Abstract
Aerobic moderately thermophilic and thermophilic methane-oxidizing bacteria make a substantial contribution in the control of global warming through biological reduction of methane emissions and have a unique capability of utilizing methane as their sole carbon and energy source. Here, we report a novel moderately thermophilic Methylococcus-like Type Ib methanotroph recovered from an alkaline thermal spring (55.4 °C and pH 8.82) in the Ethiopian Rift Valley. The isolate, designated LS7-MC, most probably represents a novel species of a new genus in the family Methylococcaceae of the class Gammaproteobacteria. The 16S rRNA gene phylogeny indicated that strain LS7-MC is distantly related to the closest described relative, Methylococcus capsulatus (92.7% sequence identity). Growth was observed at temperatures of 30–60 °C (optimal, 51–55 °C), and the cells possessed Type I intracellular membrane (ICM). The comparison of the pmoA gene sequences showed that the strain was most closely related to M. capsulatus (87.8%). Soluble methane monooxygenase (sMMO) was not detected, signifying the biological oxidation process from methane to methanol by the particulate methane monooxygenase (pMMO). The other functional genes mxaF, cbbL and nifH were detected by PCR. To our knowledge, the new strain is the first isolated moderately thermophilic methanotroph from an alkaline thermal spring of the family Methylococcaceae. Furthermore, LS7-MC represents a previously unrecognized biological methane sink in thermal habitats, expanding our knowledge of its ecological role in methane cycling and aerobic methanotrophy.
1. Introduction
Methane plays a key role in the global carbon cycle, being 34 times more powerful as a greenhouse gas than CO2, and is the most substantial contributor to climate effect [1]. Abiogenic methane from underground reservoirs is produced in catalytic reactions at high pressure and temperature. Especially in geothermal habitats, a mixture of methane and other gases (known as natural gas) enter the Earth’s atmosphere as a part of volcanic gases and hydrothermal solutions through seeps, degassing of spring water and gas venting. Moreover, anaerobic microbials (the presence of methanogenic archaea) in hyperthermal hot springs also contribute formation and releasing of biogenic methane into the atmosphere [2,3,4]. In some parts of Ethiopia (the Great Rift Valley regions), natural methane is released through thermal springs nearby the Rift Valley lakes. Such lakes and thermally heated water sediments from hot springs may affect the community structure and diversity of microorganisms and may have a major influence in the global carbon cycle.
The study of aerobic methane-oxidizing bacteria (MOB) or methanotrophs is of special interest because of their significant ecological role in the global carbon cycle and natural reduction of methane emission to the atmosphere from many different ecosystems. Moreover, these microorganisms have the ability of utilizing methane as their sole energy and have a unique multicomponent enzyme system called methane monooxygenase (MMO), of which two distinct types, a particulate membrane-bound enzyme (pMMO) and a cytoplasmic soluble, membrane-free form (sMMO) have been described. These bacteria are found worldwide in nature and have been detected and isolated from a variety of thermal and non-thermal habitats [5,6,7,8]. Until now, taxonomical and molecular diversity studies of aerobic methanotrophs comprise the three phyla of Proteobacteria, Verrucomicrobia and “Methylomirabilaeota” (candidate phylum NC10). In the phylum Proteobacteria, methanotrophs are currently reclassified and defined into five distinct families: the gammaproteobacterial Methylomonadaceae (referred to as Type Ia), Methylococcaceae (Type Ib, formerly named as Type X), Methylothermaceae (Type Ic) and the alphaproteobacterial Methylocystaceae (Type IIa) and Beijerinckiaceae (Type IIb), based on their genomic comparisons (digital DNA-DNA hybridization (dDDH)), reconstruction of genome phylogeny, average nucleotide identity (ANI) and average amino acid identity [9,10,11,12,13]. Within the phylum Verrucomicrobia (sometimes referred also to Type III methanotrophs), only one family is defined as Methylacidiphilaceae that consists of, up to now, two genera: Methylacidiphilum and Methylacidimicrobium [14,15,16].
Although the majority of reported aerobic methanotrophs are mesophilic (optimal between 10 and 35 °C) and neutrophilic, their actual physiological tolerance ranges from 0 to 72 °C, pH from 1 to 11 and salinities up to 30%. In fact, several thermotolerant (growth up to 50 °C) and moderately thermophilic methanotrophs have also been described [9,13,17]. Our knowledge of truly thermophilic or moderately thermophilic proteobacterial methanotrophs (Topt > 40 °C and Tmax < 67 °C) is still limited compared to their thermotolerant (Tmax < 50 °C), mesophilic (Tmax < 42 °C) or psychrotolerant (Tmax < 36 °C) counterparts. Only a few validly described species of thermophilic methanotrophs like Methylothermus thermalis (growth at 37−67 °C), Methylothermus subterraneus (growth at 37−65 °C) within the family Methylothermaceae and Methylocaldum szegediense (growth at 37−62 °C) within the family Methylococcaceae, could grow optimally above 55 °C [13,18,19]. The family Methylococcaceae presently comprises only seven phylogenetically associated genera: Methylococcus, Methylocaldum, Methyloparacoccus, Methylogaea, Methylomagnum, Methyloterricola and Methylotetracoccus [13,19,20,21,22,23,24,25]. In addition to these genera, the first isolated acid-tolerant moderately thermophilic (at a temperature range of 30−60 °C and at pH range 4.2−7.5) gammaproteobacterial methane oxidizers (methanotrophic isolates BFH1 and BFH2) were retrieved from a tropical topsoil with methane seeps habitat in Bangladesh, and possibly represent a novel new genus within the Type Ib methanotrophs. 16S rRNA gene phylogeny of both strains formed a cluster with the genus Methylocaldum as the closest described relative [26]. Furthermore, three novel isolates (strains GFS-K6, BRS-K6 and AK-K6) were also recovered from three different geographical regions and habitats: rice field soil, a methane seeps pond sediment from Bangladesh and a warm spring sediment from Armenia. But these microorganisms are mesophilic rather than thermotolerant and represent the members of Type Ib methanotrophs, and make, phylogenetically, a cluster together of the mesophilic genera Methylomagnum and Methyloparacoccus [27].
Methylococcus capsulatus strains, Texas and Bath, were the first reported thermotolerant methane oxidizers, growing up to 50 °C and at pH range 5.5−8.5, and were isolated from sewage sludge and geothermally heated water, respectively [21,28]. So far, strain M. capsulatus Bath is the most studied methanotroph and expanded knowledge of its ecophysiology, genetic and biochemistry. Bodrossy and colleagues, reported a bona fide novel thermophilic gammaproteobacterial methane-oxidizing bacterium (informally named as “Methylothermus” strain HB), which was recovered from underground hot springs in Hungary, and this bacterium represented the highest recorded growth temperature range at 40−72 °C, with an optimum at 62−65 °C, until now [29]. Sequence comparisons of both 16S rRNA and pmoA genes revealed that strain HB represents a novel genus of the Type Ic methanotrophs in the family Methylothermaceae, but unfortunately, this strain does not exist any longer [13]. Recently, the detection of a new gammaproteobacterial group of methanotrophs, distantly related to Methylococcus and Methylocaldum, in a Russian Far East thermal spring, provides new insights into the diversity and distribution of thermophilic methanotrophs [30]. Moreover, three thermoacidophilic strains, M. infernorum, M. fumiolicum and M. kamchatkense, were able to grow at temperatures of 37 to 65 °C and pH at up to 6.0. These were isolated from acidic geothermally heated soils and a hot spring, but they are members in the genus Methylacidiphilum of the phylum Verrucomicrobia. Verrucomicrobial methane oxidizers appear to be found only in acidic geothermal environments [14,15,16].
