Isolation of a Psychrotolerant and UV-C-Resistant Bacterium from Elephant Island, Antarctica with a Highly Thermoactive and Thermostable Catalase
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Enrichment of Strain I1P
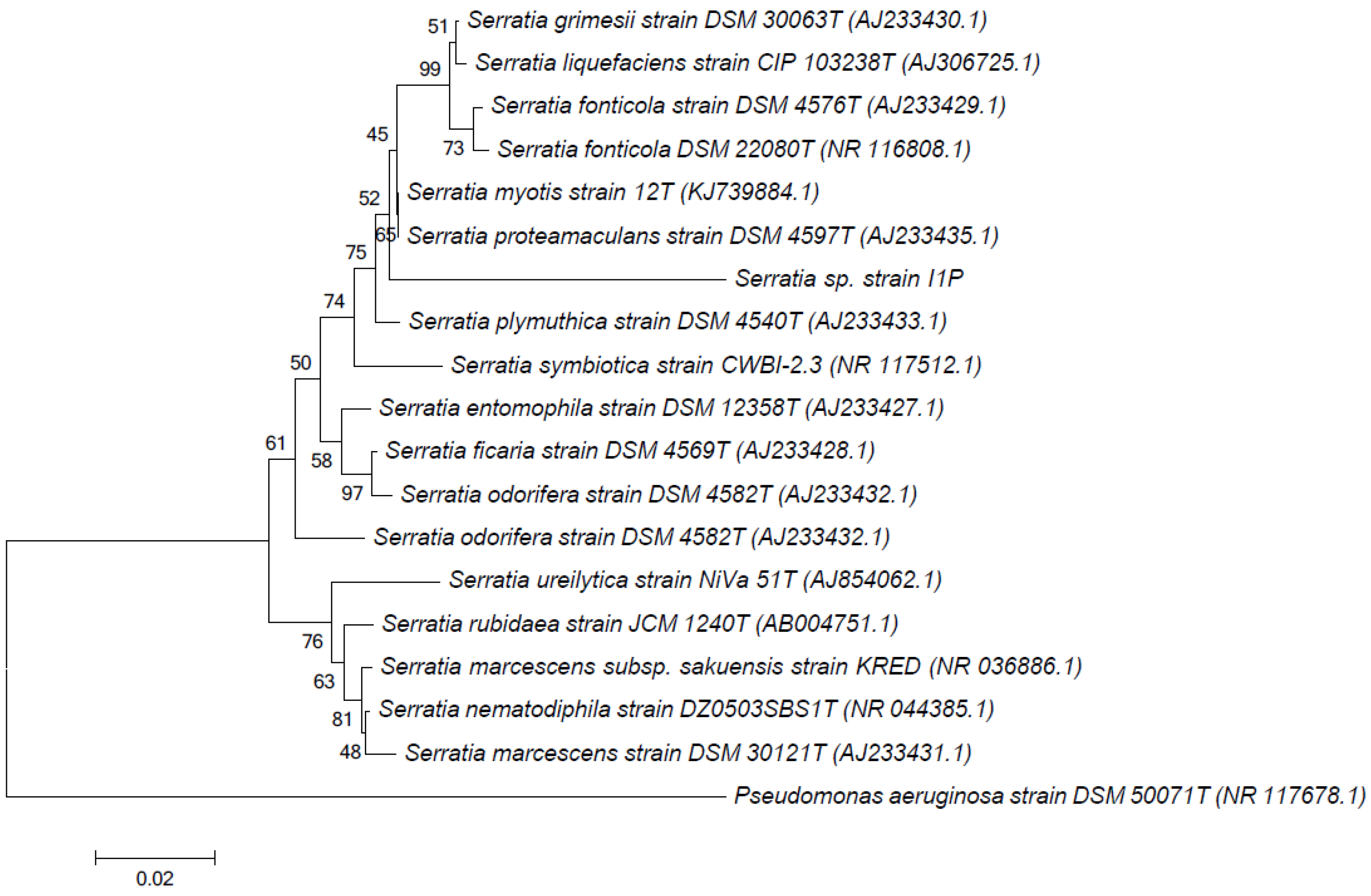
2.2. DNA Extraction, 16S rRNA Gene Amplification, and Phylogenetic Analysis
2.3. Morphological, Physiological and Biochemical Characterizations
2.4. Effect of UV Radiation on Cell Viability
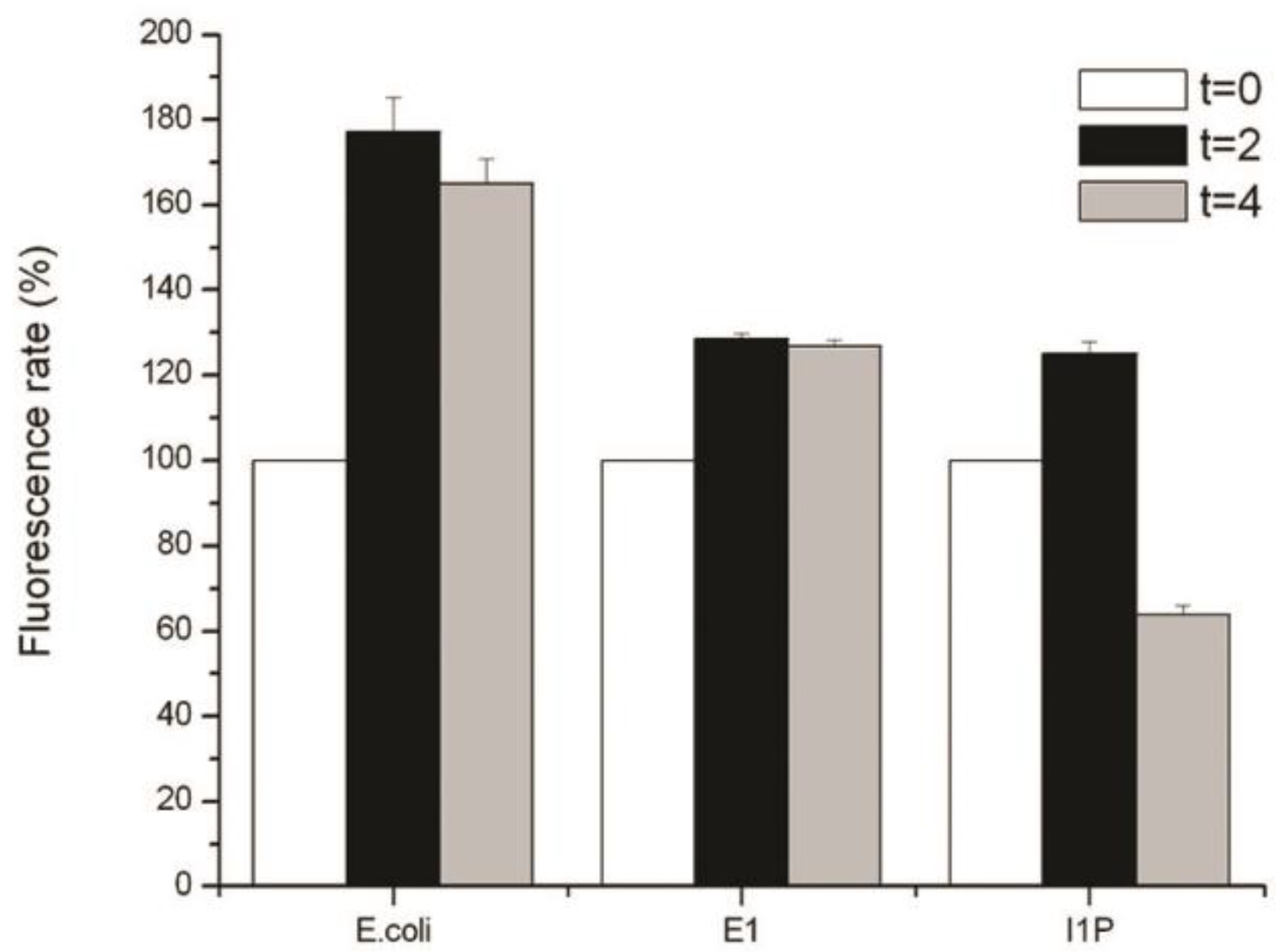
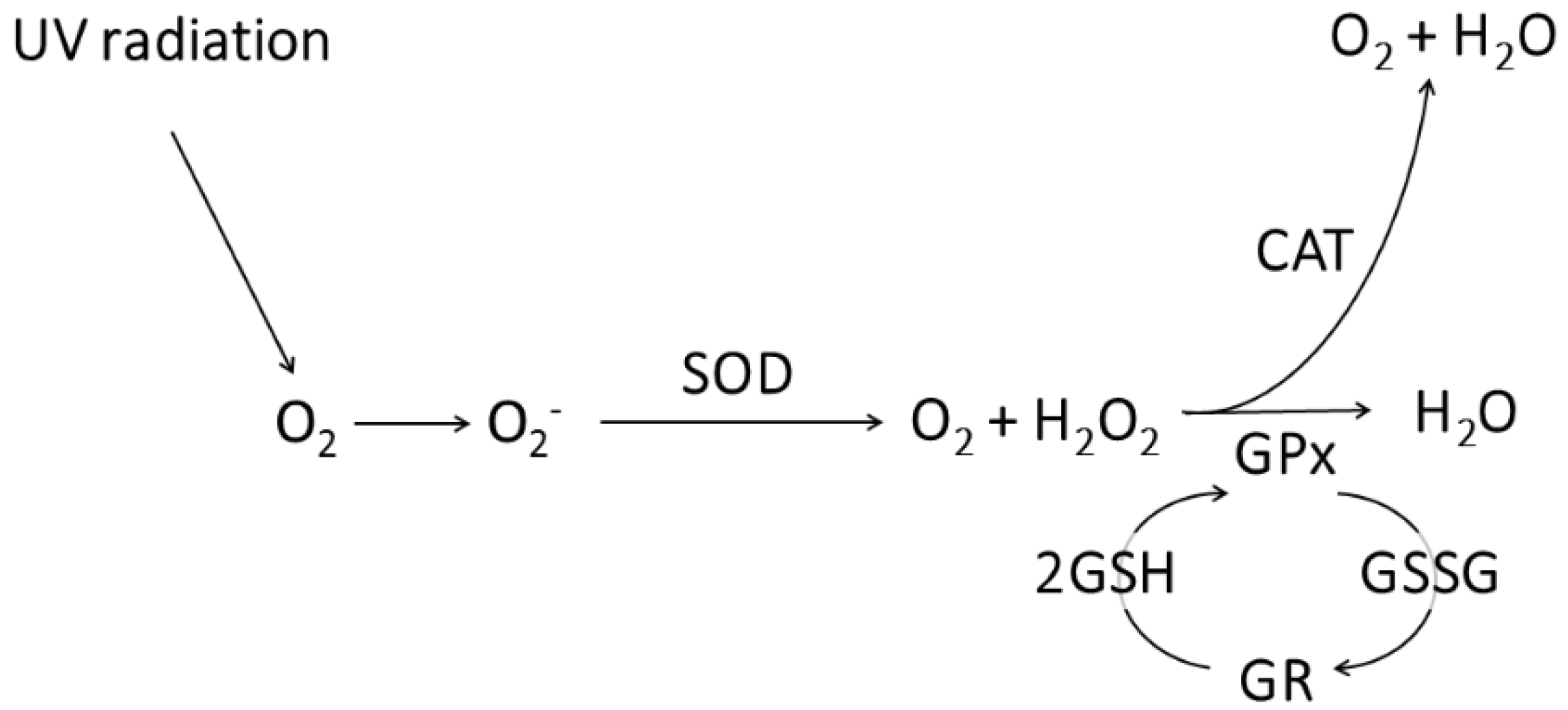
2.5. Detection of Reactive Oxygen Species (ROS)
2.6. Enzyme Assay
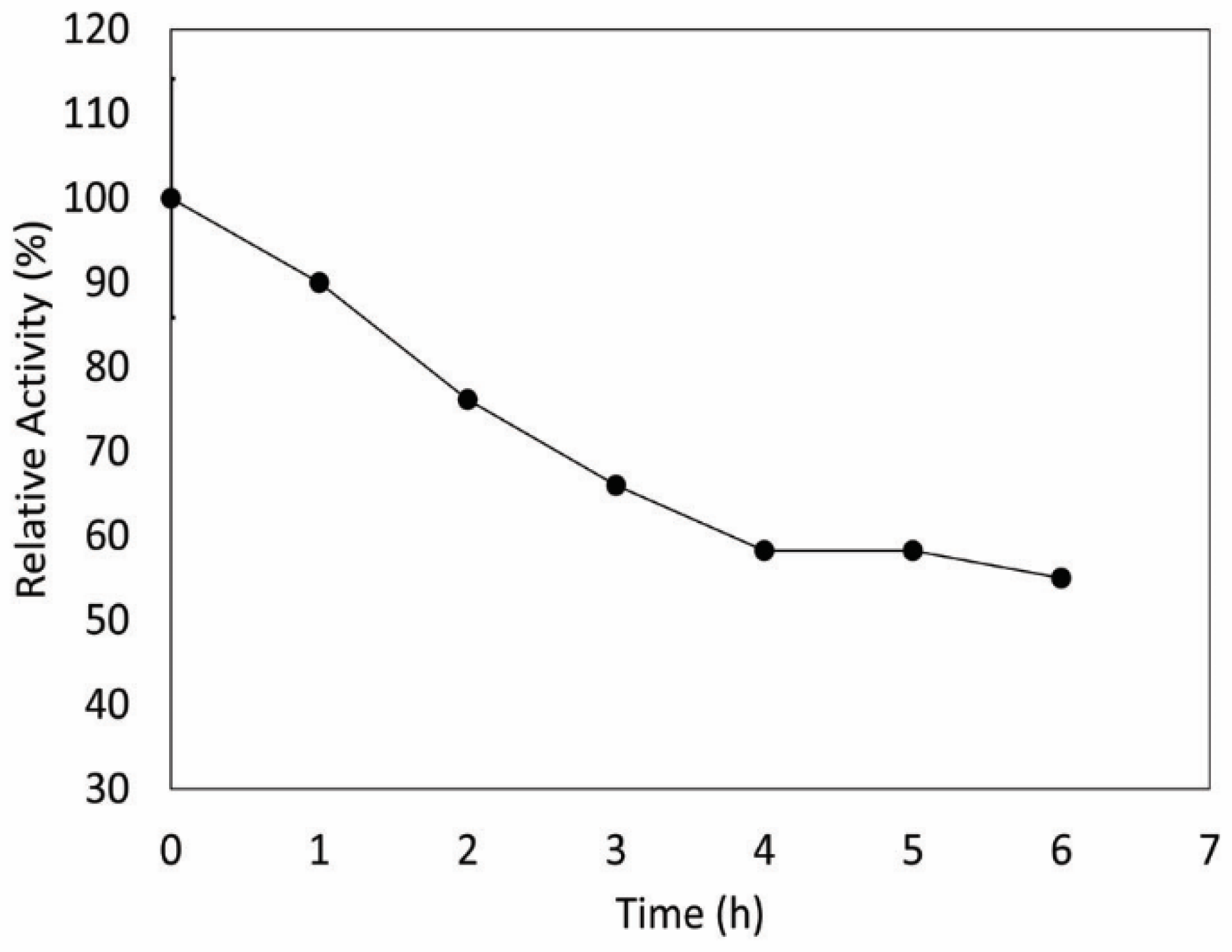
2.7. Effect of UV Radiation on Enzyme Activity
2.8. Enzyme Purification
2.9. Molecular Mass Determination
2.10. Protein Identification
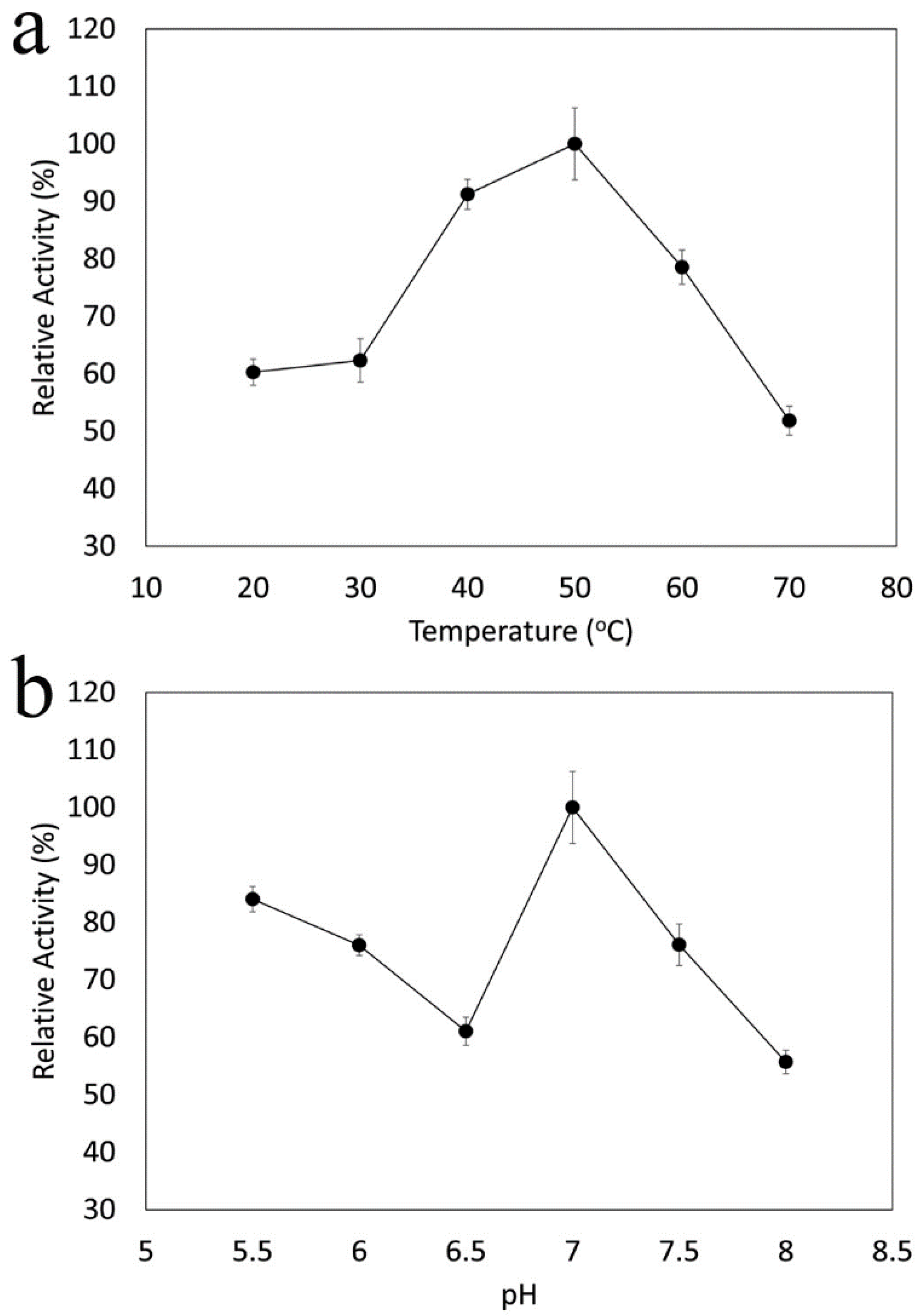
2.11. Effects of pH and Temperature on Catalase Activity
2.12. Thermostability
2.13. Effect of Inhibitors on Catalase Activity
2.14. Statistical Analyses
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Flores, P.A.; Amenábar, M.J.; Blamey, J.M. Hot Environments from Antarctica. Source of Thermophiles and Hyperthermophiles, with Potential Biotechnological Applications. In Thermophilic Microbes in Environmental and Industrial Biotechnology, 2nd ed.; Satyanarayana, T., Littlechild, J., Kawarabayasi, Y., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 99–118. [Google Scholar] [CrossRef]
- Smith, R.C.; Prézelin, B.B.; Baker, K.S.; Bidigare, R.R.; Boucher, N.; Coley, T.R.; Karentz, D.; MacIntyre, S.; Matlick, H.A.; Menzies, D. Ozone depletion: Ultraviolet radiation and phytoplankton biology in Antarctic waters. Science 1992, 255, 952–959. [Google Scholar] [CrossRef] [PubMed]
- Correa-Llantén, D.N.; Amenábar, M.J.; Blamey, J.M. Antioxidant capacity of novel pigments from an Antarctic bacterium. J. Microbiol. 2012, 50, 374–379. [Google Scholar] [CrossRef]
- Amenabar, M.J.; Flores, P.A.; Pugin, B.; Boehmwald, F.A.; Blamey, J.M. Archaeal diversity from hydrothermal systems of deception island, Antarctica. Polar Biol. 2013, 36, 373–380. [Google Scholar] [CrossRef]
