Transcriptome Analysis Reveals a Promotion of Carotenoid Production by Copper Ions in Recombinant Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Cultivation
2.2. Genetic Manipulation
2.3. Shake-Flask Fermentation and Growth Determination
2.4. Analytical Methods
2.5. Transcriptome Analysis
2.6. Quantitative PCR
3. Results and Discussion
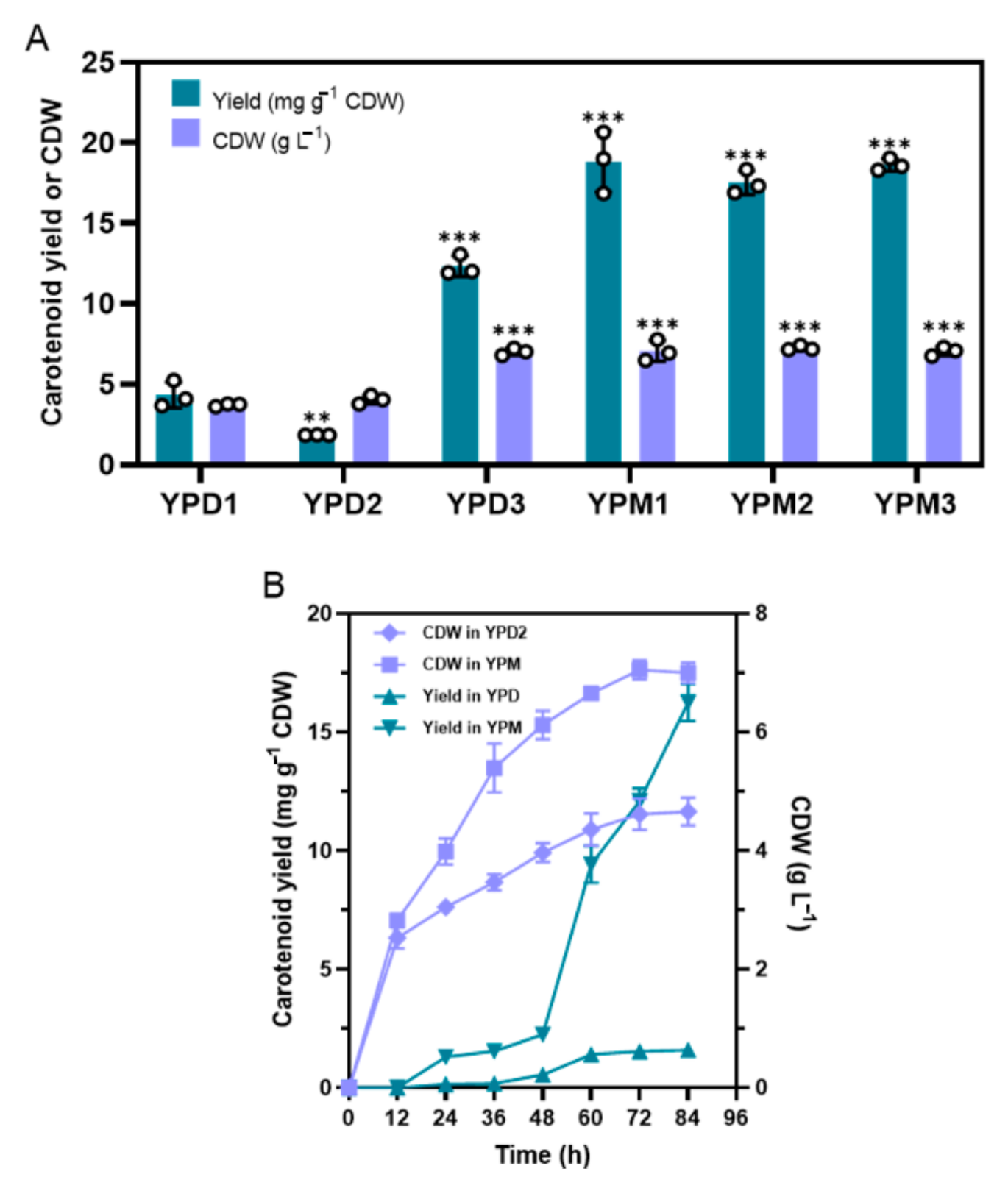
3.1. Significant Promotion of Carotenoid Accumulation is Presented in a Modified YPD Medium
3.2. Transcriptome Analysis Reveals Multiple Differentially Expressed Genes
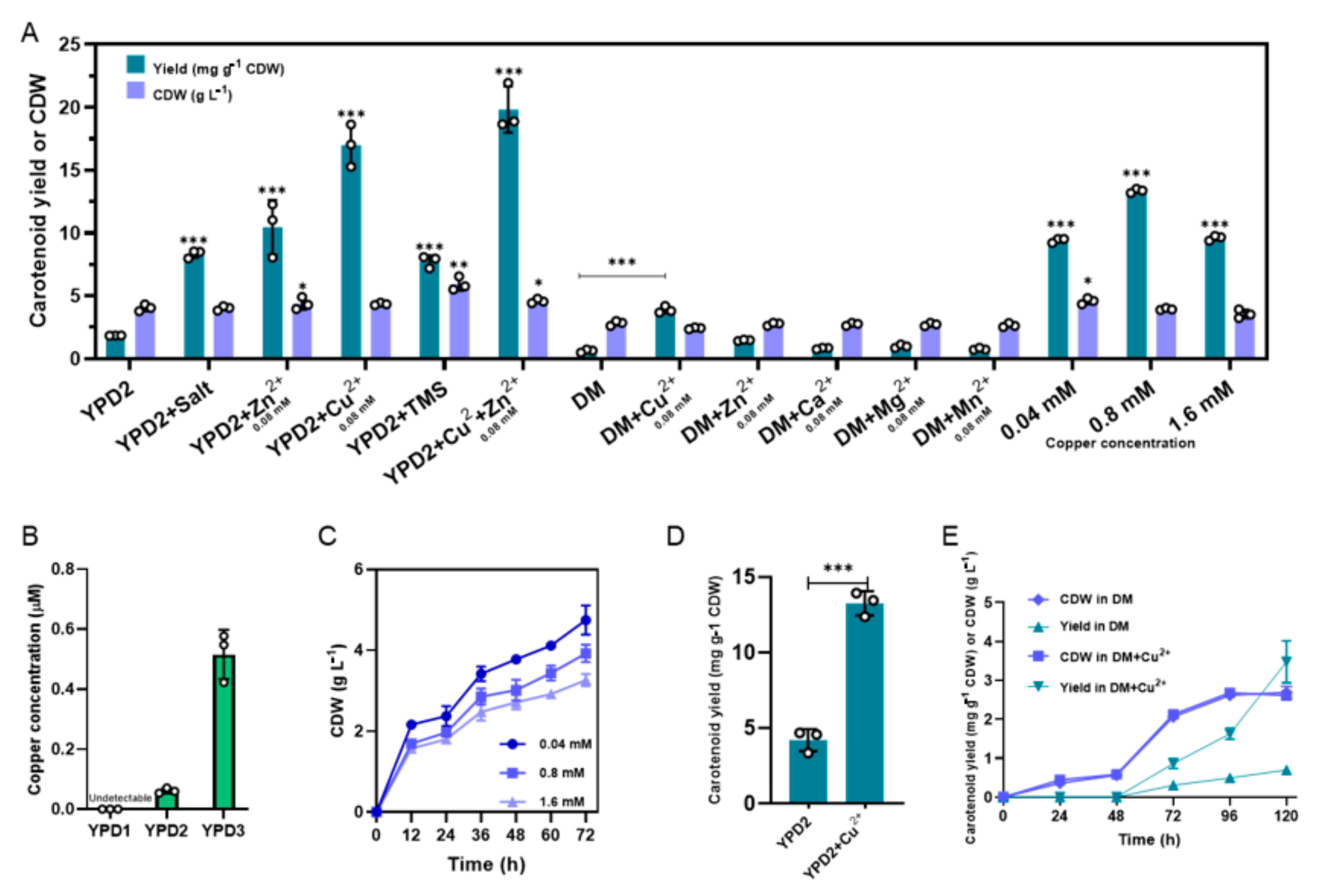
3.3. Copper Ions Play a Key Role in the Carotenoid Accumulation of BL03-D-4
3.4. ACE1 Overexpression Remarkably Improves Carotenoid Accumulation of BL03-D-4
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mussagy, C.U.; Winterburn, J.; Santos-Ebinuma, V.C.; Pereira, J.F.B. Production and extraction of carotenoids produced by microorganisms. Appl. Microbiol. Biotechnol. 2019, 103, 1095–1114. [Google Scholar] [CrossRef] [PubMed]
- Saini, R.K.; Keum, Y.S. Microbial platforms to produce commercially vital carotenoids at industrial scale: An updated review of critical issues. J. Ind. Microbiol. Biotechnol. 2019, 46, 657–674. [Google Scholar] [CrossRef]
- Sankari, M.; Rao, P.R.; Hemachandran, H.; Pullela, P.K.; Doss, C.G.; Tayubi, I.A.; Subramanian, B.; Gothandam, K.M.; Singh, P.; Ramamoorthy, S. Prospects and progress in the production of valuable carotenoids: Insights from metabolic engineering, synthetic biology, and computational approaches. J. Biotechnol. 2018, 266, 89–101. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, S.; Shao, X.; Park, J.B.; Jeong, S.H.; Park, H.J.; Kwak, W.J.; Wei, G.; Kim, S.W. Challenges and tackles in metabolic engineering for microbial production of carotenoids. Microb. Cell Fact. 2019, 18, 55. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Deng, Z.; Liu, T. Microbial production strategies and applications of lycopene and other terpenoids. World J. Microbiol. Biotechnol. 2016, 32, 15. [Google Scholar] [CrossRef] [PubMed]