Several key functional molecular gene markers like pmoA (encoding a subunit of the particulate methane monooxygenase, pMMO: a copper-dependent enzyme), mmoX (encoding a subunit of the soluble methane monooxygenase, sMMO: an iron-dependent enzyme) and mxaF (encoding the large subunit of PQQ-dependent methanol dehydrogenase, MDH: a calcium-containing enzyme) were frequently applied for detecting and diversity analysis of C1-utilizing bacteria. Especially, the pmoA gene is often applied as a phylogenetic marker for identifying aerobic methanotrophs in various habitats [6,31]. Hitherto, no thermophilic methane oxidizers have been reported to exhibit both enzymes systems, indicating that methanotrophic cytoplasmic sMMO might not be existing in cells living above 55 °C [13,18,29]. The gene cbbL, encodes the large subunit of the ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCo: an essential enzyme in the Calvin-Benson-Bossham cycle), which is responsible for autotrophic growth. This gene has been commonly utilized for analyzing marine and hypersaline microbial communities as well as studies of Type Ib methanotrophs [26,32,33,34].
Recovering of moderately thermophilic methane-oxidizing bacteria from alkaline thermal springs (pH 8.0 to 9.0) is still a challenging process. Little is known about the identity, community structure and distribution of these bacteria from such ecosystems.
In this study, we have isolated and characterized the first moderately thermophilic Type Ib methanotroph, recovered from an alkaline thermal spring sample from the Ethiopian Rift Valley. The new isolate likely represents a new genus within the family Methylococcaceae of the class Gammaproteobacteria and extends our knowledge of this group of microorganisms in these environments.
2. Materials and Methods
2.1. Sample Collection, Enrichments and Growth Conditions
The alkaline-saline Lake Shalla is situated in the eastern Ethiopian Rift Valley [35], where extensive volcanic activity has disrupted drainage and thus helped create small shallow lake basins. Lake Shalla is a terminal lake with several thermal springs close by (Figure 1). The samples were collected in November 2007 from one of the largest ponds showing diffuse venting and emitting high temperature fluids. It is located 1561 m above sea level at a position 7°28′ 666′’N and 38°38′086′’E. Water and sediment samples were collected using a sterile pitcher connected to a stick at around 50 cm depth and around 50 cm from the shore of the pond. The samples were transferred to Falcon tubes that were filled completely with sediment and water from the thermal spring outlet.
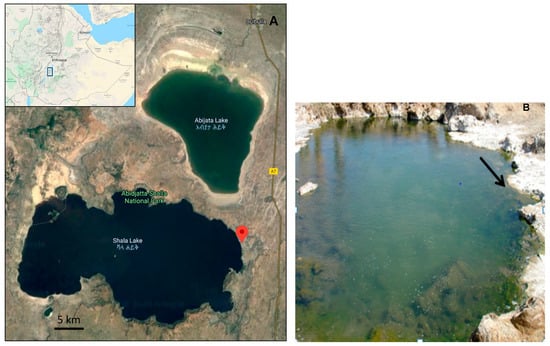
Figure 1.
Satellite image of the Ethiopian Rift Valley region, showing the Lake Shalla and the location of the hot-spring. An insert on the map shows the location of the lake in Ethiopia (Google Maps, 2020) (A). A close-up photograph of the sampling site (B).
For the enrichment and cultivation of moderately thermophilic methane oxidizers, 3 mL sediment slurry was added to 15 mL low-salt mineral medium (LMM) supplemented with KNO3 in 120 mL sterile serum flasks [27]. The pH of the LMM was adjusted to 7.0 with HCl or NaOH. In addition, a low-salt mineral medium, supplemented with either NH4 Cl (LMM-AC) or (NH4)2 SO4 (LMM-AS) adjusted to pH 7.0 was also utilized for methanotrophic enrichment cultures. The serum flasks were closed with butyl rubber stoppers with aluminum crimp seals and a mixture of 80% methane (purity 99.5%, Yara Praxair, Oslo, Norway) and 20% air aseptically was added through a syringe in the headspace. The same methane:air mixture was used in all growth experiments. The cultures were incubated at 55 °C for one week with shaking at 150 rpm in the dark.
2.2. Isolation and Purification of Strain LS7-MC
The primary enrichment culture was transferred up to five times and serial dilutions were made of the culture with LMM. 100 µL of dilutions (10−5 to 10−8) were spread onto gelrite solidified plates (20 g L−1; GelzanTM CM, Sigma-Aldrich) or agar (Difco) containing LMM. The plates were incubated for four weeks at 55 °C in jars filled with the methane and air mixture (4:1). No extra air was added in the jar. Colonies were picked and re-streaked onto fresh gelrite plates and re-incubated. Finally, one single colony was transferred to fresh liquid LMM with the methane:air mixture (4:1) in the headspace and incubated at 55 °C. These growth conditions with methane as the sole carbon source were routinely used for further cultivation of this isolate. The purity of the culture was evaluated by phase-contrast and electron microscopy, and repeated PCR amplification analysis of the partial 16S rRNA gene sequences. For further verification, the strain LS7-MC was also tested for lack of growth on acetate (10 mM), pyruvate (10 mM), succinate (10 mM), glucose (10 mM), ethanol (17 mM) and yeast extract (0.1%), streaking onto R2 A agar plates [36].
2.3. Naphthalene Assay, Acetylene Inhibition Test and Electron Microscopy
To test for the presence of sMMO, the naphthalene-oxidation assay was performed using a culture grown in liquid LMM without copper, as described by Graham and collaborators [37]. Acetylene has been applied as an inhibitor for completely blocking methane monooxygenase [38]. The effect of acetylene on strain LS7-MC was monitored by adding 4% (v/v) acetylene to the headspace after two days of incubation with methane. Three replicate flasks were used for the methane oxidation inhibition test and one flask without acetylene was used as a control. To validate the acetylene inhibition test, M. kamchatkense Kam1 and M. capsulatus Bath were used as positive controls under the same conditions [36,39]. The morphology of the strain LS7-MC was determined using phase-contrast microscopy (Nikon, Eclipse E400 microscope) and electron microscopy Jeol-1230. For electron microscopy, strain LS7-MC was grown at its optimum temperature (55 °C) and ultrathin sections were prepared as described previously [36].
