Abstract
High mobility group box B (HMGB) proteins are pivotal in the development of cancer. Although the proteomics of prostate cancer (PCa) cells has been reported, the involvement of HMGB proteins and their interactome in PCa is an unexplored field of considerable interest. We describe herein the results of the first HMGB1/HMGB2 interactome approach to PCa. Libraries constructed from the PCa cell line, PC-3, and from patients’ PCa primary tumor have been screened by the yeast 2-hybrid approach (Y2H) using HMGB1 and HMGB2 baits. Functional significance of this PCa HMGB interactome has been validated through expression and prognosis data available on public databases. Copy number alterations (CNA) affecting these newly described HMGB interactome components are more frequent in the most aggressive forms of PCa: those of neuroendocrine origin or castration-resistant PCa. Concordantly, adenocarcinoma PCa samples showing CNA in these genes are also associated with the worse prognosis. These findings open the way to their potential use as discriminatory biomarkers between high and low risk patients. Gene expression of a selected set of these interactome components has been analyzed by qPCR after HMGB1 and HMGB2 silencing. The data show that HMGB1 and HMGB2 control the expression of several of their interactome partners, which might contribute to the orchestrated action of these proteins in PCa
1. Introduction
Human high mobility group box B (HMGB) proteins HMGB1, 2, and 3 are differentially expressed in many different tissues and cell types, whereas HMGB4 expression is restricted to the testis [1]. HMGB2 has 82.3% sequence similarity with HMGB1, and both proteins have common or redundant functions in inflammation [2], chromosome remodeling activity [3], V (D) J recombination [4], and embryonic development [5].
HMGB1 has been related to the onset and progression of cancer, being involved in events such as replenishing telomeric DNA and maintaining cell immortality [6], autophagic increase, evasion of apoptosis [7,8], as well as cell proliferation and invasion [9,10]. HMGB1 is also involved in dedifferentiation during epithelial to mesenchymal transition (EMT) [11] via the receptor for advanced glycation endproducts RAGE/ nuclear factor kappaB NF-κB signaling pathways [12] and in angiogenesis [13]. The role of HMGB2 in these processes, although less well studied, has also been related to cell viability and invasion [14], EMT [10], and angiogenesis [15].
The majority of the prostate cancers (PCa) are adenocarcinomas characterized by glandular formation and the expression of androgen receptor (AR) and prostate-specific antigen (PSA). Hormonal inhibition of AR signaling is the therapeutic choice for patients with adenocarcinomas, but unfortunately, the disease usually progresses as it becomes independent of exogenous AR induction, leading to castration-resistant prostate cancer (CRPC) with a worse prognosis. In prostatic small cell neuroendocrine carcinoma (SCNC), the tumor cells are negative for AR and PSA expression and do not respond to hormonal therapy [16]. Among the most frequently used PCa cell lines, PC-3 characteristics are considered closer to a SCCN PCa model and those of DU145 (ATCC® HTB-81™) or LNCaP (lymph node carcinoma of the prostate) are considered closer to adenocarcinoma models [16]. PC-3 and DU145 are AR-independent, and LNCaP is AR-dependent [16,17]. Interestingly, upregulation of HMGB1 mRNA and protein have been detected in PCa tumors [12,18] and PCa cell lines (including PC-3 and DU145 or LNCaP) compared to the non-transformed immortalized prostate cell line RWPE-1 (prostate epithelial transformed by HPV)) [18]. Silencing of HMGB1 in LNCaP cells inhibits cell growth [19]. HMGB1 expression is notably high in PCa metastasis [12] and is positively correlated with some clinical-pathological parameters, such as Gleason score or preoperative PSA concentration, being associated with a worse prognosis [18].
Proteomic studies in relation to PCa have been reported [20,21,22], with interactome strategies being outstanding in recent developments [23,24,25]. The purpose of our study was to analyze proteins interacting with HMGB1 and HMGB2 by the yeast 2-hybrid approach (Y2H), using HMGB1 and HMGB2 baits. Results from the screening of libraries constructed from the PC-3 line, as a model of metastatic AR-independent PCa, and of libraries obtained from PCa adenocarcinoma primary tumor are presented. Analyses of copy number alterations (CNA) and mRNA levels of detected targets in public PCa databases are discussed showing that dysregulation of some HMGB1/2 targets is associated with clinical prognosis. Considering that HMGB proteins are known regulators of gene expression, we also tested whether HMGB1 and HMGB2 silencing affects the expression of their Y2H detected partners and found that this regulatory mechanism is functional in PC-3 cells.
2. Results
2.1. HMGB1 and HMGB2 Y2H Interactomes in the PCa PC-3 Cell Line and in Adenocarcinoma Primary Tumor
Human PCa cDNA libraries were constructed using total RNA from PC-3 cells and PCa adenocarcinoma primary tumor. Y2H assays were carried out as described in the Materials and Methods section, using HMGB1 and HMGB2 as baits and triple screening by 3 independent selection markers (Supplementary Figure S1). The panel of proteins interacting with HMGB1 or HMGB2 in these libraries is summarized in Table 1, Table 2, Table 3 and Table 4. The interactions of identified proteins with HMGB1 or HMGB2 have not previously been reported on Biogrid, String, or other public databases, although we have previously reported that Cytokeratin-7, the human complement subcomponent C1q (C1QPB), and zinc finger p rotein 428 (ZNF428) interact with HMGB1 and that (high density lipoprotein-binding protein (HDLBP) and ZNF428 interact with HMGB2 in noncancerous epithelial cells [26].

Table 1.
Proteins identified in the high mobility group box B 1 (HMGB1) yeast 2-hybrid (Y2H) approach interactome in PC-3 cells.

Table 2.
Proteins identified in the the high mobility group box B 2 (HMGB2) Y2H interactome in PC-3 cells.

Table 3.
Proteins identified in the HMGB1 Y2H interactome in primary tumor adenocarcinoma.

Table 4.
Proteins identified in the HMGB2 Y2H interactome in primary tumor adenocarcinoma.
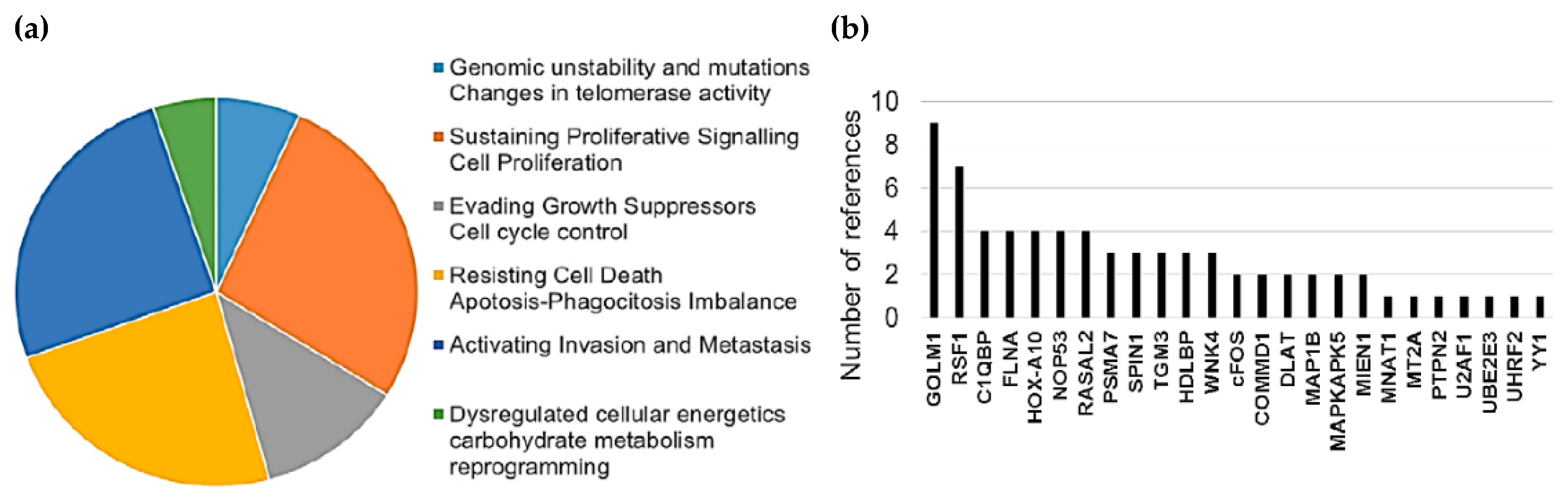
Interestingly, 43% of detected HMGB1 interactome targets (10 of a total of 28, referenced in Table 1 and Table 3) as well as 64% of HMGB2 (7 of a total of 11, referenced in Table 2 and Table 4) have previously been related to PCa, supporting the functional significance of our Y2H interactome data in PCa research. Furthermore, the detected proteins are remarkably associated with cancer hallmarks. Indeed, the oncogenic capacities of several identified proteins in our Y2H interactome had been already reported in PCa or other cancerous models by wide-ranging functional approaches, which are reviewed in Supplementary Table S1. Figure 1 summarizes the frequency distribution of the identified proteins in relation to cancer hallmarks (Figure 1a) as well as the number of references of each protein functionally related to cancer progression in diverse models (Figure 1b).
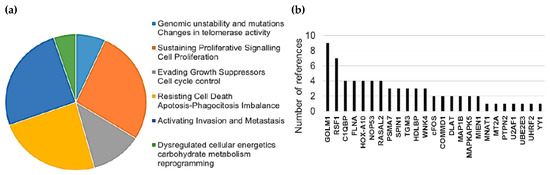
Figure 1.
Relationship between identified proteins and cancer hallmarks: (a) Distribution of HMGB1 and HMGB2 interactome targets according to cancer hallmarks and (b) number of references that associate these proteins with cancer hallmarks according to PubMed (7-31-2019).
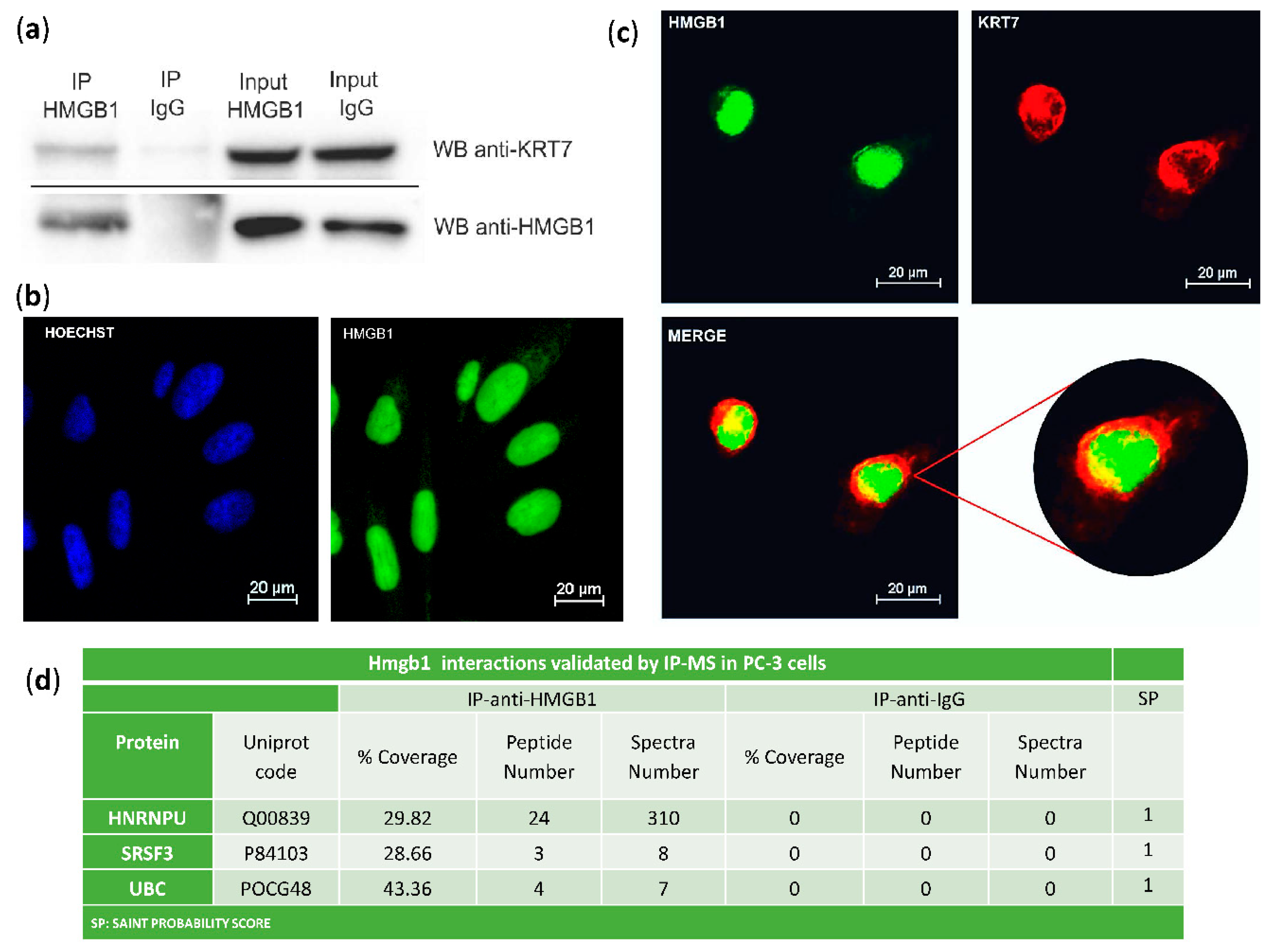
The interaction of HMGB1 with Cytokeratin-7 was validated in PC-3 cells by co-immunoprecipitation and western blot (Figure 2a). Immunodetection of HMGB1 using a green fluorescent antibody (Figure 2b,c) and of Cytokeratin-7 using a red fluorescent antibody (Figure 2c) was also assayed in PC-3 cells. Confocal microscopy showed that co-localization of HMGB1 and Cytokeratin-7 occurred principally in the perinuclear area (Figure 2c), with a Meander’s correlation coefficient of 0.87 ± 0.3. Three other interactions were also validated in PC-3 cells by immunoprecipitation and MS identification (Figure 2d).
