DeepRePath: Identifying the Prognostic Features of Early-Stage Lung Adenocarcinoma Using Multi-Scale Pathology Images and Deep Convolutional Neural Networks
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Baseline Characteristics
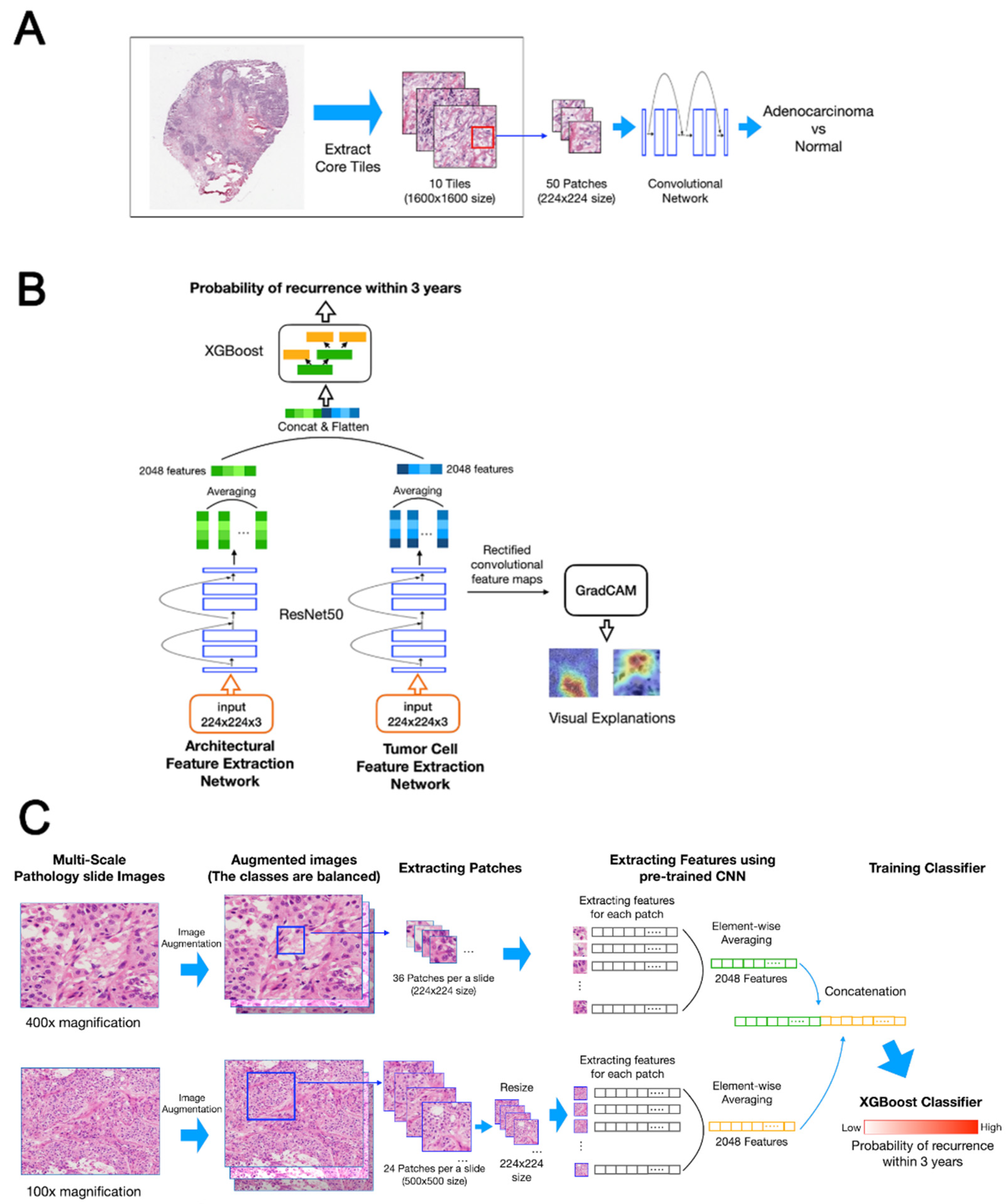
2.2. Data Preparation
2.3. Deep Learning-Based Recurrence Prediction Using Histopathology Images of LUAD in the DeepRePath Model
2.3.1. Pre-Training for Transfer Learning
2.3.2. Model Architecture
2.3.3. Visualization of the DeepRePath Model
2.4. Statistical Analysis
3. Results
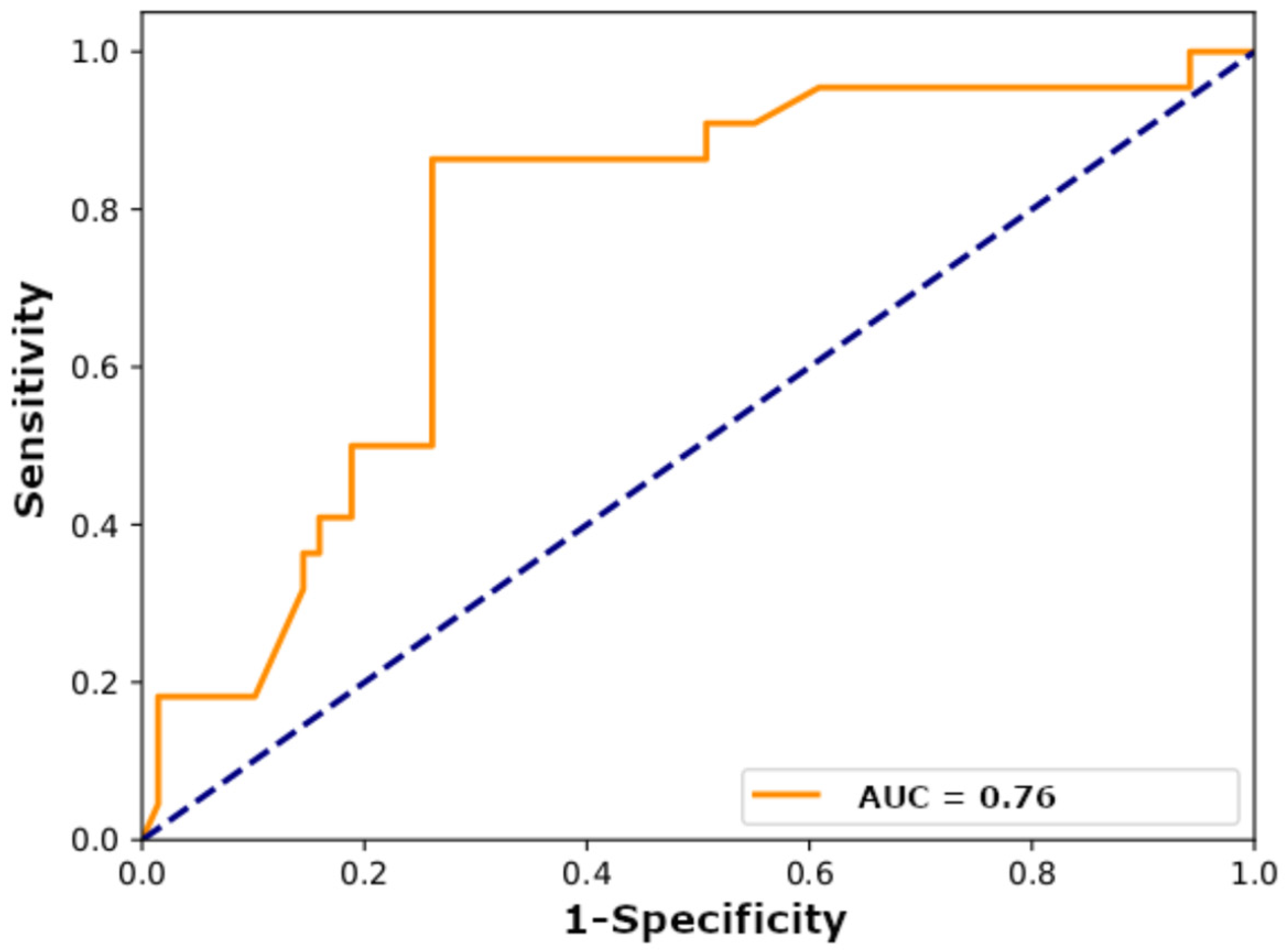
3.1. Model Performance
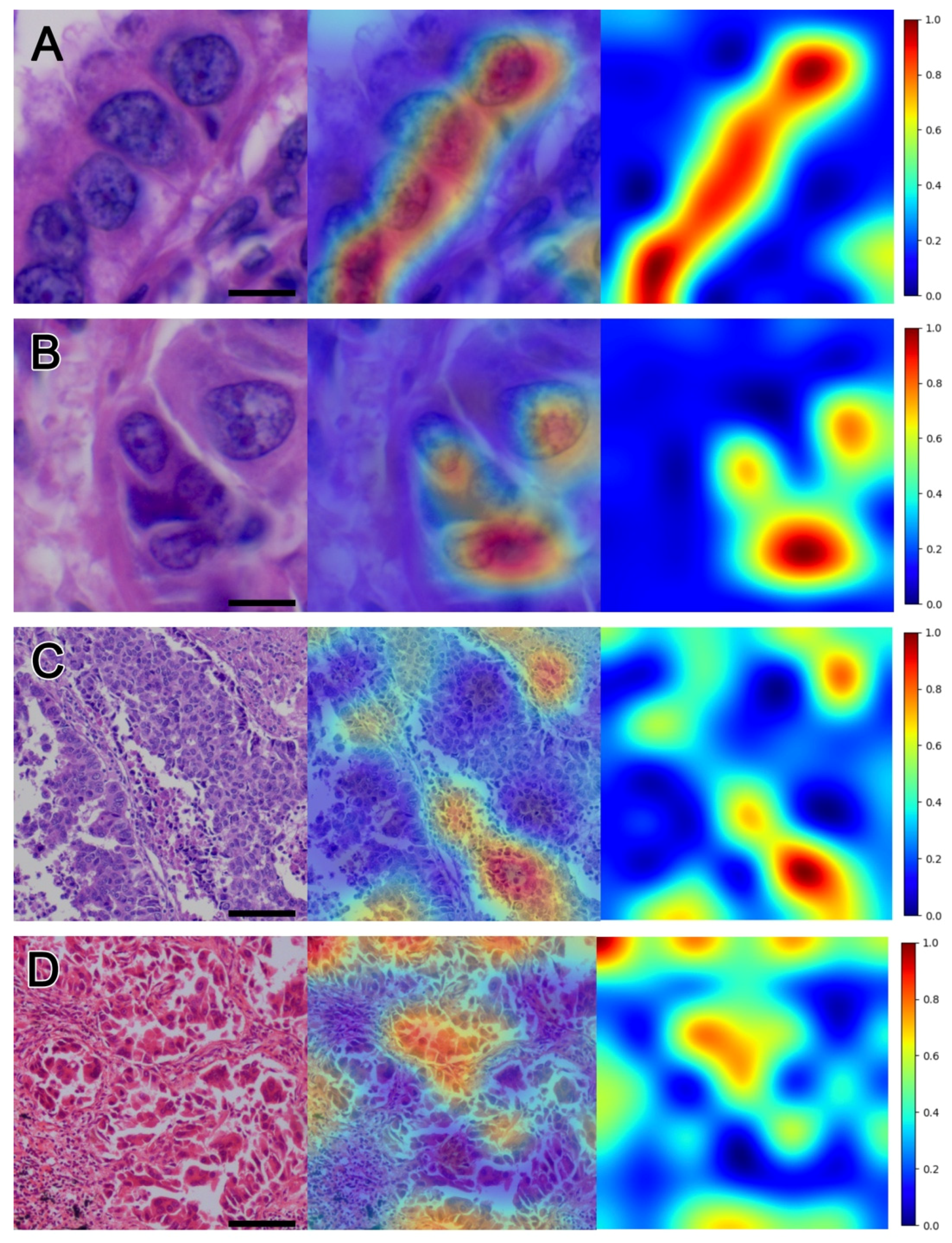
3.2. Visualization
3.3. Nuclear Morphometric Results of Hotspots and Coldspots in Heatmap Visualization
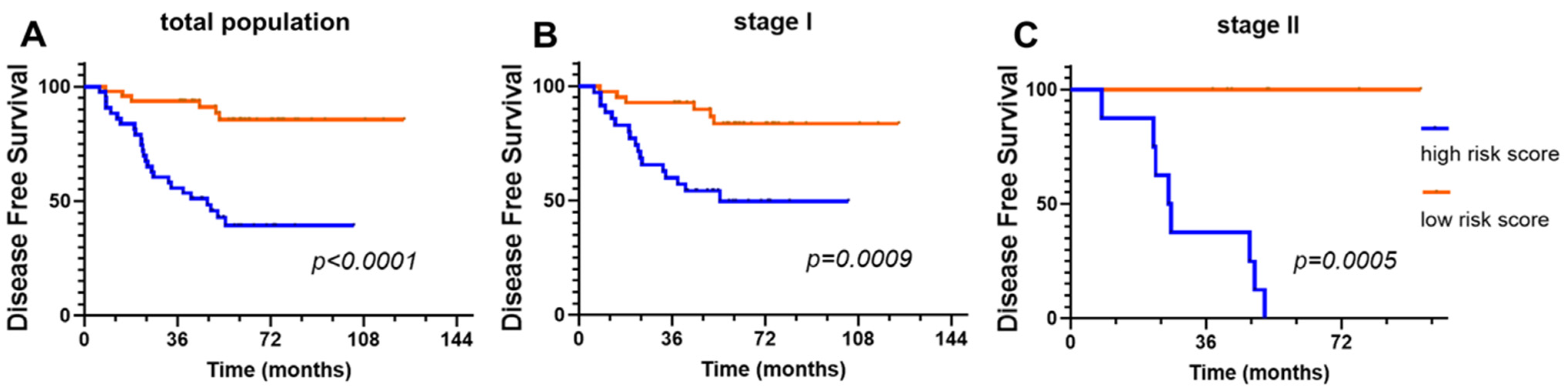
3.4. Prognostic Significance of the DeepRePath Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef]
- Goldstraw, P.; Crowley, J.; Chansky, K.; Giroux, D.J.; Groome, P.A.; Rami-Porta, R.; Postmus, P.E.; Rusch, V.; Sobin, L. The IASLC Lung Cancer Staging Project: Proposals for the Revision of the TNM Stage Groupings in the Forthcoming (Seventh) Edition of the TNM Classification of Malignant Tumours. J. Thorac. Oncol. 2007, 2, 706–714. [Google Scholar] [CrossRef] [Green Version]
- Barletta, J.A.; Yeap, B.Y.; Chirieac, L.R. Prognostic Significance of Grading in Lung Adenocarcinoma. Cancer 2009, 116, 659–669. [Google Scholar] [CrossRef] [Green Version]
- Borczuk, A.C.; Qian, F.; Kazeros, A.; Eleazar, J.; Assaad, A.; Sonett, J.R.; Ginsburg, M.; Gorenstein, L.; Powell, C.A. Invasive Size is an Independent Predictor of Survival in Pulmonary Adenocarcinoma. Am. J. Surg. Pathol. 2009, 33, 462–469. [Google Scholar] [CrossRef] [Green Version]
