Exploring the Molecular Crosstalk between Pancreatic Bud and Mesenchyme in Embryogenesis: Novel Signals Involved
Abstract
1. Introduction
2. Results
2.1. Global Gene Expression Profiling between Dorsal Pancreatic Bud and Surrounding DPB Mesenchyme
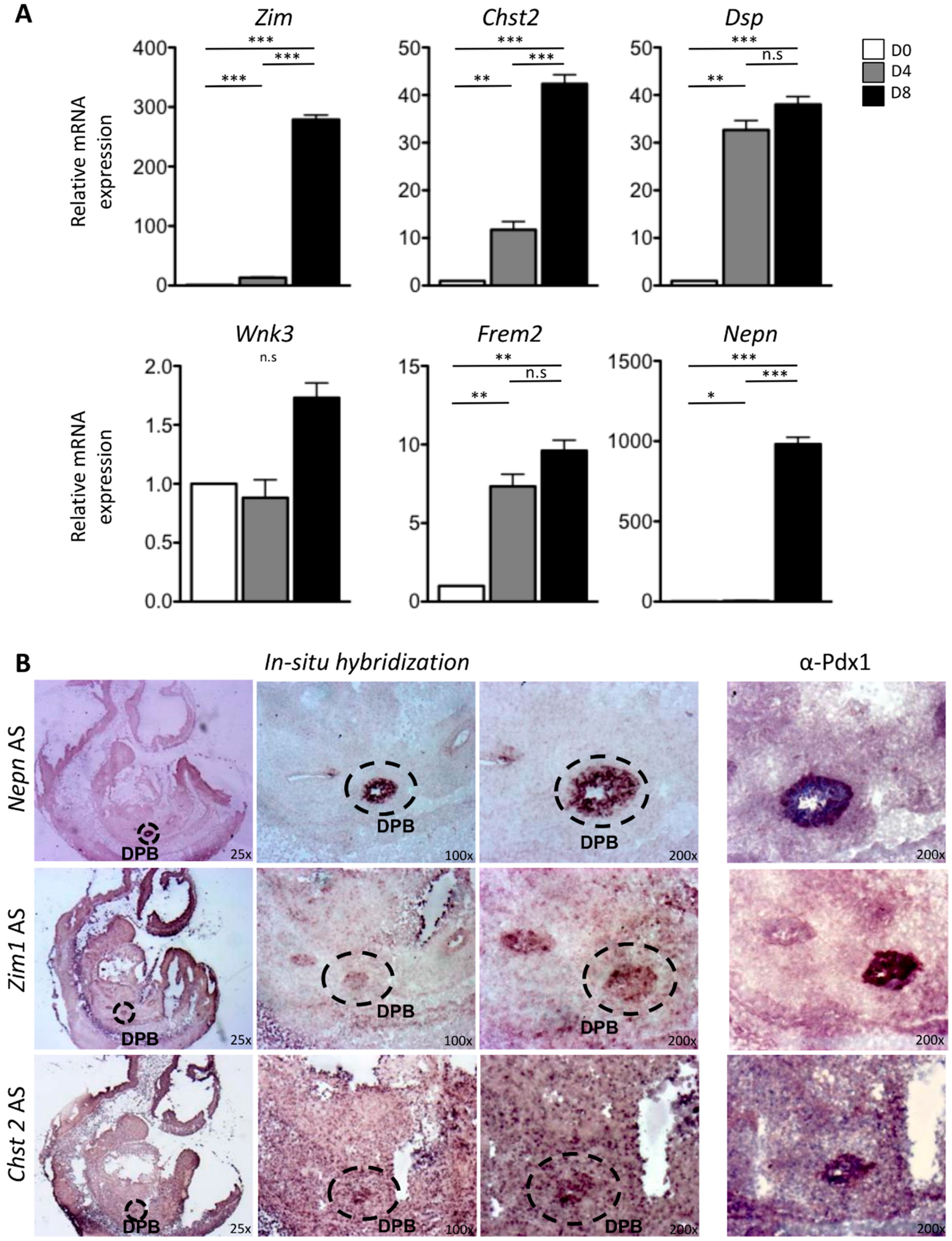
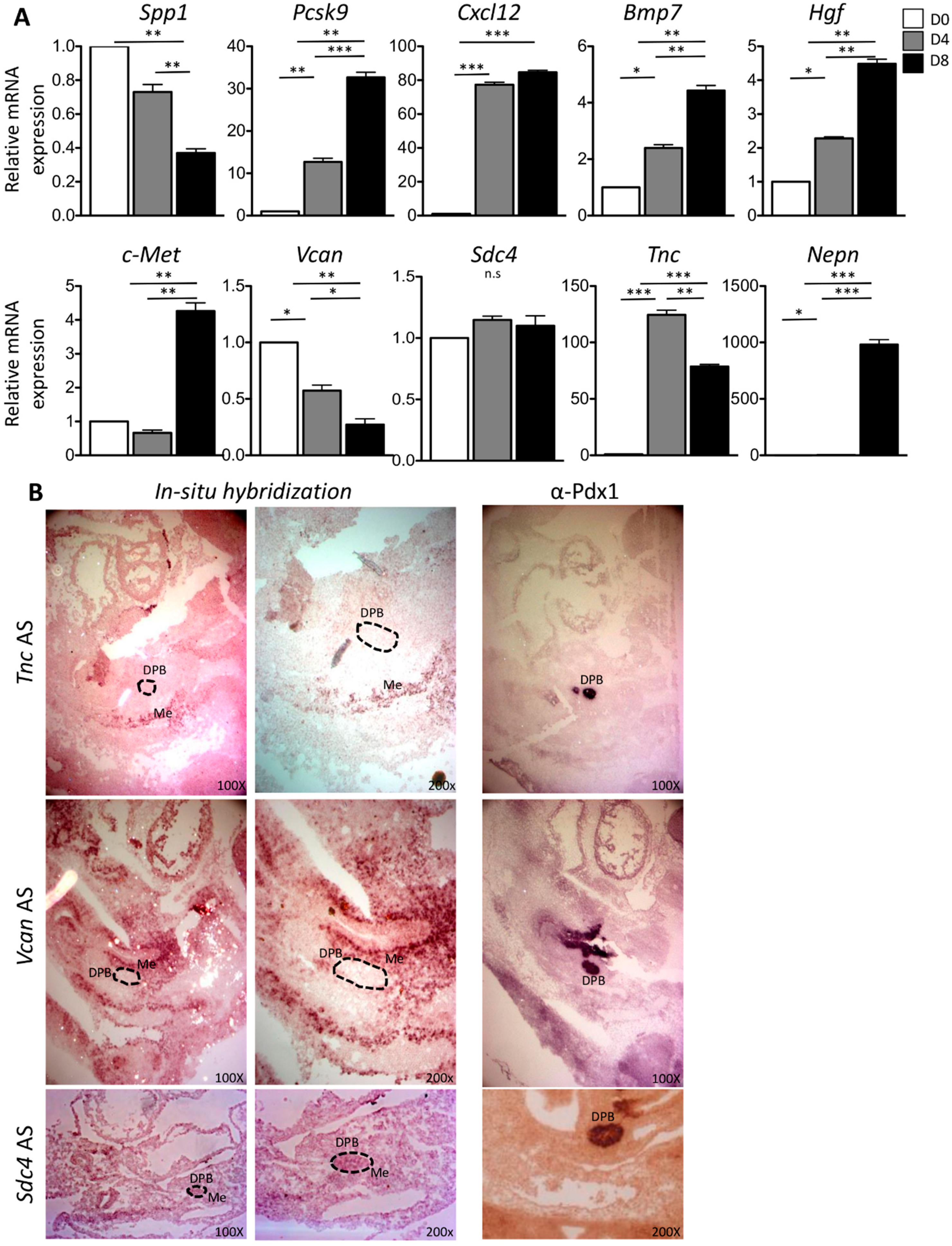
2.2. In Vitro and In Vivo Characterization of Enriched Intrinsic Factors
2.3. Validation of Enriched Intrinsic Factors and Characterization of Molecular Crosstalk between DPB and MeDPB at E10.5
3. Discussion
4. Materials and Methods
4.1. Ethics Statements
4.2. Mouse Strains and Husbandry
4.3. Embryo Dissection and Embedding for LCM
4.4. Laser Capture Microdissection (LCM)
4.5. RNA Isolation and RNAseq
4.6. RT-qPCR Analysis
4.7. ESCs Culture and Differentiation
4.8. In Situ Hybridization
4.9. Immunohistochemistry
4.10. Statistics in Biological Experiments
4.11. RNAseq Data Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| B6 | C57BL/6J |
| BP | Biological Processes |
| CC | Cellular components |
| CESA | Comitato Etico per la Sperimentazione Animale |
| Chst2 | Carbohydrate sulfotransferase 2 |
| CPM | Counts per millions |
| DE | Definitive endoderm |
| DPB | Dorsal pancreatic bud |
| Dsp | Desmoplakin |
| E10.5 | Embryonic day 10.5 |
| ESCs | Embryonic stem cells |
| EST | Expressed Sequence Tag |
| FDR | False Discovery Rate |
| Frem2 | Fras1-related extracellular matrix protein 2 |
| GO | Gene ontology |
| IHC | Immunohistochemistry |
| ISH | In situ hybridization |
| iPSCs | Induced pluripotent stem cells |
| LCM | Laser capture microdissection |
| MeDBP | DPB mesenchyme |
| mESCs | Mouse embryonic stem cells |
| MO | Molecular functions |
| PBS-DEPC | DEPC-treated phosphate-buffered saline |
| PCA | Principal component analysis |
| PCSK9 | Proproteinconvertasesubtilisin/kexin type 9 |
| PFE | Posterior foregut endoderm |
| PPCs | Pancreatic progenitor cells |
| RA | Retinoic acid |
| RIN | RNA integrity numbers |
| RNAseq | RNA high-throughput sequencing |
| Sdc4 | Syndecan 4 |
| Tnc | Tenascin C |
| Vcan | Versican |
| Wnk3 | WNK lysine-deficient protein kinase 3 |
| Zim1 | Zinc finger, imprinted 1 |