- Ma, T.; Shi, B.; Ye, Z.; Li, X.; Liu, M.; Chen, Y.; Xia, J.; Nielsen, J.; Deng, Z.; Liu, T. Lipid engineering combined with systematic metabolic engineering of Saccharomyces cerevisiae for high-yield production of lycopene. Metab. Eng. 2019, 52, 134–142. [Google Scholar] [CrossRef]
- Shi, B.; Ma, T.; Ye, Z.; Li, X.; Huang, Y.; Zhou, Z.; Ding, Y.; Deng, Z.; Liu, T. Systematic Metabolic Engineering of Saccharomyces cerevisiae for Lycopene Overproduction. J. Agric. Food Chem. 2019, 67, 11148–11157. [Google Scholar] [CrossRef]
- Hong, J.; Park, S.H.; Kim, S.; Kim, S.W.; Hahn, J.S. Efficient production of lycopene in Saccharomyces cerevisiae by enzyme engineering and increasing membrane flexibility and NAPDH production. Appl. Microbiol. Biotechnol. 2019, 103, 211–223. [Google Scholar] [CrossRef]
- Niu, F.X.; Lu, Q.; Bu, Y.F.; Liu, J.Z. Metabolic engineering for the microbial production of isoprenoids: Carotenoids and isoprenoid-based biofuels. Synth. Syst. Biotechnol. 2017, 2, 167–175. [Google Scholar] [CrossRef]
- Du, W.; Song, Y.; Liu, M.; Yang, H.; Zhang, Y.; Fan, Y.; Luo, X.; Li, Z.; Wang, N.; He, H.; et al. Gene expression pattern analysis of a recombinant Escherichia coli strain possessing high growth and lycopene production capability when using fructose as carbon source. Biotechnol. Lett. 2016, 38, 1571–1577. [Google Scholar] [CrossRef]
- Su, B.; Song, D.; Yang, F.; Zhu, H. Engineering a growth-phase-dependent biosynthetic pathway for carotenoid production in Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2020, 47, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Su, B.; Song, D.; Zhu, H. Metabolic Engineering of Saccharomyces cerevisiae for Enhanced Carotenoid Production from Xylose-Glucose Mixtures. Front. Bioeng. Biotechnol. 2020, 8, 435. [Google Scholar] [CrossRef] [PubMed]