- Amin, M.B.; Tamboli, P.; Merchant, S.H.; Ordóñez, N.G.; Ro, J.; Ayala, A.G.; Ro, J.Y. Micropapillary Component in Lung Adenocarcinoma: A Distinctive Histologic Feature with Possible Prognostic Significance. Am. J. Surg. Pathol. 2002, 26, 358–364. [Google Scholar] [CrossRef]
- Miyoshi, T.; Satoh, Y.; Okumura, S.; Nakagawa, K.; Shirakusa, T.; Tsuchiya, E.; Ishikawa, Y. Early-Stage Lung Adenocarcinomas with a Micropapillary Pattern, a Distinct Pathologic Marker for a Significantly Poor Prognosis. Am. J. Surg. Pathol. 2003, 27, 101–109. [Google Scholar] [CrossRef]
- Russell, P.A.; Wainer, Z.; Wright, G.; Daniels, M.; Conron, M.; Williams, R.A. Does Lung Adenocarcinoma Subtype Predict Patient Survival?: A Clinicopathologic Study Based on the New International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Lung Adenocarcinoma Classification. J. Thorac. Oncol. 2011, 6, 1496–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Travis, W.D.; Brambilla, E.; Noguchi, M.; Nicholson, A.G.; Geisinger, K.R.; Yatabe, Y.; Beer, D.G.; Powell, C.A.; Riely, G.J.; Van Schil, P.E.; et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J. Thorac. Oncol. 2011, 6, 244–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sica, G.; Yoshizawa, A.; Sima, C.S.; Azzoli, C.G.; Downey, R.J.; Rusch, V.W.; Travis, W.D.; Moreira, A.L. A Grading System of Lung Adenocarcinomas Based on Histologic Pattern is Predictive of Disease Recurrence in Stage I Tumors. Am. J. Surg. Pathol. 2010, 34, 1155–1162. [Google Scholar] [CrossRef]
- Higgins, K.A.; Chino, J.P.; Ready, N.; D’Amico, T.A.; Berry, M.F.; Sporn, T.; Boyd, J.; Kelsey, C.R. Lymphovascular Invasion in Non–Small-Cell Lung Cancer: Implications for Staging and Adjuvant Therapy. J. Thorac. Oncol. 2012, 7, 1141–1147. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, Y.; Ishii, G.; Kojima, M.; Yoh, K.; Otsuka, H.; Otaki, Y.; Aokage, K.; Yanagi, S.; Nagai, K.; Nishiwaki, Y.; et al. Histopathologic Features of the Tumor Budding in Adenocarcinoma of the Lung: Tumor Budding as an Index to Predict the Potential Aggressiveness. J. Thorac. Oncol. 