Symptomatology, (Co)occurrence and Differential Diagnostic PCR Identification of ‘Ca. Phytoplasma solani’ and ‘Ca. Phytoplasma convolvuli’ in Field Bindweed
Abstract
:1. Introduction
2. Results
2.1. Occurrence of ‘Ca. P. solani’ and ‘Ca. P. convolvuli’ in Symptomatic Field Bindweeds
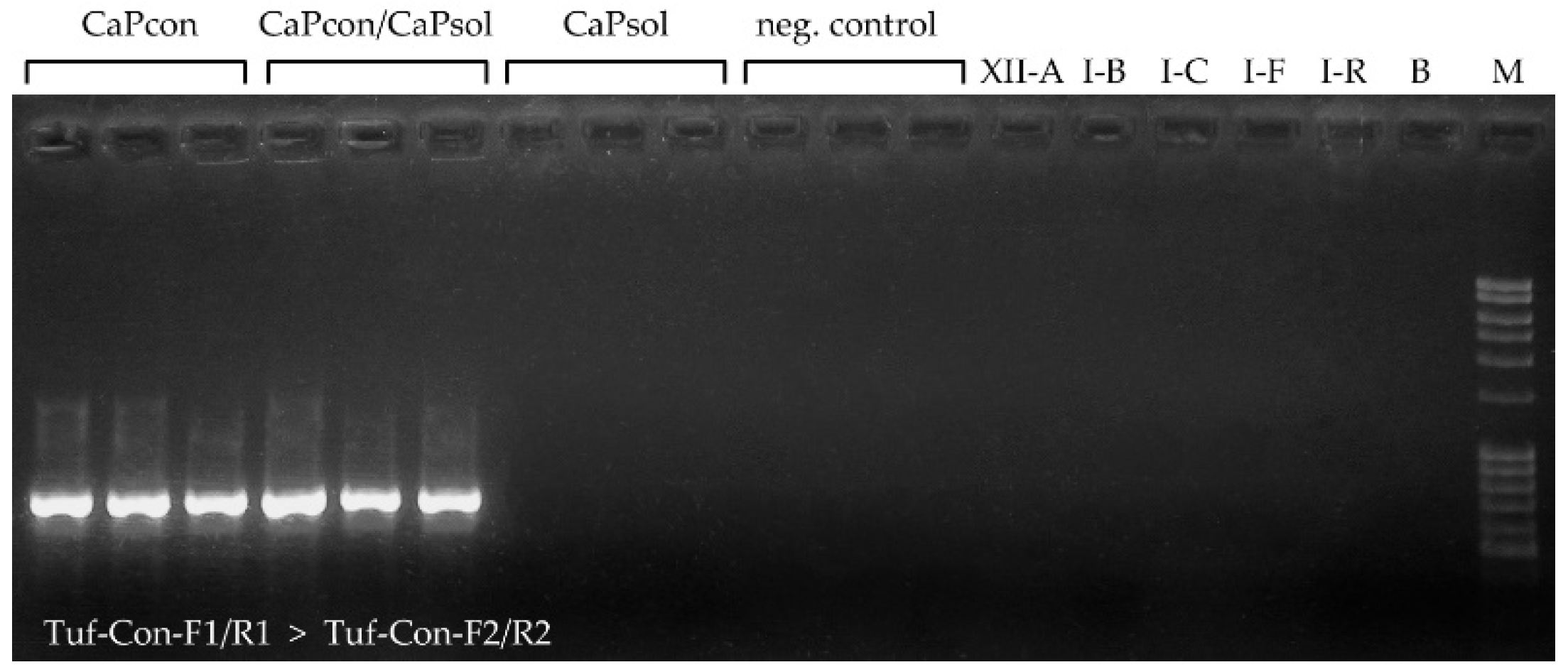
2.2. Application of Novel ‘Ca. P. convolvuli’-Specific Nested PCR Diagnostic Protocol
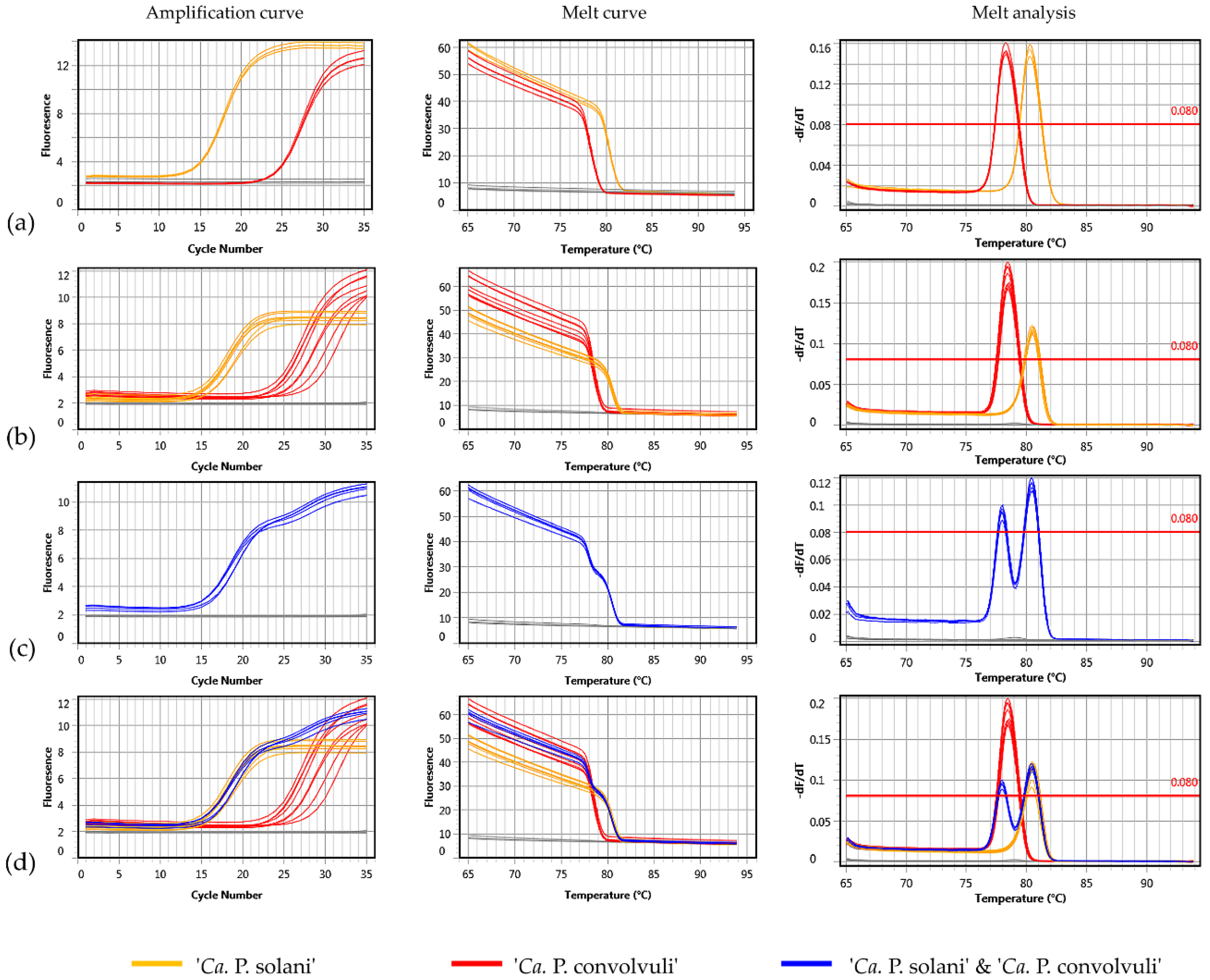
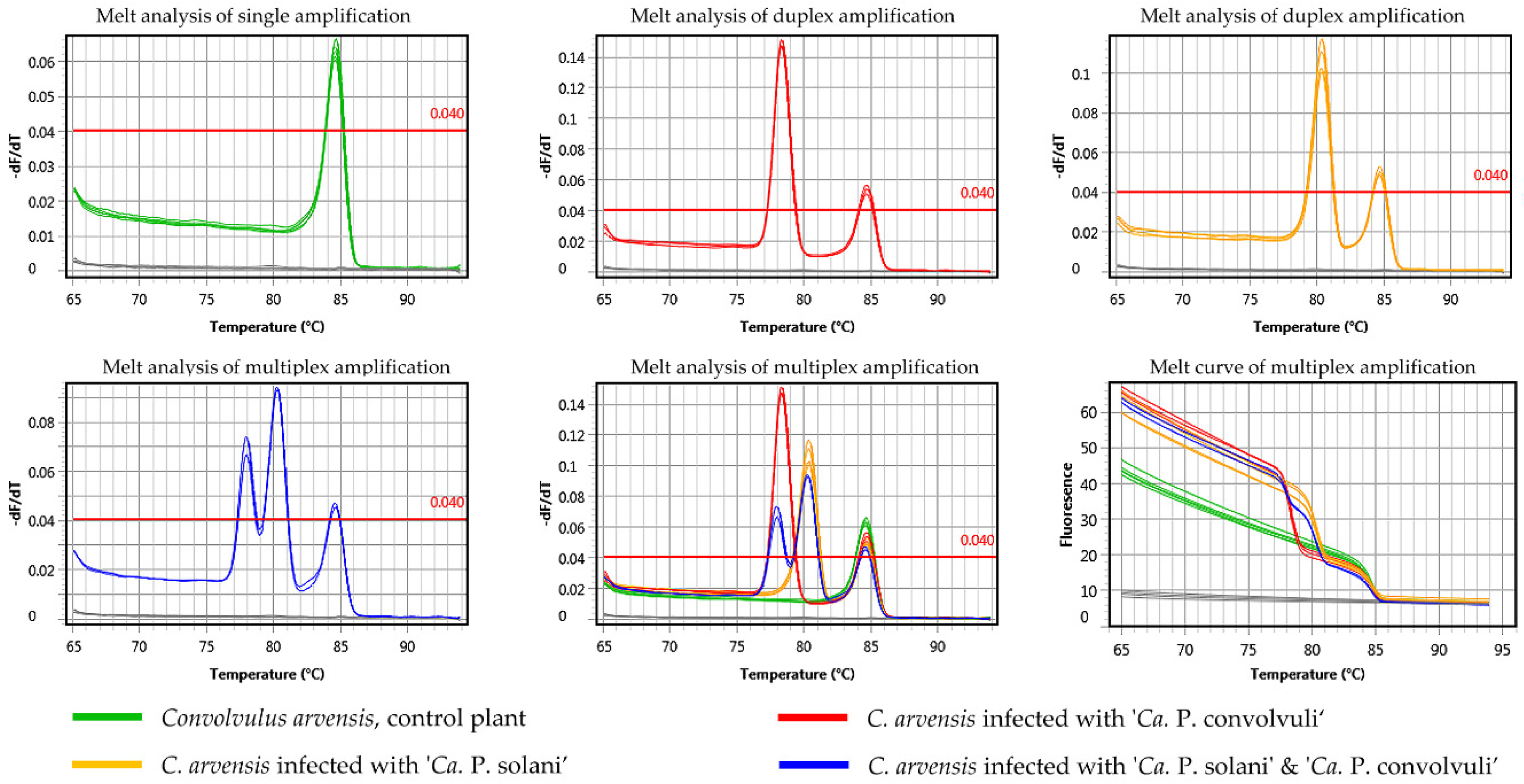
2.3. Application of Novel Duplex SYBR Green-Based Real-Time PCR for Simultaneous Detection and Differentiation of ‘Ca. P. solani’ and ‘Ca. P. convolvuli’
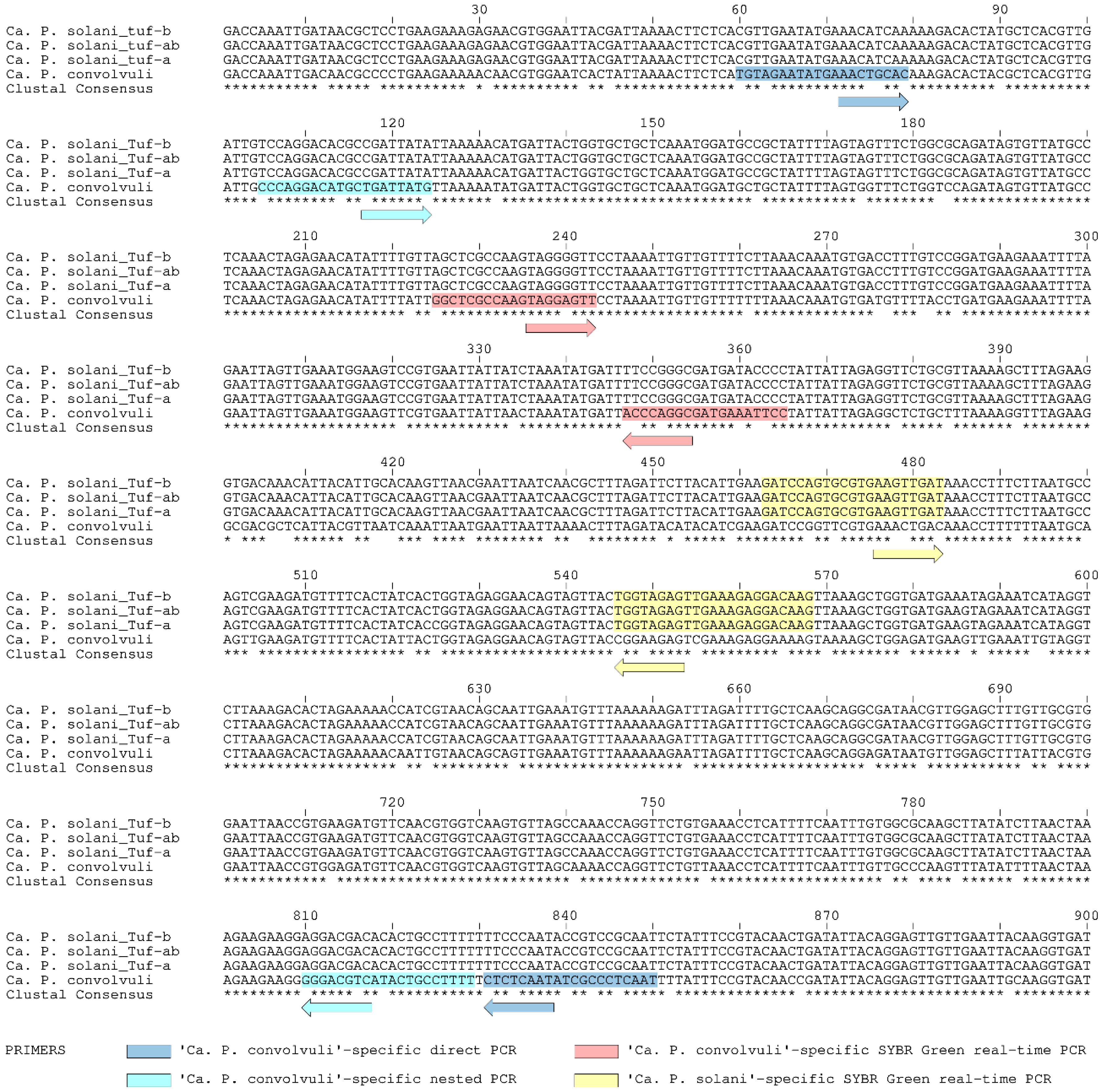
2.4. ‘Ca. P. convolvuli’ and ‘Ca. P. solani’ Genotype Strains
3. Discussion
4. Materials and Methods
4.1. Plant Sampling
4.2. DNA Extraction, Initial Phytoplasma Identification and Sequencing
4.3. Molecular Typing of ‘Ca. P. convolvuli’ Strains
4.4. Molecular Typing of ‘Ca. P. solani’ Strains
4.5. Design of ‘Ca. P. convolvuli’-Specific Nested PCR
4.6. Design of Duplex SYBR Green-Based Real-Time PCR for Simultaneous Detection of ‘Ca. P. solani’ and ‘Ca. P. convolvuli’
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cincović, T. Convolvulus, L. In Flora SR Srbije, 1st ed.; Josifović, M., Ed.; Srpska Akademija Nauka i Umetnosti: Belgrade, Serbia, 1973; Volume 5, pp. 586–589. [Google Scholar]
- Maixner, M. Transmission of German grapevine yellows (Vergilbungskrankheit) by the planthopper Hyalesthes obsoletus (Auchenorrhyncha: Cixiidae). Vitis 1994, 33, 103–104. [Google Scholar]
- Sforza, R.; Clair, D.; Daire, X.; Larrue, J.; Boudon-Padieu, E. The role of Hyalesthes obsoletus (Hemiptera: Cixiidae) in the occurrence of bois noir of grapevines in France. J. Phytopathol. 1998, 146, 549–556. [Google Scholar] [CrossRef]
- Johannesen, J.; Lux, B.; Michel, K.; Seitz, A.; Maixner, M. Invasion biology and host specificity of the grapevine yellows disease vector Hyalesthes obsoletus in Europe. Entomol. Exp. Appl. 2008, 126, 217–227. [Google Scholar] [CrossRef]
- Imo, M.; Maixner, M.; Johannesen, J. Sympatric diversification vs. immigration: Deciphering host-plant specialization in a polyphagous insect, the stolbur phytoplasma vector Hyalesthes obsoletus (Cixiidae). Mol. Ecol. 2013, 22, 2188–2203. [Google Scholar] [CrossRef] [PubMed]
- Kosovac, A.; Radonjić, S.; Hrnčić, S.; Krstić, O.; Toševski, I.; Jović, J. Molecular tracing of the transmission routes of bois noir in Mediterranean vineyards of Montenegro and experimental evidence for the epidemiological role of Vitexa gnus-castus (Lamiaceae) and associated Hyalesthes obsoletus (Cixiidae). Plant Pathol. 2016, 65, 285–298. [Google Scholar] [CrossRef] [Green Version]