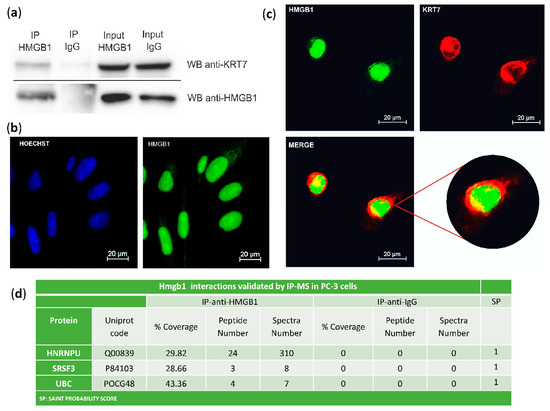
Figure 2.
Validation of HMGB1-interactions: (a) Cytokeratin-7 co-immunoprecipitation with HMGB1. PC-3 lysates were immunoprecipitated with anti-HMGB1 antibody or normal mouse IgG and immunoblotted with antibodies to Cytokeratin-7 and HMGB1; complete membranes provided as Supplementary Materials Imagen S1. Protein G horseradish peroxidase (HRP)-labelled was used as a secondary antibody to minimize the signal given by the light and heavy chains of the immunoprecipitation antibody. (b) Immunofluorescent localization of HMGB1 in PC-3 cells and comparison to Hoechst-stained nuclei. (c) Immunofluorescent co-localization of HMGB1 and Cytokeratin-7 by confocal microscopy in PC-3 cells. HMGB1 is shown in green, and Cytokeratin-7 is in red. Co-localization is seen in yellow by merging. (d) Validation of interactions with HNRNPU, SRSF3, and UBC after HMGB1 immunoprecipitation and MS peptide identification.
2.2. Mutations and Copy Number Alterations in HMGB1 and HMGB2 Interactome Targets in PCa
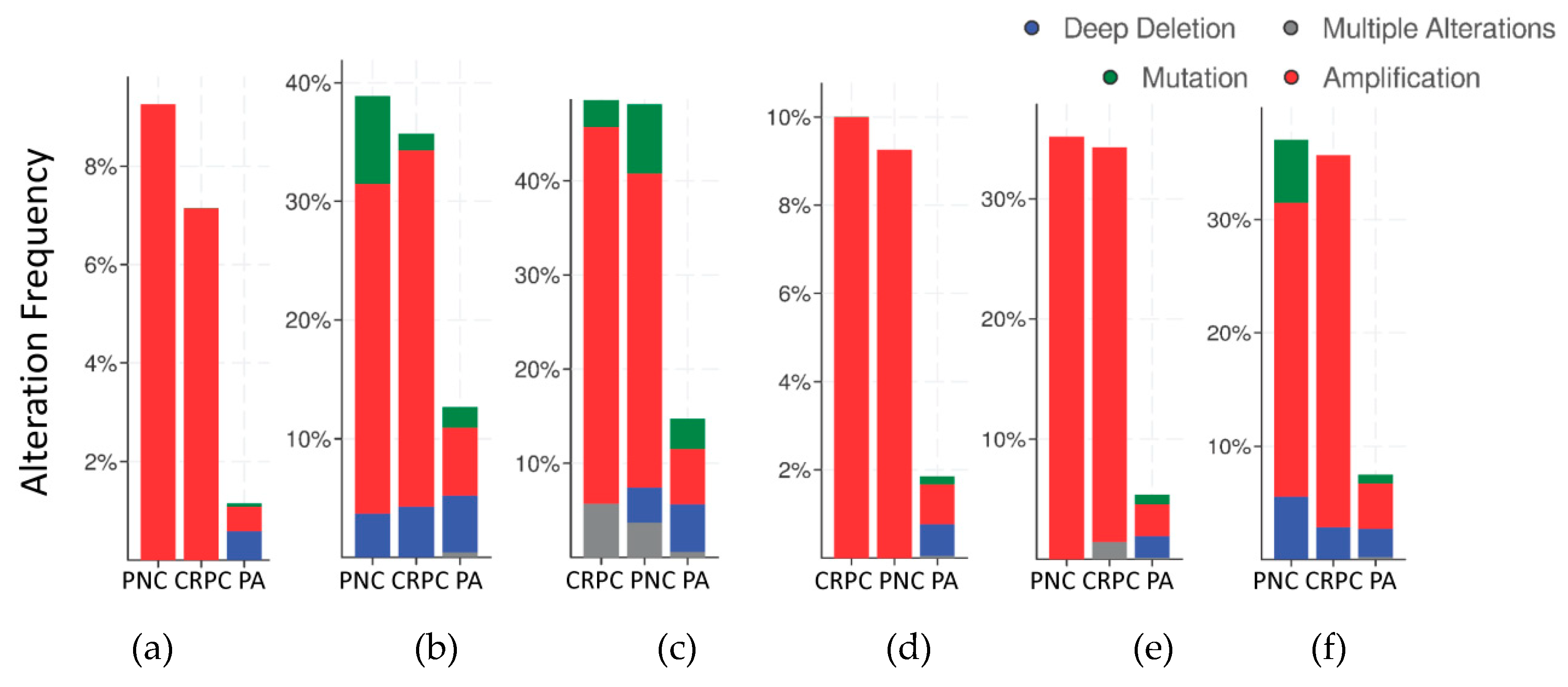
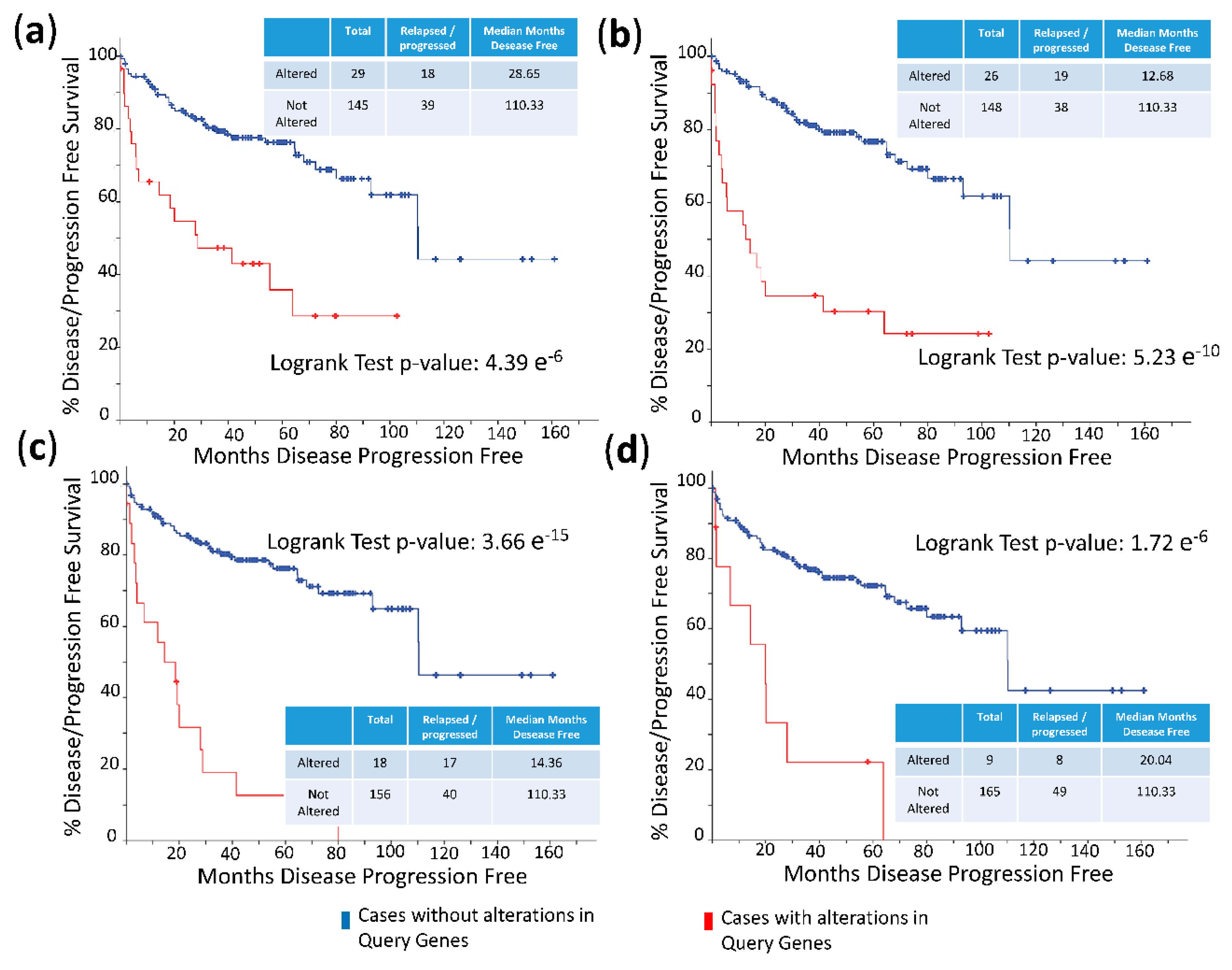
The frequency of mutations and copy number alterations (CNA) in genes encoding HMGB1 and HMGB2 proteins were analyzed as well as in those genes encoding proteins detected in the Y2H search associated with PCa, using the open platform for exploring cancer genomics data, c-Bioportal [79,80]. We included 14 PCa studies available at cBioPortal (https://www.cbioportal.org/), of which their characteristics are summarized in Supplementary Table S2. From these, 10 were adenocarcinoma studies [81,82,83,84,85,86,87,88,89,90,91], with 3218 samples; the other 3 studies corresponded to metastatic PCa [92,93,94], including 655 samples; and finally, one study corresponded to neuroendocrine PCa, which was carried out with 114 samples [95]. The data show that mutations and CNA affecting HMGB1, HMGB2, and the proteins identified in the corresponding Y2H interactome are more frequently present in neuroendocrine PCa and castration-resistant PCa than in adenocarcinoma (Figure 3). Since neuroendocrine PCa is an aggressive PCa [16], we tested whether CNA of these genes was also related to the poor prognosis in patients diagnosed with adenocarcinoma. With amplification as the most frequently detected CNA in Figure 3, we compared disease/progression-free Kaplan–Meier estimate rates calculated from the study of Taylor et al. [86] among the group of samples having gains or amplifications of these genes and the group integrated by the rest of samples. Figure 4 shows that gain or amplification of HMGB1 and HMGB2 interactome targets results in a notorious decrease of the median of months disease-free, with high significant p-values in the Logrank test.
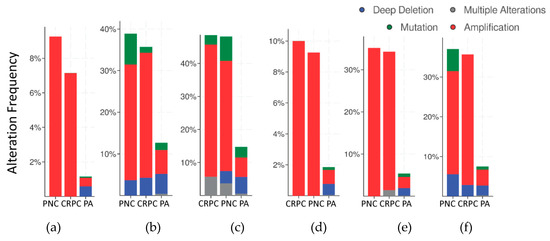
Figure 3.
