Spatial and Seasonal Analysis of Phyllosphere Bacterial Communities of the Epiphytic Gymnosperm Zamia pseudoparasitica
Abstract
1. Introduction
2. Materials and Methods
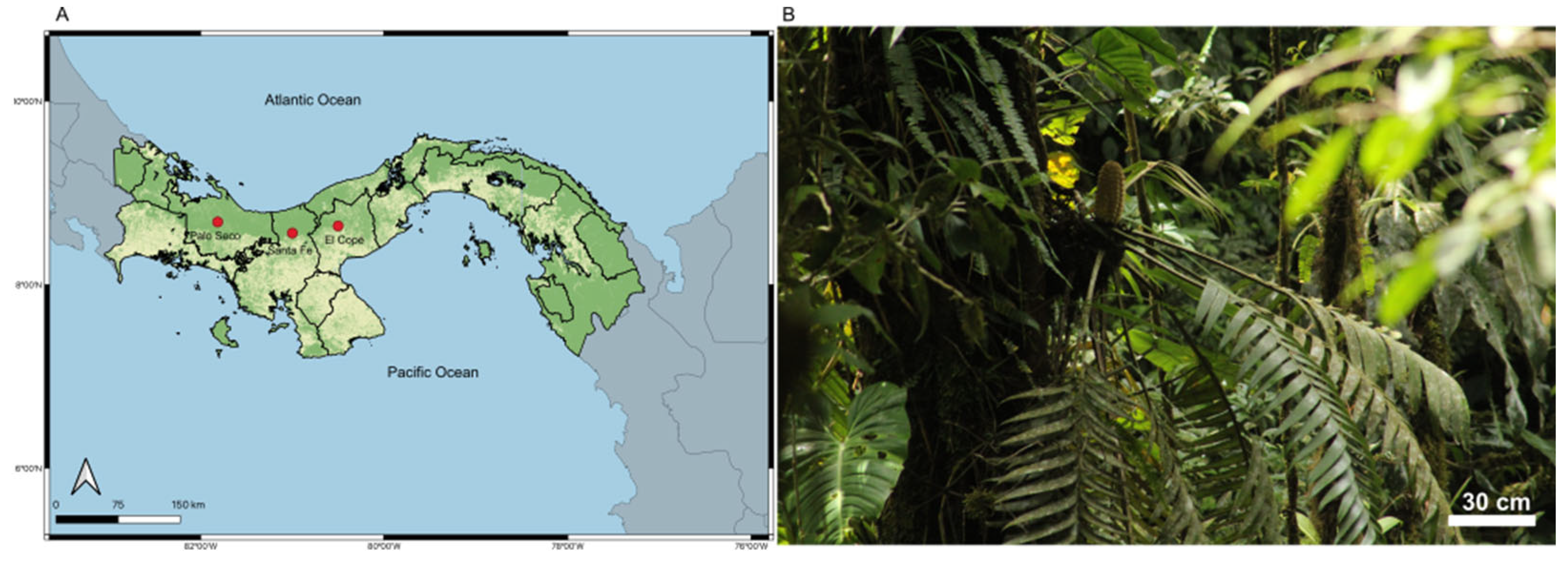
2.1. Study Sites and Field Sampling
2.2. DNA Extraction and Amplicon Sequencing
2.3. Data Analysis
2.3.1. Amplicon Sequence Data Processing
2.3.2. Diversity and Composition of the Bacterial Community
2.3.3. Beta Diversity Partitioning (Beta Net Relatedness Index, βNRI)
3. Results
3.1. Variation in Environmental Factors Between Each Sampling Site and Season
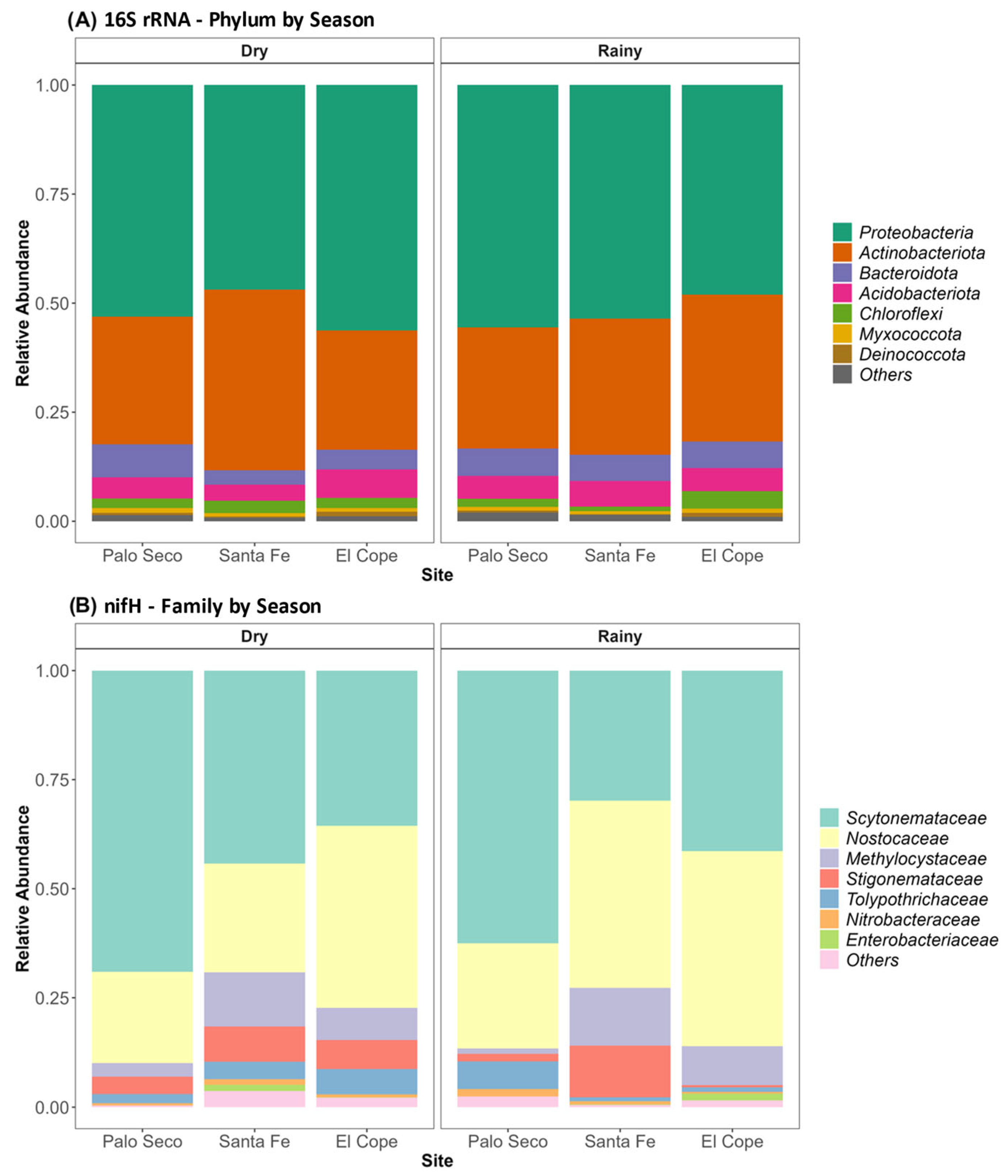
3.2. Bacterial Community Composition in the Phyllosphere of Z. pseudoparasitica
3.3. Beta Diversity Partitioning
3.4. Comparison of the Diazotrophic Communities Identified by 16S rRNA and nifH Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| βNRI | Beta Net Relatedness Index |
| ASV | Amplicon Sequence Variant |
| m.a.s.l. | Meters above sea level |
References
- Cardelús, C.L.; Colwell, R.K.; Watkins, J.E.J.R. Vascular epiphyte distribution patterns: Explaining the mid-elevation richness peak. J. Ecol. 2006, 94, 144–156. [Google Scholar] [CrossRef]
- Hietz, P.; Hietz-Seifert, U. Composition and ecology of vascular epiphyte communities along an altitudinal gradient in central Veracruz, Mexico. J. Veg. Sci. 1995, 6, 487–498. [Google Scholar] [CrossRef]
- Hietz, P.; Hietz-Seifert, U. Intra-and interspecific relations within an epiphyte community in a Mexican humid montane forest. Selbyana 1995, 16, 135–140. [Google Scholar]
- Rosa-Manzano, E.; Guerra-Pérez, A.; Mendieta-Leiva, G.; Mora-Olivo, A.; Martínez-Ávalos, J.G.; Arellano-Méndez, L.U. Vascular epiphyte diversity in two forest types of the “El Cielo” Biosphere Reserve, Mexico. Botany 2017, 95, 599–610. [Google Scholar] [CrossRef]