- Kosovac, A.; Johannesen, J.; Krstić, O.; Mitrović, M.; Cvrković, T.; Toševski, I.; Jović, J. Widespread plant specialization in the polyphagous planthopper Hyalesthes obsoletus (Cixiidae), a major vector of stolbur phytoplasma: Evidence of cryptic speciation. PLoS ONE 2018, 13, e0196969. [Google Scholar] [CrossRef]
- Quaglino, F.; Zhao, Y.; Casati, P.; Bulgari, D.; Bianco, P.A.; Wei, W.; Davis, R.E. ‘Candidatus Phytoplasma solani’, a noveltaxonassociatedwithstolbur- and boisnoir-relateddiseasesof plants. Int. J. Syst. Evol. Microbiol. 2013, 63, 2879–2894. [Google Scholar] [CrossRef] [Green Version]
- EFSA Panel on Plant Health (PLH). Scientific Opinion on the pest categorisation of Candidatus Phytoplasma solani. EFSA J. 2014, 12, 3924–3927. [Google Scholar]
- Maixner, M.; Ahrens, U.; Seemüller, E. Detection of the German grapevine yellows (Vergilbungskrankheit) MLO in grapevine, alternative hosts and a vector by a specific PCR procedure. Eur. J. Plant Pathol. 1995, 101, 241–250. [Google Scholar] [CrossRef]
- Carraro, L.; Ferrini, F.; Martini, M.; Ermacora, P.; Loi, N. A serious epidemic of stolbur on celery. J. Plant Pathol. 2008, 90, 131–135. [Google Scholar]
- Fialová, R.; Válová, P.; Balakishiyeva, G.; Danet, J.L.; Šafárová, D.; Foissac, X.; Navrátil, M. Genetic variability of stolbur phytoplasma in annual crop and wild plant species in south Moravia. J. Plant Pathol. 2009, 91, 411–416. [Google Scholar]
- Navrátil, M.; Válová, P.; Fialová, R.; Lauterer, P.; Šafářová, D.; Starý, M. The incidence of stolbur disease and associated yield losses in vegetable crops in South Moravia (Czech Republic). Crop Prot. 2009, 28, 898–904. [Google Scholar] [CrossRef]
- Murolo, S.; Marcone, C.; Prota, V.; Garau, R.; Foissac, X.; Romanazzi, G. Genetic variability of the stolbur phytoplasma vmp1 gene in grapevines, bindweeds and vegetables. J. Appl. Microbiol. 2010, 109, 2049–2059. [Google Scholar] [CrossRef] [PubMed]
- Ember, I.; Acs, Z.; Munyaneza, J.E.; Crosslin, J.M.; Kolber, M. Survey and molecular detection of phytoplasmas associated with potato in Romania and southern Russia. Eur. J. Plant Pathol. 2011, 130, 367–377. [Google Scholar] [CrossRef]
- Maixner, M.; Albert, A.; Johannesen, J. Survival relative to new and ancestral host plants, phytoplasma infection, and genetic constitution in host races of a polyphagous insect disease vector. Ecol. Evol. 2014, 4, 3082–3092. [Google Scholar] [CrossRef]
- Quaglino, F.; Maghradze, D.; Casati, P.; Chkhaidze, N.; Lobjanidze, M.; Ravasio, A.; Passera, A.; Venturini, G.; Failla, O.; Bianco, P.A. Identification and characterization of new ‘Candidatus Phytoplasma solani’ strains associated with bois noir disease in Vitis vinifera L. cultivars showing a range of symptom severity in Georgia, the Caucasus region. Plant Dis. 2016, 100, 904–915. [Google Scholar] [CrossRef] [Green Version]