2.4. Growth Conditions, Carbon and Nitrogen Sources
Growth of strain LS7-MC on various organic compounds was tested in liquid LMM supplemented with sterile substrates as potential carbon sources (acetate, pyruvate, succinate, glucose, lactate, maltose, ethanol and sorbitol) at a concentration of 10 mM. Growth on methanol, formaldehyde, methylamine, dimethylamine, formamide, glycerol, formate and yeast extract was examined using LMM containing the following variable concentration of the respective substrates: 0.01%, 0.05%, 0.1%, 0.25%, 0.5%, 0.75% and 1% (v/v). To determine the temperature range and the optimum temperature of strain LS7-MC, growth was tested at 16 different temperatures ranging from 20 to 70 °C. The effect of pH range was examined at the optimum temperature of 55 °C, with a range of 14 pH values from 5.0 to 10.0. The optical cell density was measured at 600 nm (Spectronic 21, Milton Roy Company). Nitrogen sources were tested by using liquid LMM, in which KNO3 (0.1 g L−1) was replaced by 0.1 g L−1 of NH4 Cl, (NH4)2 SO4, glycine, methylamine, dimethylamine and yeast extract. Strain LS7-MC was also tested in triplicate with nitrogen-free LMM, where N2 from the air (20% air in the headspace) was the only nitrogen source. Exponential-phase cells (1:10 dilution) were supplemented into LMM (without KNO3) and this culture was transferred three times for excluding any trace of nitrate from the original inoculum. Growth was examined after two weeks of incubation at 55 °C. The generation time and the specific growth rate were calculated from exponential phase of growth under optimal growth conditions. For testing heat-resistance, a 4-week-old culture of strain LS7-MC was heated at 80 °C for 10 min. Growth of strain LS7-MC was also tested without vitamins in LMM. The strain was examined for growth on LMM (KNO3) without added vitamins and CuSO4 · 5 H2 O (0.2 g L−1). Exponential-phase cells (1:10 dilution) were added to the medium and continued to grow until three passages. NaCl tolerance and the test of antibiotic sensitivity of strain LS7-MC were followed as delineated previously [26]. Growth was performed with methane for one week at 55 °C.
2.5. Fatty Acid Analysis
For fatty acid analysis, cultures of strain LS7-MC grown at optimum temperature (55 °C) and with cell densities 108 cells/mL were delivered to DSMZ (Deutsche Sammlung von Mikrooganismen und Zellkulturen GmbH, Germany), where the samples were processed by harvesting, saponification, methylation, extraction and base wash prior to gas chromatography analysis. The fatty acid database of the Microbial Identification System (MIS) was employed for comparing with the fatty acid patterns of strain LS7-MC.
2.6. PCR Amplification and Southern Blot Hybridization of Functional Genes
Genomic DNA was extracted using GenElute Bacterial Genomic DNA kit (Sigma). The 16S rRNA genes were amplified with the universal bacterial primers 27 f and 1492 r, using a Veriti 96-well Thermal Cycler (Applied Biosystems). The PCR was performed using DynazymeTM High-fidelity DNA polymerase (Finnzymes) and the PCR and sequencing protocols were followed as previously described [27]. The functional genes pmoA, mmoX, mxaF, cbbL and nifH (a gene responsible for nitrogen fixation) were amplified using the same PCR protocol as described above for the 16S rRNA gene. Primers used in this study are listed in Supplementary Table S1. Furthermore, amplified fragments of 16S rRNA genes, pmoA and cbbL genes were cloned using a TOPO-TA Cloning Kit (Invitrogen). The clones were screened for correct inserts and the fragments were sequenced. For confirmation of pMMO and sMMO, the Southern blotting technique was applied. Genomic DNA from strain LS7-MC, M. kamchatkense Kam1 (as a negative control) and M. capsulatus Bath and Methylococcaceae strain BFH1 (as positive controls) was extracted. Then, DNA was digested with EcoRI and HindIII. Hybridization probes and the further process were followed as previously described [32,36].
2.7. Phylogenetic Analysis and Nucleotide Sequence Accession Numbers
16S rRNA gene sequences, and PmoA, MxaF and CbbL protein sequences of strain LS7-MC were compared with available sequences in the GenBank database using the NCBI tools (Blastn and Blastp). To perform phylogenetic analysis, 16S rRNA gene sequences and deduced amino acid sequences of PmoA were aligned using the CLUSTAL W algorithm. Distances were computed and phylogenetic trees were constructed using the following methods: Neighbor Joining (NJ), Maximum Likelihood (ML), Minimum-Evolution (ME), and the following models: Maximum Composite Likelihood, Kimura 2-parameter, Tamura 3-parameter, Jukes–Cantor, Jones Taylor-Thornton (JTT), Poisson and Dayhoff, which are implemented in the MEGA7 software package. The confidence of the trees was determined by 1000 bootstrap replications [40]. The phylogenetic analysis of 16S rRNA and PmoA involved 1424 nucleotides and 167 amino acid sequences, respectively. The nearly complete 16S rRNA gene sequences and the partial sequences of the genes pmoA, mxaF, cbbL and nifH of the strain LS7-MC have been deposited in GenBank under the accession numbers KP771709, KP828775, KP843192, KP843193 and KP843194, respectively.
3. Results
3.1. Isolation of a Moderately Thermophilic Methylococcus-Like Methanotroph
The enrichments of moderately thermophilic methane oxidizers were achieved from an alkaline hydrothermal spring that has been present for the last 80 years. The in-situ temperature, the pH and the conductivity of the thermal spring were 55.4 °C, 8.82 and 8.6 S/m, respectively. Three separate enrichments, LMM (KNO3), LMM-AC (NH4 Cl) and LMM-AS ((NH4)2 SO4) were set up. After two weeks of incubation at 55 °C, the enrichment with LMM showed that cells’ turbidity and microbial growth was confirmed by phase-contrast microscopy. Growth on LMM-AC (NH4 Cl) and LMM-AS ((NH4)2 SO4) were not evident even after three weeks of incubation in the same culture conditions. Diluted LMM enrichment cultures were spread on gelrite plates and incubated for 10 days. Two different types of colonies were observed. One type was small white colonies about 0.6–0.8 mm in diameter and the other type was shiny and slightly larger, at about 1.2–1.5 mm in diameter. No such colonies appeared on agar plates. The small white colonies stopped growing after 10 days, but the cells were viable for about four weeks of incubation with methane. Using phase-contrast microscopy, the small white colonies were comprised of coccoid cells, whereas the larger and shiny colonies showed small rod-shaped cells. Physiological and taxonomic depiction of the ‘rod-shaped’ strain is currently under investigation (Islam et al., in prep.). For this current study we selected the small white colonies with coccoid cells for further characterization. The final isolate, designated LS7-MC, grew on methane or methanol as the sole carbon and energy source. It did not grow on ethanol, acetate, pyruvate, malate, succinate, methylamine, glucose, fructose or yeast extract, indicating that the isolate is an obligate aerobic methanotroph. No heterotrophic contaminants grew on these media, thus verifying the purity of the strain. Furthermore, repeated PCR amplification analysis of the partial 16S rRNA gene sequence from the methane and methanol grown cultures yielded the same sequence, confirming the purity of LS7-MC.
3.2. Growth and Physiological Characteristics of LS7-MC
The temperature range for growth was between 30 and 60 °C, and no growth was observed at 25 or 62 °C. The optimum growth temperature was 51−55 °C at pH 7.0. The cells did not appear to be heat-resistant. Growth occurred between pH 6.0–9.3 but not at pH 5 or 9.5. The optimal pH was between 7.0 and 7.5. Growth was not obtained under aerobic conditions in the absence of methane or under anaerobic conditions in the presence of methane. Furthermore, the isolate grew only in the presence of very low methanol concentrations, between 0.05% and 0.25%. Strain LS7-MC did not grow on LMM-AC (NH4 Cl), LMM-AS ((NH4)2 SO4) and nitrogen-free compounds, except LMM (KNO3). Only nitrate in the medium was used as a nitrogen source. This indicated that the addition of nitrate has strong effects on methane consumption for growth or that the strain was possibly sensitive to ammonium salts. Multicarbon compounds completely prevented growth on methane or methanol at pH 7.0. The strain was able to grow to an OD600 value of 1.2 on methane. The generation time and the specific growth rate of a culture grown on methane were estimated to 6 h and 0.115 h−1, respectively. No growth was observed after 10 days of incubation for each of these passages in LMM medium without added CuSO4 or vitamin, indicating that both vitamins and Cu+2 are essential for growth. NaCl was not required for growth, but the isolate could grow on medium supplemented with 0.1%–0.25% NaCl (w/v). No growth occurred at concentrations above 0.5% NaCl (w/v). All tested antibiotics (ampicillin 10 µg mL−1, tetracycline 10 μg mL−1, kanamycin 30 µg mL−1, streptomycin 10 µg mL−1, erythromycin 10 µg mL−1 and nalidixic acid 30 µg mL−1) suppressed growth of LS7-MC. The addition of acetylene (4%) to the headspace of exponentially growing cultures, resulted in inhibition of methane oxidation, and further growth of strain LS7-MC stopped. This demonstrated the presence of the functionally active methane oxidizing enzyme typical for MOB. Major characteristics of the strain LS7-MC with other described obligate Type Ib methanotrophic genera or species of the family Methylococcaceae are presented in Table 1.