References
- Bouwens, L.; Houbracken, I.; Mfopou, J.K. The use of stem cells for pancreatic regeneration in diabetes mellitus. Nat. Rev. Endocrinol. 2013, 9, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Melton, D.A. Pancreas regeneration. Nature 2018, 557, 351–358. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, M.T.; Russo, F.; D’Angelo, F.; Federico, A.; Gemei, M.; Del Vecchio, L.; Ceccarelli, M.; De Felice, M.; Falco, G. Novel pancreas organogenesis markers refine the pancreatic differentiation roadmap of embryonic stem cells. Stem Cell Rev. 2014, 10, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.P.; Wang, A.; Sander, M. Pancreas organogenesis: From lineage determination to morphogenesis. Annu. Rev. Cell Dev. Biol. 2013, 29, 81–105. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, M.C.; Ahnfelt-Ronne, J.; Hald, J.; Madsen, O.D.; Serup, P.; Hecksher-Sorensen, J. An illustrated review of early pancreas development in the mouse. Endocr. Rev. 2007, 28, 685–705. [Google Scholar] [CrossRef]
- Pictet, R.L.; Clark, W.R.; Williams, R.H.; Rutter, W.J. An ultrastructural analysis of the developing embryonic pancreas. Dev. Biol. 1972, 29, 436–467. [Google Scholar] [CrossRef]
- Golosow, N.; Grobstein, C. Epitheliomesenchymal interaction in pancreatic morphogenesis. Dev. Biol. 1962, 4, 242–255. [Google Scholar] [CrossRef]
- Villasenor, A.; Cleaver, O. Crosstalk between the developing pancreas and its blood vessels: An evolving dialog. Semin. Cell Dev. Biol. 2012, 23, 685–692. [Google Scholar] [CrossRef][Green Version]
- Gittes, G.K. Developmental biology of the pancreas: A comprehensive review. Dev. Biol. 2009, 326, 4–35. [Google Scholar] [CrossRef]
- Bhushan, A.; Itoh, N.; Kato, S.; Thiery, J.P.; Czernichow, P.; Bellusci, S.; Scharfmann, R. Fgf10 is essential for maintaining the proliferative capacity of epithelial progenitor cells during early pancreatic organogenesis. Development 2001, 128, 5109–5117. [Google Scholar]
- Ndlovu, R.; Deng, L.C.; Wu, J.; Li, X.K.; Zhang, J.S. Fibroblast Growth Factor 10 in Pancreas Development and Pancreatic Cancer. Front. Genet. 2018, 9, 482. [Google Scholar] [CrossRef] [PubMed]