Copy number alteration frequency of HMGB1, HMGB2, and their targets in prostate cancer: (a) HMGB1, (b) HMGB1 interactome targets from PC-3 library, (c) HMGB1 interactome targets from prostate adenocarcinoma tissue library, (d) HMGB2, (e) HMGB2 interactome targets from PC-3 library, and (f) HMGB2 interactome targets from prostate adenocarcinoma tissue library. PA, prostate adenocarcinoma; PNC, Prostate Neuroendocrine Carcinoma; CRPC, Castration Resistant Prostate Cancer. Data source: combined study from data available through c-Bioportal (detailed in Supplementary Table S2).
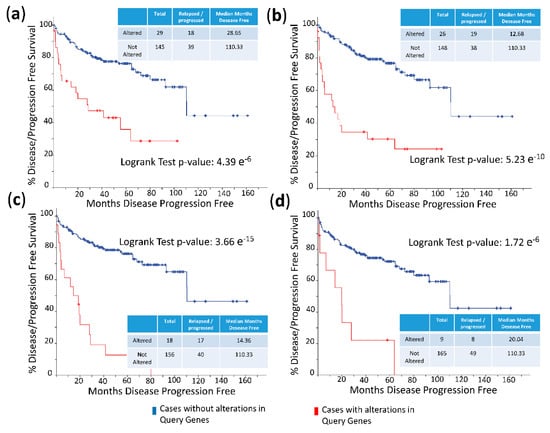
Figure 4.
Disease/progression-free Kaplan–Meier estimate: (a) Cases altered in HMGB1 interactome targets from PC-3 library; (b) cases altered in HMGB1 interactome targets from prostate adenocarcinoma tissue library; (c) cases altered in HMGB2 interactome targets from PC-3 library; and (d) cases altered in HMGB2 interactome targets from prostate adenocarcinoma tissue library. Data source: prostate adenocarcinoma study [86], including 194 patients/samples.
2.3. Expression of HMGB1 and HMGB2 Interactome Targets in PCa
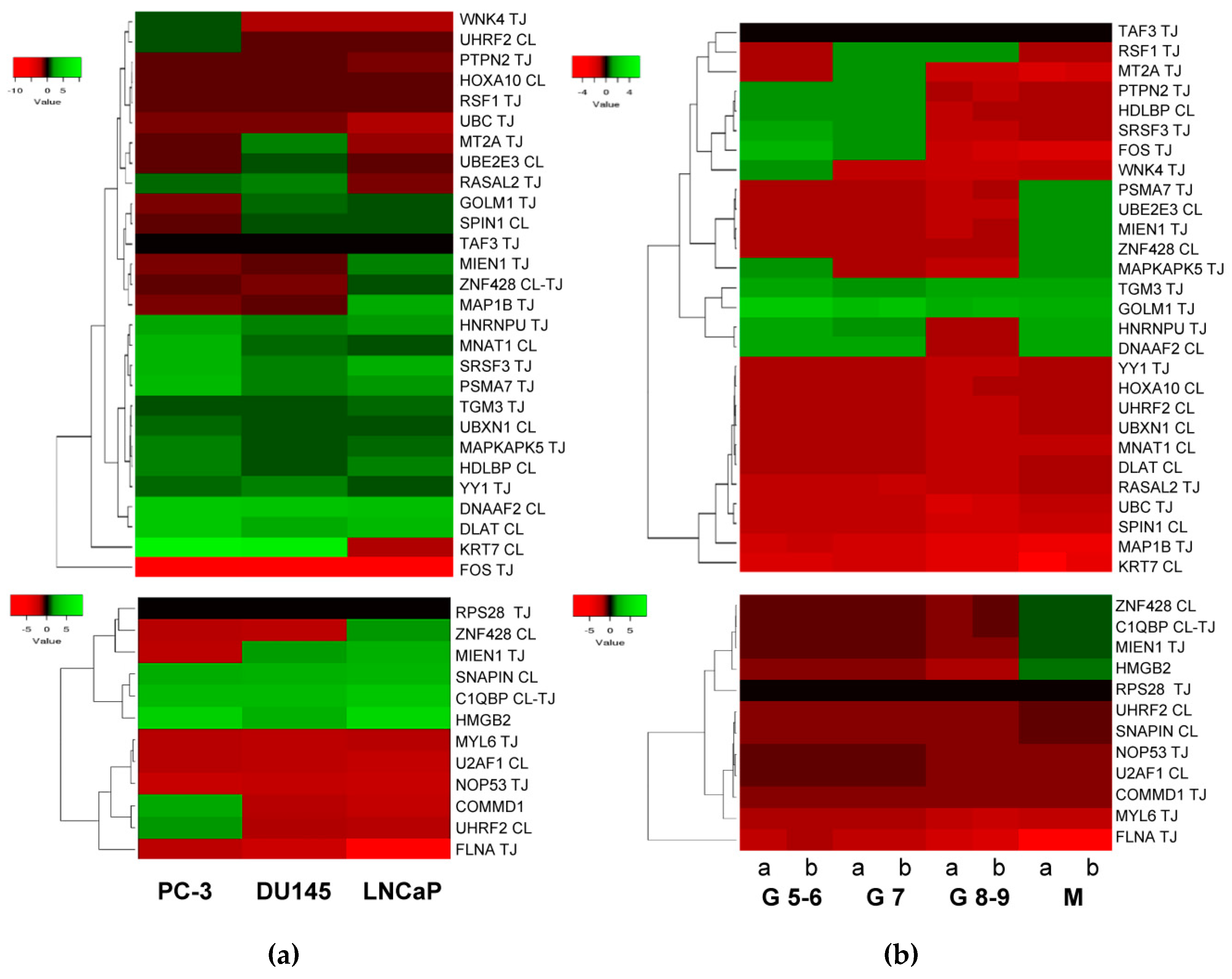
According to published data [12,18], HMGB1 expression increases in PCa cell lines and tissues from PCa, especially in metastases. With published data of RNA levels in PCa samples [86] retrieved from Geen Expression Omnibus (GEO Accession: GSE21032), the change fold expression of HMGB1 and HMGB2-interactome targets in PCa cell lines versus noncancerous cells was calculated, from which a heat map was constructed (Figure 5a). Using the same source, data was retrieved from 181 adenocarcinoma primary tumors, which were distributed in 3 groups clinically classified by Gleason scores, and in a 4th group integrated by 37 metastatic tumors. The change fold expression of HMGB1 and HMGB2-interactome targets in each group versus noncancerous cells from healthy tissues were calculated, from which the heat map shown in Figure 5b was constructed. The classification of each gene in the main clusters of the heat maps proved to be unrelated to the experimental library origin of the clone (PC-3 cell line or PCa adenocarcinoma primary tumor). The results reveal that genes encoding 11 proteins interacting with HMGB1 (Figure 4a top panel) are also upregulated in the 3 PCa cell lines (PC-3 and DU145 or LNCaP), and 8 more are upregulated in one or two PCa cell lines. Among the detected HMGB2 partners, 2 are upregulated in the 3 PCa cell lines: 1 in 2 and 3 in at least one (Figure 4a, bottom). In both HMGB1 and HMGB2 interactomes, the targets upregulated in metastatic tissue (Figure 5b) are a subset of those upregulated in one or more of the PCa cell lines. Analyzing expression in reference to Gleason score, the genes TMG3 and GOLM1 are upregulated in all the groups, whereas the others are only upregulated in groups classified with a Gleason score of less than or equal to 7 (PTPN2, HDLBP, SRF3, FOS, and WNK4). Regarding a pattern associated with the existence of metastasis, 3 genes that are not upregulated in samples from primary tumors are upregulated in metastasis: PSMA7, UBE2E3, and MIEN1 (Figure 5b).
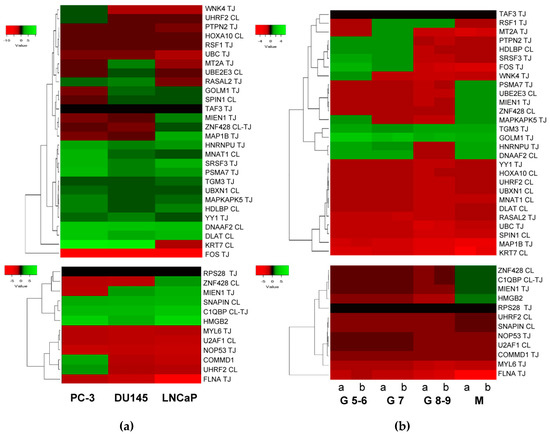
Figure 5.
Heat map expression in prostate cancer: (a) Expression of HMGB1 interactome targets (upper panel) and HMGB2 interactome targets (lower panel) in 3 prostate cancer cell lines; (b) expression of HMGB1 interactome targets (upper panel) and HMGB2 interactome targets (lower panel) in prostate adenocarcinoma cases classified by Gleason (G) score groups or metastatic (M) tumors. CL, target detected in the PC-3 library. TJ, target detected in the prostate adenocarcinoma library. Data extracted from Gene Expression Omnibus (GEO ) Accession: GSE21032.
2.4. Silencing of HMGB1 and HMGB2 Reveals Regulation of the Expression of Genes Encoding Their Interactome Targets
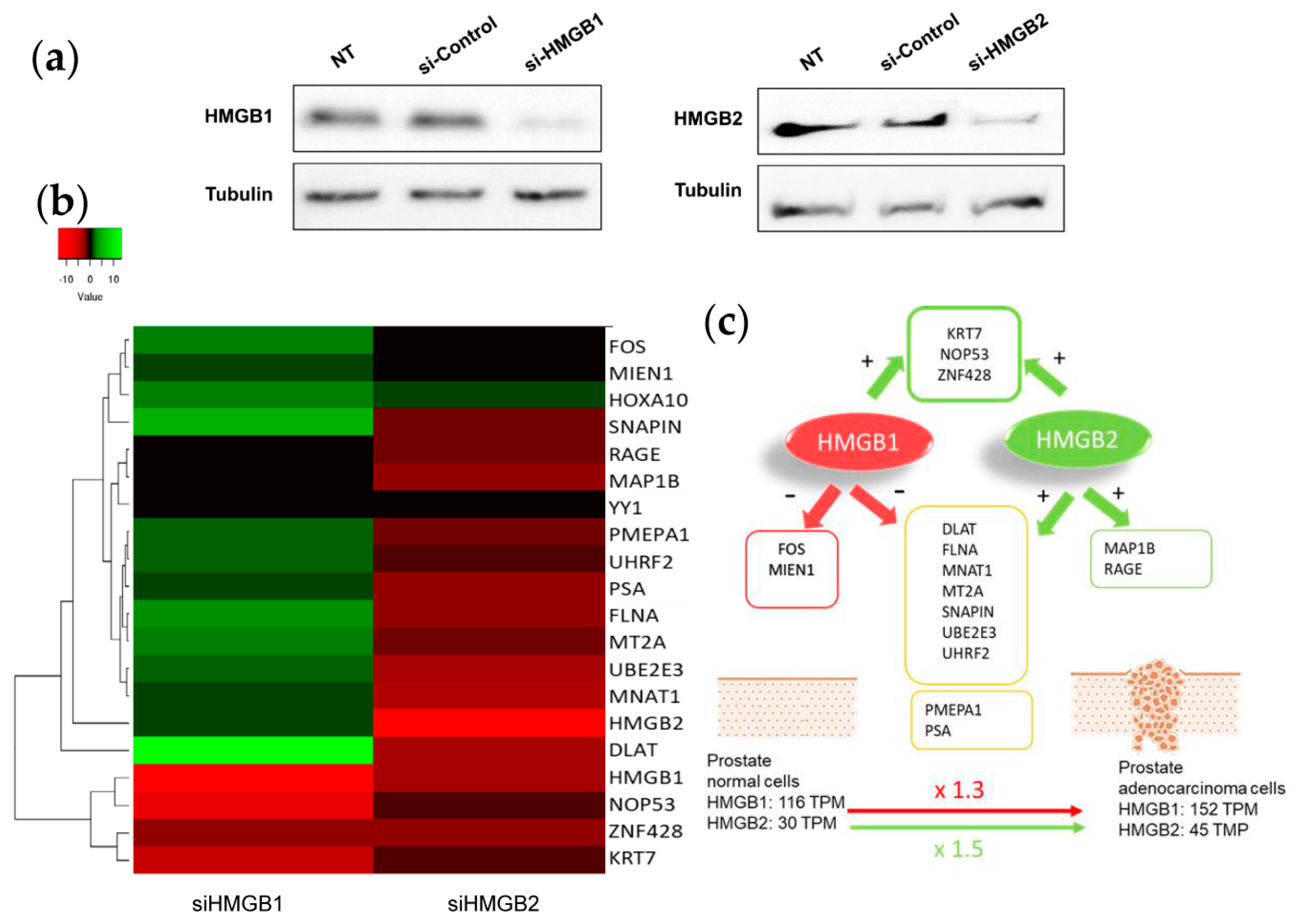
To test whether changes in HMGB1/2 protein levels in PCa cells could also be influencing the expression of their interactome targets, HMGB1 and HMGB2 in PC-3 cells were silenced by iRNA (Figure 6a). Levels of mRNA from 14 partners analyzed by qPCR and changes (siHMGB/HMGB) are summarized in the Figure 6b. This analysis also included HMGB1, HMGB2, and well-known PCa biomarkers: PSA (encoded by KLK3); PMEPA1, which is involved in downregulation of the androgen receptor, thus promoting androgen receptor-negative prostate cell proliferation [96]; and RAGE, one of the membrane receptors in the extracellular signaling function of HMGB1 [97]. Silencing of HMGB1 causes overexpression of the larger cluster of the Y2H interactome, whereas siHMGB2 has the opposite effect (Figure 6c). HMGB1 downregulates the expression of the majority of targets analyzed, and conversely, HMGB2 upregulates them. Therefore, the expression level of each regulated target would depend on the relative imbalance of HMGB1 and HMGB2 and on the differential effect of both HMGB proteins on the expression of each partner. PMEPA1 and PSA, well-known PCa biomarkers, are also oppositely regulated by HMGB1 and HMGB2 (Figure 6).