Table 1.
Comparison of the major characteristics of the strain LS7-MC with other described Type Ib methanotrophs of the family Methylococcaceae. Strains: 1, This study; 2, Methylococcus capsulatus strain Bath [20,41]; 3, Methylocaldum spp. [17,19]; 4, Methylococcaceae strain BFH1 [26]; 5, Methylotetracoccus oryzae strain C50 C1 T [25]; 6, Methylogaea oryzae E10 T [21]; 7, Methyloparacoccus murrellii R-49797 T [22]; 8, Methylomagnum ishizawai RS11 D-PrT [23]; 9, Methyloterricola oryzae 73 aT [24]; +, positive results; –, negative results; nd, not determined.
3.3. Microscopic Observations
Coccoid-type cells were observed by phase contrast microscopy (Figure 2A,B). The cells occurred individually or in pair (diplococcus) with a diameter and a length of 0.9–1.2 µm (Figure 2C,D). During cell division, an ellipsoid form appeared which turned into a diplococcus. This morphology was also seen in a thermophilic methanotrophic strain HB [29]. The strain was non-motile and multiplied by binary fission. Flagella were not apparent by transmission electron microscopy (TEM). Analysis of ultrathin sections by TEM revealed a typical Gram-negative cell wall structure and moreover, the presence of extensive intracytoplasmic membrane (ICM) (Figure 2C,D). Cells became elongated when the growth temperature was above 55 °C.
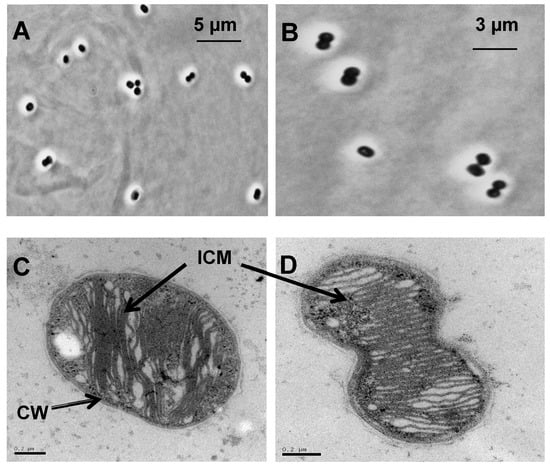
Figure 2.
Morphology of the methanotrophic strain LS7-MC. (A and B) Phase-contrast photomicrograph of cells grown in low-salt mineral medium (LMM) medium with methane at 55 °C for 5 days. Transmission electron micrograph (TEM) of the strain LS7-MC shows (C) a single cell and (D) a diplococcus. Ultrathin sections showing extensive intracytoplasmic membrane (ICM) systems and a typical Gram-negative cell wall (CW). Bars, 0.2 µm.
3.4. Phospholipids Fatty Acids (PLFA) Composition
The major fatty acids of strain LS7-MC are shown in Table 2 together with those of related Type Ib methanotrophs of the family Methylococcaceae. The fatty acid composition of this novel organism was revealed as comparable to other strains of the species M. capsulatus, Methylocaldum spp., Methylotetracoccus oryzae, Methylogaea oryzae, Methyloparacoccus murrellii, Methylomagnum ishiwaze and Metyloterricola oryzae regarding the two predominant fatty acids (C16:0 and C16:1ω7 c). In strain LS7-MC C16:0 (47.75%) and C16:1ω7 c (40.95%) accounted for 88.7% of the total amount of fatty acids. C16:0 is a major fatty acid in thermophilic, thermotolerant and mesophilic methanotrophs of the Type Ib genera like Methylocaldum, Methylococcace strain BFH1, Methylococcus, Methyloterricola and Methylogaea (30−60%), whereas mesophilic methanotroph genera (e.g., Methylotetracoccus, Methyloparacoccus and Methylomagnum) contain less than 24% of C16:0 [22]. Type Ia mesophilic or psychrotolerant genera in the family Methylomonadaceae (Methylosoma, Methylobacter and Methyloglobulus) exhibit more than 55% of C16:0 [43]. Hitherto, such high amounts of C16:1ω7 c found in strain LS7-MC have not been reported in any other described thermophilic or thermotolerant Type Ib methanotrophs. Methyloparacoccus, Methylomagnum and Methyloterricola as mesophiles, contain 54%, 47% and 27% of C16:1ω7 c, respectively. The following fatty acids were detected in less amounts (1% to 6%): C17:0 cyc, C16:0 3-OH and C18:1ω7 c, accounting for 9.0% of total fatty acids. In general, fatty acid profiles from other related methanotrophs differ significantly from that of strain LS7-MC.

Table 2.
Phospholipid fatty acids comparison of the strain LS7-MC and other related gammaproteobacterial.
3.5. Detection of Functional Genes
All amplification reactions of functional genes gave positive results except for the gene mmoX. Strain LS7-MC did not show any positive results in the naphthalene assay, verifying the absence of sMMO. Southern blotting analysis of genomic DNA also showed no positive signals with the mmoX probe, whereas the pmoA probe yielded positive signals (Supplementary Table S2). These results confirmed that strain LS7-MC does not contain the soluble form of MMO.
3.6. Phylogenetic Analysis of 16S rRNA and Functional Genes
A nearly complete sequence of the 16S rRNA gene (1424 bp) was obtained. Using Blastn search of the 16S rRNA gene sequence, strain LS7-MC showed a high sequence similarity (98.7%) to an uncultured bacterium clone (FG44 B-12) from a subterranean radioactive thermal spring in the Austrian central Alps (‘‘Franz-Josef-Quelle’’ in Bad Gastein; GenBank Accession No. FR846909) [44]. The strain showed 92.5% sequence similarity to uncultured bacterial clones from fracture water of a gold mine borehole in the USA (JX434172, JX434181, JX434257-58, JX434183, JX434188-89, JX434195, JX434201, JX434207, JX434214, JX434216, JX434220-22, JX434225), and a soil sample from Bugok geothermal in South Korea (MN726710). Low sequence similarity values were also found at 93.1% similarity to uncultured bacteria clones from industrial sugarcane bagasse feedstock piles [45], and Methylococcus sp. strain IM1 from geothermal field soils. The closest extant strains were M. capsulatus strain Bath (92.7% similarity), acid-tolerant moderately thermophilic strains BFH1 and BFH2, M. szegediense strain OR2 (92.9%), Methylocaldum sp. O-12 (92.8%) and Methylocaldum sp. E10 (92.9%). Analysis of pairwise alignment 16S rRNA gene sequence of the strain LS7-MC and the closest extant strains showed maximum sequence similarity (92.7%) with M. capsulatus strain Bath, which is a thermotolerant methanotroph of the family Methylococcaceae. Further analysis exhibited 91.2–92.4% sequence similarity to other strains (Supplementary Table S3). These results suggested that strain LS7-MC may represent a new member of the Type Ib methanotrophs rather than Type Ia (Methylomonadaceae) or Type Ic (Methylothermaceae) in the class Gammaproteobacteria. In the Neighbor-Joining tree of 16S rRNA gene (Figure 3), strain LS7-MC formed a phylogenetically separate linage within the closest genera of Methylococcus, Methylocaldum, Methylogaea and Methyloparacoccus, Methylomagnum, Methylotetracoccus and Methyloterricola. This topology was also verified with the Maximum-Likelihood (Supplementary Figure S1) and Minimum-Evolution (Supplementary Figure S2) trees, suggesting that strain LS7-MC is not a species within any of the known genera of the Type Ia, Type Ib or Type Ic methanotrophs.
