RADS® Version 2 Categories: Meta-Analysis for Prospective Studies. J. Urol. 2020, 204, 1141–1149. [Google Scholar] [CrossRef]
- Brembilla, G.; Dell’Oglio, P.; Stabile, A.; Damascelli, A.; Brunetti, L.; Ravelli, S.; Cristel, G.; Schiani, E.; Venturini, E.; Grippaldi, D.; et al. Interreader variability in prostate MRI reporting using Prostate Imaging Reporting and Data System version 2.1. Eur. Radiol. 2020, 30, 3383–3392. [Google Scholar] [CrossRef]
- Sonn, G.A.; Fan, R.E.; Ghanouni, P.; Wang, N.; Brooks, J.D.; Loening, A.; Daniel, B.L.; To’O, K.J.; Thong, A.E.; Leppert, J.T. Prostate Magnetic Resonance Imaging Interpretation Varies Substantially Across Radiologists. Eur. Urol. Focus 2019, 5, 592–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuocolo, R.; Cipullo, M.B.; Stanzione, A.; Ugga, L.; Romeo, V.; Radice, L.; Brunetti, A.; Imbriaco, M. Machine learning applications in prostate cancer magnetic resonance imaging. Eur. Radiol. Exp. 2019, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- The Royal College of Radiologists. Clinical Radiology UK Workfore Census 2018 Report; The Royal College of Radiologists: London, UK, 2018. [Google Scholar]
- Wang, S.; Burtt, K.; Turkbey, B.; Choyke, P.; Summers, R.M. Computer Aided-Diagnosis of Prostate Cancer on Multiparametric MRI: A Technical Review of Current Research. BioMed Res. Int. 2014, 2014, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Salameh, J.-P.; Bossuyt, P.M.; McGrath, T.A.; Thombs, B.D.; Hyde, C.J.; Macaskill, P.; Deeks, J.J.; Leeflang, M.; Korevaar, D.A.; Whiting, P.; et al. Preferred reporting items for systematic review and meta-analysis of diagnostic test accuracy studies (PRISMA-DTA): Explanation, elaboration, and checklist. BMJ 2020, 370, 2632. [Google Scholar] [CrossRef] [PubMed]
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.G.; Sterne, J.A.C.; Bossuyt, P.M.M.; The QUADAS-2 Group. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Whiting, P.; Davenport, C.; Deeks, J.; Mallett, S.; Takwoingi, Y.; Hyde, C.; Leeflang, M. Development of QUADAS-C, a risk of bias tool for comparative diagnostic accuracy studies. OSF 2020. [Google Scholar] [CrossRef]
- Bluemke, D.A.; Moy, L.; Bredella, M.A.; Ertl-Wagner, B.B.; Fowler, K.J.; Goh, V.J.; Halpern, E.F.; Hess, C.P.; Schiebler, M.L.; Weiss, C.R. Assessing Radiology Research on Artificial Intelligence: A Brief Guide for Authors, Reviewers, and Readers—From the Radiology Editorial Board. Radiology 2020, 294, 487–489. [Google Scholar] [CrossRef] [Green Version]
- Macaskill, P.; Gatsonis, C.; Deeks, J.; Harbord, R.; Takwoingi, Y. Cochrane Handbook for Systematci Reviews of Diagnostic Test Accuracy—Chapter 10 Analysing and Presenting Results; The Cochrane Collaboration: London, UK, 2010. [Google Scholar]
- Algohary, A.; Shiradkar, R.; Pahwa, S.; Purysko, A.; Verma, S.; Moses, D.; Shnier, R.; Haynes, A.-M.; Delprado, W.; Thompson, J.; et al. Combination of Peri-Tumoral and Intra-Tumoral Radiomic Features on Bi-Parametric MRI Accurately Stratifies Prostate Cancer Risk: A Multi-Site Study. Cancers 2020, 12, 2200. [Google Scholar] [CrossRef]
- Antonelli, M.; Johnston, E.W.; Dikaios, N.; Cheung, K.K.; Sidhu, H.S.; Appayya, M.B.; Giganti, F.; Simmons, L.A.M.; Freeman, A.; Allen, C.; et al. Machine learning classifiers can predict Gleason pattern 4 prostate cancer with greater accuracy than experienced radiologists. Eur. Radiol. 2019, 29, 4754–4764. [Google Scholar] [CrossRef] [Green Version]
- Bonekamp, D.; Kohl, S.; Wiesenfarth, M.; Schelb, P.; Radtke, J.P.; Goetz, M.; Kickingereder, P.; Yaqubi, K.; Hitthaler, B.; Gählert, N.; et al. Radiomic Machine Learning for Characterization of Prostate Lesions with MRI: Comparison to ADC Values. Radiology 2018, 289, 128–137. [Google Scholar] [CrossRef]
- Dikaios, N.; Alkalbani, J.; Sidhu, H.S.; Fujiwara, T.; Abd-Alazeez, M.; Kirkham, A.; Allen, C.; Ahmed, H.; Emberton, M.; Freeman, A.; et al. Logistic regression model for diagnosis of transition zone prostate cancer on multi-parametric MRI. Eur. Radiol. 2014, 25, 523–532. [Google Scholar] [CrossRef] [Green Version]
- Dinh, A.H.; Melodelima, C.; Souchon, R.; Moldovan, P.C.; Bratan, F.; Pagnoux, G.; Mège-Lechevallier, F.; Ruffion, A.; Crouzet, S.; Colombel, M.; et al. Characterization of Prostate Cancer with Gleason Score of at Least 7 by Using Quantitative Multiparametric MR Imaging: Validation of a Computer-aided Diagnosis System in Patients Referred for Prostate Biopsy. Radiology 2018, 287, 525–533. [Google Scholar] [CrossRef]