- Woods, C.L.; Cardelús, C.L.; DeWalt, S.J. Microhabitat associations of vascular epiphytes in a wet tropical forest canopy. J. Ecol. 2015, 103, 421430. [Google Scholar] [CrossRef]
- Nadkarni, N.M. Complex consequences of disturbance on canopy plant communities of world forests: A review and synthesis. New Phytol. 2023, 240, 1366–1380. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-G.; Xiong, C.; Wei, Z.; Chen, Q.-L.; Ma, B.; Zhou, S.-Y.; Tan, J.; Zhang, L.-M.; Cui, H.-L.; Duan, G.-L. Impacts of global change on the phyllosphere microbiome. New Phytol. 2022, 234, 1977–1986. [Google Scholar] [CrossRef]
- Bell-Doyon, P.; Laroche, J.; Saltonstall, K.; Villarreal Aguilar, J.C. Specialized bacteriome uncovered in the coralloid roots of the epiphytic gymnosperm, Zamia pseudoparasitica. Environ. DNA 2020, 2, 418–428. [Google Scholar] [CrossRef]
- Sierra, A.M.; Toupin, S.; Alonso-García, M.; Villarreal, A.J.C. Diversity of symbiotic cyanobacteria in cycad coralloid roots using a short-read rbcL-X amplicon. Symbiosis 2024, 92, 271–288. [Google Scholar] [CrossRef]
- Stone, B.W.G.; Weingarten, E.A.; Jackson, C.R. The role of the phyllosphere microbiome in plant health and function. Annu. Plant Rev. Online 2018, 1, 533–556. [Google Scholar]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef]
- Laforest-Lapointe, I.; Messier, C.; Kembel, S.W. Host species identity, site and time drive temperate tree phyllosphere bacterial community structure. Microbiome 2016, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Redford, A.J.; Fierer, N. Bacterial succession on the leaf surface, a novel system for studying successional dynamics. Microb. Ecol. 2009, 58, 189–198. [Google Scholar] [CrossRef]