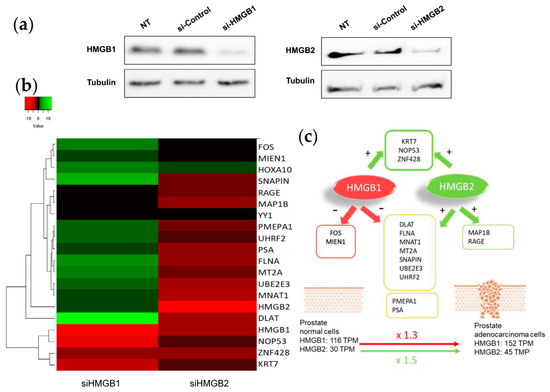
Figure 6.
HMGB1 and HMGB2 silencing: (a) Western Blot showing HMGB1 and HMGB2 silencing. Complete membranes provided as Supplementary Material, Image S2 (b) Heat map comparing the pattern of expression (siHMGB1/HMGB1 and siHMGB2/HMGB2). (c) Summary of regulatory effects of HMGB1 and HMGB2 on the selected genes.
3. Discussion
High mobility group box B (HMGB) proteins are pivotal in the development of cancer [6,8,10], and HMGB1 overexpression has been related to principal cancer hallmarks [7]. Interactome targets of HMGB1 or HMGB2 that have been identified in our Y2H study were previously found to be related to cancer hallmarks (Table S1 and Figure 1), and are also dysregulated in PCa, as confirmed by detection of changes in mRNA or protein levels. DNAAF2 [98], U2AF1 [43], C1QBP [40], Snapin, or HDLBP [99] are upregulated in prostate tumors or PCa cell lines. Others increase their expression after androgen-deprivation therapy, such as KRT7 or NOP53 [100]. Functional studies interfering the expression of several of the proteins revealed by our study also directly associated them to PCa. In this sense, selective knockdown of C1QBP through iRNA decreased cyclin D1, increased p21 expression, led to cell cycle arrest (G1/S transition) in PCa cells, and had no effect on a noncancerous cell line [40]. NOP53 acts as a tumor suppressor, and knockdown of the gene in the PCa LNCaP cell line increased the invasiveness of these cells as measured in a xenograft animal model [101].
Two already known regulatory factors have been found among the HMGB1 interactome targets, YY1 and HOXA10, and both are associated with PCa. YY1 is upregulated in human prostate cancer cell lines and tissues [66]. Inhibition of YY1 reduces expression of genes related to the Krebs cycle and electron transport chain in PCa cell lines [67], and YY1 depletion correlates with delayed progression of PCa [68]. Overexpression of YY1 can promote epithelial-mesenchymal transition by reducing hnRNPM expression [69]. YY1 can also silence tumor suppressor genes, such as XAF1 in PCa [70]. In summary, YY1 is a recognized prostate cancer driver [66] and different complexes in which YY1 takes part can induce activation or repression of gene expression, including also AR-YY1-mediated PSA transcription [102], which we found is also regulated by HMGB1 and HMGB2 silencing. HOXA10 is upregulated in PCa [31], and inverse correlations between HOXA10 expression and Gleason pattern, Gleason score, and pathological stage are found [32], although downregulation of HOXA10 gene expression may enhance lipogenesis to promote PCa cell growth and tumor progression to the castration-resistant stage [103]. Silencing of HOXA10 expression in PC-3 cells by iRNA decreased proliferation rates, whereas HOXA10 overexpression had the opposite effect [31]. Physical interaction between these PCa-associated proteins and HMGB proteins has not previously been described, and our results therefore show that there is a connection between HMGB1 and HMGB2 functions and those of their binding partners in PCa.
Considering that HMGB1, HMGB2, and a subset of their interactome partners are upregulated in PCa, we silenced HMGB1 and HMGB2 and analyzed the mRNA levels of a group of randomly selected partners in PC-3 cells (Figure 6). The data show that HMGB1 and HMGB2 control the expression of them, which might contribute to the orchestrated action of all these proteins in PCa. HMGB2 activates many of the tested targets, but unexpectedly, HMGB1 has the opposite effect. One can propose several reasons to explain upregulation of targets in these circumstances. Data from the genotype-tissue expression (GTEx) project [104] indicates that, although both HMGB1 and HMGB2 are upregulated in PCa versus noncancerous cells, the relative increase is higher for HMGB2 (×1.5) than HMGB1 (×1.3); this could explain the increased expression of several of their targets, assuming that positive regulation caused by HMGB2 predominates over negative regulation caused by HMGB1 during the onset of PCa. Alternatively, differential interaction of HMGB1 or HMGB2 with their different nuclear partners, the transcript factors detected in our Y2H analysis being among them, might condition their positive or negative regulatory roles on the expression of specific genes.
Clinically, a high frequency of CNA of the genes encoding the identified proteins is associated with the most aggressive forms of PCa: small cell neuroendocrine carcinoma (SCNC) or castration-resistant PCa (Figure 3). Their gain or amplification in the genome of the cancerous cells are positively correlated to a lesser disease-free period for PCa patients (Figure 4). The mRNA levels of a subset of these proteins are also higher in metastases than primary tumors (Figure 5). In conclusion, the set of proteins detected though our HMGB1-HMGB2 Y2H analysis are associated with the most aggressive cases of PCa. Although the PSA-based test is routinely employed for screening of PCa, it has resulted in overdiagnosis and overtreatment of nonaggressive cancers, thus reducing the quality of life of patients.
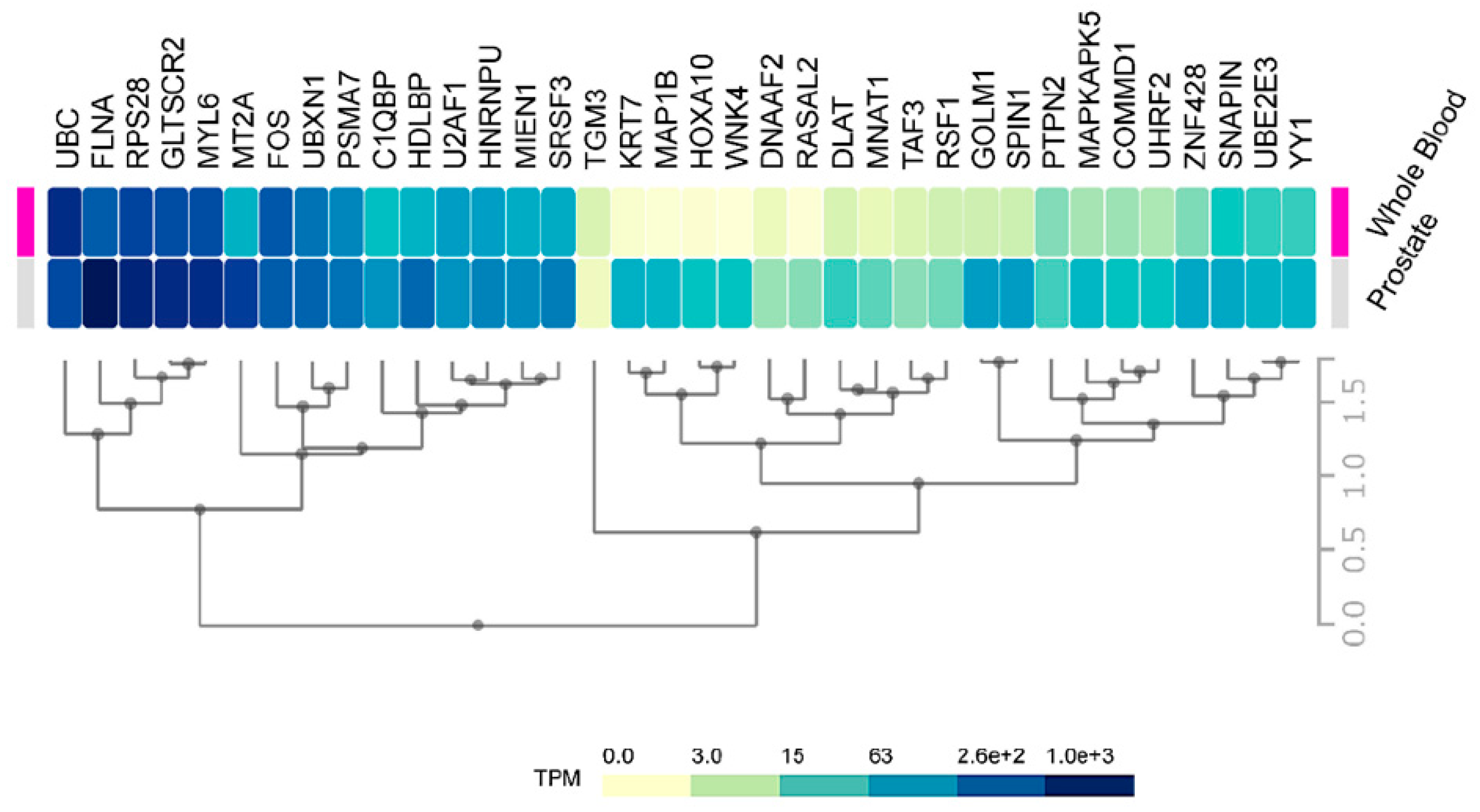
Consequently, an improvement is necessary in the initial stages to discriminate between high-risk from low risk cancers. Our data on HMGB1 and HMGB2 interactome targets, considering their correlation to high aggressiveness and bad prognosis, is a good starting point to develop new serum protein panels for improvement of PCa diagnosis. Indeed, FLNA has already been proposed in a clinical validated PCa biomarker panel in serum [74]. PSMA7 was also proposed as a PCa biomarker [55], and KRT7 is included in a whole blood mRNA 4-gene androgen regulated panel for PCa diagnosis [33]. Considering the relative expression levels of our HMGB1 and HMGB2 interactome targets in noncancerous cells or in blood of health subjects differ quite notably (Figure 7), one might anticipate that more sensitive analyses could be carried out using as biomarkers those proteins that are usually lowly expressed in noncancerous cells; thus, their levels are also low in the blood of healthy individuals. For instance, FLNA reported as a possible biomarker [74] is one of the 50 proteins most strongly expressed in normal prostate, and high levels are also detected in the blood of healthy individuals, whereas other detected HMGB1 or HMGB2 interactome targets in our study, e.g., DNAAF2, GOLM1, or TGM3, are in the lowest rank of detection in noncancerous samples and their increase should become more discriminatory.
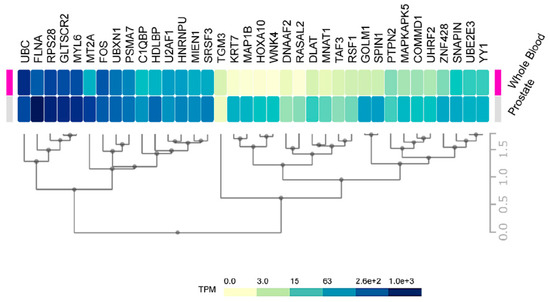
Figure 7.