- Jamshidi, E.; Murolo, S.; Ravari, S.B.; Salehi, M.; Romanazzi, G. Molecular Typing of ‘Candidatus Phytoplasma solani’ in Iranian Vineyards. Plant Dis. 2019, 103, 2412–2416. [Google Scholar] [CrossRef]
- Aryan, A.; Brader, G.; Mörtel, J.; Pastar, M.; Riedle-Bauer, M. An abundant ‘Candidatus Phytoplasma solani’ tuf b strain is associated with grapevine, stinging nettle and Hyalesthes obsoletus. Eur. J. Plant Pathol. 2014, 140, 213–227. [Google Scholar] [CrossRef] [Green Version]
- Atanasova, B.; Jakovljević, M.; Spasov, D.; Jović, J.; Mitrović, M.; Toševski, I.; Cvrković, T. The molecular epidemiology of bois noir grapevine yellows caused by ‘Candidatus Phytoplasma solani’ in the Republic of Macedonia. Eur. J. Plant Pathol. 2015, 142, 759–770. [Google Scholar] [CrossRef] [Green Version]
- Mori, N.; Quaglino, F.; Tessari, F.; Pozzebon, A.; Bulgari, D.; Casati, P.; Bianco, P.A. Investigation on ‘bois noir’ epidemiology in north-eastern Italian vineyards through a multidisciplinary approach. Ann. Appl. Biol. 2015, 166, 75–89. [Google Scholar] [CrossRef]
- Landi, L.; Riolo, P.; Murolo, S.; Romanazzi, G.; Nardi, S.; Isidoro, N. Genetic variability of stolbur phytoplasma in Hyalesthes obsoletus (Hemiptera: Cixiidae) and its main host plants in vineyard agroecosystems. J. Econ. Entomol. 2015, 108, 1506–1515. [Google Scholar] [CrossRef] [PubMed]
- Kosovac, A.; Jakovljević, M.; Krstić, O.; Cvrković, T.; Mitrović, M.; Toševski, I.; Jović, J. Role of plant-specialized Hyalesthes obsoletus associated with Convolvulus arvensis and Crepis foetida in the transmission of ‘Candidatus Phytoplasma solani’-inflicted bois noir disease of grapevine in Serbia. Eur. J. Plant Pathol. 2019, 153, 183–195. [Google Scholar] [CrossRef]
- Pierro, R.; Panattoni, A.; Passera, A.; Materazzi, A.; Luvisi, A.; Loni, A.; Ginanni, M.; Lucchi, A.; Bianco, P.A.; Quaglino, F. Proposalof A New Bois Noir Epidemiological Pattern Related to ‘Candidatus Phytoplasma Solani’ Strains Characterized by A Possible Moderate Virulence in Tuscany. Pathogens 2020, 9, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conigliaro, G.; Jamshidi, E.; Lo Verde, G.; Bella, P.; Mondello, V.; Giambra, S.; D’Urso, V.; Tsolakis, H.; Murolo, S.; Burruano, S.; et al. Epidemiological Investigations and Molecular Characterization of ‘Candidatus Phytoplasma solani’ in Grapevines, Weeds, Vectors and Putative Vectors in Western Sicily, (Southern Italy). Pathogens 2020, 9, 918. [Google Scholar] [CrossRef] [PubMed]
