Novel Sequence Feature of SecA Translocase Protein Unique to the Thermophilic Bacteria: Bioinformatics Analyses to Investigate Their Potential Roles
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Conserved Signature Indels (Insertions/Deletions) and Phylogenetic Analysis
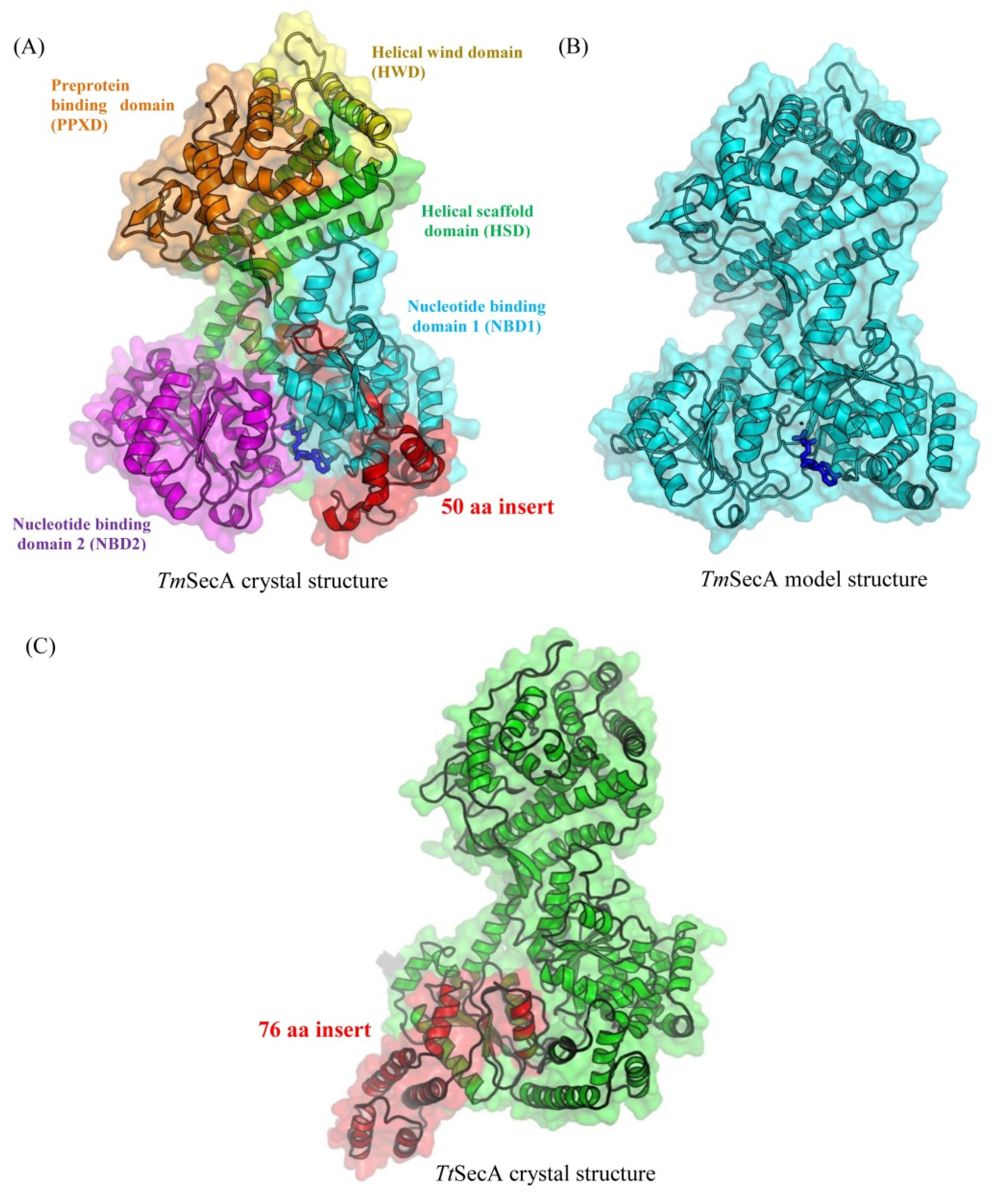
2.2. Homology Modelling of SecA Homologs and Structural Analysis of CSIs
2.3. Molecular Dynamics Simulations
3. Results
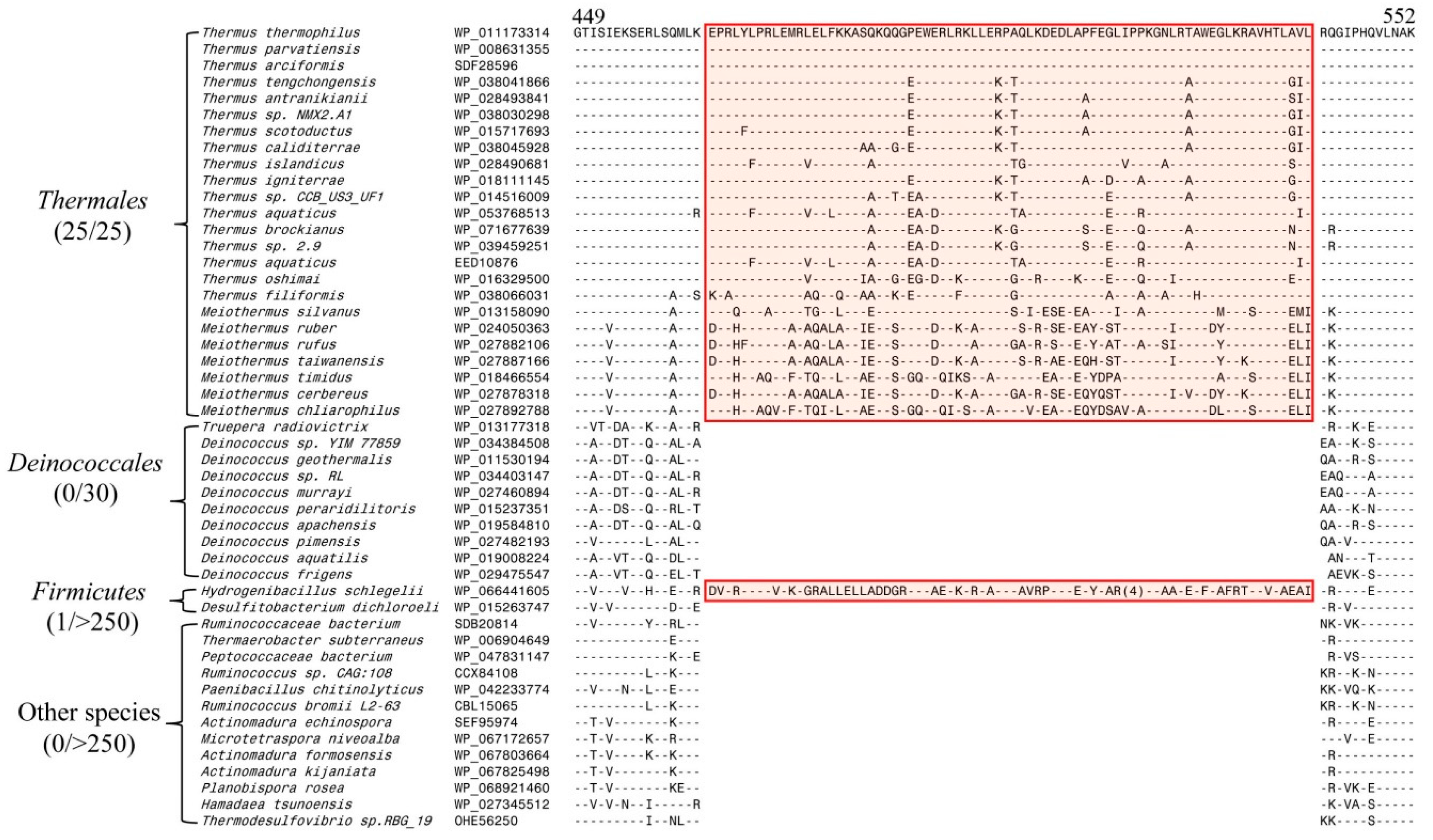
3.1. Identification of Conserved Signature Indels in SecA Homologs from Thermotogales, Aquificales, and Thermales and their Phylogenetic Implications
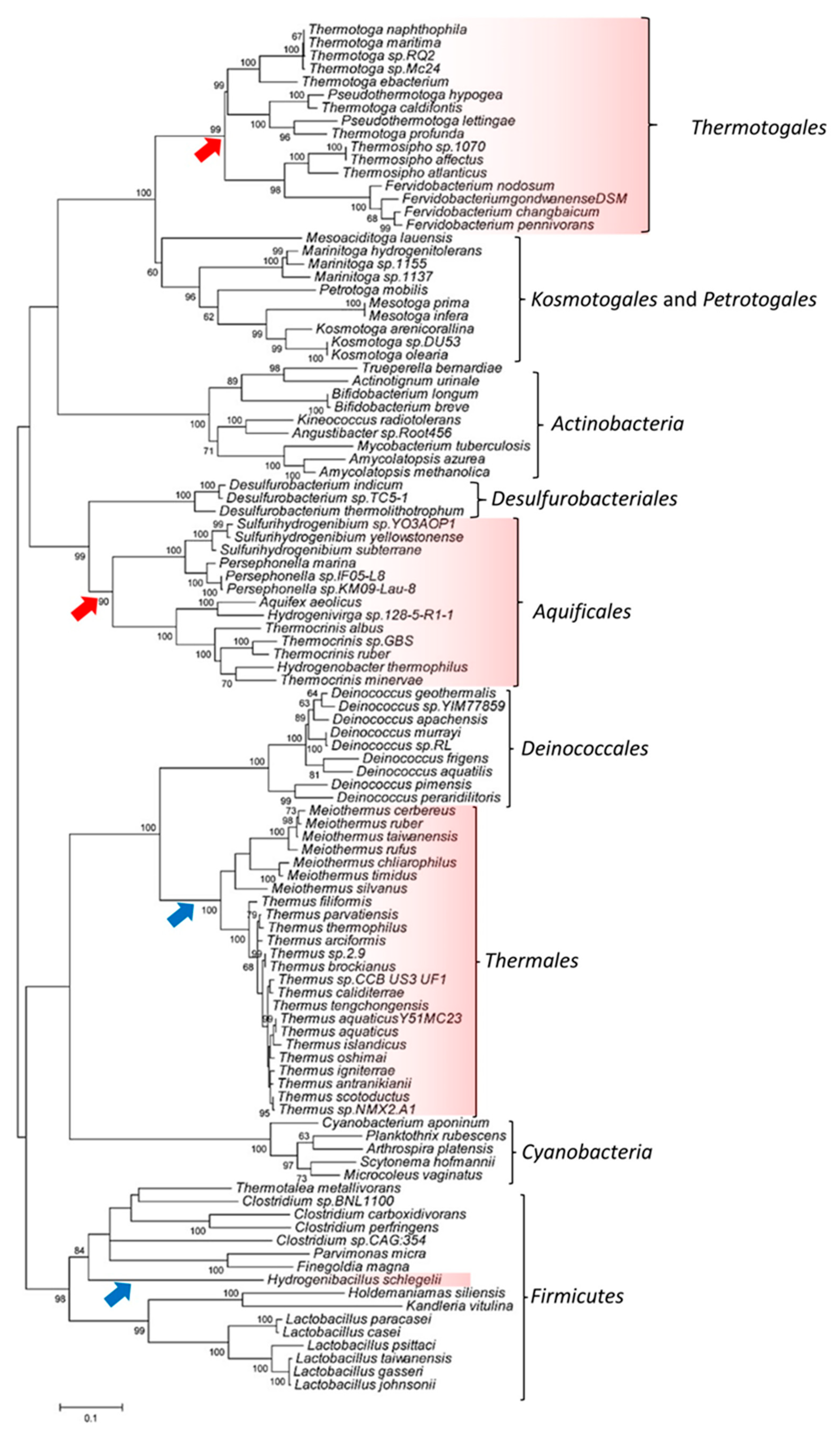
3.2. Phylogenetic Analysis of the SecA Proteins to Investigate the SHARED Presence of CSIs
3.3. Computational Analysis of the CSIs in SecA Proteins
3.4. Molecular Dynamics (MD) Simulation Studies of SecA Containing 50 aa CSI Specific for Thermotogales and Aquificales: Analysis of TmSecA Conformational Stability and Flexibility
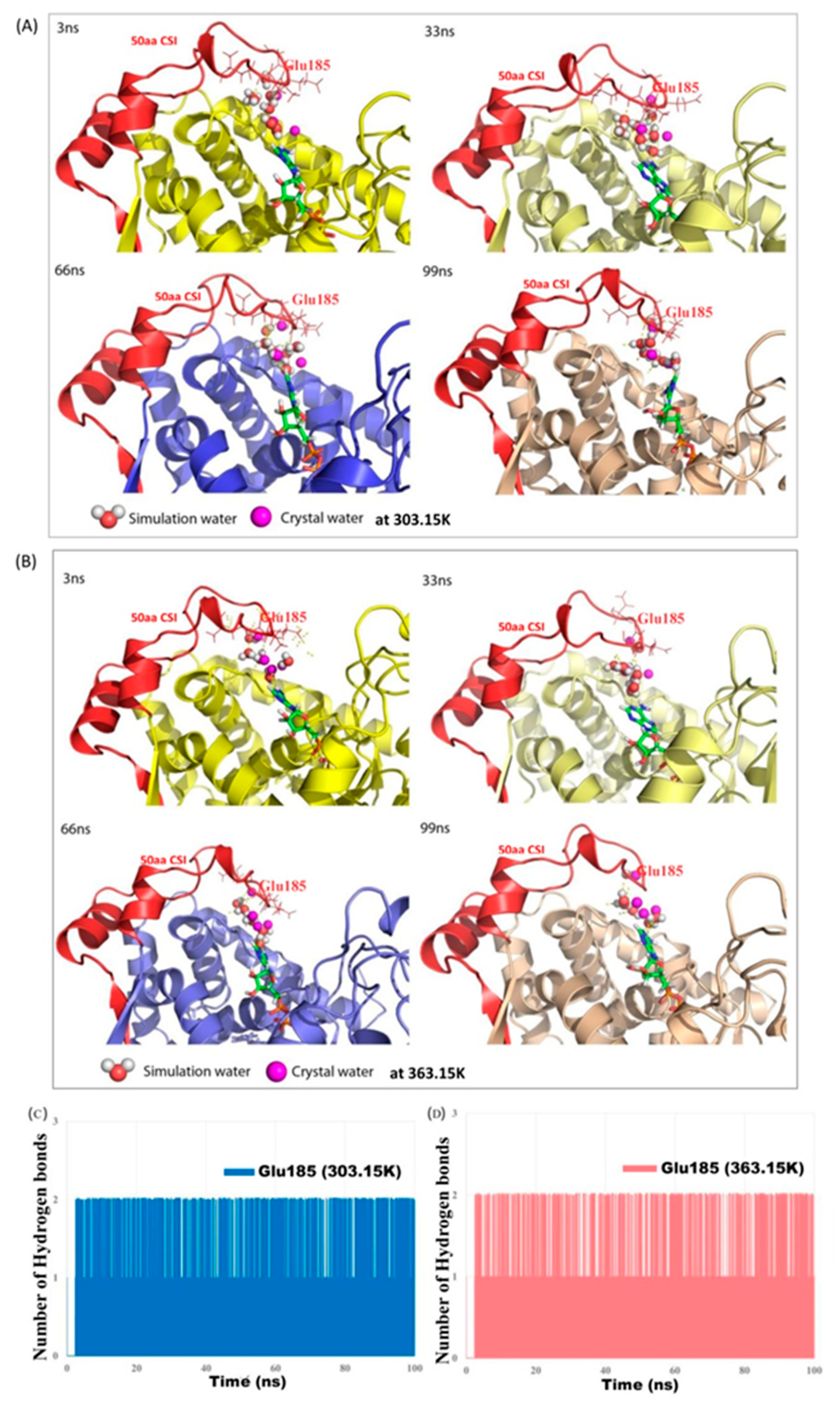
3.5. Identification of Conserved CSI-mediated Water Network in TmSecA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wiegel, J.; Ljungdhal, L.G.; Demain, A.L. The Importance of Thermophilic Bacteria in Biotechnology. Crit. Rev. Biotechnol. 1985, 3, 39–108. [Google Scholar] [CrossRef]
- Lasa, I.; Berenguer, J. Thermophilic Enzymes and Their Biotechnological Potential. Microbiologia 1993, 9, 77–89. [Google Scholar] [PubMed]
- Turner, P.; Mamo, G.; Karlsson, E.N. Potential and Utilization of Thermophiles and Thermostable Enzymes in Biorefining. Microb. Cell Fact. 2007, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, V.; Gupta, R.S. Molecular Signatures for the Phylum (class) Thermotogae and a Proposal for Its Division into Three Orders (Thermotogales, Kosmotogales ord. nov. and Petrotogales ord. nov.) Containing Four Families (Thermotogaceae, Fervidobacteriaceae fam. nov., Kosmotogaceae fam. nov. and Petrotogaceae fam. nov.) and a New Genus Pseudothermotoga gen. nov. with Five New Combinations. Antonie Van Leeuwenhoek 2014, 105, 143–168. [Google Scholar]
- Podar, M.; Reysenbach, A.L. New Opportunities Revealed by Biotechnological Explorations of Extremophiles. Curr. Opin. Biotechnol. 2006, 17, 250–255. [Google Scholar] [CrossRef]
- Huber, R.; Langworthy, T.A.; König, H.; Thomm, T.; Woese, C.R.; Sleytr, U.B.; Stetter, K.O. Thermotoga Maritima sp. nov. Represents a New Genus of Unique Extremely Thermophilic Eubacteria Growing up to 90 °C. Arch. Microbiol. 1986, 144, 324–333. [Google Scholar] [CrossRef]
- Robinson-Rechavi, M.; Godzik, A. Structural Genomics of Thermotoga Maritima Proteins Shows that Contact Order is a Major Determinant of Protein Thermostability. Structure 2005, 13, 857–860. [Google Scholar] [CrossRef]
- Sadaf, A.; Fatima, S.W.; Khare, S.K. Sporotrichum Thermophile Xylanases and Their Biotechnological Applications. In Fungi in Extreme Environments: Ecological Role and Biotechnological Significance; Tiquia-Arashiro, S., Grube, M., Eds.; Springer: Cham, Switzerland, 2019; pp. 307–328. [Google Scholar]