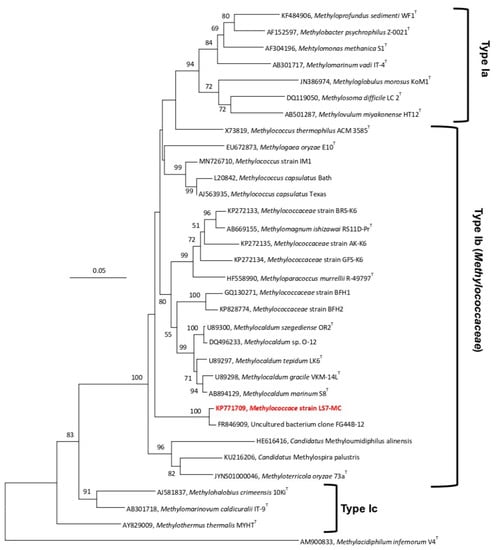
Figure 3.
16S rRNA gene-based phylogenetic tree, using the Neighbor-Joining method, showing the phylogenetic position of strain LS7-MC (highlighted in bold red) within the family Methylococcaceae (Type Ib) of the class Gammaproteobacteria. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site. Methylacidiphilum infernorum V4T (AM900833), a thermoacidophilic verrucomicrobial methanotroph, was used as an outgroup. Evolutionary analyses were conducted in MEGA7 [40]. The scale bar represents 0.05 changes per nucleotide position. Less than 50% of bootstrap values (1000 replicates) are not shown.
Based on the pmoA gene analysis and Blastn search, strain LS7-MC showed 93.8% (99.4% amino acid level) sequence identity to the partial pmoA gene sequences of a DGGE band of an uncultured bacterium clone in an Austrian radioactive thermal spring (AM749116) [46]. Lower sequence identities (88.2–91.2%) were found compared to many uncultured bacteria from different ecosystems, such as wetland soil (JQ038175), hot water (AF533666), diverse soils (MF107118), subsurface borehole water (KF901437), hot spring (KF836706), landfill cover soil (JX998182), forest soils (FN393903), coal mine soil (EU131055) and Danish soils (AF368356). The pmoA gene of the strain LS7-MC, based on pairwise sequence analysis, revealed 87.8% identity (99.4% at amino acid level) to M. capsulatus Bath, 79.7–82.7% identity (92.3–94.7% at amino acid level) to Methylocaldum spp. (M. szegediense OR2 T, M. tepidum LK6 T, M. gracile VKM 14 LT, and M. marinum S8 T), 84.6% identity (96.2%) to M. murrellii R-49797 T and 79.0% identity (95.3%) to M. oryzae E10 T (Supplementary Table S3). The phylogenetic analysis of partial derived PmoA amino acid sequences showed that the strain LS7-MC was clustered along with M. capsulatus, Methylocaldum spp., M. murrellii, M. oryzea and Methylococcaceae strains GFS-K6 and AK-K6 (Figure 4). The same topology was also found using Maximum-Likelihood (Supplementary Figure S3) and Minimum-Evolution (Supplementary Figure S4) trees, suggesting that the PmoA-based and 16S rRNA-based phylogenies generated a consistent position between strain LS7-MC and other cultivated Type Ib methanotrophs of the family Methylococcaceae. Analyses of the mxaF gene of the strain LS7-MC exhibited lower homology (83.6–85.9%) to several uncultured bacteria from coal mine soils (KF031213-15, KF031234, EU131063 and EU131061). The closest related genera, based on the mxaF gene (555 bp) and MxaF protein sequences (185 amino acids) of strain LS7-MC, were Methylococcus, Methylocaldum, Methyloparacoccus and Methylogaea, demonstrating 81.5–85.7% identity at the DNA sequences level and 96.5–97.8% similarity at the amino acid level (Supplementary Table S4). Pairwise partial-derived CbbL protein sequences (101 amino acids) comparison showed 98.1% identity with CbbL from M. capsulatus strain Bath (AF447860) and 95.0% with CbbL from M. szegediense (WP_026609010).
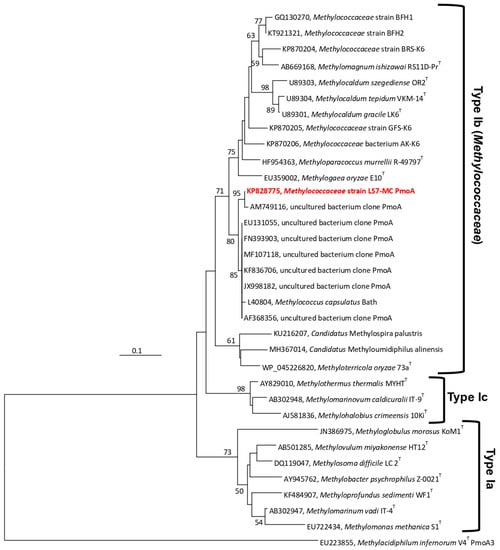
Figure 4.
Phylogenetic dendrogram based on derived deduced PmoA amino acid sequences showing the position of strain LS7-MC (highlighted in bold red) and other described gammaproteobacterial methanotrophs. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. Methylacidiphilum infernorum V4 T PmoA3 (EU223855), a thermoacidophilic verrucomicrobial methanotroph, was used as an outgroup. Evolutionary analyses were performed in MEGA7 [40]. The scale bar represents 0.1 changes per nucleotide position.
4. Discussion
Research on methane-oxidizing bacteria has mainly focused on low-temperature ecosystems. Most of the more than 50 validates species are either mesophilic or psycrophilic. Only a few thermophilic or moderately thermophilic species (8 species of proteobacterial and 3 species of verrucomicrobial methanotrophs) with optimal growth temperature between 50 and 55 °C have so far been reported [13,14,26]. In this study, enrichments for aerobic methanotrophs were established by inoculating sediment slurry from a thermal spring close to an alkaline lake (Lake Shalla) in the Ethiopian Rift Valley. From these enrichments, a novel moderately thermophilic Type Ib methanotroph, termed LS7-MC, was retrieved. This novel isolate shows an obligate aerobic methylotrophic growth with methane and methanol as the sole carbon and energy sources. According to 16S rRNA, PmoA, MxaF, CbbL and NifH sequence analyses, strain LS7-MC is most closely related to the Type Ib thermotolerant and moderately thermophilic methanotrophic bacteria within the family Methylococcaceae. Both the particulate methane monooxygenase gene pmoA and the methanol dehydrogenase gene mxaF, routinely used as functional and phylogenetic biomarkers for methanotrophic proteobacteria in natural environments [31,47], were detected in strain LS7-MC, indicating that methanotrophic proteobacteria are more widespread in thermal environments than previously thought. Furthermore, the difference in 16S rRNA gene sequences between strain LS7-MC and other related validated Type Ib methanotrophic genera ranges between 7% and 10%. The lack of soluble methane monooxygenase gene mmoX implies another significant difference between the thermotolerant genus Methylococcus and the strain LS7-MC presented here. These results ascertain that strain LS7-MC is, most probably, not a new species or subspecies of the genus Methylococcus or other genera of the families Candidate Methylomonadaceae, Methylococcaceae or Methylothermaceae. The 16S rRNA gene analyses suggested that this strain most probably represents a new genus in the methane-oxidizing bacterial family Methylococcaceae of the class Gammaproteobacteria.