- Hambrock, T.; Vos, P.C.; De Kaa, C.A.H.; Barentsz, J.O.; Huisman, H. Prostate Cancer: Computer-aided Diagnosis with Multiparametric 3-T MR Imaging—Effect on Observer Performance. Radiology 2013, 266, 521–530. [Google Scholar] [CrossRef] [Green Version]
- Iyama, Y.; Nakaura, T.; Katahira, K.; Iyama, A.; Nagayama, Y.; Oda, S.; Utsunomiya, D.; Yamashita, Y. Development and validation of a logistic regression model to distinguish transition zone cancers from benign prostatic hyperplasia on mul-ti-parametric prostate MRI. Eur. Radiol. 2017, 27, 3600–3608. [Google Scholar] [CrossRef]
- Niaf, E.; Lartizien, C.; Bratan, F.; Roche, L.; Rabilloud, M.; Mège-Lechevallier, F.; Rouviere, O. Prostate Focal Peripheral Zone Lesions: Characterization at Multiparametric MR Imaging—Influence of a Computer-aided Diagnosis System. Radiology 2014, 271, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.-K.; Chen, Z.-F.; Chen, L.; Li, J.; Peng, T.; Li, X. Clinical Application of Biparametric MRI Texture Analysis for Detection and Evaluation of High-Grade Prostate Cancer in Zone-Specific Regions. Am. J. Roentgenol. 2018, 210, 549–556. [Google Scholar] [CrossRef]
- Transin, S.; Souchon, R.; Gonindard, C.; de Rozario, R.; Walker, P.; de la Vega, M.F.; Loffroy, R.; Cormier, L.; Rouvière, O. Computer-aided diagnosis system for characterizing ISUP grade ≥ 2 prostate cancers at multiparametric MRI: A cross-vendor evaluation. Diagn. Interv. Imaging 2019, 100, 801–811. [Google Scholar] [CrossRef]
- Wang, J.; Wu, C.-J.; Bao, M.-L.; Zhang, J.; Wang, X.-N.; Zhang, Y.-D. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. Eur. Radiol. 2017, 27, 4082–4090. [Google Scholar] [CrossRef]
- Winkel, D.J.; Breit, H.-C.; Shi, B.; Boll, D.T.; Seifert, H.-H.; Wetterauer, C. Predicting clinically significant prostate cancer from quantitative image features including compressed sensing radial MRI of prostate perfusion using machine learning: Comparison with PI-RADS v2 assessment scores. Quant. Imaging Med. Surg. 2020, 10, 808–823. [Google Scholar] [CrossRef]
- Woźnicki, P.; Westhoff, N.; Huber, T.; Riffel, P.; Froelich, M.F.; Gresser, E.; Von Hardenberg, J.; Mühlberg, A.; Michel, M.S.; Schoenberg, S.O.; et al. Multiparametric MRI for Prostate Cancer Characterization: Combined Use of Radiomics Model with PI-RADS and Clinical Parameters. Cancers 2020, 12, 1767. [Google Scholar] [CrossRef]
- Zhong, X.; Cao, R.; Shakeri, S.; Scalzo, F.; Lee, Y.; Enzmann, D.R.; Wu, H.H.; Raman, S.S.; Sung, K. Deep transfer learn-ing-based prostate cancer classification using 3 Tesla multi-parametric MRI. Abdom. Radiol. 2019, 44, 2030–2039. [Google Scholar] [CrossRef]
- Cao, R.; Bajgiran, A.M.; Mirak, S.A.; Shakeri, S.; Zhong, X.; Enzmann, D.; Raman, S.; Sung, K. Joint Prostate Cancer Detection and Gleason Score Prediction in mp-MRI via FocalNet. IEEE Trans. Med. Imaging 2019, 38, 2496–2506. [Google Scholar] [CrossRef] [Green Version]
- Gaur, S.; Lay, N.; Harmon, S.A.; Doddakashi, S.; Mehralivand, S.; Argun, B.; Barrett, T.; Bednarova, S.; Girometti, R.; Karaarslan, E.; et al. Can computer-aided diagnosis assist in the identification of prostate cancer on prostate MRI? A mul-ti-center, multi-reader investigation. Oncotarget 2018, 9, 33804–33817. [Google Scholar] [CrossRef] [Green Version]
- Giannini, V.; Mazzetti, S.; Armando, E.; Carabalona, S.; Russo, F.; Giacobbe, A.; Muto, G.; Regge, D. Multiparametric magnetic resonance imaging of the prostate with computer-aided detection: Experienced observer performance study. Eur. Radiol. 2017, 27, 4200–4208. [Google Scholar] [CrossRef] [PubMed]
- Greer, M.D.; Lay, N.; Shih, J.H.; Barrett, T.; Bittencourt, L.K.; Borofsky, S.; Kabakus, I.; Law, Y.M.; Marko, J.; Shebel, H.; et al. Computer-aided diagnosis prior to conventional interpretation of prostate mpMRI: An international multi-reader study. Eur. Radiol. 2018, 28, 4407–4417. [Google Scholar] [CrossRef]
- Litjens, G.; Debats, O.; Barentsz, J.; Karssemeijer, N.; Huisman, H. Computer-Aided Detection of Prostate Cancer in MRI. IEEE Trans. Med. Imaging 2014, 33, 1083–1092. [Google Scholar] [CrossRef]