Prostate and blood levels of HMGB1 and HMGB2 interactome targets in healthy men: Expression of HMGB1 and HMGB2 interactome targets in prostate tissue and whole blood in healthy men. Data were directly obtained and processed from the GTEx Project through Expression Atlas, an integrated database of gene and protein expression in humans, animals, and plants [105], accessed through https://www.ebi.ac.uk/gxa/experiments/E-MTAB-5214/Results.
4. Materials and Methods
4.1. Biological Materials
PC-3 is an androgen-independent cell line derived from a bone metastasis [106]. The human PCa PC-3 cell line, regularly validated by DNA typing, was obtained from the American Type Culture Collection ATCC and grown in Roswell Park Memorial Institute RPMI-1640 media, supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin-streptomycin (Thermo Fisher Scientific, Inc. Waltham, MA, USA). Cells were cultured at 37 °C in 5% CO2 in air in a humidified incubator. RNA from PCa tissue, isolated after radical prostate resection of a 66-year-old man diagnosed with adenocarcinoma (Gleason score 6) and not previously treated with radiotherapy or chemotherapy, was obtained from Biobanco de Andalucía (SPAIN).
4.2. Yeast Two Hybrid Methodology
Sacchacomyces cerevisiae strains were Y187 (MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, gal80Δ, met-, and URA3::GALuas-GAL1TATA-LacZ MEL1) and Y2HGold (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1uas-GAl1TATA-His3, GAL2uas-Gal2TATA-Ade2 URA3:: MEL1UAS-Mel1TATA, and AUR1-C MEL1).
Total RNA from the PC-3 cell line obtained from the supplier (Sigma-Aldrich, Saint Louis, MO, USA) and RNA from PCa tissue (Biobanco de Andalucía, Spain) were used to construct cDNA libraries. HMGB1 and HMGB2 interacting partners were identified using Matchmaker Gold Yeast 2-Hybrid System (Clontech, Fremont, CA, USA). Library construction, bait construction, and Yeast 2-Hybrid library screening were done according to the Takara Bio USA Matchmaker® Gold Yeast 2-Hybrid System manual. In brief, the baits were cloned as fusions to the GAL4 activation domain in the plasmid pGBKT7-AD and used to transform the yeast haploid strain, Y187. cDNA libraries prepared from RNA extracted from PC-3 cells and PCa cancerous tissue were included as fusions to the GAL4 DNA-binding domain in the plasmid pGBKT7-BD and were used to transform the yeast haploid strain, Y2HGold. RNAs from human samples used to prepare the Y2H libraries were obtained from Biobanco de Andalucía (Spain). RNA was extracted from frozen tissue sections in OCT (Optimal Cutting Temperature) compound, using the Qiacube robot from QIAGEN, based on ion-exchange columns with a silica membrane. RNA was obtained with the miRNeasy mini-kit from QIAGEN that allows recovery of both total RNA and miRNAs. The samples were finally treated with RNase-free DNAase from QIAGEN. The RNA was quantified at 260 nm and 280 nm by spectrophotometry using Infinite F200 equipment of TECAN with a Nanoquant plate. Finally, the integrity of the samples was evaluated by AGILENT 2200 Tape Station apparatus, with the RIN (RNA Integrity Number) parameter being >8. Efficiency in the constructions of libraries was in the range recommended in the kit (all libraries guaranteed to have >1 × 106 independent clones). As a previous control, we confirmed that our baits (HMGB1 and HMGB2) do not autonomously activate the reporter genes in Y2HGold in the absence of a prey protein. Bait and prey fusion proteins are each expressed in different haploid yeast strains that can form diploids. The diploid yeast cell expresses both proteins, and when fusion proteins interact, the transcriptional activator GAL4 is reconstructed and brought into proximity to activate transcription of the reporter genes. For diploid formation, 1 mL of concentrated bait culture was combined with 1 mL of library culture and incubated overnight with slow shaking. A drop of the culture was checked under a phase-contrast microscope (40×) to confirm the existence of zygotes before plating on diploid-selective media. Diploids were tested for expression of the reporter genes in selective media. To reduce the appearance of false positives, a screening based on three different independent markers (ADE2, HIS3, and MEL1) was selected. pGBKT7-BD plasmids carrying the preys were rescued from confirmed positive diploids, and DNA was used to transform E. coli. The inserts were sequenced with primer T7 (5′-TAATACGACTCACTATAGGG-3′). Sequences were used for homology searches with BlastN and BlastX at the National Center for Biotechnology Information NCBI (https://blast.ncbi.nlm.nih.gov/) and proteins in the database matching the queries annotated as positives.
4.3. Expression Analysis by Quantitative Polymerase Chain Reaction (RT-qPCR)
Individual analyses of gene expression were carried out as follows. RNA samples were retro-transcribed into cDNA and labeled with the KAPPA SYBR FAST universal one-step qRT-PCR kit (Kappa Biosystems, Inc, Woburn, Massachusetts, USA). The primers for qPCR are shown in Table S3. Reaction conditions for thermal cycling were 42 °C for 5 min, 95 °C for 5 s, 40 cycles of 95 °C for 3 s, and finally 60 °C for 20 s. ECO Real-Time PCR System was used for the experiments (Illumina, Inc., San Diego, California, USA), and calculations were made by the 2−ΔΔCt method [107]. Student’s test was used to check the statistical significance of differences between samples (p < 0.05). The relative mRNA levels of the experimentally selected genes (target genes) were calculated by referring to the mRNA levels of the housekeeping gene, encoding glyceraldehyde phosphate dehydrogenase (GAPDH), which had been verified as being expressed constitutively under the assay conditions. For valid quantification using the 2−ΔΔCt method, it is crucial that target and housekeeping PCR amplification efficiencies are approximately equal: we therefore verified that the efficiencies of the 2 PCR reactions differed by <10%. At least 2 independent biological replicas and 3 technical replicas of each of them were made for all the experiments.
4.4. Immunoprecipitation
One hundred µl of Protein G Plus-Agarose immunoprecipitation-reagent (Santa Cruz Biotechnology, Dallas, TX, USA) were coupled with 4 µg of anti-HMGB1 antibody (sc-74085; Santa Cruz Biotechnology) or anti-mouse antibody (Molecular Probes, A10534) in phosphate buffered saline (PBS) for 1 h at 4 °C with rotation. PC-3 cells were lysed in 20 mM Tris/HCl, 150 mM, 1% Triton X-100, 1× phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail (Sigma-Aldrich, Saint Louis, MO, USA) and incubated for 30 min at 4 °C with rotation. Total protein (500 µg) was incubated with the antibody agarose beads overnight and eluted by incubation in 1× lithium dodecyl sulfate LDS loading buffer containing 350 mM β-mercaptoethanol at 95 °C for 10 min. Mass spectrometry and data analysis were done as previously described [26].
4.5. Western Blot Analysis
Protein samples were run on 10% SDS-PAGE gels at 80 V for 20 min followed by 200 V for 45–60 min. Proteins were transferred onto a polyvinylidene fluoride (PVDF) membrane at 0.2 A for 1 h. Membranes were blocked by incubating with 5% non-fat dry milk for 1 h at room temperature (RT) and then incubated with primary antibodies, anti-HMGB1 (sc-74085; Santa Cruz Biotechnology) or anti-Cytokeratin 7 (ab181598; Abcam, Cambridge, UK) in phosphate-buffered saline with 0.1% Tween 20® detergent PBST overnight at 4 °C. After incubation with the corresponding horseradish peroxidase-conjugated secondary antibody, enhanced chemiluminescence for high sensitivity and long-lasting signal (ECL) Anti-mouse IgG (NXA931 from GE Healthcare Sciences, Chicago, IL, USA) or ECL Anti-rabbit IgG (NA934 from GE Healthcare Sciences, Chicago, IL, USA), protein bands were detected using LuminataTMCrescendo Western HRP Substrate (Millipore Corporation, Burlington, MA, USA) and a ChemiDocTM imager (Bio-Rad laboratories Hercules, CA, USA).
4.6. Immunofluorescence and Confocal Microscopy
Cells were plated in 6-well plates, each containing 4 sterile 13-mm glass coverslips. When 80% confluent, cells were fixed in 4% paraformaldehyde in PBS for 15 min at RT. Cells were washed 3 times with PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4) and finally treated with 0.1% Triton/PBS for 15 min at RT. They were then blocked in 1% bovine serum albumin (BSA) in PBS for 1 h at RT. Primary antibodies, anti-HMGB1 (sc-74085; Santa Cruz Biotechnology) or anti-Cytokeratin 7 (ab181598; Abcam, Cambridge, UK) were diluted in 1% BSA in PBS. Cells were incubated with the corresponding primary antibodies overnight at 4 °C, followed by 3 washes with PBS and staining with the secondary antibodies, modified with Alexa Fluor 488 and 568 (Invitrogen, Carlsbad, CA, USA) previously diluted in 1% BSA in PBS for 1 h at RT in the dark. For nuclear staining, after secondary antibody incubation, wells were washed 3 times and stained with Hoechst (Life Technologies, Carlsbad, CA, USA) for 5 min at RT in the dark. Cells were washed once with PBS and once with sterile distilled water. Each coverslip was mounted on a clean slide using ProLong™ Gold Antifade Mountant (Invitrogen). After drying, the slides were stored at 4 °C in the dark until they were examined by confocal microscopy (Nikon A1R). Meander’s correlation coefficient was calculated using Nis Elements software from Nikon.
4.7. HMGB1 and HMGB2 Silencing by siRNA
The PC-3 cell line was transfected with small interfering (si)RNA oligonucleotides using Lipofectamine 2000 (Invitrogen). siRNA and Lipofectamine 2000 were each diluted separately with Opti-MEM (Gibco), mixed together, and incubated for 5 min at RT. The mixture was added to cells plated in 3 mL RPMI 1610 medium (final concentration of siRNA, 50 nM). Cells were collected at 48 h post transfection for further analysis. The following siRNAs (Life Technologies) were used for the silencing of each gene: s20254 Silencer Select for HMGB1, s6650 for HMGB2, and AS02A5Z3 for the siRNA negative control.
Total RNA was extracted from different conditions (siHMGB1, siHMGB2, and siCtrl#2) of the PC-3 cell line using GeneJET RNA Purification Kit (#K0731, Thermo Scientific). The remaining DNA was removed by incubating with DNase I, RNase-free (#EN0521, Thermo Scientific). DNA-free RNA was finally purified using GeneJET RNA Cleanup and Concentration Micro Kit (#K0842, Thermo Scientific). qPCR reactions were run in triplicate in an Eco Real-Time PCR System (Illumina) using 1 ng per reaction. PC-3 lysates of each condition were extracted with lysis buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 0.1% NP40, 1 mM ethylenediaminetetraacetic acid disodium salt (EDTA), and 2 mM MgCl2), and protein concentration was quantified using Bradford Reagent (Bio-Rad). Protein samples of 25–40 µg were loaded for western blotting. PVDF membranes were incubated overnight at 4 °C with primary antibodies, anti-HMGB1 (ab18256, Abcam), anti-HMGB2 (ab67282, Abcam), or anti-α-tubulin (sc53646, Santa Cruz Biotechnology).
4.8. Heat Maps
Heat maps from change-fold ratios (Figure 5 and Figure 6) were drawn with Heatmapper (http://heatmapper.ca/expression/), using complete linkage as clustering method and Euclidean distance as the measurement method [108].
4.9. Statistical Analysis
Analyses were carried out using GraphPad Prism 6 (GraphPad Software, San Diego, CA, USA). Continuous variables were expressed as mean ± SE. Relative gene expression assays were tested using independent t-tests. A 2-tailed p-value test was used with p < 0.05 considered significant.
5. Conclusions
We have carried out the first HMGB1/HMGB2 interactome approach in prostate cancer (PCa) using both the PC-3 cell line and adenocarcinoma tissue. Gene or protein expressions of the majority of targets are dysregulated in PCa, and functional relationships between these proteins and PCa had also previously been confirmed by different laboratories using different models and technical approaches. We have shown by interference analysis that several HMGB1 and HMGB2 partners are regulated by HMGB1 and HMGB2 themselves, which might contribute to the coordination of their cellular action in PCa. Copy number alterations in the detected HMGB1 and HMGB2 partners are associated with aggressive forms of PCa and a poor prognosis. These characteristics can potentially be used as discriminatory biomarkers between high and low risk patients.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/11/11/1729/s1, Table S1: Association of proteins that interact with HMGB1 or HMGB2 to cancer hallmarks, Table S2: Prostate cancer studies available through cBioportal, Table S3: List of oligonucleotides used in this work. Figure S1: Y2H triple screening by 3 independent selection markers. Image S1: Blots corresponding to Figure 2a. Image S2: Blots corresponding to Figure 6a.