- Vieille, C.; Zeikus, G.J. Hyperthermophilic Enzymes: Sources, Uses, and Molecular Mechanisms for Thermostability. Microbiol. Mol. Biol. Rev. 2001, 65, 1–43. [Google Scholar] [CrossRef]
- Chien, A.; Edgar, D.B.; Trela, J.M. Deoxyribonucleic acid Polymerase from the Extreme Thermophile Thermus aquaticus. J. Bacteriol. 1976, 127, 1550–1557. [Google Scholar]
- Sterner, R.; Liebl, W. Thermophilic Adaptation of Proteins. Crit. Rev. Biochem. Mol. Biol. 2001, 36, 39–106. [Google Scholar] [CrossRef]
- Zhou, X.X.; Wang, Y.B.; Pan, Y.J.; Li, W.F. Differences in Amino acids Composition and Coupling Patterns between Mesophilic and Thermophilic Proteins. Amino. Acids 2008, 34, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Szilagyi, A.; Zavodszky, P. Structural Differences between Mesophilic, Moderately Thermophilic and Extremely Thermophilic Protein Subunits: Results of a Comprehensive Survey. Structure 2000, 8, 493–504. [Google Scholar] [CrossRef]
- Vogt, G.; Woell, S.; Argos, P. Protein Thermal Stability, Hydrogen Bonds, and Ion Pairs. J. Mol. Biol. 1997, 269, 631–643. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Gerike, U.; Danson, M.J.; Hough, D.W.; Taylor, G.L. Structural Adaptations of the Cold-Active Citrate Synthase from an Antarctic Bacterium. Structure 1998, 6, 351–361. [Google Scholar] [CrossRef]
- Glyakina, A.V.; Garbuzynskiy, S.O.; Lobanov, M.Y.; Galzitskaya, O.V. Different Packing of External Residues Can Explain Differences in the Thermostability of Proteins from Thermophilic and Mesophilic Organisms. Bioinformatics 2007, 23, 2231–2238. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Siso, M.I. Editorial for the Special Issue: Thermophiles and Thermozymes. Microorganisms 2019, 7, 62. [Google Scholar] [CrossRef] [PubMed]
- Reysenbach, A.-L. Phylum BI. Aquificae phy. nov. In Bergey’s Manual of Systematic Bacteriology; Boone, D.R., Castenholz, R.W., Eds.; Springer: Berlin, Germany, 2001; pp. 359–367. [Google Scholar]
- Reysenbach, A.-L. Phylum BII. Thermotogae phy. nov. In Bergey’s Manual of Systematic Bacteriology; Boone, D.R., Castenholz, R.W., Eds.; Springer: Berlin, Germany, 2001; pp. 369–387. [Google Scholar]
- Gupta, R.S. The Phylum Aquificae. In The Prokaryotes: Prokaryotic Biology and Symbiotic Associations; Rosenberg, E., DeLong, E.F., Thompson, F., Lory, S., Stackebrand, E., Eds.; Springer: Berlin, Germany, 2014; pp. 418–445. [Google Scholar]
- Albuquerque, L.; Costa, M.S. The Family Thermaceae. In The Prokaryotes-Other Major Lineages of Bacteria and the Archaea; Rosenberg, E., DeLong, E., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: New York, NY, USA, 2014; pp. 955–987. [Google Scholar]
- Escuder-Rodriguez, J.J.; DeCastro, M.E.; Cerdan, M.E.; Rodriguez-Belmonte, E.; Becerra, M.; Gonzalez-Siso, M.I. Cellulases from Thermophiles Found by Metagenomics. Microorganisms 2018, 6, 66. [Google Scholar] [CrossRef]
- Bhandari, V.; Gupta, R.S. Phylum Thermotogae. In The Prokaryotes—Other Major Lineages of Bacteria and the Archea; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 989–1015. [Google Scholar] [CrossRef]