- Marcone, C.; Ragozzino, A.; Seemüller, E. Detection and identification of phytoplasmas in yellows-diseased weeds in Italy. Plant Pathol. 1997, 46, 530–537. [Google Scholar] [CrossRef]
- Seemüller, E.; Marcone, C.; Lauer, U.; Ragozzino, A.; Göschl, M. Current status of molecular classification of the phytoplasmas. J. Plant Pathol. 1998, 80, 3–26. [Google Scholar]
- Martini, M.; Marcone, C.; Mitrović, J.; Maixner, M.; Delić, D.; Myrta, A.; Ermacora, P.; Bertaccini, A.; Duduk, B. ‘Candidatus Phytoplasma convolvuli’, a new phytoplasma taxon associated with bindweed yellows in four European countries. Int. J. Syst. Evol. Microbiol. 2012, 62, 2910–2915. [Google Scholar] [CrossRef] [Green Version]
- Quaglino, F.; Maghradze, D.; Chkhaidze, N.; Casati, P.; Failla, O.; Bianco, P.A. First report of ‘Candidatus Phytoplasma solani’ and ‘Ca. P. convolvuli’ associated with grapevine Bois Noir and bindweed yellows, respectively, in Georgia. Plant Dis. 2014, 98, 1151. [Google Scholar] [CrossRef]
- Zwolińska, A.; Krawczyk, K.; Borodynko-Filas, N.; Pospieszny, H. Non-crop sources of Rapeseed Phyllody phytoplasma (‘Candidatus Phytoplasma asteris’: 16SrI-B and 16SrI-(B/L) L), and closely related strains. Crop Prot. 2019, 119, 59–68. [Google Scholar] [CrossRef]
- Langer, M.; Maixner, M. Molecular characterisation of grapevine yellows associated phytoplasmas of the stolbur group based on RFLP analysis of non-ribosomal DNA. Vitis 2004, 43, 191–199. [Google Scholar]
- Bressan, A.; Sémétey, O.; Nusillard, B.; Clair, D.; Boudon-Padieu, E. Insect vectors (Hemiptera: Cixiidae) and pathogens associated with the disease syndrome “basses richesses” of sugarbeet in France. Plant Dis. 2008, 92, 113–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kessler, S.; Schaerer, S.; Delabays, N.; Turlings, T.C.; Trivellone, V.; Kehrli, P. Host plant preferences of Hyalesthes obsoletus, the vector of the grapevine yellows disease ‘bois noir’, in Switzerland. Entomol. Exp. Appl. 2011, 139, 60–67. [Google Scholar] [CrossRef]
- Johannesen, J.; Foissac, X.; Kehrli, P.; Maixner, M. Impact of vector dispersal and host-plant fidelity on the dissemination of an emerging plant pathogen. PLoS ONE 2012, 7, e51809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trivellone, V.; Filippin, L.; Narduzzi-Wicht, B.; Angelini, E. A regional-scale survey to define the known and potential vectors of grapevine yellow phytoplasmas in vineyards South of Swiss Alps. Eur. J. Plant Pathol. 2016, 145, 915–927. [Google Scholar] [CrossRef]
- Chuche, J.; Danet, J.L.; Rivoal, J.B.; Arricau-Bouvery, N.; Thiéry, D. Minor cultures as hosts for vectors of extensive crop diseases: Does Salvia sclarea actas a pathogen and vector reservoir for lavender decline? J. Pest Sci. 2018, 91, 145–155. [Google Scholar] [CrossRef]