- Sies, H. Oxidative stress: Oxidants and antioxidants. Exp. Physiol. Trans. Integr. 1997, 82, 291–295. [Google Scholar] [CrossRef]
- Briviba, K.; Klotz, L.O.; Sies, H. Toxic and signaling effects of photochemically or chemically generated singlet oxygen in biological systems. Biol. Chem. 1997, 378, 1259–1265. [Google Scholar]
- Wu, H.; Gao, K.; Villafañe, V.E.; Watanabe, T.; Helbling, E.W. Effects of solar UV radiation on morphology and photosynthesis of filamentous cyanobacterium Arthrospira platensis. Appl. Environ. Microbiol. 2005, 71, 5004–5013. [Google Scholar] [CrossRef]
- Imlay, J.A. Pathways of oxidative damage. Annu. Rev. Microbiol. 2003, 57, 395–418. [Google Scholar] [CrossRef]
- Correa-Llantén, D.N.; Amenábar, M.J.; Muñoz, P.A.; Monsalves, M.T.; Castro, M.E.; Blamey, J.M. Alicyclobacillus sp. strain CC2, a thermo-acidophilic bacterium isolated from deception island (Antarctica) containing a thermostable superoxide dismutase enzyme. Adv. Polar Sci. 2014, 25, 92–96. [Google Scholar] [CrossRef]
- Mondino, L.J.; Asao, M.; Madigan, M.T. Cold-active halophilic bacteria from the ice-sealed Lake Vida, Antarctica. Arch. Microbiol. 2009, 191, 785–790. [Google Scholar] [CrossRef]
- Cabiscol, E.; Tamarit, J.; Ros, J. Oxidative stress in bacteria and protein damage by reactive oxygen species. Int. Microbiol. 2000, 3, 3–8. [Google Scholar]
- Correa-Llantén, D.N.; Amenabar, M.J.; Blamey, J.M. Resistance to hypoosmotic shock of liposomes containing novel pigments from an Antarctic bacterium. Microbiol. Biotechnol. Lett. 2012, 40, 215–219. [Google Scholar] [CrossRef]
- Dieser, M.; Greenwood, M.; Foreman, C.M. Carotenoid Pigmentation in Antarctic Heterotrophic Bacteria as a Strategy to Withstand Environmental Stresses. Arct. Antarct. Alp. Res. 2010, 42, 396–405. [Google Scholar] [CrossRef]
- Monsalves, M.T.; Amenabar, M.J.; Ollivet-Besson, G.P.; Blamey, J.M. Effect of UV radiation on a thermostable superoxide dismutase purified from a thermophilic bacterium isolated from a sterilization drying oven. Protein Pept. Lett. 2013, 20, 749–754. [Google Scholar] [CrossRef]