- Mehralivand, S.; Harmon, S.A.; Shih, J.H.; Smith, C.P.; Lay, N.; Argun, B.; Bednarova, S.; Baroni, R.H.; Canda, A.E.; Ercan, K.; et al. Multicenter Multireader Evaluation of an Artificial Intelligence–Based Attention Mapping System for the Detection of Prostate Cancer With Multiparametric MRI. Am. J. Roentgenol. 2020, 215, 903–912. [Google Scholar] [CrossRef]
- Schelb, P.; Kohl, S.; Radtke, J.P.; Wiesenfarth, M.; Kickingereder, P.; Bickelhaupt, S.; Kuder, T.A.; Stenzinger, A.; Hohenfellner, M.; Schlemmer, H.-P.; et al. Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology 2019, 293, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Schelb, P.; Wang, X.; Radtke, J.P.; Wiesenfarth, M.; Kickingereder, P.; Stenzinger, A.; Hohenfellner, M.; Schlemmer, H.-P.; Maier-Hein, K.H.; Bonekamp, D. Simulated clinical deployment of fully automatic deep learning for clinical prostate MRI assessment. Eur. Radiol. 2021, 31, 302–313. [Google Scholar] [CrossRef]
- Thon, A.; Teichgräber, U.; Tennstedt-Schenk, C.; Hadjidemetriou, S.; Winzler, S.; Malich, A.; Papageorgiou, I. Computer aided detection in prostate cancer diagnostics: A promising alternative to biopsy? A retrospective study from 104 lesions with histological ground truth. PLoS ONE 2017, 12, e0185995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.; Gao, G.; Liu, Y.; Han, C.; Liu, J.; Zhang, X.; Wang, X. Feasibility of integrating computer-aided diagnosis with structured reports of prostate multiparametric MRI. Clin. Imaging 2020, 60, 123–130. [Google Scholar] [CrossRef]
- Deniffel, D.; Abraham, N.; Namdar, K.; Dong, X.; Salinas, E.; Milot, L.; Khalvati, F.; Haider, M.A. Using decision curve analysis to benchmark performance of a magnetic resonance imaging–based deep learning model for prostate cancer risk assessment. Eur. Radiol. 2020, 30, 6867–6876. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.J.S.; Barentsz, J.O.; Karssemeijer, N.; Huisman, H.J. Clinical evaluation of a computer-aided diagnosis system for determining cancer aggressiveness in prostate MRI. Eur. Radiol. 2015, 25, 3187–3199. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Yang, L.; Yue, Y.; Xu, J.; Huang, C.; Song, B. Use of Radiomics to Improve Diagnostic Performance of PI-RADS v2.1 in Prostate Cancer. Front. Oncol. 2021, 10, 631831. [Google Scholar] [CrossRef]
- Wang, N.N.; Fan, R.E.; Leppert, J.T.; Ghanouni, P.; Kunder, C.A.; Brooks, J.D.; Chung, B.I.; Sonn, G.A. Performance of multiparametric MRI appears better when measured in patients who undergo radical prostatectomy. Res. Rep. Urol. 2018, ume 10, 233–235. [Google Scholar] [CrossRef] [Green Version]
- Miah, S.; Ahmed, H.U.; Freeman, A.; Emberton, M. Does true Gleason pattern 3 merit its cancer descriptor? Nat. Rev. Urol. 2016, 13, 541–548. [Google Scholar] [CrossRef]
- Park, S.H.; Han, K. Methodologic Guide for Evaluating Clinical Performance and Effect of Artificial Intelligence Technology for Medical Diagnosis and Prediction. Radiology 2018, 286, 800–809. [Google Scholar] [CrossRef]
- Stavrinides, V.; Syer, T.; Hu, Y.; Giganti, F.; Freeman, A.; Karapanagiotis, S.; Bott, S.R.; Brown, L.C.; Burns-Cox, N.; Dudderidge, T.J.; et al. False Positive Multiparametric Magnetic Resonance Imaging Phenotypes in the Biopsy-naïve Prostate: Are They Distinct from Significant Cancer-associated Lesions? Lessons from PROMIS. Eur. Urol. 2021, 79, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Distler, F.A.; Radtke, J.P.; Bonekamp, D.; Kesch, C.; Schlemmer, H.-P.; Wieczorek, K.; Kirchner, M.; Pahernik, S.; Hohenfellner, M.; Hadaschik, B.A. The Value of PSA Density in Combination with PI-RADS™ for the Accuracy of Prostate Cancer Prediction. J. Urol. 2017, 198, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.A.; Brittenden, J.; Lenton, J.; Lambie, H.; Goldstone, A.; Wylie, P.N.; Tolan, D.; Burling, D.; Honeyfield, L.; Bassett, P.; et al. Influence of Computer-Aided Detection False-Positives on Reader Performance and Diagnostic Confidence for CT Colonography. Am. J. Roentgenol. 2009, 192, 1682–1689. [Google Scholar] [CrossRef]
- National Cancer Imaging Translational Accelerator Novel and Multi-Parametric (mp) MRI PROSTATE Image Repository for Development of Artificial Intelligence Automated Reporting and Multi-Centre Clinical Trials. Available online: https://ncita.org.uk/exemplar-4 (accessed on 1 May 2021).