- Moussa, A.; Mori, N.; Faccincani, M.; Pavan, F.; Bianco, P.A.; Quaglino, F. Vitex agnus-castus cannot be used as trap plant for the vector Hyalesthes obsoletus to prevent infections by ‘Candidatus Phytoplasma solani’ in northern Italian vineyards: Experimental evidence. Ann. Appl. Biol. 2019, 175, 302–312. [Google Scholar] [CrossRef]
- Mori, N.; Cargnus, E.; Martini, M.; Pavan, F. Relationships between Hyalesthes obsoletus, Its Herbaceous Hosts and Bois Noir Epidemiology in Northern Italian Vineyards. Insects 2020, 11, 606. [Google Scholar] [CrossRef]
- Cvrković, T.; Jović, J.; Mitrović, M.; Krstić, O.; Toševski, I. Experimental and molecular evidence of Reptalus panzeri as a natural vector of bois noir. Plant Pathol. 2014, 63, 42–53. [Google Scholar] [CrossRef] [Green Version]
- Mitrović, M.; Jakovljević, M.; Jović, J.; Krstić, O.; Kosovac, A.; Trivellone, V.; Jermini, M.; Toševski, I.; Cvrković, T. ‘Candidatus Phytoplasma solani’ genotypes associated with potato stolbur in Serbia and the role of Hyalesthes obsoletus and Reptalus panzeri (Hemiptera, Cixiidae) as natural vectors. Eur. J. Plant Pathol. 2016, 144, 619–630. [Google Scholar] [CrossRef]
- Chuche, J.; Danet, J.L.; Salar, P.; Foissac, X.; Thiéry, D. Transmission of ‘Candidatus Phytoplasma solani’ by Reptalus quinquecostatus (Hemiptera: Cixiidae). Ann. Appl. Biol. 2016, 169, 214–223. [Google Scholar] [CrossRef]
- Jakovljević, M.; Jović, J.; Krstić, O.; Mitrović, M.; Marinković, S.; Toševski, I.; Cvrković, T. Diversity of phytoplasmas identified in the polyphagous leafhopper Euscelis incisus (Cicadellidae, Deltocephalinae) in Serbia: Pathogen inventory, epidemiological significance and vectoring potential. Eur. J. Plant Pathol. 2020, 156, 201–221. [Google Scholar] [CrossRef]
- Fabre, A.; Danet, J.-L.; Foissac, X. The stolbur phytoplasma antigenic membrane protein gene stamp is submitted to diversifying positive selection. Gene 2011, 472, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Jović, J.; Cvrković, T.; Mitrović, M.; Krnjanjić, S.; Petrović, A.; Redinbaugh, M.G.; Pratt, R.C.; Hogenhout, S.A.; Toševski, I. Stolbur phytoplasma transmission to maize by Reptalus panzeri and the disease cycle of maize redness in Serbia. Phytopathology 2009, 99, 1053–1061. [Google Scholar] [CrossRef] [Green Version]