- Doan, H.K.; Ngassam, V.N.; Gilmore, S.F.; Tecon, R.; Parikh, A.N.; Leveau, J.J.H. Topography-Driven shape, Spread, and Retention of Leaf Surface Water Impacts Microbial Dispersion and Activity in the Phyllosphere. Phytobiomes J. 2020, 4, 268–280. [Google Scholar] [CrossRef]
- Remus-Emsermann, M.N.; De Oliveira, S.; Schreiber, L.; Leveau, J.J.H. Quantification of lateral heterogeneity in carbohydrate permeability of isolated plant leaf cuticles. Front. Microbiol. 2011, 2, 12467. [Google Scholar] [CrossRef] [PubMed]
- Brader, G.; Compant, S.; Vescio, K.; Mitter, B.; Trognitz, F.; Ma, L.-J.; Sessitsch, A. Ecology and Genomic Insights into Plant-Pathogenic and Plant-Nonpathogenic Endophytes. Annu. Rev. Phytopath. 2017, 55, 61–83. [Google Scholar] [CrossRef]
- Truchado, P.; Gil, M.I.; Moreno-Candel, M.; Allende, A. Impact of weather conditions, leaf age and irrigation water disinfection on the major epiphytic bacterial genera of baby spinach grown in an open field. Food Microbiol. 2019, 78, 46–52. [Google Scholar] [CrossRef]
- Hacquard, S.; Martin, F.M.; Wang, E. Impact of global chance on the plant microbiome. New Phytol. 2022, 234, 181–187. [Google Scholar] [CrossRef]
- Hirano, S.S.; Upper, C.D. Bacteria in the Leaf Ecosystem with Emphasis on Pseudomonas syringae—A Pathogen, Ice Nucleus, and Epiphyte. Microbiol. Molec. Biol. Rev. 2000, 64, 624–653. [Google Scholar] [CrossRef]
- Lindow, S.E.; Leveau, J.H.J. Phyllosphere microbiology. Curr. Opin. Biotechnol. 2002, 13, 238–243. [Google Scholar] [CrossRef]
- Boutin, S.; Lussier, E.; Laforest-Lapointe, I. Investigating the spatiotemporal dynamics of apple tree phyllosphere bacterial and fungal communities across cultivars in orchards. Can. J. Microbiol. 2024, 70, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Redford, A.J.; Bowers, R.M.; Knight, R.; Linhart, Y.; Fierer, N. The ecology of the phyllosphere: Geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 2010, 12, 2885–2893. [Google Scholar] [CrossRef] [PubMed]