- Halliwell, B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef]
- Boehmwald, F.; Muñoz, P.; Flores, P.; Blamey, J.M. Functional screening for the discovery of new extremophilic enzymes. In Biotechnology of Extremophiles: Grand Challenges in Biology and Biotechnology; Rampelotto, P., Ed.; Springer: Cham, Switzerland, 2016; pp. 321–350. [Google Scholar] [CrossRef]
- Zheng, Z.; Jiang, Y.; Miao, J.; Wang, Q.F.; Zhang, B.T.; Li, G.Y. Purification and characterization of a cold-active iron superoxide dismutase from a psychrophilic bacterium, Marinomonas sp. NJ522. Biotechnol. Lett. 2006, 28, 85–88. [Google Scholar] [CrossRef]
- Ji, M.; Barnwell, C.; Grunden, A. Characterization of recombinant glutathione reductase from the psychrophilic Antarctic bacterium Colwellia psychrerythraea. Extremophiles 2015, 19, 863–874. [Google Scholar] [CrossRef]
- Kaushal, J.; Mehandia, S.; Singh, G.; Raina, A.; Arya, S.K. Catalase enzyme: Application in bioremediation and food industry. Biocatal. Agric. Biotechnol. 2018, 16, 192–199. [Google Scholar] [CrossRef]
- Liu, X.; Kokare, C. Microbial Enzymes of Use in Industry. In Biotechnology of Microbial Enzymes; Brahmachari, G., Demain, A.L., Adrio, J.L., Eds.; Elsevier: London, UK, 2017; pp. 267–298. [Google Scholar] [CrossRef]
- Lončar, N.; Fraaije, M.W. Catalases as biocatalysts in technical applications: Current state and perspectives. Appl. Microbiol. Biotechnol. 2015, 99, 3351–3357. [Google Scholar] [CrossRef]
- Fruhwirth, G.O.; Paar, A.; Gudelj, M.; Cavaco-Paulo, A.; Robra, K.H.; Gubitz, G.M. An immobilized catalase peroxidase from the alkalothermophilic Bacillus SF for the treatment of textile bleaching effluents. Appl. Microbiol. Biotechnol. 2002, 60, 313–319. [Google Scholar] [CrossRef]
- Yu, Z.; Zheng, H.; Zhao, X.; Li, S.; Xu, J.; Song, H. High level extracellular production of a recombinant alkaline catalase in E. coli BL21 under ethanol stress and its application in hydrogen peroxide removal after cotton fabrics bleaching. Bioresour. Technol. 2016, 214, 303–310. [Google Scholar] [CrossRef]