2010, 5, 1361–1368. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.B.; Kim, H.; Mino-Kenudson, M.; Cho, S.; Kwon, H.J.; Lee, K.R.; Kwon, S.; Lee, J.; Kim, K.; Jheon, S. Tumor spread through air spaces (STAS): Prognostic significance of grading in non-small cell lung cancer. Mod. Pathol. 2020, 34, 549–561. [Google Scholar] [CrossRef]
- Lee, M.A.; Kang, J.; Lee, H.Y.; Kim, W.; Shon, I.; Hwang, N.Y.; Kim, H.K.; Choi, Y.S.; Kim, J.; Zo, J.I.; et al. Spread Through Air Spaces (STAS) in Invasive Mucinous Adenocarcinoma of the Lung: Incidence, Prognostic Impact, and Prediction Based on Clinicoradiologic Factors. Thorac. Cancer 2020, 11, 3145–3154. [Google Scholar] [CrossRef]
- Van den Bent, M.J. Interobserver Variation of the Histopathological Diagnosis in Clinical Trials on Glioma: A Clinician’s Perspective. Acta Neuropathol. 2010, 120, 297–304. [Google Scholar] [CrossRef] [Green Version]
- Cooper, L.A.; Kong, J.; Gutman, D.A.; Dunn, W.D.; Nalisnik, M.; Brat, D.J. Novel Genotype-Phenotype Associations in Human Cancers Enabled by Advanced Molecular Platforms and Computational Analysis of Whole Slide Images. Lab. Investig. 2015, 95, 366–376. [Google Scholar] [CrossRef] [Green Version]
- Dimitriou, N.; Arandjelović, O.; Caie, P.D. Deep Learning for Whole Slide Image Analysis: An Overview. Front. Med. 2019, 6, 264. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Zang, X.; Yang, L.; Huang, J.; Liang, F.; Rodriguez-Canales, J.; Wistuba, I.I.; Gazdar, A.; Xie, Y.; Xiao, G. Comprehensive Computational Pathological Image Analysis Predicts Lung Cancer Prognosis. J. Thorac. Oncol. 2017, 12, 501–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Chen, A.; Yang, L.; Cai, L.; Xie, Y.; Fujimoto, J.; Gazdar, A.; Xiao, G. Comprehensive Analysis of Lung Cancer Pathology Images to Discover Tumor Shape and Boundary Features that Predict Survival Outcome. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Wang, T.; Yang, L.; Yang, D.M.; Fujimoto, J.; Yi, F.; Luo, X.; Yang, Y.; Yao, B.; Lin, S.; et al. ConvPath: A Software Tool for Lung Adenocarcinoma Digital Pathological Image Analysis Aided by a Convolutional Neural Network. EBioMedicine 2019, 50, 103–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Rong, R.; Yang, D.M.; Fujimoto, J.; Yan, S.; Cai, L.; Yang, L.; Luo, D.; Behrens, C.; Parra, E.R.; et al. Computational Staining of Pathology Images to Study the Tumor Microenvironment in Lung Cancer. Cancer Res. 2020, 80, 2056–2066. [Google Scholar] [CrossRef] [Green Version]
- Wulczyn, E.; Steiner, D.F.; Xu, Z.; Sadhwani, A.; Wang, H.; Flament-Auvigne, I.; Mermel, C.H.; Chen, P.-H.C.; Liu, Y.; Stumpe, M.C. Deep learning-based survival prediction for multiple cancer types using histopathology images. PLoS ONE 2020, 15, e0233678. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Janowczyk, A.; Zhou, Y.; Thawani, R.; Fu, P.; Schalper, K.; Velcheti, V.; Madabhushi, A. Prediction of Recurrence in Early Stage Non-Small Cell Lung Cancer Using Computer Extracted Nuclear Features from Digital H&E Images. Sci. Rep. 2017, 7, 13543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, K.-H.; Zhang, C.; Berry, G.J.; Altman, R.B.; Ré, C.; Rubin, D.L.; Snyder, M. Predicting Non-Small Cell Lung Cancer Prognosis by Fully Automated Microscopic Pathology Image Features. Nat. Commun. 2016, 7, 12474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, F.; Yang, L.; Wang, S.; Guo, L.; Huang, C.; Xie, Y.; Xiao, G. Microvessel Prediction in H&E Stained Pathology Images Using Fully Convolutional Neural Networks. BMC Bioinform. 2018, 19, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Wang, L.; Li, C.; Cai, Y.; Liang, Y.; Mo, X.; Lu, Q.; Dong, L.; Liu, Y. DeepLRHE: A Deep Convolutional Neural Network Framework to Evaluate the Risk of Lung Cancer Recurrence and Metastasis from Histopathology Images. Front. Genet. 2020, 11, 768. [Google Scholar] [CrossRef]