- ReIMAGINE Prostate Cancer Risk Study. Available online: https://www.reimagine-pca.org (accessed on 1 May 2021).
| Study | Year | Country | No. of Patients | Age, Years | PSA, ng/mL | Patient Population | Pre- or Post- Biopsy MRI | Field Strength, T | n° Centers/Vendors | Reference Standard |
|---|---|---|---|---|---|---|---|---|---|---|
| ROI Classification (ROI-C) | ||||||||||
| Algohary [19] | 2020 | USA | 115 | NR | NR | Biopsy proven | Pre-biopsy | 3 | 4/2 | TRUS (12-core) |
| Antonelli [20] | 2019 | UK | 134 | 64 (43–83) | 7.4 (2.5–30.3) | Clinically suspected | Pre-biopsy | 3 | 1/1 | TTMB +/− TB |
| Bonekamp [21] | 2018 | Germany | 133 | 63 (58–71) | 7.5 (5.4–11) | Clinically suspected | Pre-biopsy | 3 | 1/1 | TB |
| Dikaios [22] | 2015 | UK | 85 | 63 * (45–77) | 8.7 * (0.2–39) | Clinically suspected | Pre-biopsy | 1.5 | 1/1 | TTMB |
| Dinh [23] | 2018 | France | 129 | 67 (IQR 61–70) | 7.3 (IQR 5.1–11.5) | Clinically suspected | Pre-biopsy | 3 | 1/2 | TRUS (4-core) + TB |
| Hambrock [24] | 2013 | Netherlands | 34 | 64 * (53–74) | 7.5 * (3.4–21.8) | Biopsy proven | Post-biopsy | 3 | 1/1 | RP |
| Iyama [25] | 2017 | Japan | 60 | 70 * (55–81) | 10.4 * (5–160) | Biopsy proven | Unclear | 3 | 1/1 | RP |
| Li [45] | 2021 | China | 62 | 65 * (SD 9.4) | NR | Clinically suspected | Pre-biopsy | 3 | 1/1 | TRUS (12-core)/RP |
| Litjens [44] | 2015 | Netherlands | 107 | 66 (48–83) | 13 (1–56) | Clinically suspected ‡ | Pre-biopsy | 3 | 1/1 | TB |
| Niaf [26] | 2014 | France | 30 | 62 (45–70) | 7.4 (4.6–40) | Biopsy proven | Post-biopsy | 1.5 | 1/1 | RP |
| Niu [27] | 2017 | China | 184 | 59 (42–79) | 12.0 (4–98.3) | Clinically suspected | Pre-biopsy | 1.5 | 1/1 | TRUS (12-core) + TB |
| Transin [28] | 2019 | France | 74 | 66 (IQR, 62–69) | 7.4 (IQR 5.7–11) | Biopsy proven | Post-biopsy | 3/1.5 § | 1/1 | RP |
| Wang [29] | 2017 | China | 54 | 74 (IQR, 66–78) | 23.6 (IQR, 12.5–56.1) | Biopsy proven | Post-biopsy | 3 | 1/1 | RP |
| Winkel [30] | 2020 | Switzerland | 40 ** | 69 * (SD 8.7) † | 7 * (SD 11.2) † | Biopsy proven | Pre-biopsy | 3 | 1/1 | TRUS (12/18 core) + TB |
| Woźnicki [31] | 2020 | Germany | 40 | 69 (IQR 63–72) | 8.2 (IQR 6.8–11.9) | Clinically suspected | Pre-biopsy | 3 | 1/1 | TRUS (12-core) + TB |
| Zhong [32] | 2019 | USA | 30 | NR (43–80) † | 7.9 * (SD 12.5) † | Biopsy proven | Mix | 3 | 1/1 | RP |
| Lesion Localization and Classification (LL&C) | ||||||||||
| Cao [33] | 2019 | USA | 417 | NR | NR | Biopsy proven | Pre-biopsy | 3 | 1/1 | RP |
| Gaur [34] | 2018 | USA | 216 | 62 * (42–79) | 8.1 * (0.3–31.9) | Clinically suspected | Pre-biopsy | 3 | 5/3 | RP/TRUS (12/24-core) |
| Giannini [35] | 2017 | Italy | 89 | 67 (63–73) | 7.5 (6.2–11.0) | Clinically suspected | Pre-biopsy | 1.5 | 1/1 | TB/Saturation biopsy ^ |
| Greer [36] | 2018 | USA | 163 | 62 * (43–83) | 9.8 * (1.5–84.6) | Clinically suspected | Pre-biopsy | 3 | 1/1 | RP/TRUS (12-core) |
| Litjens [37] | 2014 | Netherlands | 347 | NR | NR | Clinically suspected | Pre-biopsy | 3 | 1/1 | TB |
| Mehralivand [38] | 2020 | Multiple | 236 | NR | NR | Clinically suspected | Pre-biopsy | 3 | 5/NR | RP/TRUS (12-core) |
| Schelb [39] | 2019 | Germany | 62 | 64 (IQR 60–69) | 6.9 (IQR 5.1–8.9) | Clinically suspected | Pre-biopsy | 3 | 1/1 | TRUS + TB |
| Schelb [40] | 2020 | Germany | 259 | 61 (IQR 61–72) | 7.2 (IQR 5.2–10.0) | Clinically suspected | Pre-biopsy | 3 | 1/1 | TB + Saturation biopsy |
| Thon [41] | 2017 | Germany | 79 | 65 * (48–80) | NR | Clinically suspected ‡ | Pre-biopsy | 3 | 1/1 | TB |
| Zhu [42] | 2019 | China | 153 | 66 (IQR 30–73) | 12.3 (IQR 7.9–21) | Clinically suspected | Pre-biopsy | 3 | 1/1 | TRUS (12/13-core) + TB |
| Patient Classification (PAT-C) | ||||||||||
| Deniffel [43] | 2020 | Canada | 50 | 64 * (SD 8.4) | 7.2 (IQR 5.2–11.2) | Clinically suspected | Pre-biopsy | 3 | 1/1 | TB +/− TRUS (12-core) |
| Study | Reader Characteristics | CAD System Characteristics | |||||
|---|---|---|---|---|---|---|---|