- Gudelj, M.; Fruhwirth, G.O.; Paar, A.; Lottspeich, F.; Robra, K.; Cavaco-Paulo, A.; Gübitz, G.P. A catalase-peroxidase from a newly isolated thermoalkaliphilic Bacillus sp. with potential for the treatment of textile bleaching effluents. Extremophiles 2001, 5, 423–429. [Google Scholar] [CrossRef]
- Amenabar, M.J.; Colman, D.R.; Poudel, S.; Roden, E.E.; Boyd, E.S. Electron acceptor availability alters carbon and energy metabolism in a thermoacidophile. Environ. Microbiol. 2018, 20, 2523–2537. [Google Scholar] [CrossRef]
- Amenabar, M.J.; Boyd, E.S. Mechanisms of mineral substrate acquisition in a thermoacidophile. Appl. Environ. Microbiol. 2018, 84, e00334-18. [Google Scholar] [CrossRef]
- Boyd, E.S.; Cummings, D.E.; Geesey, G.G. Mineralogy influences structure and diversity of bacterial communities associated with geological substrata in a pristine aquifer. Microb. Ecol. 2007, 54, 170–182. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acid Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Altschul, S.; Madden, T.; Schaffer, A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acid Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef]
- Correa-Llantén, D.; Larraín-Linton, J.; Muñoz, P.A.; Castro, M.; Boehmwald, F.; Blamey, J.M. Characterization of the thermophilic bacterium Geobacillus sp. strain GWE1 isolated from a sterilization oven. Microbiol. Biotechnol. Lett. 2013, 41, 278–283. [Google Scholar] [CrossRef]
- Beers, R.F.; Sizer, I.W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 1952, 195, 133–140. [Google Scholar]
- Chance, B.; Maehly, A.C. Assay of Catalases and Peroxidases. In Methods of Biochemical Analysis; Glick, D., Ed.; Interscience Publishers: New York, NY, USA, 1955; pp. 764–775. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Amenabar, M.J.; Blamey, J.M. Purification and characterization of a thermostable glutamate dehydrogenase from a thermophilic bacterium isolated from a sterilization drying oven. Biochem. Mol. Biol. 2012, 45, 91–95. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Allgood, G.S.; Perry, J.J. Characterization of a manganese-containing catalase from the obligate thermophile Thermoleophilum album. J. Bacteriol. 1986, 168, 563–567. [Google Scholar] [CrossRef]