Author Contributions
Conceptualization, M.-E.C. and M.L.-M.; methodology, A.B.-A., M.C.-Q., and M.S.-M.; software, Á.V.-V.; validation, A.B.-A., O.M.-I., Á.V.-V., and A.F.; formal analysis, M.Q.-V. and E.R.-B.; investigation, A.B.-A., M.C.-Q., and M.S.-M.; data curation, A.B.-A. and M.-E.C.; Writing—Original Draft preparation, A.B.-A., M.L.-M., and M.-E.C.; Writing—Review and Editing, all authors; project administration, M.-E.C.; funding acquisition, M.-E.C. and M.L.-M.
Funding
This work has been supported by Plan Estatal I + D + I cofunded by the Instituto Carlos III (ISCIII, Spain) under grant agreement AES no. PI14/01031 by Fondo Europeo de Desarrollo Regional-FEDER (The European Regional Development Fund-ERDF) “A way of Making Europe”, and by Xunta de Galicia (Consolidación Grupos Referencia Competitiva contract no. ED431C 2016-012. Aida Barreiro-Alonso was funded by a predoctoral fellowship from Xunta de Galicia-2013 (Spain).
Acknowledgments
We thank the Biobanco of Andalucía (Spain) for the RNA samples from cancerous prostate tissue and adjacent healthy tissue. We also thank technical support for confocal microscopy to the microscopy unit from “Servicio de Apoyo a la Investigación” (SAI) at University of a Coruña (SPAIN). Collaboration of the postgrad students Ana Feijó and Carlos Coppel in library screening during their laboratory stages is acknowledged. The English text was edited by Biomedes (http://www.biomedes.co.uk/).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Catena, R.; Escoffier, E.; Caron, C.; Khochbin, S.; Martianov, I.; Davidson, I. HMGB4, a Novel Member of the HMGB Family, is Preferentially Expressed in the Mouse Testis and Localizes to the Basal Pole of Elongating Spermatids. Biol. Reprod. 2009, 80, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Pusterla, T.; de Marchis, F.; Palumbo, R.; Bianchi, M.E. High Mobility Group B2 is Secreted by Myeloid Cells and has Mitogenic and Chemoattractant Activities Similar to High Mobility Group B1. Autoimmunity 2009, 42, 308–310. [Google Scholar] [CrossRef] [PubMed]
- Ugrinova, I.; Pashev, I.G.; Pasheva, E.A. Nucleosome Binding Properties and Co-Remodeling Activities of Native and in Vivo Acetylated HMGB-1 and HMGB-2 Proteins. Biochemistry 2009, 48, 6502–6507. [Google Scholar] [CrossRef] [PubMed]
- Swanson, P.C. Fine Structure and Activity of Discrete RAG-HMG Complexes on V (D)J Recombination Signals. Mol. Cell. Biol. 2002, 22, 1340–1351. [Google Scholar] [CrossRef]
- Bagherpoor, A.J.; Dolezalova, D.; Barta, T.; Kucirek, M.; Sani, S.A.; Esner, M.; Kunova Bosakova, M.; Vinarsky, V.; Peskova, L.; Hampl, A.; et al. Properties of Human Embryonic Stem Cells and their Differentiated Derivatives Depend on Nonhistone DNA-Binding HMGB1 and HMGB2 Proteins. Stem Cells Dev. 2017, 26, 328–340. [Google Scholar] [CrossRef]
- Ke, S.; Zhou, F.; Yang, H.; Wei, Y.; Gong, J.; Mei, Z.; Wu, L.; Yu, H.; Zhou, Y. Downregulation of High Mobility Group Box 1 Modulates Telomere Homeostasis and Increases the Radiosensitivity of Human Breast Cancer Cells. Int. J. Oncol. 2015, 46, 1051–1058. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Zeh, H.J., 3rd; Lotze, M.T. High-Mobility Group Box 1 and Cancer. Biochim. Biophys. Acta 2010, 1799, 131–140. [Google Scholar] [CrossRef]
- Liu, K.; Huang, J.; Xie, M.; Yu, Y.; Zhu, S.; Kang, R.; Cao, L.; Tang, D.; Duan, X. MIR34A Regulates Autophagy and Apoptosis by Targeting HMGB1 in the Retinoblastoma Cell. Autophagy 2014, 10, 442–452. [Google Scholar] [CrossRef]
- Chandrasekaran, K.S.; Sathyanarayanan, A.; Karunagaran, D. Downregulation of HMGB1 by miR-34a is Sufficient to Suppress Proliferation, Migration and Invasion of Human Cervical and Colorectal Cancer Cells. Tumor Biol. 2016, 37, 13155–13166. [Google Scholar] [CrossRef]
- Tang, C.; Yang, Z.; Chen, D.; Xie, Q.; Peng, T.; Wu, J.; Qi, S. Downregulation of miR-130a Promotes Cell Growth and Epithelial to Mesenchymal Transition by Activating HMGB2 in Glioma. Int. J. Biochem. Cell Biol. 2017, 93, 25–31. [Google Scholar] [CrossRef]
- Liu, P.L.; Liu, W.L.; Chang, J.M.; Chen, Y.H.; Liu, Y.P.; Kuo, H.F.; Hsieh, C.C.; Ding, Y.S.; Chen, W.W.; Chong, I.W. MicroRNA-200c Inhibits Epithelial-Mesenchymal Transition, Invasion, and Migration of Lung Cancer by Targeting HMGB1. PLoS ONE 2017, 12, e0180844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shao, S.; Han, D.; Xu, Y.; Jiao, D.; Wu, J.; Yang, F.; Ge, Y.; Shi, S.; Li, Y.; et al. High Mobility Group Box 1 Promotes the Epithelial-to-Mesenchymal Transition in Prostate Cancer PC3 Cells Via the RAGE/NF-kappaB Signaling Pathway. Int. J. Oncol. 2018, 53, 659–671. [Google Scholar] [CrossRef] [PubMed]
- van Beijnum, J.R.; Nowak-Sliwinska, P.; van den Boezem, E.; Hautvast, P.; Buurman, W.A.; Griffioen, A.W. Tumor Angiogenesis is Enforced by Autocrine Regulation of High-Mobility Group Box 1. Oncogene 2013, 32, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.B.; Cai, L.; Lin, S.J.; Xiong, Z.K.; Lu, J.L.; Mao, Y.; Yao, Y.; Zhou, L.F. High-Mobility Group Box 2 is Associated with Prognosis of Glioblastoma by Promoting Cell Viability, Invasion, and Chemotherapeutic Resistance. Neuro Oncol. 2013, 15, 1264–1275. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Jiang, H.; Zhu, H.; Zhang, H.; Gong, J.; Zhang, L.; Ding, Q. Overexpression of High Mobility Group Box 1 and 2 is Associated with the Progression and Angiogenesis of Human Bladder Carcinoma. Oncol. Lett. 2013, 5, 884–888. [Google Scholar] [CrossRef]
- Tai, S.; Sun, Y.; Squires, J.M.; Zhang, H.; Oh, W.K.; Liang, C.Z.; Huang, J. PC3 is a Cell Line Characteristic of Prostatic Small Cell Carcinoma. Prostate 2011, 71, 1668–1679. [Google Scholar] [CrossRef]
- Elangovan, I.; Thirugnanam, S.; Chen, A.; Zheng, G.; Bosland, M.C.; Kajdacsy-Balla, A.; Gnanasekar, M. Targeting Receptor for Advanced Glycation End Products (RAGE) Expression Induces Apoptosis and Inhibits Prostate Tumor Growth. Biochem. Biophys. Res. Commun. 2012, 417, 1133–1138. [Google Scholar] [CrossRef]
- Li, T.; Gui, Y.; Yuan, T.; Liao, G.; Bian, C.; Jiang, Q.; Huang, S.; Liu, B.; Wu, D. Overexpression of High Mobility Group Box 1 with Poor Prognosis in Patients After Radical Prostatectomy. BJU Int. 2012, 110, E1125–E1130. [Google Scholar] [CrossRef]
- Gnanasekar, M.; Thirugnanam, S.; Ramaswamy, K. Short Hairpin RNA (shRNA) Constructs Targeting High Mobility Group Box-1 (HMGB1) Expression Leads to Inhibition of Prostate Cancer Cell Survival and Apoptosis. Int. J. Oncol. 2009, 34, 425–431. [Google Scholar] [CrossRef]
- Flores-Morales, A.; Iglesias-Gato, D. Quantitative Mass Spectrometry-Based Proteomic Profiling for Precision Medicine in Prostate Cancer. Front. Oncol. 2017, 7, e267. [Google Scholar] [CrossRef]
- Muller, A.K.; Foll, M.; Heckelmann, B.; Kiefer, S.; Werner, M.; Schilling, O.; Biniossek, M.L.; Jilg, C.A.; Drendel, V. Proteomic Characterization of Prostate Cancer to Distinguish Nonmetastasizing and Metastasizing Primary Tumors and Lymph Node Metastases. Neoplasia 2018, 20, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, D.; Li, M.; Wei, X.; Liu, S.; Zhao, M.; Liu, C.; Wang, X.; Jiang, X.; Li, X.; et al. Quantitative Proteomics of TRAMP Mice Combined with Bioinformatics Analysis Reveals that PDGF-B Regulatory Network Plays a Key Role in Prostate Cancer Progression. J. Proteome Res. 2018, 17, 2401–2411. [Google Scholar] [CrossRef] [PubMed]
- Stelloo, S.; Nevedomskaya, E.; Kim, Y.; Hoekman, L.; Bleijerveld, O.B.; Mirza, T.; Wessels, L.F.A.; van Weerden, W.M.; Altelaar, A.F.M.; Bergman, A.M.; et al. Endogenous Androgen Receptor Proteomic Profiling Reveals Genomic Subcomplex Involved in Prostate Tumorigenesis. Oncogene 2018, 37, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.; Brady, N.J.; Bareja, R.; Robinson, B.D.; Conteduca, V.; Augello, M.A.; Puca, L.; Ahmed, A.; Dardenne, E.; Lu, X.; et al. N-Myc-Mediated Epigenetic Reprogramming Drives Lineage Plasticity in Advanced Prostate Cancer. J. Clin. Investig. 2019, 130, 3924–3940. [Google Scholar] [CrossRef]
- Zhang, Z.; Chng, K.R.; Lingadahalli, S.; Chen, Z.; Liu, M.H.; Do, H.H.; Cai, S.; Rinaldi, N.; Poh, H.M.; Li, G.; et al. An AR-ERG Transcriptional Signature Defined by Long-Range Chromatin Interactomes in Prostate Cancer Cells. Genome Res. 2019, 29, 223–235. [Google Scholar] [CrossRef]
- Barreiro-Alonso, A.; Lamas-Maceiras, M.; García-Díaz, R.; Rodríguez-Belmonte, E.; Yu, L.; Pardo, M.; Choudhary, J.S.; Cerdán, M.E. Delineating the HMGB1 and HMGB2 interactome in prostate and ovary epithelial cells and its relationship with cancer. Oncotarget 2018, 9, 19050–19064. [Google Scholar] [CrossRef]
- Chen, R.; Xiao, M.; Gao, H.; Chen, Y.; Li, Y.; Liu, Y.; Zhang, N. Identification of a Novel Mitochondrial Interacting Protein of C1QBP using Subcellular Fractionation Coupled with CoIP-MS. Anal. Bioanal. Chem. 2016, 408, 1557–1564. [Google Scholar] [CrossRef]
- Li, C.; He, C.; Xu, Y.; Xu, H.; Tang, Y.; Chavan, H.; Duan, S.; Artigues, A.; Forrest, M.L.; Krishnamurthy, P.; et al. Alternol Eliminates Excessive ATP Production by Disturbing Krebs Cycle in Prostate Cancer. Prostate 2019, 79, 628–639. [Google Scholar] [CrossRef]
- Omran, H.; Kobayashi, D.; Olbrich, H.; Tsukahara, T.; Loges, N.T.; Hagiwara, H.; Zhang, Q.; Leblond, G.; O’Toole, E.; Hara, C.; et al. Ktu/PF13 is Required for Cytoplasmic Pre-Assembly of Axonemal Dyneins. Nature 2008, 456, 611–616. [Google Scholar] [CrossRef]