Thermotolerant and moderately thermophilic proteobacterial methanotrophs have been found in various ecosystems from thermal spring sediments, subsurface hot aquifer, tropical landfill wetlands, methane seeps topsoil, compost and marine sediments [17,18,26,42]. The existence of these bacteria was supported by phylogenetic analyses of genes (like 16S rRNA, pmoA, mxaF and mmoX), through cultivation efforts and bacterial community analyses (using DNA-SIP, metagenomics, metatranscriptomics and next-generation sequencing) [48,49,50,51]. The comparative analysis of the 16S rRNA gene showed a relatively high sequence identity (>98%) to a clone of subsurface water sample (average pH 7.88) from an Austrian radioactive thermal spring [44]. The high sequence similarity of this clone to strain LS7-MC may indicate common physiology and metabolism properties between the strain LS7-MC and the uncultured thermal spring bacterium. Moderately thermophilic methanotrophs related to strain LS7-MC could be present in the central Austrian radioactive thermal spring and possibly in other related habitations of radioactive geothermal, and pmoA gene sequences obtained from the same thermal spring showed 93.8% sequence identity to pmoA of the strain LS7-MC. This is an indication that moderately thermophilic gammaproteobacterial methanotrophs may play a significant role in the carbon cycle in such subsurface thermal spring ecosystems. A relatively lower percentage identity of 16S rRNA, pmoA and mxaF sequences have also been detected in coal mine, diverse soils, wetland and landfill cover soils as well as hot spring water [19,52,53,54], suggesting that Methylococcus-like mesophilic or moderately thermophilic methanotrophs might be found in these various environments.
Strain LS7-MC required vitamins and copper for consistent growth and was negative with the naphthalene-oxidation assay. The lack of sMMO was also verified using the Southern blotting technique and PCR amplification. These central observations have also been described in other thermophilic proteobacterial isolates such as ‘Methylothermus’ strain HB [29], Methylothermus spp. [18,48] and thermophilic Methylocaldum spp. [17], as well as the verrucomicrobial M. kamchatkense Kam1 [36], which suggests that genes encoding soluble methane monooxygenase most probably do not exist in moderately thermophilic or thermophilic proteobacterial methanotrophs or verrucomicrobial thermoacidophilic methanotrophs [14]. On the other hand, within Type Ib methane-oxidizers, thermotolerants such as M. capsulatus and M. marinum, and mesophilic M. ishizawai (growth range 4–37 °C) possess both sMMO and pMMO enzyme systems [13,42,55].
5. Conclusions
We have retrieved an obligate moderately thermophilic Type Ib methanotroph that belongs to the family Methylococcaceae of the class Gammaproteobacteria. This new isolate is a Methylococcus-like bacterium that contains the particulate methane monooxygenase (pMMO) but does not contain soluble methane monooxygenase (sMMO) in the methane oxidation process. Based on the physiological, biochemical and genotypic properties, strain LS7-MC most probably represents a novel genus within the family Methylococcaceae. This strain also denotes a previously unrecognized biological methane sink, diversity of methane oxidation and on the adaptation of this process to alkaline thermal habitats. Furthermore, this finding will increase our knowledge of methanotroph ecology and its involvement to global cycles of carbon and nitrogen, and the thermophilic nature of this strain possibly makes a considerable candidate for potential biotechnological applications [56]. Additional studies regarding the molecular biology, biochemistry and whole genome of LS7-MC are needed to provide insight into how biological methane oxidation processes and mechanisms are regulated in alkaline thermal ecosystems.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/2/250/s1. Table S1: Primers for amplification of functional genes of the strain LS7-MC. Table S2: Results of Southern blot analysis of radioactively labeled pmoA and mmoX probes. Table S3: Pairwise sequence (EMBOSS) alignment analysis of 16S rRNA gene sequences, pmoA gene and partial-derived PmoA amino acid sequences shows similarity between strain LS7-MC and other cultivated gammaproteobacterial methanotrophs. Table S4: Pairwise MxaF protein sequences similarity comparisons (EMBOSS) between LS7-MC and other related methanotrophs of the family Methylococcaceae. Figure S1: Phylogenetic tree based on 16S rRNA gene sequences. Figure S2: Phylogenetic tree based on 16S rRNA gene sequences. Figure S3: Phylogenetic tree of PmoA protein sequences. Figure S4: Phylogenetic tree of PmoA protein sequences.
Author Contributions
T.I., A.G., A.G.-M., J.C.M. and L.Ø. designed the experiments, analyzed the data and wrote and edited the manuscript. T.I. performed the isolation of strain LS7-MC, and carried out morphological, biochemical characterization and phylogenetic analysis. L.Ø. collected samples and contributed reagents/materials/TEM analysis. All authors have read and agreed to the published version of the manuscript.
Funding
The Norwegian Council of Universities Committee for Developmental Research and Education (NUFU) funded this project through grant number NUFUPRO-2007 10069 to University of Addis Ababa and University of Bergen.