- Rosenberg, E. The family Deinococcaceae. In The Prokaryotes- Other Major Lineages of Bacteria and the Archaea; Rosenberg, E., DeLong, E., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: New York, NY, USA, 2014; pp. 613–615. [Google Scholar]
- Huber, R.; Hannig, M. Thermotogales. Prokaryotes 2006, 7, 899–922. [Google Scholar]
- Pollo, S.M.; Zhaxybayeva, O.; Nesbo, C.L. Insights into Thermoadaptation and the Evolution of Mesophily from the Bacterial Phylum Thermotogae. Can. J. Microbiol. 2015, 61, 655–670. [Google Scholar] [CrossRef]
- Griffiths, E.; Gupta, R.S. Molecular Signatures in Protein Sequences that are Characteristics of the Phylum Aquificae. Int. J. Syst. Evol. Microbiol. 2006, 56, 99–107. [Google Scholar] [CrossRef]
- Gupta, R.S.; Bhandari, V. Phylogeny and Molecular Signatures for the Phylum Thermotogae and Its Subgroups. Antonie Van Leeuwenhoek 2011, 100, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Lali, R. Molecular Signatures for the Phylum Aquificae and Its Different Clades: Proposal for Division of the phylum Aquificae into the Emended Order Aquificales, Containing the Families Aquificaceae and Hydrogenothermaceae, and a New Order Desulfurobacteriales ord. nov., containing the family Desulfurobacteriaceae. Antonie Van Leeuwenhoek 2013, 104, 349–368. [Google Scholar] [PubMed]
- Ho, J.; Adeolu, M.; Khadka, B.; Gupta, R.S. Identification of Distinctive Molecular Traits that are Characteristic of the Phylum “Deinococcus-Thermus” and Distinguish Its Main Constituent Groups. Syst. Appl. Microbiol. 2016, 39, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Rapoport, T.A.; Jungnickel, B.; Kutay, U. Protein Transport across the Eukaryotic Endoplasmic Reticulum and Bacterial Inner Membranes. Annu. Rev. Biochem. 1996, 65, 271–303. [Google Scholar] [CrossRef] [PubMed]
- Fekkes, P.; Driessen, A.J. Protein Targeting to the Bacterial Cytoplasmic Membrane. Microbiol. Mol. Biol. Rev. 1999, 63, 161–173. [Google Scholar] [PubMed]
- Lill, R.; Cunningham, K.; Brundage, L.A.; Ito, K.; Oliver, D.; Wickner, W. SecA Protein Hydrolyzes ATP and is an Essential Component of the Protein Translocation ATPase of Escherichia Coli. EMBO J. 1989, 8, 961–966. [Google Scholar] [CrossRef]
- Collinson, I. SecA-a New Twist in the Tale. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef]
- Chen, Y.; Bauer, B.W.; Rapoport, T.A.; Gumbart, J.C. Conformational Changes of the Clamp of the Protein Translocation ATPase SecA. J. Mol. Biol. 2015, 427, 2348–2359. [Google Scholar] [CrossRef]
- Vassylyev, D.G.; Mori, H.; Vassylyeva, M.N.; Tsukazaki, T.; Kimura, Y.; Tahirov, T.H.; Ito, K. Crystal Structure of the Translocation ATPase SecA from Thermus Thermophilus Reveals a Parallel, Head-To-Head Dimer. J. Mol. Biol. 2006, 364, 248–258. [Google Scholar] [CrossRef]
- Zimmer, J.; Rapoport, T.A. Conformational Flexibility and Peptide Interaction of the Translocation ATPase SecA. J. Mol. Biol. 2009, 394, 606–612. [Google Scholar] [CrossRef][Green Version]