- Massafra, V.; Milona, A.; Vos, H.R.; Burgering, B.M.; van Mil, S.W. Quantitative Liver Proteomics Identifies FGF19 Targets that Couple Metabolism and Proliferation. PLoS ONE 2017, 12, e0171185. [Google Scholar] [CrossRef]
- Li, B.; Cao, X.; Weng, C.; Wu, Y.; Fang, X.; Zhang, X.; Liu, G. HoxA10 Induces Proliferation in Human Prostate Carcinoma PC-3 Cell Line. Cell Biochem. Biophys. 2014, 70, 1363–1368. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, Y.; de Velasco, M.A.; Oki, T.; Shimizu, N.; Nozawa, M.; Yoshimura, K.; Yoshikawa, K.; Nishio, K.; Uemura, H. HOXA10 Expression Profiling in Prostate Cancer. Prostate 2019, 79, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Thomas, B.C.; Kay, J.D.; Menon, S.; Vowler, S.L.; Dawson, S.N.; Bucklow, L.J.; Luxton, H.J.; Johnston, T.; Massie, C.E.; Pugh, M.; et al. Whole Blood mRNA in Prostate Cancer Reveals a Four-Gene Androgen Regulated Panel. Endocr. Relat. Cancer 2016, 23, 797–812. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Lu, J.; Li, Y.; Chen, C.; Cai, Y.; Tan, G.; Peng, Z.; Zhang, Z.; Dong, Z.; Kang, T.; et al. MNAT1 is Overexpressed in Colorectal Cancer and Mediates p53 Ubiquitin-Degradation to Promote Colorectal Cancer Malignance. J. Exp. Clin. Cancer Res. 2018, 37, e284. [Google Scholar] [CrossRef]
- Wang, W.; Chen, Z.; Mao, Z.; Zhang, H.; Ding, X.; Chen, S.; Zhang, X.; Xu, R.; Zhu, B. Nucleolar Protein Spindlin1 Recognizes H3K4 Methylation and Stimulates the Expression of rRNA Genes. EMBO Rep. 2011, 12, 1160–1166. [Google Scholar] [CrossRef]
- Plafker, K.S.; Plafker, S.M. The Ubiquitin-Conjugating Enzyme UBE2E3 and its Import Receptor Importin-11 Regulate the Localization and Activity of the Antioxidant Transcription Factor NRF2. Mol. Biol. Cell 2015, 26, 327–338. [Google Scholar] [CrossRef]
- Huang, K.; Yang, C.; Wang, Q.X.; Li, Y.S.; Fang, C.; Tan, Y.L.; Wei, J.W.; Wang, Y.F.; Li, X.; Zhou, J.H.; et al. The CRISPR/Cas9 System Targeting EGFR Exon 17 Abrogates NF-kappaB Activation Via Epigenetic Modulation of UBXN1 in EGFRwt/vIII Glioma Cells. Cancer Lett. 2017, 388, 269–280. [Google Scholar] [CrossRef]
- Lu, H.; Hallstrom, T.C. The Nuclear Protein UHRF2 is a Direct Target of the Transcription Factor E2F1 in the Induction of Apoptosis. J. Biol. Chem. 2013, 288, 23833–23843. [Google Scholar] [CrossRef]
- McGee, A.M.; Douglas, D.L.; Liang, Y.; Hyder, S.M.; Baines, C.P. The Mitochondrial Protein C1qbp Promotes Cell Proliferation, Migration and Resistance to Cell Death. Cell Cycle 2011, 10, 4119–4127. [Google Scholar] [CrossRef]
- Amamoto, R.; Yagi, M.; Song, Y.; Oda, Y.; Tsuneyoshi, M.; Naito, S.; Yokomizo, A.; Kuroiwa, K.; Tokunaga, S.; Kato, S.; et al. Mitochondrial p32/C1QBP is Highly Expressed in Prostate Cancer and is Associated with Shorter Prostate-Specific Antigen Relapse Time After Radical Prostatectomy. Cancer Sci. 2011, 102, 639–647. [Google Scholar] [CrossRef]
- Quintero, I.B.; Herrala, A.M.; Araujo, C.L.; Pulkka, A.E.; Hautaniemi, S.; Ovaska, K.; Pryazhnikov, E.; Kulesskiy, E.; Ruuth, M.K.; Soini, Y.; et al. Transmembrane Prostatic Acid Phosphatase (TMPAP) Interacts with Snapin and Deficient Mice Develop Prostate Adenocarcinoma. PLoS ONE 2013, 8, e73072. [Google Scholar] [CrossRef] [PubMed]
- Fei, D.L.; Motowski, H.; Chatrikhi, R.; Prasad, S.; Yu, J.; Gao, S.; Kielkopf, C.L.; Bradley, R.K.; Varmus, H. Wild-Type U2AF1 Antagonizes the Splicing Program Characteristic of U2AF1-Mutant Tumors and is Required for Cell Survival. PLoS Genet. 2016, 12, e1006384. [Google Scholar] [CrossRef] [PubMed]
- Daures, M.; Idrissou, M.; Judes, G.; Rifai, K.; Penault-Llorca, F.; Bignon, Y.J.; Guy, L.; Bernard-Gallon, D. A New Metabolic Gene Signature in Prostate Cancer Regulated by JMJD3 and EZH2. Oncotarget 2018, 9, 23413–23425. [Google Scholar] [CrossRef] [PubMed]
- Shankar, E.; Song, K.; Corum, S.L.; Bane, K.L.; Wang, H.; Kao, H.Y.; Danielpour, D. A Signaling Network Controlling Androgenic Repression of c-Fos Protein in Prostate Adenocarcinoma Cells. J. Biol. Chem. 2016, 291, 5512–5526. [Google Scholar] [CrossRef]
- Yan, G.; Ru, Y.; Wu, K.; Yan, F.; Wang, Q.; Wang, J.; Pan, T.; Zhang, M.; Han, H.; Li, X.; et al. GOLM1 Promotes Prostate Cancer Progression through Activating PI3K-AKT-mTOR Signaling. Prostate 2018, 78, 166–177. [Google Scholar] [CrossRef]
- Zhang, L.; Song, D.; Zhu, B.; Wang, X. The Role of Nuclear Matrix Protein HNRNPU in Maintaining the Architecture of 3D Genome. Semin. Cell Dev. Biol. 2019, 90, 161–167. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, J.W.; Jeong, M.H.; An, J.H.; Jang, S.M.; Song, K.H.; Choi, K.H. Microtubule-Associated Protein 1B Light Chain (MAP1B-LC1) Negatively Regulates the Activity of Tumor Suppressor p53 in Neuroblastoma Cells. FEBS Lett. 2008, 582, 2826–2832. [Google Scholar] [CrossRef]
- Zheng, M.; Wang, Y.H.; Wu, X.N.; Wu, S.Q.; Lu, B.J.; Dong, M.Q.; Zhang, H.; Sun, P.; Lin, S.C.; Guan, K.L.; et al. Inactivation of Rheb by PRAK-Mediated Phosphorylation is Essential for Energy-Depletion-Induced Suppression of mTORC1. Nat. Cell Biol. 2011, 13, 263–272. [Google Scholar] [CrossRef]
- Dwyer, S.F.; Gelman, I.H. Cross-Phosphorylation and Interaction between Src/FAK and MAPKAP5/PRAK in Early Focal Adhesions Controls Cell Motility. J. Cancer Biol. Res. 2014, 2, e1045. [Google Scholar]
- Dasgupta, S.; Wasson, L.M.; Rauniyar, N.; Prokai, L.; Borejdo, J.; Vishwanatha, J.K. Novel Gene C17orf37 in 17q12 Amplicon Promotes Migration and Invasion of Prostate Cancer Cells. Oncogene 2009, 28, 2860–2872. [Google Scholar] [CrossRef]
- Chen, S.H.; Chen, L.; Russell, D.H. Metal-Induced Conformational Changes of Human Metallothionein-2A: A Combined Theoretical and Experimental Study of Metal-Free and Partially Metalated Intermediates. J. Am. Chem. Soc. 2014, 136, 9499–9508. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, M.; Nomura, T.; Sato, F.; Mimata, H. Metallothionein is Up-Regulated Under Hypoxia and Promotes the Survival of Human Prostate Cancer Cells. Oncol. Rep. 2007, 18, 1145–1153. [Google Scholar] [PubMed]
- Lin, H.K.; Altuwaijri, S.; Lin, W.J.; Kan, P.Y.; Collins, L.L.; Chang, C. Proteasome Activity is Required for Androgen Receptor Transcriptional Activity Via Regulation of Androgen Receptor Nuclear Translocation and Interaction with Coregulators in Prostate Cancer Cells. J. Biol. Chem. 2002, 277, 36570–36576. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Choi, Y.J.; Kim, J.M.; Jeong, S.T.; Kim, J.H.; Kim, S.H.; Ryu, S.E. Binding and Regulation of HIF-1alpha by a Subunit of the Proteasome Complex, PSMA7. FEBS Lett. 2001, 498, 62–66. [Google Scholar] [CrossRef]
- Romanuik, T.L.; Ueda, T.; Le, N.; Haile, S.; Yong, T.M.; Thomson, T.; Vessella, R.L.; Sadar, M.D. Novel Biomarkers for Prostate Cancer Including Noncoding Transcripts. Am. J. Pathol. 2009, 175, 2264–2276. [Google Scholar] [CrossRef]
- Kim, M.; Morales, L.D.; Jang, I.S.; Cho, Y.Y.; Kim, D.J. Protein Tyrosine Phosphatases as Potential Regulators of STAT3 Signaling. Int. J. Mol. Sci. 2018, 19, 2708. [Google Scholar] [CrossRef]
- Hui, K.; Gao, Y.; Huang, J.; Xu, S.; Wang, B.; Zeng, J.; Fan, J.; Wang, X.; Yue, Y.; Wu, S.; et al. RASAL2, a RAS GTPase-Activating Protein, Inhibits Stemness and Epithelial-Mesenchymal Transition Via MAPK/SOX2 Pathway in Bladder Cancer. Cell Death Dis. 2017, 8, e2600. [Google Scholar] [CrossRef] [PubMed]
- Min, S.; Kim, K.; Kim, S.G.; Cho, H.; Lee, Y. Chromatin-Remodeling Factor, RSF1, Controls p53-Mediated Transcription in Apoptosis upon DNA Strand Breaks. Cell Death Dis. 2018, 9, e1079. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Y.; Zhang, Y.; Bai, X.; Peng, Y.; He, P. Rsf-1 Overexpression in Human Prostate Cancer, Implication as a Prognostic Marker. Tumor Biol. 2014, 35, 5771–5776. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Ajiro, M.; Yu, L.; McCoy, P., Jr.; Zheng, Z.M. Oncogenic Splicing Factor SRSF3 Regulates ILF3 Alternative Splicing to Promote Cancer Cell Proliferation and Transformation. RNA 2019, 25, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Bowler, E.; Porazinski, S.; Uzor, S.; Thibault, P.; Durand, M.; Lapointe, E.; Rouschop, K.M.A.; Hancock, J.; Wilson, I.; Ladomery, M. Hypoxia Leads to Significant Changes in Alternative Splicing and Elevated Expression of CLK Splice Factor Kinases in PC3 Prostate Cancer Cells. BMC Cancer 2018, 18, e355. [Google Scholar] [CrossRef] [PubMed]
- Bereczki, O.; Ujfaludi, Z.; Pardi, N.; Nagy, Z.; Tora, L.; Boros, I.M.; Balint, E. TATA Binding Protein Associated Factor 3 (TAF3) Interacts with p53 and Inhibits its Function. BMC Mol. Biol. 2008, 9, e57. [Google Scholar] [CrossRef] [PubMed]
- Fesus, L.; Thomazy, V.; Falus, A. Induction and Activation of Tissue Transglutaminase during Programmed Cell Death. FEBS Lett. 1987, 224, 104–108. [Google Scholar] [CrossRef]
- Moniz, S.; Jordan, P. Emerging Roles for WNK Kinases in Cancer. Cell. Mol. Life Sci. 2010, 67, 1265–1276. [Google Scholar] [CrossRef] [PubMed]
- Seligson, D.; Horvath, S.; Huerta-Yepez, S.; Hanna, S.; Garban, H.; Roberts, A.; Shi, T.; Liu, X.; Chia, D.; Goodglick, L.; et al. Expression of Transcription Factor Yin Yang 1 in Prostate Cancer. Int. J. Oncol. 2005, 27, 131–141. [Google Scholar] [CrossRef]
- Kashyap, V.; Bonavida, B. Role of YY1 in the Pathogenesis of Prostate Cancer and Correlation with Bioinformatic Data Sets of Gene Expression. Genes Cancer 2014, 5, 71–83. [Google Scholar] [PubMed]