- Heiser, P.W.; Lau, J.; Taketo, M.M.; Herrera, P.L.; Hebrok, M. Stabilization of beta-catenin impacts pancreas growth. Development 2006, 133, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Bravo, J.L.; Flores-Martinez, A.; Herrero-Martin, G.; Puri, S.; Taketo, M.M.; Rojas, A.; Hebrok, M.; Cano, D.A. Loss of Pancreas upon Activated Wnt Signaling Is Concomitant with Emergence of Gastrointestinal Identity. PLoS ONE 2016, 11, e0164714. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Gallego-Llamas, J.; Ribes, V.; Kedinger, M.; Niederreither, K.; Chambon, P.; Dolle, P.; Gradwohl, G. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev. Biol. 2005, 284, 399–411. [Google Scholar] [CrossRef]
- Molotkov, A.; Molotkova, N.; Duester, G. Retinoic acid generated by Raldh2 in mesoderm is required for mouse dorsal endodermal pancreas development. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2005, 232, 950–957. [Google Scholar]
- Kumar, M.; Jordan, N.; Melton, D.; Grapin-Botton, A. Signals from lateral plate mesoderm instruct endoderm toward a pancreatic fate. Dev. Biol. 2003, 259, 109–122. [Google Scholar] [CrossRef]
- Harmon, E.B.; Apelqvist, A.A.; Smart, N.G.; Gu, X.; Osborne, D.H.; Kim, S.K. GDF11 modulates NGN3+ islet progenitor cell number and promotes beta-cell differentiation in pancreas development. Development 2004, 131, 6163–6174. [Google Scholar] [CrossRef]
- Gnatenko, D.A.; Kopantzev, E.P.; Sverdlov, E.D. [Fibroblast growth factors and their effects in pancreas organogenesis]. Biomeditsinskaia Khimiia 2017, 63, 211–218. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Li, X.Y.; Zhai, W.J.; Teng, C.B. Notch Signaling in Pancreatic Development. Int. J. Mol. Sci. 2015, 17, 48. [Google Scholar] [CrossRef]
- Belo, J.; Krishnamurthy, M.; Oakie, A.; Wang, R. The role of SOX9 transcription factor in pancreatic and duodenal development. Stem Cells Dev. 2013, 22, 2935–2943. [Google Scholar] [CrossRef] [PubMed]
- Marty-Santos, L.; Cleaver, O. Pdx1 regulates pancreas tubulogenesis and E-cadherin expression. Development 2016, 143, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.ncbi.nlm.nih.gov/unigene (accessed on 15 June 2018).