- Griffiths, E.; Gupta, R.S. Signature Sequences in Diverse Proteins Provide Evidence for the Late Divergence of the Order Aquificales. Int. Microbiol. 2004, 7, 41–52. [Google Scholar]
- Gupta, R.S. Identification of Conserved Indels that are Useful for Classification and Evolutionary Studies. In Methods in Microbiology New Approaches to Prokaryotics Systematics; Goodfellow, M., Sutcliffe, I.C., Chun, J., Eds.; Elsevier: London, UK, 2014; pp. 153–182. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Goujon, M.; McWilliam, H.; Li, W.; Valentin, F.; Squizzato, S.; Paern, J.; Lopez, R. A New Bioinformatics Analysis Tools Framework at EMBL-EBI. Nucleic Acids Res. 2010, 38, W695–W699. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Comput. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Alnajar, S.; Khadka, B.; Gupta, R.S. Ribonucleotide Reductases from Bifidobacteria Contain Multiple Conserved Indels Distinguishing Them from All Other Organisms: In Silico Analysis of the Possible Role of a 43 aa Bifidobacteria-Specific Insert in the Class III RNR Homolog. Front. Microbiol. 2017, 8, 1409. [Google Scholar] [CrossRef]
- Gupta, R.S.; Nanda, A.; Khadka, B. Novel Molecular, Structural and Evolutionary Characteristics of the Phosphoketolases from Bifidobacteria and Coriobacteriales. PLoS ONE 2017, 12, e0172176. [Google Scholar] [CrossRef]
- Khadka, B.; Gupta, R.S. Identification of a Conserved 8 aa Insert in the PIP5K Protein in the Saccharomycetaceae Family of Fungi and the Molecular Dynamics Simulations and Structural Analysis to Investigate Its Potential Functional Role. Proteins 2017, 85, 1454–1467. [Google Scholar] [CrossRef]
- Khadka, B.; Gupta, R.S. Novel Molecular Signatures in the PIP4K/PIP5K Family of Proteins Specific for Different Isozymes and Subfamilies Provide Important Insights into the Evolutionary Divergence of this Protein Family. Genes 2019, 10, 312. [Google Scholar] [CrossRef]
- Shen, M.Y.; Sali, A. Statistical Potential for Assessment and Prediction of Protein Structures. Protein Sci. 2006, 15, 2507–2524. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.R.; Heo, L.; Seok, C. Effective Protein Model Structure Refinement by Loop Modeling and Overall Relaxation. Proteins 2016, 84 (Suppl. 1), 293–301. [Google Scholar] [CrossRef]
- Lovell, S.C.; Davis, I.W.; Arendall, W.B., III; de Bakker, P.I.; Word, J.M.; Prisant, M.G.; Richardson, J.S.; Richardson, D.C. Structure Validation by Calpha Geometry: Phi, psi and Cbeta deviation. Proteins 2003, 50, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of Protein Structures: Patterns of Nonbonded Atomic Interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Sippl, M.J. Recognition of Errors in Three-Dimensional Structures of Proteins. Proteins 1993, 17, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Wiederstein, M.; Sippl, M.J. ProSA-Web: Interactive Web Service for the Recognition of Errors in Three-Dimensional Structures of Proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Eisenberg, D.; Luthy, R.; Bowie, J.U. VERIFY3D: Assessment of Protein Models with Three-Dimensional Profiles. Methods Enzym. 1997, 277, 396–404. [Google Scholar]