- Park, A.; Lee, J.; Mun, S.; Kim, D.J.; Cha, B.H.; Moon, K.T.; Yoo, T.K.; Kang, H.G. Identification of Transcription Factor YY1 as a Regulator of a Prostate Cancer-Specific Pathway using Proteomic Analysis. J. Cancer 2017, 8, 2303–2311. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Tao, T.; Liu, C.; Guan, H.; Zhang, G.; Ling, Z.; Zhang, L.; Lu, K.; Chen, S.; Xu, B.; et al. Upregulation of miR-146a by YY1 Depletion Correlates with Delayed Progression of Prostate Cancer. Int. J. Oncol. 2017, 50, 421–431. [Google Scholar] [CrossRef]
- Yang, T.; An, Z.; Zhang, C.; Wang, Z.; Wang, X.; Liu, Y.; Du, E.; Liu, R.; Zhang, Z.; Xu, Y. HnRNPM, a Potential Mediator of YY1 in Promoting the Epithelial-Mesenchymal Transition of Prostate Cancer Cells. Prostate 2019, 79, 1199–1210. [Google Scholar] [CrossRef]
- Camacho-Moctezuma, B.; Quevedo-Castillo, M.; Melendez-Zajgla, J.; Aquino-Jarquin, G.; Martinez-Ruiz, G.U. YY1 Negatively Regulates the XAF1 Gene Expression in Prostate Cancer. Biochem. Biophys. Res. Commun. 2019, 508, 973–979. [Google Scholar] [CrossRef]
- Riera-Romo, M. COMMD1: A Multifunctional Regulatory Protein. J. Cell. Biochem. 2018, 119, 34–51. [Google Scholar] [CrossRef] [PubMed]
- Zoubeidi, A.; Ettinger, S.; Beraldi, E.; Hadaschik, B.; Zardan, A.; Klomp, L.W.; Nelson, C.C.; Rennie, P.S.; Gleave, M.E. Clusterin Facilitates COMMD1 and I-kappaB Degradation to Enhance NF-kappaB Activity in Prostate Cancer Cells. Mol. Cancer. Res. 2010, 8, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Loy, C.J.; Sim, K.S.; Yong, E.L. Filamin-A Fragment Localizes to the Nucleus to Regulate Androgen Receptor and Coactivator Functions. Proc. Natl. Acad. Sci. USA 2003, 100, 4562–4567. [Google Scholar] [CrossRef] [PubMed]
- Ravipaty, S.; Wu, W.; Dalvi, A.; Tanna, N.; Andreazi, J.; Friss, T.; Klotz, A.; Liao, C.; Garren, J.; Schofield, S.; et al. Clinical Validation of a Serum Protein Panel (FLNA, FLNB and KRT19) for Diagnosis of Prostate Cancer. J. Mol. Biomark. Diagn. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Yeh, D.W.; Chen, Y.S.; Lai, C.Y.; Liu, Y.L.; Lu, C.H.; Lo, J.F.; Chen, L.; Hsu, L.C.; Luo, Y.; Xiang, R.; et al. Downregulation of COMMD1 by miR-205 Promotes a Positive Feedback Loop for Amplifying Inflammatory- and Stemness-Associated Properties of Cancer Cells. Cell Death Differ. 2016, 23, 841–852. [Google Scholar] [CrossRef]
- Park, I.; Han, C.; Jin, S.; Lee, B.; Choi, H.; Kwon, J.T.; Kim, D.; Kim, J.; Lifirsu, E.; Park, W.J.; et al. Myosin Regulatory Light Chains are Required to Maintain the Stability of Myosin II and Cellular Integrity. Biochem. J. 2011, 434, 171–180. [Google Scholar] [CrossRef]
- Lee, S.; Kim, J.Y.; Kim, Y.J.; Seok, K.O.; Kim, J.H.; Chang, Y.J.; Kang, H.Y.; Park, J.H. Nucleolar Protein GLTSCR2 Stabilizes p53 in Response to Ribosomal Stresses. Cell Death Differ. 2012, 19, 1613–1622. [Google Scholar] [CrossRef]
- Kim, H.K.; Fuchs, G.; Wang, S.; Wei, W.; Zhang, Y.; Park, H.; Roy-Chaudhuri, B.; Li, P.; Xu, J.; Chu, K.; et al. A Transfer-RNA-Derived Small RNA Regulates Ribosome Biogenesis. Nature 2017, 552, 57–62. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Baca, S.C.; Prandi, D.; Lawrence, M.S.; Mosquera, J.M.; Romanel, A.; Drier, Y.; Park, K.; Kitabayashi, N.; MacDonald, T.Y.; Ghandi, M.; et al. Punctuated Evolution of Prostate Cancer Genomes. Cell 2013, 153, 666–677. [Google Scholar] [CrossRef]
- Barbieri, C.E.; Baca, S.C.; Lawrence, M.S.; Demichelis, F.; Blattner, M.; Theurillat, J.P.; White, T.A.; Stojanov, P.; Van Allen, E.; Stransky, N.; et al. Exome Sequencing Identifies Recurrent SPOP, FOXA1 and MED12 Mutations in Prostate Cancer. Nat. Genet. 2012, 44, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Fraser, M.; Sabelnykova, V.Y.; Yamaguchi, T.N.; Heisler, L.E.; Livingstone, J.; Huang, V.; Shiah, Y.J.; Yousif, F.; Lin, X.; Masella, A.P.; et al. Genomic Hallmarks of Localized, Non-Indolent Prostate Cancer. Nature 2017, 541, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Leyh-Bannurah, S.R.; Gazdovich, S.; Budaus, L.; Zaffuto, E.; Briganti, A.; Abdollah, F.; Montorsi, F.; Schiffmann, J.; Menon, M.; Shariat, S.F.; et al. Local Therapy Improves Survival in Metastatic Prostate Cancer. Eur. Urol. 2017, 72, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Coleman, I.; Morrissey, C.; Zhang, X.; True, L.D.; Gulati, R.; Etzioni, R.; Bolouri, H.; Montgomery, B.; White, T.; et al. Substantial Interindividual and Limited Intraindividual Genomic Diversity among Tumors from Men with Metastatic Prostate Cancer. Nat. Med. 2016, 22, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative Genomic Profiling of Human Prostate Cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Armenia, J.; Wankowicz, S.A.M.; Liu, D.; Gao, J.; Kundra, R.; Reznik, E.; Chatila, W.K.; Chakravarty, D.; Han, G.C.; Coleman, I.; et al. The Long Tail of Oncogenic Drivers in Prostate Cancer. Nat. Genet. 2018, 50, 645–651. [Google Scholar] [CrossRef]
- Rosario, S.R.; Long, M.D.; Affronti, H.C.; Rowsam, A.M.; Eng, K.H.; Smiraglia, D.J. Pan-Cancer Analysis of Transcriptional Metabolic Dysregulation using the Cancer Genome Atlas. Nat. Commun. 2018, 9, e5330. [Google Scholar] [CrossRef]
- Hieronymus, H.; Schultz, N.; Gopalan, A.; Carver, B.S.; Chang, M.T.; Xiao, Y.; Heguy, A.; Huberman, K.; Bernstein, M.; Assel, M.; et al. Copy Number Alteration Burden Predicts Prostate Cancer Relapse. Proc. Natl. Acad. Sci. USA 2014, 111, 11139–11144. [Google Scholar] [CrossRef]
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid Cultures Derived from Patients with Advanced Prostate Cancer. Cell 2014, 159, 176–187. [Google Scholar] [CrossRef]
- Abida, W.; Armenia, J.; Gopalan, A.; Brennan, R.; Walsh, M.; Barron, D.; Danila, D.; Rathkopf, D.; Morris, M.; Slovin, S.; et al. Prospective Genomic Profiling of Prostate Cancer Across Disease States Reveals Germline and Somatic Alterations that may Affect Clinical Decision Making. JCO Precis. Oncol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Grasso, C.S.; Wu, Y.M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The Mutational Landscape of Lethal Castration-Resistant Prostate Cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Abida, W.; Cyrta, J.; Heller, G.; Prandi, D.; Armenia, J.; Coleman, I.; Cieslik, M.; Benelli, M.; Robinson, D.; Van Allen, E.M.; et al. Genomic Correlates of Clinical Outcome in Advanced Prostate Cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 11428–11436. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef] [PubMed]
- Beltran, H.; Prandi, D.; Mosquera, J.M.; Benelli, M.; Puca, L.; Cyrta, J.; Marotz, C.; Giannopoulou, E.; Chakravarthi, B.V.; Varambally, S.; et al. Divergent Clonal Evolution of Castration-Resistant Neuroendocrine Prostate Cancer. Nat. Med. 2016, 22, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhou, Z.; Huang, J.; Chen, C. PMEPA1 Promotes Androgen Receptor-Negative Prostate Cell Proliferation through Suppressing the Smad3/4-c-Myc-p21 Cip1 Signaling Pathway. J. Pathol. 2011, 223, 683–694. [Google Scholar] [CrossRef]
- Zhao, C.B.; Bao, J.M.; Lu, Y.J.; Zhao, T.; Zhou, X.H.; Zheng, D.Y.; Zhao, S.C. Co-Expression of RAGE and HMGB1 is Associated with Cancer Progression and Poor Patient Outcome of Prostate Cancer. Am. J. Cancer Res. 2014, 4, 369–377. [Google Scholar] [CrossRef][Green Version]
- Bull, J.H.; Ellison, G.; Patel, A.; Muir, G.; Walker, M.; Underwood, M.; Khan, F.; Paskins, L. Identification of Potential Diagnostic Markers of Prostate Cancer and Prostatic Intraepithelial Neoplasia using cDNA Microarray. Br. J. Cancer 2001, 84, 1512–1519. [Google Scholar] [CrossRef]
- Iglesias-Gato, D.; Wikstrom, P.; Tyanova, S.; Lavallee, C.; Thysell, E.; Carlsson, J.; Hagglof, C.; Cox, J.; Andren, O.; Stattin, P.; et al. The Proteome of Primary Prostate Cancer. Eur. Urol. 2016, 69, 942–952. [Google Scholar] [CrossRef]
- Rajan, P.; Sudbery, I.M.; Villasevil, M.E.; Mui, E.; Fleming, J.; Davis, M.; Ahmad, I.; Edwards, J.; Sansom, O.J.; Sims, D.; et al. Next-Generation Sequencing of Advanced Prostate Cancer Treated with Androgen-Deprivation Therapy. Eur. Urol. 2014, 66, 32–39. [Google Scholar] [CrossRef]
- Kim, J.Y.; Cho, Y.E.; Kim, G.Y.; Lee, H.L.; Lee, S.; Park, J.H. Down-Regulation and Aberrant Cytoplasmic Expression of GLTSCR2 in Prostatic Adenocarcinomas. Cancer Lett. 2013, 340, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Cao, P.; Wan, M.M.; Sui, G. Yin Yang 1: A Multifaceted Protein Beyond a Transcription Factor. Transcription 2010, 1, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.; Li, Y.; Gan, Y.; Zhao, D.; Wang, G.; Xie, N.; Lovnicki, J.M.; Fazli, L.; Cao, Q.; Chen, K.; et al. Roles of the HOXA10 Gene during Castrate-Resistant Prostate Cancer Progression. Endocr. Relat. Cancer 2019, 26, 279–292. [Google Scholar] [CrossRef] [PubMed]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Petryszak, R.; Keays, M.; Tang, Y.A.; Fonseca, N.A.; Barrera, E.; Burdett, T.; Fullgrabe, A.; Fuentes, A.M.; Jupp, S.; Koskinen, S.; et al. Expression Atlas Update, an Integrated Database of Gene and Protein Expression in Humans, Animals and Plants. Nucleic Acids Res. 2016, 44, D746–D752. [Google Scholar] [CrossRef] [PubMed]
- Kaighn, M.E.; Narayan, K.S.; Ohnuki, Y.; Lechner, J.F.; Jones, L.W. Establishment and Characterization of a Human Prostatic Carcinoma Cell Line (PC-3). Investig. Urol. 1979, 17, 16–23. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR and the 2(-Delta Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-Enabled Heat Mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).