- Chiotaki, R.; Petrou, P.; Giakoumaki, E.; Pavlakis, E.; Sitaru, C.; Chalepakis, G. Spatiotemporal distribution of Fras1/Frem proteins during mouse embryonic development. Gene Expr. Patterns Gep 2007, 7, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Hale, M.A.; Swift, G.H.; Hoang, C.Q.; Deering, T.G.; Masui, T.; Lee, Y.K.; Xue, J.; MacDonald, R.J. The nuclear hormone receptor family member NR5A2 controls aspects of multipotent progenitor cell formation and acinar differentiation during pancreatic organogenesis. Development 2014, 141, 3123–3133. [Google Scholar] [CrossRef] [PubMed]
- Gallicano, G.I.; Bauer, C.; Fuchs, E. Rescuing desmoplakin function in extra-embryonic ectoderm reveals the importance of this protein in embryonic heart, neuroepithelium, skin and vasculature. Development 2001, 128, 929–941. [Google Scholar]
- Gallicano, G.I.; Kouklis, P.; Bauer, C.; Yin, M.; Vasioukhin, V.; Degenstein, L.; Fuchs, E. Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J. Cell Biol. 1998, 143, 2009–2022. [Google Scholar] [CrossRef] [PubMed]
- Ye, A.; He, H.; Kim, J. Paternally expressed Peg3 controls maternally expressed Zim1 as a trans factor. PLoS ONE 2014, 9, e108596. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Landsman, L.; Nijagal, A.; Whitchurch, T.J.; Vanderlaan, R.L.; Zimmer, W.E.; Mackenzie, T.C.; Hebrok, M. Pancreatic mesenchyme regulates epithelial organogenesis throughout development. PLoS Biol. 2011, 9, e1001143. [Google Scholar] [CrossRef]
- Guo, T.; Landsman, L.; Li, N.; Hebrok, M. Factors expressed by murine embryonic pancreatic mesenchyme enhance generation of insulin-producing cells from hESCs. Diabetes 2013, 62, 1581–1592. [Google Scholar] [CrossRef]
- Wali, J.A.; Thomas, H.E. Pancreatic Alpha Cells Hold the Key to Survival. EBioMedicine 2015, 2, 368–369. [Google Scholar] [CrossRef] [PubMed]
- Murtaugh, L.C.; Keefe, M.D. Regeneration and repair of the exocrine pancreas. Annu. Rev. Physiol. 2015, 77, 229–249. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, M.; Hruban, R.H.; Wood, L.D. New Developments in the Molecular Mechanisms of Pancreatic Tumorigenesis. Adv. Anat. Pathol. 2018, 25, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Shih, H.P.; Panlasigui, D.; Cirulli, V.; Sander, M. ECM Signaling Regulates Collective Cellular Dynamics to Control Pancreas Branching Morphogenesis. Cell Rep. 2016, 14, 169–179. [Google Scholar] [CrossRef]
- Kilic, G.; Wang, J.; Sosa-Pineda, B. Osteopontin is a novel marker of pancreatic ductal tissues and of undifferentiated pancreatic precursors in mice. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2006, 235, 1659–1667. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Katsumoto, K.; Kume, S. The role of CXCL12-CXCR4 signaling pathway in pancreatic development. Theranostics 2013, 3, 11–17. [Google Scholar] [CrossRef]