- Moyes, A.B.; Kueppers, L.M.; Pett-Ridge, J.; Carper, D.L.; Vandehey, N.; O’Neil, J.; Frank, C. Evidence for foliar endophytic nitrogen fixation in a widely distributed subalpine conifer. New Phytol. 2016, 210, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Dorokhov, Y.L.; Sheshukova, E.V.; Komarova, T.V. Methanol in Plant Life. Front. Plant Sci. 2018, 9, 1623. [Google Scholar] [CrossRef]
- Lajoie, G.; Maglione, R.; Kembel, S.W. Adaptive matching between phyllosphere bacteria and their tree hosts in a neotropical forest. Microbiome 2020, 8, 1–10. [Google Scholar] [CrossRef]
- Fürnkranz, M.; Wanek, W.; Richter, A.; Abell, G.; Rasche, F.; Sessitsch, A. Nitrogen fixation by phyllosphere bacteria associated with higher plants and their colonizing epiphytes of a tropical lowland rainforest of Costa Rica. ISME J. 2008, 2, 561–570. [Google Scholar] [CrossRef]
- Camelo, C.; Vera, S.; Bonilla, R. Mecanismos de acción de las rizobacterias promotoras del crecimiento vegetal. Corpoica. Cienc. Y Tecnol. Agropecu. 2011, 12, 159–166. Available online: http://www.redalyc.org/articulo.oa?id=449945031010 (accessed on 21 April 2021). [CrossRef]
- Spicer, M.E.; Woods, C.L. A case for studying biotic interactions in epiphyte ecology and evolution. Perspect. Plant Ecol. Evol. Syst. 2022, 54, 125658. [Google Scholar] [CrossRef]
- Taylor, A.S.; Haynes, J.L.; Stevenson, D.W.; Holzman, G.; Mendieta, J. Biogeographic Insights in Central American Cycad Biology. In Global Advances in Biogeography; Stevens, L., Ed.; InTech: Rijeka, Croatia, 2012; pp. 73–98. Available online: www.intechopen.com (accessed on 13 September 2022).