- Verwaal, R.; Buiting-Wiessenhaan, N.; Dalhuijsen, S.; Roubos, J.A. CRISPR/Cpf1 enables fast and simple genome editing of Saccharomyces cerevisiae. Yeast 2018, 35, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Letchworth, G.J. High efficiency transformation by electroporation of Pichia pastoris pretreated with lithium acetate and dithiothreitol. BioTechniques 2004, 36, 152–154. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, Q.; Sun, T.; Zhu, X.; Xu, H.; Tang, J.; Zhang, X.; Ma, Y. Engineering central metabolic modules of Escherichia coli for improving beta-carotene production. Metab. Eng. 2013, 17, 42–50. [Google Scholar] [CrossRef]
- Pan, X.; Wang, B.; Duan, R.; Jia, J.; Li, J.; Xiong, W.; Ling, X.; Chen, C.; Huang, X.; Zhang, G.; et al. Enhancing astaxanthin accumulation in Xanthophyllomyces dendrorhous by a phytohormone: Metabolomic and gene expression profiles. Microb. Biotechnol. 2020, 13, 1446–1460. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, K.; Mehmood, M.A.; Zhao, Z.K.; Bai, F.; Zhao, X. Deletion of acetate transporter gene ADY2 improved tolerance of Saccharomyces cerevisiae against multiple stresses and enhanced ethanol production in the presence of acetic acid. Bioresour. Technol. 2017, 245, 1461–1468. [Google Scholar] [CrossRef]
- Qi, Y.; Liu, H.; Yu, J.; Chen, X.; Liu, L. Med15B Regulates Acid Stress Response and Tolerance in Candida glabrata by Altering Membrane Lipid Composition. Appl. Environ. Microbiol. 2017, 83, e01128-17. [Google Scholar] [CrossRef]
- Caspeta, L.; Chen, Y.; Ghiaci, P.; Feizi, A.; Buskov, S.; Hallstrom, B.M.; Petranovic, D.; Nielsen, J. Biofuels. Altered sterol composition renders yeast thermotolerant. Science 2014, 346, 75–78. [Google Scholar] [CrossRef]
- Guo, L.; Pang, Z.; Gao, C.; Chen, X.; Liu, L. Engineering microbial cell morphology and membrane homeostasis toward industrial applications. Curr. Opin. Biotechnol. 2020, 66, 18–26. [Google Scholar] [CrossRef]
- Walker, C.; Ryu, S.; Trinh, C.T. Exceptional solvent tolerance in Yarrowia lipolytica is enhanced by sterols. Metab. Eng. 2019, 54, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Liu, H.; Chen, X.; Liu, L. Engineering microbial membranes to increase stress tolerance of industrial strains. Metab. Eng. 2019, 53, 24–34. [Google Scholar] [CrossRef]