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R.; et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 2018, 23, 181–193.e7. [Google Scholar] [CrossRef] [Green Version]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Norimatsu, Y.; Irino, S.; Maeda, Y.; Yanoh, K.; Kurokawa, T.; Hirai, Y.; Kobayashi, T.K.; Fulciniti, F. Nuclear Morphometry as An Adjunct to Cytopathologic Examination of Endometrial Brushings on LBC Samples: A Prospective Approach to Combined Evaluation in Endometrial Neoplasms and Look Alikes. Cytopathology 2021, 32, 65–74. [Google Scholar] [CrossRef]
- Kashyap, A.; Jain, M.; Shukla, S.; Andley, M. Study of Nuclear Morphometry on Cytology Specimens of Benign and Malignant Breast Lesions: A study of 122 cases. J. Cytol. 2017, 34, 10–15. [Google Scholar] [CrossRef]
- Coudray, N.; Ocampo, P.S.; Sakellaropoulos, T.; Narula, N.; Snuderl, M.; Fenyö, D.; Moreira, A.L.; Razavian, N.; Tsirigos, A. Classification and Mutation Prediction from Non–Small Cell Lung Cancer Histopathology Images Using Deep Learning. Nat. Med. 2018, 24, 1559–1567. [Google Scholar] [CrossRef]
- Gertych, A.; Swiderska-Chadaj, Z.; Ma, Z.; Ing, N.; Markiewicz, T.; Cierniak, S.; Salemi, H.; Guzman, S.; Walts, A.E.; Knudsen, B.S. Convolutional Neural Networks Can Accurately Distinguish Four Histologic Growth Patterns of Lung Adenocarcinoma in Digital Slides. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Yip, S.S.F.; Sha, L.; Osinski, B.L.; Ho, I.Y.; Tan, T.L.; Willis, C.; Weiss, H.; Beaubier, N.; Mahon, B.M.; Taxter, T.J. Multi-Field-of-View Deep Learning Model Predicts Nonsmall Cell Lung Cancer Programmed Death-Ligand 1 status from whole-slide hematoxylin and eosin images. J. Pathol. Inform. 2019, 10, 24. [Google Scholar] [CrossRef]