- Van Der, S.D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Abrahama, M.J.; Schulzb, R.; Pálla, S.; Smith, J.C.; Hessa, B.; Lindahla, E. GROMACS: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 2016, 1–2, 19–25. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical Sampling through Velocity Rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef] [PubMed]
- Nosé, S.; Klein, M.L. Constant Pressure Molecular Dynamics for Molecular Systems. Mol. Phys. 1983, 50, 1055–1076. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic Transitions in Single crYstals: A New Molecular Dynamics Method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Kutzner, C.; Pall, S.; Fechner, M.; Esztermann, A.; de Groot, B.L.; Grubmuller, H. Best Bang for Your Buck: GPU Nodes for GROMACS Biomolecular Simulations. J. Comput. Chem. 2015, 36, 1990–2008. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual Molecular Dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Nesbo, C.L.; Bradnan, D.M.; Adebusuyi, A.; Dlutek, M.; Petrus, A.K.; Foght, J.; Doolittle, W.F.; Noll, K.M. Mesotoga Prima gen. nov., sp. nov., the First Described Mesophilic Species of the Thermotogales. Extremophiles 2012, 16, 387–393. [Google Scholar] [CrossRef]
- L’Haridon, S.; Reysenbach, A.L.; Tindall, B.J.; Schonheit, P.; Banta, A.; Johnsen, U.; Schumann, P.; Gambacorta, A.; Stackebrandt, E.; Jeanthon, C. Desulfurobacterium Atlanticum sp. nov., Desulfurobacterium Pacificum sp. nov. and Thermovibrio Guaymasensis sp. nov., Three Thermophilic Members of the Desulfurobacteriaceae fam. nov., a deep Branching Lineage within the Bacteria. Int. J. Syst. Evol. Microbiol. 2006, 56, 2843–2852. [Google Scholar] [CrossRef]
- Hassan, F.M.N.; Gupta, R.S. Novel Sequence Features of DNA Repair Genes/Proteins from Deinococcus Species Implicated in Protection from Oxidatively Generated Damage. Genes 2018, 9, 149. [Google Scholar] [CrossRef]
- Bonjour, F.; Graber, A.; Aragno, M. Isolation of Bacillus schlegelii, a Thermophilic, Hydrogen Oxidizing, Aerobic Autotroph, from Geothermal and Nongeothermal Environments. Microb. Ecol. 1988, 16, 331–337. [Google Scholar] [CrossRef]
- Kampfer, P.; Glaeser, S.P.; Busse, H.J. Transfer of Bacillus Schlegelii to a Novel Genus and Proposal of Hydrogenibacillus Schlegelii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 2013, 63, 1723–1727. [Google Scholar] [CrossRef]
- Maker, A.; Hemp, J.; Pace, L.A.; Ward, L.M.; Fischer, W.W. Draft Genome Sequence of Hydrogenibacillus schlegelii MA48, a Deep-Branching Member of the Bacilli Class of Firmicutes. Genome Announc. 2017, 5, e00380-16. [Google Scholar] [CrossRef] [PubMed]
- Zhaxybayeva, O.; Swithers, K.S.; Lapierre, P.; Fournier, G.P.; Bickhart, D.M.; Deboy, R.T.; Nelson, K.E.; Nesbo, C.L.; Doolittle, W.F.; Gogarten, J.P.; et al. On the Chimeric Nature, Thermophilic Origin, and Phylogenetic Placement of the Thermotogales. Proc. Natl. Acad. Sci. USA 2009, 106, 5865–5870. [Google Scholar] [CrossRef] [PubMed]