- Jorda, T.; Puig, S. Regulation of Ergosterol Biosynthesis in Saccharomyces cerevisiae. Genes 2020, 11, 795. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Ohta, A.; Horiuchi, H.; Fukuda, R. Oxysterol-binding protein homologs mediate sterol transport from the endoplasmic reticulum to mitochondria in yeast. J. Biol. Chem. 2018, 293, 5636–5648. [Google Scholar] [CrossRef] [PubMed]
- Vaquero, I.; Ruiz-Domínguez, M.C.; Márquez, M.; Vílchez, C. Cu-mediated biomass productivity enhancement and lutein enrichment of the novel microalga Coccomyxa onubensis. Process Biochem. 2012, 47, 694–700. [Google Scholar] [CrossRef]
- Reider, A.A.; d’Espaux, L.; Wehrs, M.; Sachs, D.; Li, R.A.; Tong, G.J.; Garber, M.; Nnadi, O.; Zhuang, W.; Hillson, N.J.; et al. A Cas9-based toolkit to program gene expression in Saccharomyces cerevisiae. Nucleic Acids Res. 2017, 45, 496–508. [Google Scholar]
- Shi, H.; Jiang, Y.; Yang, Y.; Peng, Y.; Li, C. Copper metabolism in Saccharomyces cerevisiae: An update. BioMetals 2020. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, B.; Li, A.; Deng, M.-R.; Zhu, H. Transcriptome Analysis Reveals a Promotion of Carotenoid Production by Copper Ions in Recombinant Saccharomyces cerevisiae. Microorganisms 2021, 9, 233. https://doi.org/10.3390/microorganisms9020233
Su B, Li A, Deng M-R, Zhu H. Transcriptome Analysis Reveals a Promotion of Carotenoid Production by Copper Ions in Recombinant Saccharomyces cerevisiae. Microorganisms. 2021; 9(2):233. https://doi.org/10.3390/microorganisms9020233
Chicago/Turabian StyleSu, Buli, Anzhang Li, Ming-Rong Deng, and Honghui Zhu. 2021. "Transcriptome Analysis Reveals a Promotion of Carotenoid Production by Copper Ions in Recombinant Saccharomyces cerevisiae" Microorganisms 9, no. 2: 233. https://doi.org/10.3390/microorganisms9020233
APA StyleSu, B., Li, A., Deng, M.-R., & Zhu, H. (2021). Transcriptome Analysis Reveals a Promotion of Carotenoid Production by Copper Ions in Recombinant Saccharomyces cerevisiae. Microorganisms, 9(2), 233. https://doi.org/10.3390/microorganisms9020233