- Stevenson, D.W. The Zamiaceae in Panama with comments on phytogeography and species relationships. Brittonia 1993, 45, 1–16. [Google Scholar] [CrossRef]
- Bell-Doyon, P.; Villarreal, A.J.C. New Notes on the Ecology of the Epiphytic Gymnosperm and Panamanian Endemic Zamia pseudoparasitica. Neotrop. Nat. 2020, 2, 1–7. [Google Scholar]
- Monteza-Moreno, C.M.; Rodriguez-Castro, L.; Castillo-Caballero, P.L.; Toribio, E.; Saltonstall, K. Arboreal camera trapping sheds light on seed dispersal of the world’s only epiphytic gymnosperm: Zamia pseudoparasitica. Ecol. Evol. 2022, 12, e8769. [Google Scholar] [CrossRef]
- Sierra, A.M.; Meléndez, O.; Bethancourt, R.; Bethancourt, A.; Rodríguez-Castro, L.; López, C.A.; Sedio, B.E.; Saltonstall, K.; Villarreal, A.J.C. Leaf Endophytes Relationship with Host Metabolome Expression in Tropical Gymnosperms. J. Chem. Ecol. 2024, 50, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Instituto de Meteorología e Hidrología de Panamá (IMHPA). Available online: https://www.imhpa.gob.pa/es/ (accessed on 7 January 2025).
- Tichý, L. Field test of canopy cover estimation by hemispherical photographs taken with a smartphone. J. Veg. Sci. 2015, 27, 427–435. [Google Scholar] [CrossRef]
- Rueden, C.; Dietz, C.; Horn, M.; Schindelin, J.; Northan, B.; Berthold, M.; Eliceiri, Y.K. ImageJ Ops, version 2.2.0; ImageJ Project: Madison, WI, USA, 2021. Available online: https://imagej.net/Ops (accessed on 25 October 2024).
- Kembel, S.W.; O’Connor, T.K.; Arnold, H.K.; Hubbell, S.P.; Wright, S.J.; Green, J.L. Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc. Natl. Acad. Sci. USA 2014, 111, 13715–13720. [Google Scholar] [CrossRef]
- Hanshew, A.S.; Mason, C.M.; Raffa, K.F.; Currie, C.R. Minimization of chloroplast contamination in 16S rRNA gene pyrosequencing of insect herbivore bacterial communities. J. Microbiol. Meth. 2013, 95, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Wong, J.; Heiner, C.; Steve, O.H.; Theriot, C.M.; Gulati, A.S.; McGill, S.K.; Dougherty, M.K. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 2019, 47, e103. [Google Scholar] [CrossRef]
- Moynihan, M.A.; Reeder, C.F. moyn413/nifHdada2, version 2.0.5; Zenodo: Geneva, Switzerland, 2023. [CrossRef]
- Callahan, B.; McMurdle, P.; Rosen, M.; Han, A.; Johnson, A.; Holmes, S. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Meth. 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. In R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 14 October 2024).
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- Gaby, J.C.; Buckley, D.H. A comprehensive, aligned nifH gene database: A multi-purpose tool for studies of nitrogen-fixing bacteria. Database. 2014, 2014, bauoo1. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef] [PubMed]
- Schliep, K.P. phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. vegan: Community Ecology Package, version 2.5-6; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Chase, J.M.; Kraft, N.J.B.; Smith, K.G.; Vellend, M.; Inouye, B.D. Using null models to disentangle variation in community dissimilarity from variation in α-diversity. Ecosphere 2011, 2, art24. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Kembel, S.W.; Cowan, P.D.; Helmus, M.R.; Cornwell, W.K.; Morlon, H.; Ackerly, D.D.; Blomberg, S.P.; Webb, C.O. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010, 26, 1463–1464. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef]
- Rastogi, G.; Sbodio, A.; Tech, J.J.; Suslow, T.V.; Coaker, G.L.; Leveau, J.H. Leaf microbiota in an agroecosystem: Spatiotemporal variation in bacterial community composition on field-grown lettuce. Int. Soc. Microb. Ecol. J. 2012, 6, 1812–1822. [Google Scholar] [CrossRef]