- Yellowley, C. CXCL12/CXCR4 signaling and other recruitment and homing pathways in fracture repair. Bonekey Rep. 2013, 2, 300. [Google Scholar] [CrossRef]
- Paron, I.; Berchtold, S.; Voros, J.; Shamarla, M.; Erkan, M.; Hofler, H.; Esposito, I. Tenascin-C enhances pancreatic cancer cell growth and motility and affects cell adhesion through activation of the integrin pathway. PLoS ONE 2011, 6, e21684. [Google Scholar] [CrossRef]
- Zhang, Z.; Rankin, S.A.; Zorn, A.M. Syndecan4 coordinates Wnt/JNK and BMP signaling to regulate foregut progenitor development. Dev. Biol. 2016, 416, 187–199. [Google Scholar] [CrossRef]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Kato, T. Biological roles of hepatocyte growth factor-Met signaling from genetically modified animals. Biomed. Rep. 2017, 7, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Sahlberg, C.; Aukhil, I.; Thesleff, I. Tenascin-C in developing mouse teeth: Expression of splice variants and stimulation by TGFbeta and FGF. Eur. J. Oral Sci. 2001, 109, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Midwood, K.S.; Orend, G. The role of tenascin-C in tissue injury and tumorigenesis. J. Cell Commun. Signal 2009, 3, 287–310. [Google Scholar] [CrossRef] [PubMed]
- Lau, T.L.; Kim, C.; Ginsberg, M.H.; Ulmer, T.S. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 2009, 28, 1351–1361. [Google Scholar] [CrossRef] [PubMed]
- Yamagata, M.; Yamada, K.M.; Yoneda, M.; Suzuki, S.; Kimata, K. Chondroitin sulfate proteoglycan (PG-M-like proteoglycan) is involved in the binding of hyaluronic acid to cellular fibronectin. J. Biol. Chem. 1986, 261, 13526–13535. [Google Scholar]
- Woods, A.; Couchman, J.R. Syndecan 4 heparan sulfate proteoglycan is a selectively enriched and widespread focal adhesion component. Mol. Biol. Cell 1994, 5, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Lee, E.; Kwon, S.; Park, H.; Yi, J.Y.; Kim, S.; Han, I.O.; Yun, Y.; Oh, E.S. Transmembrane domain-induced oligomerization is crucial for the functions of syndecan-2 and syndecan-4. J. Biol. Chem. 2005, 280, 42573–42579. [Google Scholar] [CrossRef]
- Sheng, W.; Wang, G.; La Pierre, D.P.; Wen, J.; Deng, Z.; Wong, C.K.; Lee, D.Y.; Yang, B.B. Versican mediates mesenchymal-epithelial transition. Mol. Biol. Cell 2006, 17, 2009–2020. [Google Scholar] [CrossRef][Green Version]