- Wei, J.W.; Tafe, L.J.; Linnik, Y.A.; Vaickus, L.J.; Tomita, N.; Hassanpour, S. Pathologist-Level Classification of Histologic Patterns on Resected Lung Adenocarcinoma Slides with Deep Neural Networks. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Roxanis, I.; Chow, J. Cellular cohesion as a prognostic factor in malignant melanoma: A Retrospective Study with Up to 12 Years Follow-Up. Mod. Pathol. 2010, 23, 502–510. [Google Scholar] [CrossRef] [Green Version]
- Park, S.Y.; Lee, H.-S.; Jang, H.-J.; Lee, G.K.; Chung, K.Y.; Zo, J.I. Tumor Necrosis as a Prognostic Factor for Stage IA Non-Small Cell Lung Cancer. Annal. Thorac. Surg. 2011, 91, 1668–1673. [Google Scholar] [CrossRef]
- Cortés, Á.A.; Urquizu, L.C.; Cubero, J.H. Adjuvant Chemotherapy in Non-Small Cell Lung Cancer: State-of-The-Art. Transl. Lung Cancer Res. 2015, 4, 191–197. [Google Scholar]
- Uramoto, H.; Tanaka, F. Recurrence After Surgery in Patients with NSCLC. Transl. Lung Cancer Res. 2014, 3, 242–249. [Google Scholar] [PubMed]
- Zarella, M.D.; Bowman, D.; Aeffner, F.; Farahani, N.; Xthona, A.; Absar, S.F.; Parwani, A.; Bui, M.; Hartman, D.J. A Practical Guide to Whole Slide Imaging: A White Paper from the Digital Pathology Association. Arch. Pathol. Lab. Med. 2019, 143, 222–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
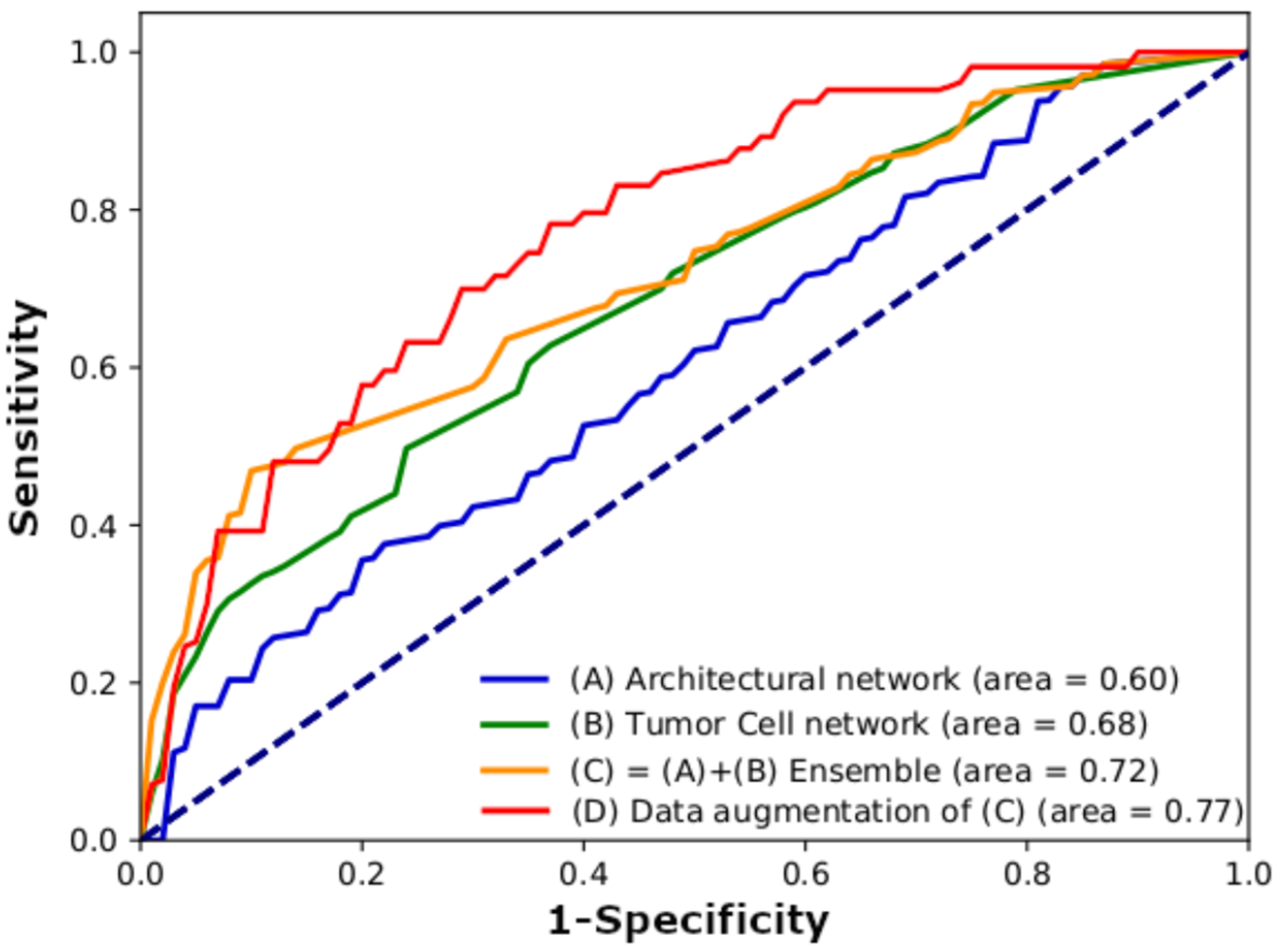
| Input Images | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | AUC |
|---|---|---|---|---|---|---|
| Architectural network (100× magnification) | 65 | 59 | 47 | 86 | 62 | 0.6 |
| Tumor cell network (400× magnification) | 52 | 78 | 56 | 84 | 71 | 0.68 |
| Architectural + tumor cell ensemble | 46 | 94 | 78 | 83 | 82 | 0.72 |
| Architectural + tumor cell ensemble (data augmentation) | 74 | 78 | 59 | 89 | 77 | 0.77 |