- Yumoto, I.; Ichihashi, D.; Iwata, H.; Istokovics, A.; Ichise, N.; Matsuyama, H.; Okuyama, H.; Kawasaki, K. Purification and characterization of a catalase from the facultatively psychrophilic bacterium Vibrio rumoiensis S-1T exhibiting high catalase activity. J. Bacteriol. 2000, 182, 1903–1909. [Google Scholar] [CrossRef]
- Kang, Y.S.; Lee, D.H.; Yoon, B.J.; Oh, D.C. Purification and characterization of a catalase from photosynthetic bacterium Rhodospirillum rubrum S1 grown under anaerobic conditions. J. Microbiol. 2006, 44, 185–191. [Google Scholar]
- Chattopadhyay, M.; Raghu, G.; Sharma, Y.; Biju, A.; Rajasekharan, M.; Shivaji, S. Increase in oxidative stress at low temperature in an Antarctic bacterium. Curr. Microbiol. 2011, 62, 544–546. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Ebers, J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 33, 152–155. [Google Scholar]
- Janda, J.M.; Abbott, S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007, 45, 2761–2764. [Google Scholar] [CrossRef]
- Grimont, P.A.; Grimont, F.; Starr, M.P. Serratia proteamaculans (Paine and Stansfield) comb. nov., a senior subjective synonym of Serratia liquefaciens (Grimes and Hennerty) Bascomb et al. Int. J. Syst. Evol. Microbiol. 1978, 28, 503–510. [Google Scholar] [CrossRef]
- Amara, U. Molecular and Biochemical Characaterization of Plant Growth Promoting Rhizobacteria for Enhancing Crop Yield. Ph.D. Thesis, Arid Agriculture University, Rawalpindi, Pakistan, 2015. [Google Scholar]
- Grimont, F.; Grimont, P.A. The Genus Serratia. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; Volume 6, pp. 219–244. [Google Scholar]
- Sánchez, L.A.; Gómez, F.F.; Delgado, O.D. Cold-adapted microorganisms as a source of new antimicrobials. Extremophiles 2009, 13, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Shivaji, S.; Reddy, G.; Aduri, R.; Kutty, R.; Ravenschlag, K. Bacterial diversity of a soil sample from Schirmacher Oasis, Antarctica. Cell. Mol. Biol. 2004, 50, 525–536. [Google Scholar] [PubMed]