- Wang, X.; He, J.; Zhao, X.; Qi, T.; Zhang, T.; Kong, C. Syndecan-1 suppresses epithelial-mesenchymal transition and migration in human oral cancer cells. Oncol. Rep. 2018, 39, 1835–1842. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Falco, G.; Stanghellini, I.; Ko, M.S. Use of Chuk as an internal standard suitable for quantitative RT-PCR in mouse preimplantation embryos. Reprod. Biomed. Online 2006, 13, 394–403. [Google Scholar] [CrossRef]
- Fontana, R.; Guidone, D.; Sangermano, F.; Calabro, V.; Pollice, A.; La Mantia, G.; Vivo, M. PKC Dependent p14ARF Phosphorylation on Threonine 8 Drives Cell Proliferation. Sci. Rep. 2018, 8, 7056. [Google Scholar] [CrossRef] [PubMed]
- Fagman, H.; Amendola, E.; Parrillo, L.; Zoppoli, P.; Marotta, P.; Scarfo, M.; De Luca, P.; de Carvalho, D.P.; Ceccarelli, M.; De Felice, M.; et al. Gene expression profiling at early organogenesis reveals both common and diverse mechanisms in foregut patterning. Dev. Biol. 2011, 359, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Credendino, S.C.; Lewin, N.; de Oliveira, M.; Basu, S.; D’Andrea, B.; Amendola, E.; Di Guida, L.; Nardone, A.; Sanges, R.; De Felice, M.; et al. Tissue- and Cell Type-Specific Expression of the Long Noncoding RNA Klhl14-AS in Mouse. Int. J. Genom. 2017, 2017, 9769171. [Google Scholar] [CrossRef] [PubMed]
- Marotta, P.; Amendola, E.; Scarfo, M.; De Luca, P.; Zoppoli, P.; Amoresano, A.; De Felice, M.; Di Lauro, R. The paired box transcription factor Pax8 is essential for function and survival of adult thyroid cells. Mol. Cell. Endocrinol. 2014, 396, 26–36. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, M.; Risselada, H.J.; Endter, L.J.; Comte-Miserez, V.; Mayer, A. SNARE-mediated membrane fusion arrests at pore expansion to regulate the volume of an organelle. EMBO J. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Chan, C.K. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol. Biol. 2016, 1374, 339–361. [Google Scholar]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Nikolayeva, O.; Robinson, M.D. edgeR for differential RNA-seq and ChIP-seq analysis: An application to stem cell biology. Methods Mol. Biol. 2014, 1150, 45–79. [Google Scholar]
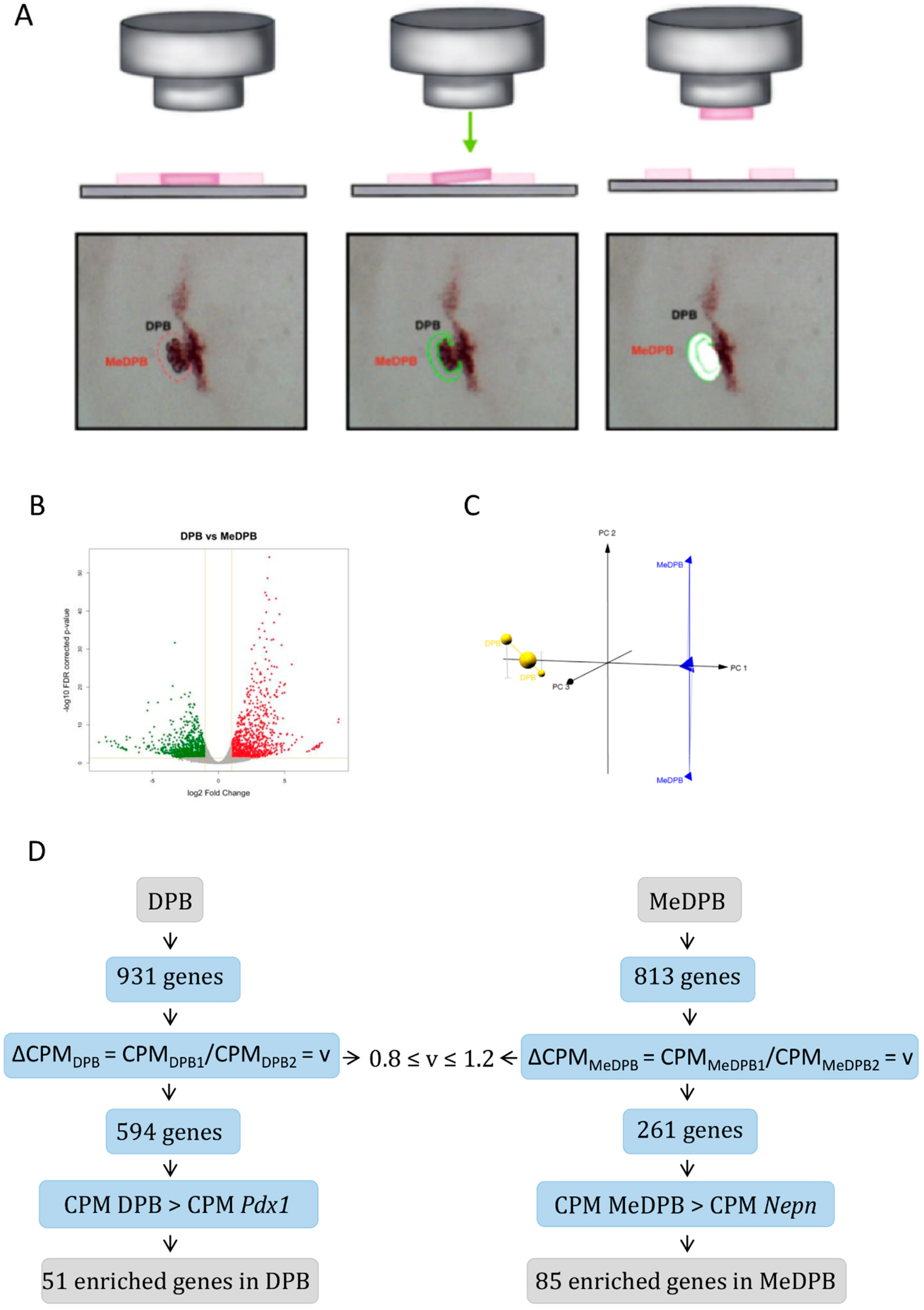
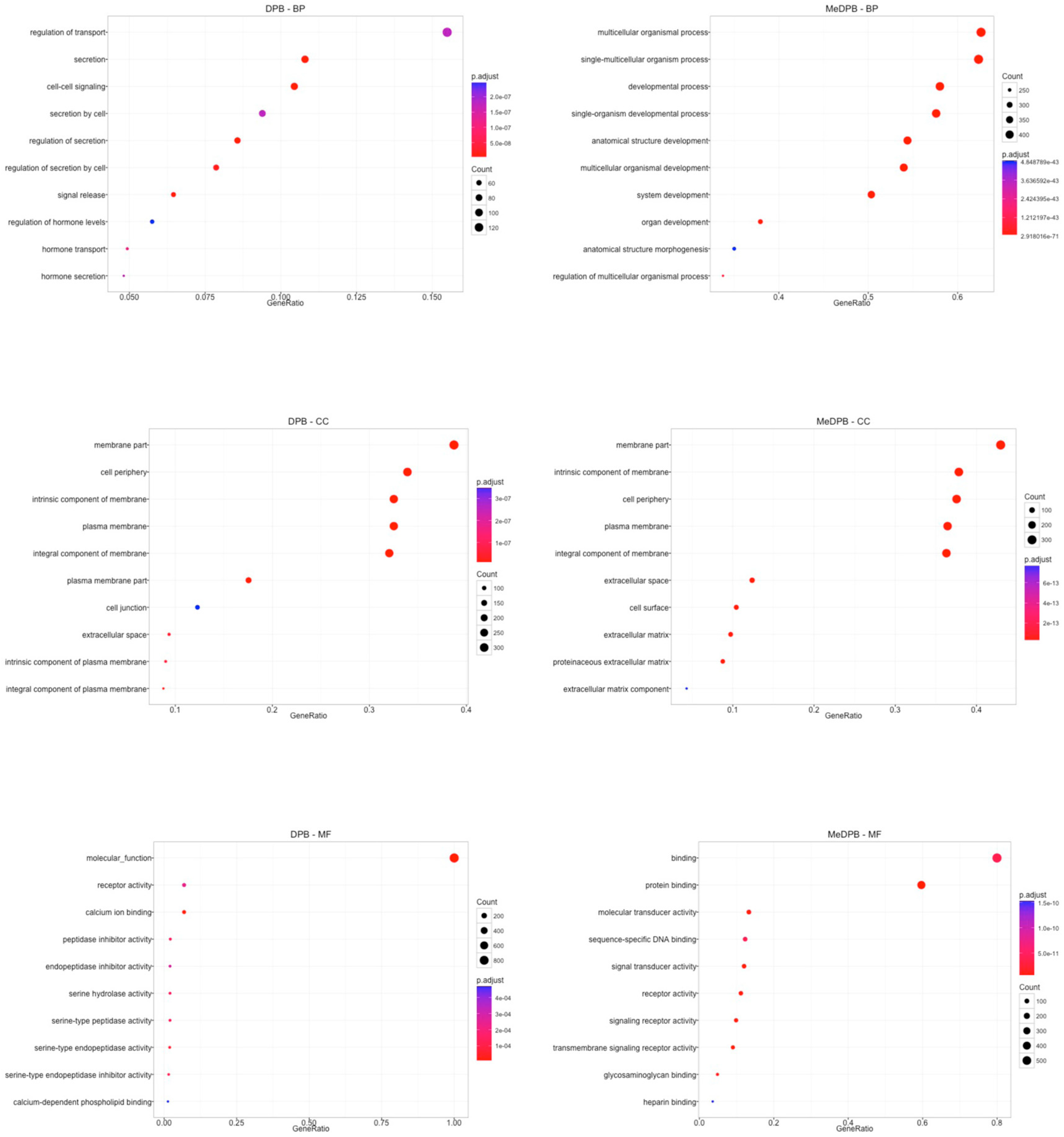
| Gene Name | CPM DPB Mean | Gene Name | CPM DPB Mean |
|---|---|---|---|
| Peg3 | 3769.3 | Fryl | 187.7 |
| Rian | 728.7 | Zim1 | 167.5 |
| Nepn | 686.2 | Rap1gap2 | 167.0 |
| Ptprf | 638.4 | Etl4 | 161.1 |
| Gcg | 571.5 | Itga6 | 159.2 |
| Meg3 | 567.0 | Lama5 | 157.4 |
| Myh9 | 376.5 | Epcam | 155.1 |
| Frem2 | 369.1 | Nav2 | 154.0 |
| Fras1 | 356.4 | Appl2 | 153.5 |
| Ccnd1 | 322.5 | Nr5a2 | 152.6 |
| Prox1 | 308.0 | Svil | 143.9 |
| Spon1 | 303.3 | Parm1 | 135.8 |
| Cdh1 | 291.1 | Itpr3 | 135.7 |
| Ptpn13 | 281.9 | Abcc8 | 135.5 |