| Input Images | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | AUC |
|---|---|---|---|---|---|---|
| Architectural network (100× magnification) | 91 | 14 | 25 | 83 | 33 | 0.43 |
| Tumor cell network (400× magnification) | 73 | 62 | 38 | 88 | 65 | 0.65 |
| Architectural+tumor cell ensemble | 59 | 87 | 59 | 87 | 80 | 0.75 |
| Architectural+tumor cell ensemble (data augmentation) | 86 | 74 | 51 | 94 | 77 | 0.76 |
| Cohort | Model | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | AUC |
|---|---|---|---|---|---|---|---|
| I | DeepRePath with TL | 46 | 94 | 78 | 83 | 82 | 0.72 |
| DeepRePath without TL | 80 | 89 | 75 | 93 | 87 | 0.87 | |
| II | DeepRePath with TL | 59 | 87 | 59 | 87 | 80 | 0.75 |
| DeepRePath without TL | 64 | 62 | 35 | 84 | 63 | 0.58 |
| Histopathology | Hotspot (107 Nuclei) | Coldspot (107 Nuclei) | p |
|---|---|---|---|
| Area (μm2) | 16.83 ± 8.39 | 12.97 ± 5.15 | <0.001 |
| Primary axis (μm) | 5.59 ± 1.38 | 4.89 ± 1.06 | <0.001 |
| Secondary axis (μm) | 3.68 ± 1.04 | 3.29 ± 0.72 | 0.001 |
| Maximum Feret (μm) | 5.81 ± 1.43 | 5.09 ± 1.04 | <0.001 |
| Minimum Feret (μm) | 3.83 ± 1.05 | 3.39 ± 0.73 | <0.001 |
| Perimeter (μm) | 15.69 ± 3.73 | 13.66 ± 2.70 | <0.001 |
| Shape factor * | 0.821 ± 0.098 | 0.846 ± 0.070 | 0.036 |
| Roughness † | 0.942 ± 0.037 | 0.949 ± 0.020 | 0.101 |
| Aspect ratio ‡ | 1.584 ± 0.446 | 1.519 ± 0.326 | 0.221 |
| Roundness § | 0.672 ± 0.155 | 0.685 ± 0.128 | 0.526 |
| Effect | Univariate | p | Multivariate | p |
|---|---|---|---|---|
| Hazard Ratio (95% CI) | Hazard Ratio (95% CI) | |||
| Gender (female vs. male) | 0.650 (0.306–1.381) | 0.262 | ||
| Age (≥60 vs. <60) | 1.360 (0.640–2.889) | 0.424 | ||
| ECOG (≥1 vs. 0) | 3.285 (1.604–6.727) | 0.001 | 2.631 (1.267–5.467) | 0.009 |
| Tumor grade (moderate to poor vs. well) | 2.483 (1.110–5.555) | 0.027 | 1.296 (0.563–2.987) | 0.542 |