- Xiao, X.; Li, M.; You, Z.; Wang, F. Bacterial communities inside and in the vicinity of the Chinese Great Wall Station, King George Island, Antarctica. Antarct. Sci. 2007, 19, 11–16. [Google Scholar] [CrossRef]
- Santos, A.L.; Gomes, N.C.; Henriques, I.; Almeida, A.; Correia, A.; Cunha, Â. Contribution of reactive oxygen species to UV-B-induced damage in bacteria. J. Photochem. Photobiol. B 2012, 117, 40–46. [Google Scholar] [CrossRef]
- Krisko, A.; Radman, M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc. Natl. Acad. Sci. USA 2010, 107, 14373–14377. [Google Scholar] [CrossRef]
- Salma, K.B.; Lobna, M.; Sana, K.; Kalthoum, C.; Imene, O.; Abdelwaheb, C. Antioxidant enzymes expression in Pseudomonas aeruginosa exposed to UV-C radiation. J. Basic Microbiol. 2016, 56, 736–740. [Google Scholar] [CrossRef]
- Meng, J.Y.; Zhang, C.Y.; Fen, Z.; Wang, X.P.; Lei, C.L. Ultraviolet light-induced oxidative stress: Effects on antioxidant response of Helicoverpa armigera adults. J. Insect Physiol. 2009, 55, 588–592. [Google Scholar] [CrossRef]
- Arenas, F.A.; Díaz, W.A.; Leal, C.A.; Pérez-Donoso, J.M.; Imlay, J.A.; Vásquez, C.C. The Escherichia coli btuE gene, encodes a glutathione peroxidase that is induced under oxidative stress conditions. Biochem. Biophys. Res. Commun. 2010, 398, 690–694. [Google Scholar] [CrossRef]
- Alquéres, S.; Meneses, C.; Rouws, L.; Rothballer, M.; Baldani, I.; Schmid, M.; Hartmann, A. The bacterial superoxide dismutase and glutathione reductase are crucial for endophytic colonization of rice roots by Gluconacetobacter diazotrophicus PAL5. Mol. Plant Microbe Interact. 2013, 26, 937–945. [Google Scholar] [CrossRef]
- Wang, W.; Sun, M.; Liu, W.; Zhang, B. Purification and characterization of a psychrophilic catalase from antarctic bacillus. Can. J. Microbiol. 2008, 54, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Danson, M.J.; Hough, D.W.; Russell, R.J.; Taylor, G.L.; Pearl, L. Enzyme thermostability and thermoactivity. Protein Eng. 1996, 9, 629–630. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Switala, J.; Loewen, P.C. Diversity of properties among catalases. Arch. Biochem. Biophys. 2002, 401, 145–154. [Google Scholar] [CrossRef]