| MRI | No. of Readers § | Reader Experience, Years | Algorithm | Input Sequences | Discriminative Features | Evaluation Strategy | |
| ROI Classification (ROI-C) | |||||||
| Algohary [19] | bp | 1 | 7–25 | QDA | T2WI, ADC | ROI radiomic features (intra-tumoral and peri-tumoral) | Randomly split test cohort |
| Antonelli [20] | mp | 1 | 10 (>100 MRIs/year) | PZ: LinR, TZ: NB | ADC, DCE, PSAd | ROI radiomic features, PSAd | Five-fold CV |
| Bonekamp [21] | mp | 1 | >3 * | RF | T2WI, ADC, DWI (b1500) | ROI radiomic features | Temporally split test cohort |
| Dikaios [22] | mp | 1 | 7 (300–400 MRIs/year) | LogR | T2WI, ADC, DCE | ROI radiomic features | Temporally split test cohort |
| Dinh [23] | mp | 1 | 0.25–17 | GLMM | ADC, DCE | ROI radiomic features | Temporally split test cohort |
| Hambrock [24] | mp | 4 ‡ | NR (>100 MRIs) | LDA | ADC, DCE | ROI radiomic features | LOPO CV |
| Iyama [25] | mp | 2 | 7 | LogR | T2WI, ADC | ROI radiomic features | LOPO CV |
| Li [45] | mp | 2 | >5 | LogR | T2WI, ADC, DWI (b1000), DCE | ROI radiomic features, PI-RADS v2.1 score, PSA | Temporally split test cohort |
| Litjens [44] | mp | 1 | 2–20 | LogR | PDI, T2WI, ADC, DWI (b800), DCE | RF likelihood, PI-RADS v1 score | Internal independent test cohort |
| Niaf [26] | mp | 5 ‡ | 1–7 | SVM | T2WI, DWI (b600), DCE | ROI radiomic features | LOPO CV |
| Niu [27] | mp | 2 | 1–4 | LogR | T2WI, ADC | ROI radiomic features | NR |
| Transin [28] | mp | 1 | 20 | GLMM | ADC, DCE | ROI radiomic features | External test cohort |
| Wang [29] | mp | 2 | >10 | SVM | T2WI, ADC, DWI (b1500), DCE | ROI radiomic features, PI-RADS v2 score | LOPO CV |
| Winkel [30] | mp | 2 | >5 | RF | T2WI, ADC, DCE | ROI radiomic features | Randomly split test cohort |
| Woźnicki [31] | mp | 2 | >7 | Ensemble (SVM, LogR) | T2WI, ADC | ROI/WG radiomic features, PI-RADS v2 score, PSAd, DRE findings | Randomly split test cohort |
| Zhong [32] | mp | 1 | >10 (>500 MRIs/year) | CNN | T2WI, ADC | CNN learned features | Randomly split test cohort |
| Lesion Localization and Classification (LL&C) | |||||||
| Cao [33] | mp | 1 | >10 (>1000 MRIs/year) | CNN | T2WI, ADC | CNN learned features | Five-fold CV |
| Gaur [34] | mp | 2 | NR (500–2000 MRIs/year) | RF | T2WI, ADC, DWI (b1500) | Voxel radiomic features | Multicenter external test cohort |
| Giannini [35] | mp | 3 | 2–4 (120–200 MRIs/year) | SVM | T2WI, ADC, DCE | Voxel radiomic features | Internal independent test cohort |
| Greer [36] | mp | 2 | NR (<500–2000 MRIs/year) | RF | T2WI, ADC, DWI (b2000) | Voxel radiomic features | Internal independent test cohort |
| Litjens [37] | mp | 1 | 20 † | RF | PDI, T2WI, ADC, DWI (b800), DCE | Stage 1: voxel radiomic features, Stage 2: candidate radiomic features | LOPO CV |
| Mehralivand [38] | mp | 1 | <1–>3 or <100–>300 MRIs/year | RF | T2WI, ADC, DWI (b1500) | Patch-based radiomic features | Multicenter external test cohort |
| Schelb [39] | mp | 1 | >3 * | CNN | T2WI, ADC, DWI (b1500) | CNN learned features | Randomly split test cohort |
| Schelb [40] | mp | 1 | >3 * | CNN | T2WI, ADC, DWI (b1500) | CNN learned features | Temporally split test cohort |
| Thon [41] | mp | 2 | NR | Watson ElementaryTM | T2WI, ADC, DCE | Voxel radiomic features | External test cohort |
| Zhu [42] | mp | 1 | 1–2 (200 MRIs/year) | ANN | T2WI, ADC, DCE | Voxel radiomic features | Internal independent test cohort |
| Patient Classification (PAT-C) | |||||||
| Deniffel [43] | mp | 1 | 3–15 | CNN | T2WI, ADC, DWI (b1600) | CNN learned features | Randomly split test cohort |
| Study | Are All Applicable Image Sets Defined? | Is an External Test Set Used for Final Statistical Reporting? | Have Multivendor Images Been Used to Evaluate the AI Algorithm? | Are the Size of the Training, Validation and Test Sets Justified? | Was the Algorithm Trained Using a Standard of Reference That Is Widely Accepted in the Field? | Was the Preparation of Images for the AI Algorithm Adequately Described? | Were the Results of the AI Algorithm Compared with Expert Radiologists? | Was the Manner in Which the AI Algorithm Makes Decisions Demonstrated? | Is the AI Algorithm Publicly Available? |