- Hunt, J.F.; Weinkauf, S.; Henry, L.; Fak, J.J.; McNicholas, P.; Oliver, D.B.; Deisenhofer, J. Nucleotide Control of Interdomain Interactions in the Conformational Reaction Cycle of SecA. Science 2002, 297, 2018–2026. [Google Scholar] [CrossRef] [PubMed]
- Papanikolau, Y.; Papadovasilaki, M.; Ravelli, R.B.; McCarthy, A.A.; Cusack, S.; Economou, A.; Petratos, K. Structure of Dimeric SecA, the Escherichia Coli Preprotein Translocase Motor. J. Mol. Biol. 2007, 366, 1545–1557. [Google Scholar] [CrossRef]
- Sharma, V.; Arockiasamy, A.; Ronning, D.R.; Savva, C.G.; Holzenburg, A.; Braunstein, M.; Jacobs, W.R., Jr.; Sacchettini, J.C. Crystal Structure of Mycobacterium Tuberculosis SecA, a Preprotein Translocating ATPase. Proc. Natl. Acad. Sci. USA 2003, 100, 2243–2248. [Google Scholar]
- Vrontou, E.; Economou, A. Structure and Function of SecA, the Preprotein Translocase Nanomotor. Biochim. Biophys Acta 2004, 1694, 67–80. [Google Scholar]
- Zimmer, J.; Nam, Y.; Rapoport, T.A. Structure of a Complex of the ATPase SecA and the Protein-Translocation Channel. Nature 2008, 455, 936–943. [Google Scholar] [CrossRef]
- Barillari, C.; Taylor, J.; Viner, R.; Essex, J.W. Classification of Water Molecules in Protein Binding Sites. J. Am. Chem. Soc. 2007, 129, 2577–2587. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, R.; Yang, C.Y.; Wang, S. Analysis of Ligand-Bound Water Molecules in High-Resolution Crystal Structures of Protein-Ligand Complexes. J. Chem. Inf. Model. 2007, 47, 668–675. [Google Scholar] [CrossRef]
- Homans, S.W. Water, Water Everywhere--Except Where it Matters? Drug Discov. Today 2007, 12, 534–539. [Google Scholar] [CrossRef]
- Singh, N.; Briggs, J.M. Molecular Dynamics Simulations of Factor Xa: Insight into Conformational Transition of Its Binding Subsites. Biopolymers 2008, 89, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Schiebel, J.; Gaspari, R.; Wulsdorf, T.; Ngo, K.; Sohn, C.; Schrader, T.E.; Cavalli, A.; Ostermann, A.; Heine, A.; Klebe, G. Intriguing Role of Water in Protein-Ligand Binding Studied by Neutron Crystallography on Trypsin Complexes. Nat. Commun. 2018, 9, 3559. [Google Scholar] [CrossRef] [PubMed]
- Vieille, C.; Burdette, D.S.; Zeikus, J.G. Thermozymes. Biotechnol. Annu. Rev. 1996, 2, 1–83. [Google Scholar] [PubMed]
- Kumar, S.; Tsai, C.J.; Nussinov, R. Factors Enhancing Protein Thermostability. Protein Eng. 2000, 13, 179–191. [Google Scholar] [CrossRef]
- Berezovsky, I.N.; Shakhnovich, E.I. Physics and Evolution of Thermophilic Adaptation. Proc. Natl. Acad. Sci. USA 2005, 102, 12742–12747. [Google Scholar] [CrossRef]
- Mizuguchi, K.; Sele, M.; Cubellis, M.V. Environment Specific Substitution Tables for Thermophilic Proteins. BMC Bioinform. 2007, 8 (Suppl. 1), S15. [Google Scholar] [CrossRef]
- Taylor, T.J.; Vaisman, I.I. Discrimination of Thermophilic and Mesophilic Proteins. BMC Struct. Biol. 2010, 10 (Suppl. 1), S5. [Google Scholar] [CrossRef]
- Singh, B.; Gupta, R.S. Conserved Inserts in the Hsp60 (GroEL) and Hsp70 (DnaK) Proteins are Essential for Cellular Growth. Mol. Genet. Genom. 2009, 281, 361–373. [Google Scholar] [CrossRef]