- Lorentzen, M.S.; Moe, E.; Jouve, H.M.; Willassen, N.P. Cold adapted features of Vibrio salmonicida catalase: Characterisation and comparison to the mesophilic counterpart from Proteus mirabilis. Extremophiles 2006, 10, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Díaz, A.; Loewen, P.C.; Fita, I.; Carpena, X. Thirty years of heme catalases structural biology. Arch. Biochem. Biophys. 2012, 525, 102–110. [Google Scholar] [CrossRef]
- Chelikani, P.; Fita, I.; Loewen, P.C. Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 2004, 61, 192–208. [Google Scholar] [CrossRef]

| Characteristics | I1P |
|---|---|
| Morphology | rod |
| Gram stain | negative |
| Temperature range (optimum) | 4–37 °C (22 °C) |
| pH range (optimum) | 5–10 (7–9) |
| Salinity range (optimum) | 2–19% (2–6%) |
| API 20 E tests | |
| β-galactosidase | − |
| Arginine dihydrolase | − |
| Lysine decarboxylase | − |
| Ornithine decarboxylas | − |
| Citrate utilization | − |
| H2S production | − |
| Urease | − |
| l-tryptophane deaminase | + |
| Indol production | − |
| Acetoin production | − |
| Gelatinase | + |
| d-glucose (fermentation-oxidation) | − |
| d-mannitol (fermentation-oxidation) | − |
| Inositol (fermentation-oxidation) | − |
| d-sorbitol (fermentation-oxidation) | − |
| l-rhamnose (fermentation-oxidation) | − |
| d-sucrose (fermentation-oxidation) | − |
| d-melibiose (fermentation-oxidation) | − |
| Amygdalin (fermentation-oxidation) | − |
| l-arabinose (fermentation-oxidation) | − |
| Nitrate reduction to NO2 | + |
| Nitrate reduction to N2 | + |
| E. coli | GEW1 | I1P | ||||
|---|---|---|---|---|---|---|
| Time (min) | Viability * | UV Dose # | Viability * | UV Dose # | Viability * | UV Dose # |
| 0 | 100 | 0 | 100 | 0 | 100 | 0 |
| 5 | 0 | 0.19 | 4.5 | 0.19 | 75.0 | 0.19 |
| 30 | 0 | 1.26 | 0.08 | 1.26 | 18.5 | 1.26 |
| 60 | 0 | 2.63 | 0.02 | 2.63 | 5.0 | 2.63 |
| 120 | 0 | 6.33 | 0.01 | 6.33 | 2.0 | 6.33 |
| Purification Step | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Purification (Fold) |