- Schneider, B.; Gibb, K.S.; Seemüller, E. Sequence and RFLP analysis of the elongation factor Tu gene used in differentiation and classification of phytoplasmas. Microbiology 1997, 143, 3381–3389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.H.; Karlen, Y.; Bakker, O.; Van den Hoff, M.J.B.; Moorman, A.F.M. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakers, C.; Ruijter, J.M.; LekanneDeprez, R.H.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Mittelberger, C.; Obkircher, L.; Oberkofler, V.; Ianeselli, A.; Kerschbamer, C.; Gallmetzer, A.; Reyes-Dominguez, Y.; Letschka, T.; Janik, K. Development of a universal endogenous qPCR control for eukaryotic DNA samples. Plant Methods 2020, 16, 1–14. [Google Scholar] [CrossRef]
- Cimerman, A.; Pacifico, D.; Salar, P.; Marzachi, C.; Foissac, X. Striking diversity of vmp1, a variable geneencoding a putative membrane protein of the stolbur phytoplasma. Appl. Environ. Microbiol. 2009, 75, 2951–2957. [Google Scholar] [CrossRef] [Green Version]
- Sharon, R.; Soroker, V.; Wesley, S.D.; Zahavi, T.; Harari, A.; Weintraub, P.G. Vitex agnus-castus is a preferred host plant for Hyalesthes obsoletus. J. Chem. Ecol. 2005, 31, 1051–1063. [Google Scholar] [CrossRef]
- Lee, I.M.; Gundersen-Rindal, D.E.; Davis, R.E.; Bottner, K.D.; Marcone, C.; Seemüller, E. ‘Candidatus Phytoplasma asteris’, a novel phytoplasma taxon associated with aster yellows and related diseases. Int. J. Syst. Evol. Microbiol. 2004, 54, 1037–1048. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.M.; Gundersen-Rindal, D.E.; Davis, R.E.; Bartoszyk, I.M. Revised classification scheme of phytoplasmas based on RFLP analysesof 16S rRNA and ribosomal protein gene sequences. Int. J. Syst. Evol. Microbiol. 1998, 48, 1153–1169. [Google Scholar]
- Landi, L.; Murolo, S.; Romanazzi, G. Detection of ‘Candidatus Phytoplasma solani’ in roots from Bois noir symptomatic and recovered grapevines. Sci. Rep. 2019, 9, 2013. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aleksić, Ž.; Šutić, D.; Aleksić, D. Transmission intensity of stolbur virus by means of Hyalesthes obsoletus Sign. on some host plants. Plant Prot. 1967, 93–95, 67–73. [Google Scholar]
- Kosovac, A. The Influence of Host-Plant Use on Cryptic Differentiation of Vector Hyalesthes obsoletus Signoret, 1865 (Hemiptera: Cixiidae) and on Epidemiological Transmission Routes of ‘Candidatus Phytoplasma solani’. Ph.D. Thesis, University of Belgrade, Belgrade, Serbia, 2018. [Google Scholar]
- Angelini, E.; Clair, D.; Borgo, M.; Bertaccini, A.; Boudon-Padieu, E. Flavescence dorée in France and Italy—Occurrence of closely related phytoplasma isolates and their near relationships to Palatinate grapevine yellows and an alder yellows phytoplasma. Vitis 2001, 40, 79–86. [Google Scholar]
- Deng, S.; Hiruki, C. Amplification of 16S rRNA genes from culturable and non culturable mollicutes. J. Microbiol. Methods 1991, 14, 53–61. [Google Scholar] [CrossRef]