- Huber, R.; Eder, W. Aquificales. In the Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, E., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Bonch-Osmolovskaya, E. Aquificales. In Encylopedia of Life Sciences; Wieley: Hoboken, NJ, USA, 2008; pp. 1–7. [Google Scholar]
- Knight, J.D.; Hamelberg, D.; McCammon, J.A.; Kothary, R. The Role of Conserved Water Molecules in the Catalytic Domain of Protein Kinases. Proteins 2009, 76, 527–535. [Google Scholar] [CrossRef]
- Nygaard, R.; Valentin-Hansen, L.; Mokrosinski, J.; Frimurer, T.M.; Schwartz, T.W. Conserved Water-Mediated Hydrogen Bond Network between TM-I, -II, -VI, and -VII in 7TM Receptor Activation. J. Biol. Chem. 2010, 285, 19625–19636. [Google Scholar] [CrossRef]
- Kaur, M.; Bahia, M.S.; Silakari, O. Exploring the Role of Water Molecules for Docking and Receptor Guided 3D-QSAR Analysis of Naphthyridine Derivatives as Spleen Tyrosine Kinase (Syk) Inhibitors. J. Chem. Inf. Model. 2012, 52, 2619–2630. [Google Scholar] [CrossRef] [PubMed]
- Jeszenoi, N.; Balint, M.; Horvath, I.; Van Der, S.D.; Hetenyi, C. Exploration of Interfacial Hydration Networks of Target-Ligand Complexes. J. Chem. Inf. Model. 2016, 56, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Lett, C.M.; Berghuis, A.M.; Frey, H.E.; Lepock, J.R.; Guillemette, J.G. The Role of a Conserved Water Molecule in the Redox-Dependent Thermal Stability of Iso-1-Cytochrome c. J. Biol. Chem. 1996, 271, 29088–29093. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Lin, S.X. Cofactor Hydrogen Bonding onto the Protein Main Chain is Conserved in the Short Chain Dehydrogenase/Reductase Family and Contributes to Nicotinamide Orientation. J. Biol. Chem. 2004, 279, 16778–16785. [Google Scholar]
- Sterpone, F.; Stirnemann, G.; Hynes, J.T.; Laage, D. Water Hydrogen-Bond Dynamics around Amino Acids: The Key Role of Hydrophilic Hydrogen-Bond Acceptor Groups. J. Phys. Chem. B 2010, 114, 2083–2089. [Google Scholar] [CrossRef]
- Huggins, D.J.; Tidor, B. Systematic Placement of Structural Water Molecules for Improved Scoring of Protein-Ligand Interactions. Protein Eng. Des. Sel. 2011, 24, 777–789. [Google Scholar] [CrossRef]
- Milenkovic, S.; Bondar, A.N. Mechanism of Conformational Coupling in SecA: Key Role of Hydrogen-Bonding Networks and Water Interactions. Biochim. Biophys. Acta 2016, 1858, 374–385. [Google Scholar] [CrossRef]
- Geszvain, K.; Gruber, T.M.; Mooney, R.A.; Gross, C.A.; Landick, R. A Hydrophobic Patch on the Flap-Tip Helix of E.coli RNA Polymerase Mediates Sigma(70) Region 4 Function. J. Mol. Biol. 2004, 343, 569–587. [Google Scholar] [CrossRef]
- Akiva, E.; Itzhaki, Z.; Margalit, H. Built-In Loops Allow Versatility in Domain-Domain Interactions: Lessons from Self-Interacting Domains. Proc. Natl. Acad. Sci. USA 2008, 105, 13292–13297. [Google Scholar]
- Hashimoto, K.; Panchenko, A.R. Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states. Proc. Natl. Acad. Sci. USA 2010, 107, 20352–20357. [Google Scholar] [CrossRef]