Acknowledgments
This study was supported through the Ethiopian–Norwegian collaboration project “Biotechnology and microbial diversity of Ethiopian Soda Lakes”. The electron microscopy imaging was performed at the Molecular Imaging Center (MIC), Department of Biomedicine, University of Bergen.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- IPCC. Climate Change:The Physical Science Basis, Contribution of Working Group I to the Fith Assessment Report of the IPCC.; Cambridge University Press: Cambridge, UK, 2013. [Google Scholar]
- Etiope, G.; Lassey, K.R.; Klusman, R.W.; Boschi, E. Reappraisal of the fossil methane budget and related emission from geologic sources. Geophy. Res. Lett. 2008, 35, L09307. [Google Scholar] [CrossRef]
- Nazaries, L.; Murrell, J.C.; Millard, P.; Baggs, L.; Singh, B.K. Methane, microbes and models: Fundamental understanding of the soil methane cycle for future predictions. Environ. Microbiol. 2013, 15, 2395–2417. [Google Scholar] [CrossRef] [PubMed]
- Tikhonova, E.N.; Kravchenko, I.K. Activity and Diversity of Aerobic Methanotrophs in Thermal Springs of the Russian Far East. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–30. [Google Scholar]
- Hanson, R.S.; Hanson, T.E. Methanotrophic bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef] [PubMed]
- McDonald, I.R.; Bodrossy, L.; Chen, Y.; Murrell, J.C. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 2008, 74, 1305–1315. [Google Scholar] [CrossRef]
- Semrau, J.D.; Dispirito, A.A.; Murrell, J.C. Life in the extreme: Thermoacidophilic methanotrophy. Trends Microbiol. 2008, 16, 190–193. [Google Scholar] [CrossRef]
- Saidi-Mehrabad, A.; He, Z.; Tamas, I.; Sharp, C.E.; Brady, A.L.; Rochman, F.F.; Bodrossy, L.; Abell, G.C.; Penner, T.; Dong, X.; et al. Methanotrophic bacteria in oilsands tailings ponds of northern Alberta. ISME J. 2013, 7, 908–921. [Google Scholar] [CrossRef]
- Bowman, J. The Methanotrophs—The Families Methylococcaceae and Methylocystaceae, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Eds.; Springer: New York, NY, USA, 2006; pp. 266–289. [Google Scholar]
- Trotsenko, Y.A.; Murrell, J.C. Metabolic Aspects of Aerobic Obligate Methanotrophy. Adv. Appl. Microbiol. 2008, 63, 183–229. [Google Scholar]
- Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 2015, 6, 1346. [Google Scholar] [CrossRef]
- Orata, F.D.; Meier-Kolthoff, J.P.; Sauvageau, D.; Stein, L.Y. Phylogenomic analysis of the gammaproteobacterial methanotrophs (Order Methylococcales) calls for the reclassification of members at the genus and species levels. Front. Microbiol. 2018, 9, 3162. [Google Scholar] [CrossRef]
- Houghton, K.M.; Carere, C.R.; Stott, M.B.; McDonald, I.R. Thermophilic methanotrophs: In hot pursuit. FEMS Microbiol. Ecol. 2019, 95, fiz125. [Google Scholar] [CrossRef]
- Op den Camp, H.J.; Islam, T.; Stott, M.B.; Harhangi, H.R.; Hynes, A.; Schouten, S.; Jetten, M.S.; Birkeland, N.K.; Pol, A.; Dunfield, P.F. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 2009, 1, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Sharp, C.E.; Smirnova, A.V.; Graham, J.M.; Stott, M.B.; Khadka, R.; Moore, T.R.; Grasby, S.E.; Strack, M.; Dunfield, P.F. Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ. Microbiol. 2014, 16, 1867–1878. [Google Scholar] [CrossRef] [PubMed]
- Van Teeseling, M.C.; Pol, A.; Harhangi, H.R.; van der Zwart, S.; Jetten, M.S.; Op den Camp, H.J.; van Niftrik, L. Expanding the verrucomicrobial methanotrophic world: Description of three novel species of Methylacidimicrobium gen. nov. Appl. Environ. Microbiol. 2014, 80, 6782–6791. [Google Scholar] [CrossRef] [PubMed]
- Eshinimaev, B.T.; Medvedkova, K.A.; Khmelenina, V.N.; Suzina, N.E.; Osipov, G.A.; Lysenko, A.M.; Trotsenko, Y.A. New thermophilic methanotrophs of the genus Methylocaldum. Mikrobiologiia 2004, 73, 530–539. [Google Scholar] [CrossRef]
- Hirayama, H.; Suzuki, Y.; Abe, M.; Miyazaki, M.; Makita, H.; Inagaki, F.; Uematsu, K.; Takai, K. Methylothermus subterraneus sp. nov., a moderately thermophilic methanotroph isolated from a terrestrial subsurface hot aquifer. Int. J. Syst. Evol. Microbiol. 2011, 61, 2646–2653. [Google Scholar] [CrossRef]
- Bodrossy, L.; Holmes, E.M.; Holmes, A.J.; Kovacs, K.L.; Murrell, J.C. Analysis of 16 S rRNA and methane monooxygenase gene sequences reveals a novel group thermotolerant and thermophilic methanotrophs, Methylocaldum gen. nov. Arch. Microbiol. 1997, 168, 493–503. [Google Scholar] [CrossRef]
- Foster, J.W.; Davis, R.H. A methane-dependent coccus, with notes on classification and nomenclature of obligate, methane-utilizing bacteria. J Bacteriol. 1966, 91, 1924–1931. [Google Scholar] [CrossRef]
- Geymonat, E.; Ferrando, L.; Tarlera, S.E. Methylogaea oryzae gen. nov., sp. nov., a mesophilic methanotroph isolated from a rice paddy field. Int. J. Syst. Evol. Microbiol. 2011, 61, 2568–2572. [Google Scholar] [CrossRef]
- Hoefman, S.; van der Ha, D.; Iguchi, H.; Yurimoto, H.; Sakai, Y.; Boon, N.; Vandamme, P.; Heylen, K.; de Vos, P. Methyloparacoccus murrellii gen. nov., sp. nov., a methanotroph isolated from pond water. Int. J. Syst. Evol. Microbiol. 2014, 64, 2100–2107. [Google Scholar] [CrossRef]
- Khalifa, A.; Lee, C.G.; Ogiso, T.; Ueno, C.; Dianou, D.; Demachi, T.; Katayama, A.; Asakawa, S. Methylomagnum ishizawai gen. nov., sp. nov., a mesophilic type I methanotroph isolated from rice rhizosphere. Int. J. Syst. Evol. Microbiol. 2015, 65, 3527–3534. [Google Scholar] [CrossRef]
- Frindte, K.; Maarastawi, S.A.; Lipski, A.; Hamacher, J.; Knief, C. Characterization of the first rice paddy cluster I isolate, Methyloterricola oryzae gen. nov., sp. nov. and amended description of Methylomagnum ishizawai. Int. J. Syst. Evol. Microbiol. 2017, 67, 4507–4514. [Google Scholar] [CrossRef] [PubMed]
- Ghashghavi, M.; Belova, S.E.; Bodelier, P.L.; Dedysh, S.N.; Kox, M.A.; Speth, D.R.; Frenzel, P.; Jetten, M.S.M.; Lüker, S.; Lüke, C. Methylotetracoccus oryzae Strain C50 C1 Is a Novel Type Ib Gammaproteobacterial Methanotroph Adapted to Freshwater Environments. Msphere 2019, 4, e00631-18. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Torsvik, V.; Larsen, Ø.; Bodrossy, L.; Øvreås, L.; Birkeland, N.K. Acid-tolerant moderately thermophilic methanotrophs of the class Gammaproteobacteria isolated from tropical topsoil with methane seeps. Front. Microbiol. 2016, 7, 851. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Larsen, Ø.; Torsvik, V.; Øvreås, L.; Panosyan, H.; Murrell, J.C.; Birkeland, N.K.; Bodrossy, L. Novel methanotrophs of the family Methylococcaceae from different geographical regions and habitats. Microorganisms 2015, 3, 484–499. [Google Scholar] [CrossRef] [PubMed]
- Whittenbury, R.; Phillips, K.C.; Wilkinson, J.F. Enrichment, isolation and some properties of methane-utilizing bacteria. J. Gen. Microbiol. 1970, 61, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Bodrossy, L.; Kovacs, K.L.; McDonald, I.R.; Murrell, J.C. A novel thermophilic methane-oxidising γ-Proteobacterium. FEMS Microbiol. Lett. 1999, 170, 335–341. [Google Scholar] [CrossRef]