|---|---|---|---|---|---|---|---|---|---|
| ROI Classification (ROI-C) | |||||||||
| Algohary [19] | ✗ | ✗ | ✓ | ✗ | ✗ | ✓ | ✓ | ✗ | ✗ |
| Antonelli [20] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ |
| Bonekamp [21] | ✓ | ✗ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✗ |
| Dikaios [22] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ |
| Dinh [23] | ✓ | ✗ | ✓ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ |
| Hambrock [24] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ |
| Iyama [25] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✗ | ✗ |
| Li [45] | ✓ | ✗ | ✗ | ✗ | ? | ✓ | ✓ | ✗ | ✗ |
| Litjens [44] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ? | ✗ |
| Niaf [26] | ✓ | ✗ | ✗ | ✗ | ✓ | ✗ | ✓ | ✗ | ✗ |
| Niu [27] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✗ | ✗ |
| Transin [28] | ✓ | ✓ | ✗ | ✗ | ✓ | ✗ | ✓ | ✓ | ✗ |
| Wang [29] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✗ | ✗ |
| Winkel [30] | ✗ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✗ | ✗ |
| Woźnicki [31] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✗ | ✗ |
| Zhong [32] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✗ | ✗ |
| Lesion Localization and Classification (LL&C) | |||||||||
| Cao [33] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ |
| Gaur [34] | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✗ |
| Giannini [35] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ |
| Greer [36] | ✓ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ | ✗ |
| Litjens [37] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ |
| Mehralivand [38] | ✓ | ✓ | ✓ | ✓ | ? | ? | ✓ | ✓ | ✗ |
| Schelb [39] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Schelb [40] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Thon [41] | ✓ | ✓ | ✗ | ✗ | ? | ✓ | ✓ | ✓ | ✓ * |
| Zhu [42] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ |
| Patient Classification (PAT-C) | |||||||||
| Deniffel [43] | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✓ | ✓ | ✗ |
| Study | Endpoint | Level | Zone | Readers(s) Alone | CAD System Alone | Combination | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cut-Off | SN % | SP % | AUC | Chosen Threshold | SN % | SP % | AUC | Interaction | SN % | SP % | AUC | ||||
| ROI Classification (ROI-C) | |||||||||||||||
| Algohary [19] | D’Amico ≥ Intermediate | Lesion | WP | PI-RADSv2, ≥3 | 71 (61–80) | 67 (52–80) | NR | NR | 63 (52–72) | 91 (79–98) | 0.75 | NA | NA | NA | NA |
| Antonelli [20] | GS 3 + 3 vs. 4 component | Index Lesion | PZ | Suspected GS ≥ 3 + 4 | 72 | 40 | NR | Matched to reader SP in training set | 90 | 65 | 0.83 | NA | NA | NA | NA |
| TZ | 82 | 44 | NR | 92 | 56 | 0.75 | NA | NA | NA | NA | |||||
| Bonekamp [21] | GS ≥ 3 + 4 | Lesion | WP | PI-RADSv2, ≥4 | 88 (77–95) | 50 (42–58) | NR | Matched to reader SN in training set | 97 (88–100) | 58 (50–66) | 0.88 | NA | NA | NA | NA |
| Dinh [23] | GS ≥ 3 + 4 | Lesion | WP | Likert (1–5), ≥3 | 100 (100–100) | 14 (8–19) | 0.84 (0.77–0.89) | CAD SN of 95% in training set | 96 (91–100) | 44 (36–52) | 0.88 (0.82–0.93) | NA | NA | NA | NA |
| Dikaios [22] | GS ≥ 3 + 4 or CCL ≥ 4 mm | Lesion | TZ | PI-RADSv1, ≥3 | 92 (74–99) | 37 (25–50) | 0.74 (0.63–0.86) | Probability threshold > 0.5 | 60 | 73 | 0.67 (0.55–0.79) | NA | NA | NA | NA |
| Hambrock [24] | GS ≥ 3 + 3 and >0.5 cm3 | Lesion | WP | Likelihood scale (0–100), no cut-off | NR | NR | 0.88 (0.85–0.93) | NA | NR | NR | 0.90 (0.83–0.96) | CAD scores available to radiologist for interpretation | NR | NR | 0.91 (0.86–0.97) |
| Iyama [25] | GS ≥ 3 + 4 and >10 mm vs. BPH | Lesion | TZ | PI-RADSv2, no cut-off | NR | NR | 0.87 (0.81–0.93) | NA | NR | NR | 0.97 (0.94–0.99) | NA | NA | NA | NA |