- Smart, C.D.; Schneider, B.; Blomquist, C.L.; Guerra, L.J.; Harrison, N.A.; Ahrens, U.; Lorenz, K.H.; Seemüller, E.; Kirkpatrick, B.C. Phytoplasma-specific PCR primers based on sequences of the 16S-23S rRNA spacer region. Appl. Environ. Microbiol. 1996, 62, 2988–2993. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.M.; Martini, M.; Marcone, C.; Zhu, S.F. Classificati on of phytoplasma strains in the elm yellows group (16SrV) and proposal of ‘Candidatus Phytoplasma ulmi’ for the phytoplasma associated with elm yellows. Int. J. Syst. Evol. Microbiol. 2004, 54, 337–347. [Google Scholar] [CrossRef] [Green Version]
- Gundersen, D.E.; Lee, I.M. Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathol. Mediterr. 1996, 35, 144–151. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dwight, Z.; Palais, R.; Wittwer, C.T. uMELT: Prediction of high-resolution melting curves and dynamic melting profiles of PCR products in a rich web application. Bioinformatics 2011, 27, 1019–1020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]

| Region | Location | GPS Coordinates | Date | Symptoms Occurrence | Number of Samples |
|---|---|---|---|---|---|
| Eastern Serbia | Donji Milanovac | 44°31.501′ N 22°02.500′ E | August 2018 | symptomatic | 6 |
| July 2019 | symptomatic | 10 | |||
| August 2019 | symptomatic | 6 | |||
| June 2020 | symptomatic | 6 | |||
| non-symptomatic | 6 | ||||
| August 2020 | non-symptomatic | 24 | |||
| Štubik | 44°17.237′ N 22°21.713′ E | July 2019 | symptomatic | 6 | |
| August 2019 | symptomatic | 8 | |||
| non-symptomatic | 12 | ||||
| Kladovo | 44°36.850′ N 22°36.233′ E | September 2020 | symptomatic | 12 | |
| non-symptomatic | 12 | ||||
| Central Serbia | Deč | 44°49.292′ N 20°09.354′ E | August 2020 | symptomatic | 8 |
| non-symptomatic | 12 | ||||
| September 2020 | symptomatic | 6 | |||
| Total | 134 |
| Location | Date | Symptoms a | Number of Samples | Number (%) of Phytoplasma Positive Samples b,c | CaPsol stamp Genotype | ||
|---|---|---|---|---|---|---|---|
| CaPsol | CaPcon | MIX | |||||
| Donji Milanovac | August 2018 | ba, el, dy | 6 | 0 | 6 | 0 | / |
| July 2019 | ba, ul, mrl | 10 | 4 | 0 | 6 | Rqg50 (10) | |
| August 2019 | ba | 6 | 0 | 6 | 0 | / | |
| June 2020 | ba, y, lvr, sp | 6 | 0 | 6 | 0 | / | |
| control plants | 6 | 0 | 0 | 0 | / | ||
| August 2020 | control plants | 24 | 0 | 0 | 0 | / | |
| Štubik | July 2019 | ba, lvr, sp | 6 | 0 | 3 | 3 | Rqg50 (3) |
| August 2019 | ba, y | 8 | 0 | 3 | 5 | Rqg50 (5) | |
| control plants | 12 | 0 | 0 | 0 | / | ||
| Kladovo | September 2020 | ba, y, ul, is, ssp | 12 | 12 | 0 | 0 | Rqg50 (8) GGY (2) Rpm35 (2) |
| control plants | 12 | 0 | 0 | 0 | / | ||
| Deč | August 2020 | ba, ul, y | 8 | 1 | 2 | 5 | Rqg31 (6) |