| Tumor size (≥2.4 vs. <2.4 cm) | 1.938 (0.925–4.062) | 0.080 | 2.065 (0.968–4.405) | 0.061 |
| LVI (yes vs. no) | 2.332 (1.064–5.113) | 0.035 | 1.681 (0.739–3.826) | 0.215 |
| pT stage (≥T3 vs. T1–2) | 1.490 (0.733–3.030) | 0.271 | ||
| pN stage (≥N1 vs. N0) | 1.763 (0.673–4.621) | 0.249 | ||
| High vs. low score for recurrence * | 6.358 (2.599–15.554) | <0.001 | 5.564 (2.245–13.789) | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shim, W.S.; Yim, K.; Kim, T.-J.; Sung, Y.E.; Lee, G.; Hong, J.H.; Chun, S.H.; Kim, S.; An, H.J.; Na, S.J.; et al. DeepRePath: Identifying the Prognostic Features of Early-Stage Lung Adenocarcinoma Using Multi-Scale Pathology Images and Deep Convolutional Neural Networks. Cancers 2021, 13, 3308. https://doi.org/10.3390/cancers13133308
Shim WS, Yim K, Kim T-J, Sung YE, Lee G, Hong JH, Chun SH, Kim S, An HJ, Na SJ, et al. DeepRePath: Identifying the Prognostic Features of Early-Stage Lung Adenocarcinoma Using Multi-Scale Pathology Images and Deep Convolutional Neural Networks. Cancers. 2021; 13(13):3308. https://doi.org/10.3390/cancers13133308
Chicago/Turabian StyleShim, Won Sang, Kwangil Yim, Tae-Jung Kim, Yeoun Eun Sung, Gyeongyun Lee, Ji Hyung Hong, Sang Hoon Chun, Seoree Kim, Ho Jung An, Sae Jung Na, and et al. 2021. "DeepRePath: Identifying the Prognostic Features of Early-Stage Lung Adenocarcinoma Using Multi-Scale Pathology Images and Deep Convolutional Neural Networks" Cancers 13, no. 13: 3308. https://doi.org/10.3390/cancers13133308
APA StyleShim, W. S., Yim, K., Kim, T.-J., Sung, Y. E., Lee, G., Hong, J. H., Chun, S. H., Kim, S., An, H. J., Na, S. J., Kim, J. J., Moon, M. H., Moon, S. W., Park, S., Hong, S. A., & Ko, Y. H. (2021). DeepRePath: Identifying the Prognostic Features of Early-Stage Lung Adenocarcinoma Using Multi-Scale Pathology Images and Deep Convolutional Neural Networks. Cancers, 13(13), 3308. https://doi.org/10.3390/cancers13133308