- Kizilova, A.K.; Sukhacheva, M.V.; Pimenov, N.V.; Yurkov, A.M.; Kravchenko, I.K. Methane oxidation activity and diversity of aerobic methanotrophs in pH-neutral and semi-neutral thermal springs of the Kunashir Island, Russian Far East. Extremophiles 2014, 18, 207–218. [Google Scholar] [CrossRef]
- Lau, E.; Fisher, M.C.; Steudler, P.A.; Cavanaugh, C.M. The methanol dehydrogenase gene, mxaF, as a functional and phylogenetic marker for proteobacterial methanotrophs in natural environments. PLoS ONE 2013, 8, e56993. [Google Scholar] [CrossRef]
- Baxter, N.J.; Hirt, R.P.; Bodrossy, L.; Kovacs, K.L.; Embley, T.M.; Prosser, J.I.; Murrell, J.C. The ribulose-1,5-bisphosphate carboxylase/oxygenase gene cluster of Methylococcus capsulatus (Bath). Arch. Microbiol. 2002, 177, 279–289. [Google Scholar] [CrossRef]
- Tourova, T.P.; Kovaleva, O.L.; Sorokin, D.Y.; Muyzer, G. Ribulose-1, 5-bisphosphate carboxylase/oxygenase genes as a functional marker for chemolithoautotrophic halophilic sulfur-oxidizing bacteria in hypersaline habitats. Microbiology 2010, 156, 2016–2025. [Google Scholar] [CrossRef]
- Danilova, O.V.; Suzina, N.E.; Van De Kamp, J.; Svenning, M.M.; Bodrossy, L.; Dedysh, S.N. A new cell morphotype among methane oxidizers: A spiral-shaped obligately microaerophilic methanotroph from northern low-oxygen environments. ISME J. 2016, 10, 2734. [Google Scholar] [CrossRef]
- Lanzén, A.; Simmachew, A.; Gessesse, A.; Chmolowska, D.; Jonassen, I.; Øvreås, L. Surprising Prokaryotic and Eukaryotic diversity, community structure and biogeography of Ehiopian soda lakes. PLoS ONE 2013, 8, e72577. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Jensen, S.; Reigstad, L.J.; Larsen, O.; Birkeland, N.K. Methane oxidation at 55 degrees C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. USA 2008, 105, 300–304. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.; Korich, D.; LeBlanc, R.; Sinclair, N.; Arnold, R. Applications of a colorimetric plate assay for soluble methane monooxygenase activity. Appl. Environ. Microbiol. 1992, 58, 2231–2236. [Google Scholar] [CrossRef] [PubMed]
- Prior, S.; Dalton, H. Acetylene as a suicide substrate and active site probe for methane monooxygenase from Methylococcus capsulatus (Bath). FEMS Microbiol. Lett. 1985, 29, 105–109. [Google Scholar] [CrossRef]
- Bédard, C.; Knowles, R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol. Rev. 1989, 53, 68–84. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Takeuchi, M.; Kamagata, Y.; Oshima, K.; Hanada, S.; Tamaki, H.; Marumo, K.; Maeda, H.; Nedachi, M.; Hattori, M.; Iwasaki, W.; et al. Methylocaldum marinum sp. nov., a thermotolerant, methane-oxidizing bacterium isolated from marine sediments, and emended description of the genus Methylocaldum. Int. J. Syst. Evol. Microbiol. 2014, 64, 3240–3246. [Google Scholar] [CrossRef]
- Bowman, J.P.; Sly, L.I.; Nichols, P.D.; Hayward, A.C. Revised taxonomy of the methanotrophs: Description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int. J. Syst. Bacteriol. 1993, 43, 735–753. [Google Scholar] [CrossRef]
- Deutzmann, J.S.; Hoppert, M.; Schink, B. Characterization and phylogeny of a novel methanotroph, Methyloglobulus morosus gen. nov., spec. nov. Syst. Appl. Microbiol. 2014, 37, 165–169. [Google Scholar] [CrossRef]
- Weidler, G.W.; Gerbl, F.W.; Stan-Lotter, H. Crenarchaeota and Their Role in the Nitrogen Cycle in a Subsurface Radioactive Thermal Spring in the Austrian Central Alps. Appl. Environ. Microbiol. 2008, 74, 5934–5942. [Google Scholar] [CrossRef] [PubMed]
- Rattanachomsri, U.; Kanokratana, P.; Eurwilaichitr, L.; Igarashi, Y.; Champreda, V. Culture-Independent Phylogenetic Analysis of the Microbial Community in Industrial Sugarcane Bagasse Feedstock Piles. Biosci. Biotechnol. Biochem. 2011, 75, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Weidler, G.W.; Dornmayr-Pfaffenhuemer, M.; Gerbl, F.W.; Heinen, W.; Stan-Lotter, H. Communities of archaea and bacteria in a subsurface radioactive thermal spring in the Austrian Central Alps, and evidence of ammonia-oxidizing Crenarchaeota. Appl. Environ. Microbiol. 2007, 73, 259–270. [Google Scholar] [CrossRef] [PubMed]
- McDonald, I.R.; Murrell, J.C. The particulate methane monooxygenase gene pmoA and its use as a functional gene probe for methanotrophs. FEMS Microbiol. Lett. 1997, 156, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Osaka, T.; Ebie, Y.; Tsuneda, S.; Inamori, Y. Identification of the bacterial community involved in methane-dependent denitrification in activated sludge using DNA stable-isotope probing. FEMS Microbiol. Ecol. 2008, 64, 494–506. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, M.; Abraham, W.R.; Shrestha, P.M.; Noll, M.; Conrad, R. Activity and composition of methanotrophic bacterial communities in planted rice soil studied by flux measurements, analyses of pmoA gene and stable isotope probing of phospholipid fatty acids. Environ. Microbiol. 2008, 10, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Dumont, M.G.; Pommerenke, B.; Casper, P. Using stable isotope probing to obtain a targeted metatranscriptome of aerobic methanotrophs in lake sediment. Environ. Microbiol. Rep. 2013, 5, 757–764. [Google Scholar] [CrossRef]
- Lau, E.; Iv, E.J.; Dillard, Z.W.; Dague, R.D.; Semple, A.L.; Wentzell, W.L. High Throughput Sequencing to Detect Differences in Methanotrophic Methylococcaceae and Methylocystaceae in Surface Peat, Forest Soil, and Sphagnum Moss in Cranesville Swamp Preserve, West Virginia, USA. Microorganisms 2015, 3, 113–136. [Google Scholar] [CrossRef]
- Knief, C.; Dunfield, P.F. Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environ. Microbiol. 2005, 7, 1307–1317. [Google Scholar] [CrossRef]
- Kong, J.Y.; Su, Y.; Zhang, Q.Q.; Bai, Y.; Xia, F.F.; Fang, C.R.; He, R. Vertical profiles of community and activity of methanotrophs in landfill cover soils of different age. J. Appl. Microbiol. 2013, 115, 756–765. [Google Scholar] [CrossRef]
- Sengupta, A.; Dick, W.A. Methanotrophic bacterial diversity in two diverse soils under varying land-use practices as determined by high-throughput sequencing of the pmoA gene. Appl. Soil Ecol. 2017, 119, 35–45. [Google Scholar] [CrossRef]
- Ward, N.; Larsen, Ø.; Sakwa, J.; Bruseth, L.; Khouri, H.; Durkin, A.S.; Dimitrov, G.; Jiang, L.; Scanlan, D.; Kang, K.H.; et al. Genomic insights into methanotrophy: The complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2004, 2, e303. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.; Ho, A.; Yoon, S. Novel approaches and reasons to isolate methanotrophic bateria with biotechnological potentials: Recent achievements and perspectives. Appl. Microbiol. Biotechnol. 2019, 103, 1–8. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).