| control plants | 12 | 0 | 0 | 0 | / | ||
| September 2020 | ba, ul, y | 6 | 0 | 2 | 4 | Rqg31 (4) | |
| Total | 134 | 17 (25%) | 28 (41%) | 23 (34%) | Rqg50 (26) Rqg31 (10) GGY (2) Rpm35 (2) | ||
| symptomatic/control plants | 68/66 | 17/0 | 28/0 | 23/0 | |||
| Application | Primer Name | Primer Sequence (5′→3′) | Amplicon | ||
|---|---|---|---|---|---|
| Length (bp) | GC (%) | Tm (°C) a | |||
| ‘Ca. P. convolvuli’-specific direct PCR | Tuf-Con-F1 | TGTAGAATATGAAACTGCAC | 791 | n.a. | n.a. |
| Tuf-Con-R1 | ATTGAGGGCGATATTGAGAG | ||||
| ‘Ca. P. convolvuli’-specific nested PCR | Tuf-Con-F2 | CCCAGGACATGCTGATTATG | 725 | n.a. | n.a. |
| Tuf-Con-R2 | AAAAGGCAGTATGACGTCCC | ||||
| ‘Ca. P. convolvuli’-specific real-time PCR | Tuf-Con-Fq | GGCTCGCCAAGTAGGAGTT | 141 | 31 | 78.0/78.26 ± 0.11 |
| Tuf-Con-Rq | GGAATTTCATCGCCTGGGT | ||||
| ‘Ca. P. solani’-specific real-time PCR | Tuf-Sol-Fq | GATCCAGTGCGTGAAGTTGAT | 106 | 41 | 81.0/80.38 ± 0.12 |
| Tuf-Sol-Rq | CTTGTCCTCTTTCAACTCTACCA | ||||
| DNA Concentration (ng/reaction) | ‘Ca. P. convolvuli’ | ‘Ca. P. solani’ | ||||
|---|---|---|---|---|---|---|
| Ct Mean ± SD a | Efficiency (E) b | Value of fit (R2) c | Ct Mean ± SD a | Efficiency (E) b | Value of Fit (R2) c | |
| 25 | 13.70 ± 0.06 | 0.95–0.98 | >0.999 | 14.14 ± 0.03 | 0.96–0.98 | >0.999 |
| 2.5 | 17.00 ± 0.01 | 0.91–0.98 | >0.999 | 17.43 ± 0.03 | 095–0.96 | >0.999 |
| 0.25 | 20.56 ± 0.06 | 0.90–0.95 | >0.999 | 20.92 ± 0.06 | 0.90–0.97 | >0.999 |
| 0.025 | 24.08 ± 0.04 | 091–0.97 | >0.999 | 24.36 ± 0.04 | 0.88–0.92 | >0.999 |
| 0.0025 | 27.48 ± 0.21 | 0.91–0.95 | >0.999 | 28.67 ± 0.64 | 0.87–0.89 | >0.999 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jović, J.; Marinković, S.; Jakovljević, M.; Krstić, O.; Cvrković, T.; Mitrović, M.; Toševski, I. Symptomatology, (Co)occurrence and Differential Diagnostic PCR Identification of ‘Ca. Phytoplasma solani’ and ‘Ca. Phytoplasma convolvuli’ in Field Bindweed. Pathogens 2021, 10, 160. https://doi.org/10.3390/pathogens10020160
Jović J, Marinković S, Jakovljević M, Krstić O, Cvrković T, Mitrović M, Toševski I. Symptomatology, (Co)occurrence and Differential Diagnostic PCR Identification of ‘Ca. Phytoplasma solani’ and ‘Ca. Phytoplasma convolvuli’ in Field Bindweed. Pathogens. 2021; 10(2):160. https://doi.org/10.3390/pathogens10020160
Chicago/Turabian StyleJović, Jelena, Slavica Marinković, Miljana Jakovljević, Oliver Krstić, Tatjana Cvrković, Milana Mitrović, and Ivo Toševski. 2021. "Symptomatology, (Co)occurrence and Differential Diagnostic PCR Identification of ‘Ca. Phytoplasma solani’ and ‘Ca. Phytoplasma convolvuli’ in Field Bindweed" Pathogens 10, no. 2: 160. https://doi.org/10.3390/pathogens10020160