| Li [45] | GS ≥ 3 + 3 | Index Lesion | WP | PI-RADSv2.1, ≥4 | 91 | 68 | 0.85 | NR | 82 | 82 | 0.86 (0.75–0.94) | LR model of PI-RADS, CAD score and PSA | 79 | 96 | 0.94 |
| Litjens [44] | GS ≥ 3 + 4 | Lesion | WP | PI-RADSv1, ≥3 | 100 (98–100) | 9 (0–19) | 0.78 (0.70–0.85) | NR | NA | NA | NA | LR model of PI-RADS and CAD score | 99 (98–100) | 26 (0–60) | 0.87 (0.81–0.93) |
| Niaf [26] | GS ≥ 3 + 3 and >2 × 2 mm in-plane | Lesion | PZ | Likelihood score (0–4), no cut-off | NR | NR | 0.87 (0.81–0.92) | NA | NR | NR | 0.82 (0.73–0.90) | CAD scores available to radiologist for interpretation | NR | NR | 0.89 (0.83–0.94) |
| Niu [27] | GS ≥ 3 + 4 | Lesion | PZ | PI-RADSv2, ≥4 | 79 | 75 | 0.76 (0.74–0.83) | NR | 87 | 89 | 0.89 (0.82–0.94) | NA | NA | NA | NA |
| TZ | 73 | 77 | 0.73 (0.69–0.81) | NR | 88 | 81 | 0.87 (0.81–0.92) | NA | NA | NA | NA | ||||
| Transin [28] | GS ≥ 3 + 4 | Lesion | PZ | PI-RADSv2, ≥3 | 97 (93–100) | 37 (22–52) | 0.74 (0.62–0.86) | CAD SN of 95% in training set | 89 (82–97) | 42 (26–58) | 0.78 (0.69–0.87) | NA | NA | NA | NA |
| Wang [29] | GS ≥ 3 + 3 and >0.5 cm3 | Index Lesion | WP | PI-RADSv2, ≥3 | 76 (67–84) | 91 (87–94) | 0.86 (0.83–0.90) | Youden statistics on test set | 90 (84–95) | 88 (85–93) | 0.95 (0.93–0.97) | SVM model of PI-RADS and CAD score | 92 (87–96) | 95 (93–99) | 0.98 (0.95–0.99) |
| Winkel [30] | GS ≥ 3 + 4 | Lesion | PZ | PI-RADSv2, ≥3 | 100 | 53 | 0.60 | NR | 100 | 58 | 0.90 | NA | NA | NA | NA |
| Woźnicki [31] | GS ≥ 3 + 4 | Index Lesion | WP | PI-RADSv2, ≥4 | NR | NR | 0.69 (0.43–0.89) | NA | NA | NA | NA | Radiomics model ensembled with PI-RADS, PSAd and DRE models | NR | NR | 0.84 (0.60–1.00) |
| Zhong [32] | GS ≥ 3 + 4 | Lesion | WP | PI-RADSv2, ≥4 | 86 | 48 | 0.71 (0.58–0.85) | Point of best accuracy in test set | 64 | 80 | 0.73 (0.58–0.88) | NA | NA | NA | NA |
| Lesion Localization and Classification (LL&C) | |||||||||||||||
| Cao [33] | GS ≥ 3 + 4 | Lesion | WP | PI-RADSv2, ≥3 | 81 | NR | NR | FP per patient in test set matched to radiologist (0.62) | 79 | NR | 0.81 | NA | NA | NA | NA |
| Gaur [34] | GS ≥ 3 + 3 | Index Lesion | WP | PI-RADSv2, ≥3 | 78 | NR | NR | NR | NA | NA | NA | CAD identified lesions reviewed by radiologist | 68 | NR | NR |
| Giannini [35] | GS ≥ 3 + 4 | Lesion | WP | PI-RADSv2, ≥3 and max diameter ≥7 mm | 72 (61–81) | NR | NR | Voxel likelihood of malignancy ≥60% and lesion candidate ≥ 100 voxels in size | 81 (61–93) | NR | NR | CAD identified lesions reviewed by radiologist | 76 (65–85) | NR | NR |
| Greer [36] | GS ≥ 3 + 3 | Index Lesion | WP | PI-RADSv2, ≥3 | 78 (69–85) | NR | NR | NR | NA | NA | NA | CAD identified lesions reviewed by radiologist | 78 (69–86) | NR | NR |
| Mehralivand [38] | GS ≥ 3 + 4 | Lesion | WP | PI-RADSv2, ≥3 | 51 (46–57) | NR | 0.75 | NR | NA | NA | NA | CAD identified lesions reviewed by radiologist | 52 (45–61) | NR | 0.78 |
| Schelb [39] | GS ≥ 3 + 4 | Sextant | WP | Mix of PI-RADSv1/v2, ≥3 | 67 (55–78) | 68 (62–73) | NR | Point that most closely matched PI-RADS ≥ 3 performance in training set | 59 (47–70) | 66 (61–72) | NR | NA | NA | NA | NA |
| Schelb [40] | GS ≥ 3 + 4 | Sextant | WP | PI-RADSv2, ≥3 | 71 (65–76) | 62 (60–65) | NR | Iterative dynamic threshold that most closely matches PI-RADS ≥ 3 performance in most recent cases | 70 (64–75) | 66 (63–69) | NR | NA | NA | NA | NA |
| Thon [41] | GS ≥ 2 + 3 | Lesion | WP | PI-RADSv2, no cut-off | NR | NR | 0.68 (0.59–0.76) | Youden statistics on test set | 47 | 75 | 0.64 (0.53–0.75) | NA | NA | NA | NA |
| Zhu [42] | GS ≥ 3 + 4 | Lesion | WP | PI-RADSv2, ≥3 | 77 (68–84) | NR | NR | NR | NA | NA | NA | Radiologist reported with but not limited by CAD probability map | 89 (82–94) | NR | NR |
| Study | Endpoint | Zone | Reader(s) Alone | CAD System Alone | Combination | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cut-Off | SN % | SP % | AUC | Chosen Threshold | SN % | SP % | AUC | Interaction | SN % | SP % | AUC | |||