| Slc38a5 | 275.5 | Plk2 | 129.4 |
| Dlk1 | 266.3 | Fbxo21 | 128.0 |
| Chst2 | 243.9 | Wnk3 | 127.6 |
| Adamts1 | 230.2 | Lrba | 120.0 |
| Dlg5 | 229.1 | Arg1 | 119.5 |
| Dsp | 225.8 | Wnk2 | 118.5 |
| Notch1 | 214.6 | Jmy | 114.6 |
| Sox9 | 210.0 | Cep170b | 113.2 |
| Ankrd50 | 189.5 | Rnf213 | 111.9 |
| Pam | 189.4 | Fndc3b | 110.4 |
| Aes | 188.9 | Gatsl2 | 109.5 |
| Gene Name | CPM MeDPB Mean | Gene Name | CPM MeDPB Mean |
|---|---|---|---|
| Mest | 589.9 | Akap12 | 85.67 |
| Fbn2 | 401.2 | Efnb2 | 83.16 |
| Zfp462 | 352.5 | Oxct1 | 80.50 |
| Zfhx4 | 217.0 | Epha4 | 78.03 |
| Sulf2 | 164.1 | Zfp423 | 77.27 |
| Cdh11 | 157.5 | Fbn1 | 75.36 |
| Dpysl3 | 157.0 | Sacs | 75.36 |
| Nefm | 144.0 | Arhgef40 | 71.37 |
| Fndc3c1 | 140.0 | Aff3 | 70.69 |
| Slit2 | 131.7 | Runx1t1 | 67.79 |
| Basp1 | 122.1 | Lin28b | 67.29 |
| Gli3 | 117.3 | Dennd5b | 66.06 |
| Nid1 | 116.7 | Cachd1 | 64.35 |
| Cald1 | 111.8 | Kdr | 64.18 |
| Amot | 108.9 | Phf6 | 64.12 |
| Msn | 105.8 | Nefl | 62.98 |
| Zeb2 | 101.9 | Foxp2 | 62.41 |
| 2810417H13Rik | 95.3 | Map1a | 60.62 |
| Hdgfrp3 | 92.8 | Ets1 | 60.32 |
| Nhsl2 | 90.4 | Far1 | 59.52 |
| Flnc | 88.6 | Col11a1 | 55.89 |
| Col5a1 | 88.5 | Mmp2 | 55.79 |
| Phactr2 | 87.8 | Cdh5 | 55.58 |
| Hmcn1 | 86.9 | Atp11c | 55.41 |
| Pcdh18 | 86.7 | Adam19 | 55.25 |
| Genes Expressed in Pancreatic Bud | |
|---|---|
| Gene Name | Description |
| Gcg | glucagon [Source:MGI Symbol;Acc:MGI:95674] |
| Bmp7 | bone morphogenetic protein 7 [Source:MGI Symbol;Acc:MGI:103302] |
| Met | met proto-oncogene [Source:MGI Symbol;Acc:MGI:96969] |
| Spp1 | secreted phosphoprotein 1 [Source:MGI Symbol;Acc:MGI:98389] |
| Pcsk9 | proproteinconvertasesubtilisin/kexin type 9 [Source:MGI Symbol;Acc:MGI:2140260] |
| Clu | clusterin [Source:MGI Symbol;Acc:MGI:88423] |
| Pyy | peptide YY [Source:MGI Symbol;Acc:MGI:99924] |
| Edn3 | endothelin 3 [Source:MGI Symbol;Acc:MGI:95285] |
| Sdc4 | syndecan 4 [Source:MGI Symbol;Acc:MGI:1349164] |
| Serpinf2 | serine (or cysteine) peptidase inhibitor, clade F, member 2 [Source:MGI Symbol;Acc:MGI:107173] |
| Pcsk6 | proproteinconvertasesubtilisin/kexin type 6 [Source:MGI Symbol;Acc:MGI:102897] |
| F5 | coagulation factor V [Source:MGI Symbol;Acc:MGI:88382] |
| Iapp | islet amyloid polypeptide [Source:MGI Symbol;Acc:MGI:96382] |
| Casr | calcium-sensing receptor [Source:MGI Symbol;Acc:MGI:1351351] |
| Cck | cholecystokinin [Source:MGI Symbol;Acc:MGI:88297] |
| Sct | secretin [Source:MGI Symbol;Acc:MGI:99466] |
| Gene Name | Description |
| Vcan | versican [Source:MGI Symbol;Acc:MGI:102889] |
| Tnc | tenascin C [Source:MGI Symbol;Acc:MGI:101922] |
| Igf1 | insulin-like growth factor 1 [Source:MGI Symbol;Acc:MGI:96432] |