|---|---|---|---|---|
| Crude Extract | 2320 | 59,143 | 25 | 1 |
| DEAE- Sepharose FF | 26 | 82,141 | 318 | 13 |
| Superdex-200 | 13 | 81,321 | 6265 | 251 |
| Q-HiTrap FF | 2 | 12,511 | 7447 | 298 |
| Manufacturer | Commercial Name | Source | Applications | Specific Activity (U/mg) |
|---|---|---|---|---|
| Novozyme | Terminox Ultra | No data | Textile industry | 10 |
| Catazyme | Aspergillus niger * | Food and textile industry | 4960 | |
| Sigma-Aldrich | Catalase | A. niger | Research market | >4000 |
| Catalase | Corynebacterium glutamicum | >71,428 | ||
| Catalase | Micrococcus luteus | 65–150 | ||
| Biocatalysts | Catalase 929 L | A. niger | Food industry | 16.5 |
| Calzyme | Catalase | A. niger | No data | 2000 |
| This study | Catalase | Serratia sp. I1P | Research market | 7447 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monsalves, M.T.; Ollivet-Besson, G.P.; Amenabar, M.J.; Blamey, J.M. Isolation of a Psychrotolerant and UV-C-Resistant Bacterium from Elephant Island, Antarctica with a Highly Thermoactive and Thermostable Catalase. Microorganisms 2020, 8, 95. https://doi.org/10.3390/microorganisms8010095
Monsalves MT, Ollivet-Besson GP, Amenabar MJ, Blamey JM. Isolation of a Psychrotolerant and UV-C-Resistant Bacterium from Elephant Island, Antarctica with a Highly Thermoactive and Thermostable Catalase. Microorganisms. 2020; 8(1):95. https://doi.org/10.3390/microorganisms8010095
Chicago/Turabian StyleMonsalves, María T., Gabriela P. Ollivet-Besson, Maximiliano J. Amenabar, and Jenny M. Blamey. 2020. "Isolation of a Psychrotolerant and UV-C-Resistant Bacterium from Elephant Island, Antarctica with a Highly Thermoactive and Thermostable Catalase" Microorganisms 8, no. 1: 95. https://doi.org/10.3390/microorganisms8010095
APA StyleMonsalves, M. T., Ollivet-Besson, G. P., Amenabar, M. J., & Blamey, J. M. (2020). Isolation of a Psychrotolerant and UV-C-Resistant Bacterium from Elephant Island, Antarctica with a Highly Thermoactive and Thermostable Catalase. Microorganisms, 8(1), 95. https://doi.org/10.3390/microorganisms8010095