| ROI Classification (ROI-C) | ||||||||||||||
| Bonekamp [21] | GS ≥ 3 + 4 | WP | PI-RADSv2, ≥4 | 89 (76–96) | 43 (33–54) | NR | Matched to reader SN in training set | 96 (85–99) | 51 (40–62) | NR | NA | NA | NA | NA |
| Dinh [23] | GS ≥ 3 + 4 | WP | Likert (1–5), ≥3 | 100 (100–100) | 9 (2–15) | 0.88 (0.68–0.96) | CAD SN of 95% in training set | 100 (100–100) | 40 (28–51) | 0.95 (0.90–0.98) | NA | NA | NA | NA |
| Woźnicki [31] | GS ≥ 3 + 4 | WP | PI-RADSv2, ≥4 | 91 (82–98) | 28 (13–46) | NR | NR | NA | NA | NA | Radiomics model ensembled with PI-RADS, PSAd and DRE models | 91 (81–98) | 57 (38–74) | NR |
| Lesion Localization and Classification (LL&C) | ||||||||||||||
| Gaur [34] | GS ≥ 3 + 3 | WP | PI-RADSv2, ≥3 | 94 (91–96) | 45 (38–52) | 0.82 | NR | NA | NA | NA | CAD identified lesions reviewed by radiologist | 82 (75–88) | 72 (63–80) | 0.83 |
| Giannini [35] | GS ≥ 3 + 4 | WP | PI-RADSv2, ≥3 and max diameter ≥7 mm | 81 (70–90) | 75 (68–92) | NR | NR | 96 (78–100) | NR | NR | CAD identified lesions reviewed by radiologist | 91 (82–97) | 78 (71–85) | NR |
| Greer [36] | GS ≥ 3 + 3 | WP | PI-RADSv2, ≥3 | 91 (87–95) | 70 (62–79) | 0.88 (0.83–0.92) | NR | NA | NA | NA | CAD identified lesions reviewed by radiologist | 90 (85–95) | 57 (47–66) | 0.85 (0.79–0.90) |
| Litjens [37] | GS ≥ 3 + 4 * | WP | PI-RADSv1, ≥3 | ≈100 † | ≈52 † | NR | NA | NR | NR | 0.83 | NA | NA | NA | NA |
| Mehralivand [38] | GS ≥ 3 + 4 | WP | PI-RADSv2, ≥3 | 82 | NR | 0.82 | NR | NA | NA | NA | CAD identified lesions reviewed by radiologist | 84 | NR | 0.78 |
| Schelb [39] | GS ≥ 3 + 4 | WP | Mix of PI-RADSv1/v2, ≥3 | 96 (80–100) | 22 (10–39) | NR | Point that most closely matched PI-RADS ≥3 performance in training set | 96 (80–100) | 31 (16–48) | NR | NA | NA | NA | NA |
| Schelb [40] | GS ≥ 3 + 4 | WP | PI-RADSv2, ≥3 | 98 (94–100) | 17 (11–24) | NR | Iterative dynamic threshold that most closely matches PI-RADS ≥ 3 performance in most recent cases | 99 (95–100) | 24 (17–31) | NR | NA | NA | NA | NA |
| Zhu [42] | GS ≥ 3 + 4 | WP | PI-RADSv2, ≥3 | 84 (75–91) | 56 (43–69) | 0.83 (0.76–0.88) | NR | NA | NA | NA | Radiologist reported with but not limited by CAD probability map | 93 (86–98) | 66 (53–77) | 0.89 (0.83–0.94) |
| Patient Classification (PAT-C) | ||||||||||||||
| Deniffel [43] | GS ≥ 3 + 4 | WP | PI-RADSv2, ≥3 and PSAd ≥0.15 ng/mL2 | 95 (84–100) | 35 (19–52) | NR | CSPCa likelihood ≥ 0.2 | 100 (100–100) | 52 (32–68) | 0.85 (0.76–0.97) | NA | NA | NA | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syer, T.; Mehta, P.; Antonelli, M.; Mallett, S.; Atkinson, D.; Ourselin, S.; Punwani, S. Artificial Intelligence Compared to Radiologists for the Initial Diagnosis of Prostate Cancer on Magnetic Resonance Imaging: A Systematic Review and Recommendations for Future Studies. Cancers 2021, 13, 3318. https://doi.org/10.3390/cancers13133318
Syer T, Mehta P, Antonelli M, Mallett S, Atkinson D, Ourselin S, Punwani S. Artificial Intelligence Compared to Radiologists for the Initial Diagnosis of Prostate Cancer on Magnetic Resonance Imaging: A Systematic Review and Recommendations for Future Studies. Cancers. 2021; 13(13):3318. https://doi.org/10.3390/cancers13133318
Chicago/Turabian StyleSyer, Tom, Pritesh Mehta, Michela Antonelli, Sue Mallett, David Atkinson, Sébastien Ourselin, and Shonit Punwani. 2021. "Artificial Intelligence Compared to Radiologists for the Initial Diagnosis of Prostate Cancer on Magnetic Resonance Imaging: A Systematic Review and Recommendations for Future Studies" Cancers 13, no. 13: 3318. https://doi.org/10.3390/cancers13133318
APA StyleSyer, T., Mehta, P., Antonelli, M., Mallett, S., Atkinson, D., Ourselin, S., & Punwani, S. (2021). Artificial Intelligence Compared to Radiologists for the Initial Diagnosis of Prostate Cancer on Magnetic Resonance Imaging: A Systematic Review and Recommendations for Future Studies. Cancers, 13(13), 3318. https://doi.org/10.3390/cancers13133318