- Schoeffler, A.J.; May, A.P.; Berger, J.M. A Domain Insertion in Escherichia Coli GyrB Adopts a Novel Fold that Plays a Critical Role in Gyrase Function. Nucleic Acids Res. 2010, 38, 7830–7844. [Google Scholar] [CrossRef] [PubMed]
- Gouridis, G.; Karamanou, S.; Sardis, M.F.; Scharer, M.A.; Capitani, G.; Economou, A. Quaternary Dynamics of the SecA Motor Drive Translocase Catalysis. Mol. Cell 2013, 52, 655–666. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Kraft, C.; Jaiswal, R.; Sejwal, K.; Kasaragod, V.B.; Kuper, J.; Burger, J.; Mielke, T.; Luirink, J.; Bhushan, S. Cryo-Electron Microscopic Structure of SecA Protein Bound to the 70S Ribosome. J. Biol. Chem. 2014, 289, 7190–7199. [Google Scholar] [CrossRef] [PubMed]
- Wowor, A.J.; Yan, Y.; Auclair, S.M.; Yu, D.; Zhang, J.; May, E.R.; Gross, M.L.; Kendall, D.A.; Cole, J.L. Analysis of SecA Dimerization in Solution. Biochemistry 2014, 53, 3248–3260. [Google Scholar] [CrossRef] [PubMed]
- Collinson, I.; Corey, R.A.; Allen, W.J. Channel Crossing: How are Proteins Shipped across the Bacterial Plasma Membrane? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370. [Google Scholar] [CrossRef]
- Fekkes, P.; de Wit, J.G.; van der Wolk, J.P.; Kimsey, H.H.; Kumamoto, C.A.; Driessen, A.J. Preprotein Transfer to the Escherichia coli Translocase Requires the Co-Operative Binding of SecB and the Signal Sequence to SecA. Mol. Microbiol. 1998, 29, 1179–1190. [Google Scholar] [CrossRef]
- Pretz, M.G.; Remigy, H.; Swaving, J.; Albers, S.V.; Garrido, V.G.; Chami, M.; Engel, A.; Driessen, A.J. Functional and Structural Characterization of the Minimal Sec Translocase of the Hyperthermophile Thermotoga maritima. Extremophiles 2005, 9, 307–316. [Google Scholar] [CrossRef]
- Sala, A.; Calderon, V.; Bordes, P.; Genevaux, P. TAC from Mycobacterium Tuberculosis: A Paradigm for Stress-Responsive Toxin-Antitoxin Systems Controlled by SecB-like Chaperones. Cell Stress Chaperones 2013, 18, 129–135. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khadka, B.; Persaud, D.; Gupta, R.S. Novel Sequence Feature of SecA Translocase Protein Unique to the Thermophilic Bacteria: Bioinformatics Analyses to Investigate Their Potential Roles. Microorganisms 2020, 8, 59. https://doi.org/10.3390/microorganisms8010059
Khadka B, Persaud D, Gupta RS. Novel Sequence Feature of SecA Translocase Protein Unique to the Thermophilic Bacteria: Bioinformatics Analyses to Investigate Their Potential Roles. Microorganisms. 2020; 8(1):59. https://doi.org/10.3390/microorganisms8010059
Chicago/Turabian StyleKhadka, Bijendra, Dhillon Persaud, and Radhey S. Gupta. 2020. "Novel Sequence Feature of SecA Translocase Protein Unique to the Thermophilic Bacteria: Bioinformatics Analyses to Investigate Their Potential Roles" Microorganisms 8, no. 1: 59. https://doi.org/10.3390/microorganisms8010059
APA StyleKhadka, B., Persaud, D., & Gupta, R. S. (2020). Novel Sequence Feature of SecA Translocase Protein Unique to the Thermophilic Bacteria: Bioinformatics Analyses to Investigate Their Potential Roles. Microorganisms, 8(1), 59. https://doi.org/10.3390/microorganisms8010059