| Cxcl12 | chemokine (C-X-C motif) ligand 12 [Source:MGI Symbol;Acc:MGI:103556] |
| Tgfb2 | transforming growth factor, beta 2 [Source:MGI Symbol;Acc:MGI:98726] |
| Thbs1 | thrombospondin 1 [Source:MGI Symbol;Acc:MGI:98737] |
| Itga8 | integrin alpha 8 [Source:MGI Symbol;Acc:MGI:109442] |
| Lpar1 | lysophosphatidic acid receptor 1 [Source:MGI Symbol;Acc:MGI:108429] |
| Pcsk5 | proproteinconvertasesubtilisin/kexin type 5 [Source:MGI Symbol;Acc:MGI:97515] |
| Apob | apolipoprotein B [Source:MGI Symbol;Acc:MGI:88052] |
| Adcyap1r1 | adenylate cyclase activating polypeptide 1 receptor 1 [Source:MGI Symbol;Acc:MGI:108449] |
| Cxcr4 | chemokine (C-X-C motif) receptor 4 [Source:MGI Symbol;Acc:MGI:109563] |
| Dcn | decorin [Source:MGI Symbol;Acc:MGI:94872] |
| Thbd | thrombomodulin [Source:MGI Symbol;Acc:MGI:98736] |
| Hgf | hepatocyte growth factor [Source:MGI Symbol;Acc:MGI:96079] |
| Ntsr1 | neurotensin receptor 1 [Source:MGI Symbol;Acc:MGI:97386] |
| Bmp2 | bonemorphogeneticprotein2 [Source:MGI Symbol;Acc:MGI:88177] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guerriero, I.; De Angelis, M.T.; D’Angelo, F.; Leveque, R.; Savignano, E.; Roberto, L.; Lucci, V.; Mazzone, P.; Laurino, S.; Storto, G.; et al. Exploring the Molecular Crosstalk between Pancreatic Bud and Mesenchyme in Embryogenesis: Novel Signals Involved. Int. J. Mol. Sci. 2019, 20, 4900. https://doi.org/10.3390/ijms20194900
Guerriero I, De Angelis MT, D’Angelo F, Leveque R, Savignano E, Roberto L, Lucci V, Mazzone P, Laurino S, Storto G, et al. Exploring the Molecular Crosstalk between Pancreatic Bud and Mesenchyme in Embryogenesis: Novel Signals Involved. International Journal of Molecular Sciences. 2019; 20(19):4900. https://doi.org/10.3390/ijms20194900
Chicago/Turabian StyleGuerriero, Ilaria, Maria Teresa De Angelis, Fulvio D’Angelo, Rita Leveque, Eleonora Savignano, Luca Roberto, Valeria Lucci, Pellegrino Mazzone, Simona Laurino, Giovanni Storto, and et al. 2019. "Exploring the Molecular Crosstalk between Pancreatic Bud and Mesenchyme in Embryogenesis: Novel Signals Involved" International Journal of Molecular Sciences 20, no. 19: 4900. https://doi.org/10.3390/ijms20194900
APA StyleGuerriero, I., De Angelis, M. T., D’Angelo, F., Leveque, R., Savignano, E., Roberto, L., Lucci, V., Mazzone, P., Laurino, S., Storto, G., Nardelli, A., Sgambato, A., Ceccarelli, M., De Felice, M., Amendola, E., & Falco, G. (2019). Exploring the Molecular Crosstalk between Pancreatic Bud and Mesenchyme in Embryogenesis: Novel Signals Involved. International Journal of Molecular Sciences, 20(19), 4900. https://doi.org/10.3390/ijms20194900