- Bao, L.; Gu, L.; Sun, B.; Cai, W.; Zhang, S.; Zhuang, G.; Bai, Z.; Zhuang, X. Seasonal variation of epiphytic bacteria in the phyllosphere of Gingko biloba, Pinus bungeana and Sabina chinensis. FEMS Microbiol. Ecol. 2020, 96, fiaa017. [Google Scholar] [CrossRef]
- Copeland, J.K.; Yuan, L.; Layeghifard, M.; Wang, P.W.; Guttman, D.S. Seasonal community succession of the phyllosphere microbiome. Mol. Plant Microbe Interact. 2015, 28, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Peñuelas, J.; Rico, J.; Ogaya, R.; Jump, A.S.; Terradas, J. Summer season and long-term drought increase the richness of bacteria and fungi in the foliar phyllosphere of Quercus ilex in a mixed Mediterranean forest. Plant Biol. 2012, 14, 565–575. [Google Scholar] [CrossRef]
- Finkel, O.M.; Burch, A.Y.; Lindow, S.E.; Post, A.F.; Belkin, S. Geographical location determines the population structure in phyllosphere microbial communities of a salt-excreting desert tree. Appl. Environ. Microbiol. 2011, 77, 7647–7655. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Konopka, A.E.; Fredrickson, J.K. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J. 2012, 6, 1653–1664. [Google Scholar] [CrossRef]
- Tripathi, B.M.; Stegen, J.C.; Kim, M.; Dong, K.; Adams, J.M.; Lee, Y.K. Soil pH mediates the balance between stochastic and deterministic assembly of bacteria. ISME J. 2018, 12, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Delmotte, N.; Knief, C.; Chaffron, S.; Innerebner, G.; Roschitzki, B.; Schlapbach, R.; von Mering, C.; Vorholt, J.A. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 16428–16433. [Google Scholar] [CrossRef] [PubMed]
- Martiny, J.B.H.; Bohannan, B.J.; Brown, J.H.; Colwell, R.K.; Fuhrman, J.A.; Green, J.L.; Horner-Devine, M.C.; Kane, M.; Krumins, J.A.; Kuske, C.R.; et al. Microbial biogeography: Putting microorganisms on the map. Nat. Rev. Microbiol. 2006, 4, 102–112. [Google Scholar] [CrossRef]
- Kecskeméti, E.; Berkelmann-Löhnertz, B.; Reineke, A. Are Epiphytic Microbial Communities in the Carposphere of Ripening Grape Clusters (Vitis vinifera L.) Different between Conventional, Organic, and Biodynamic Grapes? PLoS ONE 2016, 11, e0160852. [Google Scholar] [CrossRef] [PubMed]
- Grady, K.L.; Sorensen, J.W.; Stopnisek, N.; Guittar, J.; Shade, A. Assembly and seasonality of core phyllosphere microbiota on perennial biofuel crops. Nat. Commun. 2019, 10, 4135. [Google Scholar] [CrossRef]
- Atamna-Ismaeel, N.; Finkel, O.; Glaser, F.; von Mering, C.; Vorholt, J.A.; Koblížek, M.; Belkin, S.; Béjà, O. Bacterial anoxygenic photosynthesis on plant leaf surfaces. Environ. Microbiol. Rep. 2012, 4, 209–216. [Google Scholar] [CrossRef]
- Watanabe, K.; Kohzu, A.; Suda, W.; Yamamura, S.; Takamatsu, T.; Takenaka, A.; Koshikawa, M.K.; Hayashi, S.; Watanabe, M. Microbial nitrification in throughfall of a Japanese cedar associated with archaea from the tree canopy. SpringerPlus 2016, 5, 1596. [Google Scholar] [CrossRef]
- Romero, F.M.; Marina, M.; Pieckenstain, F.L. Novel components of leaf bacterial communities of field-grown tomato plants and their potential for plant growth promotion and biocontrol of tomato diseases. Res. Microbiol. 2016, 167, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, W.; Liu, Y.; Zhu, W.; Yuan, Z.; Su, X.; Ding, C. Differences in phyllosphere microbiomes among different Populus spp. in the same habitat. Front. Plant Sci. 2023, 14, 1143878. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, J.; Sun, P.; Chen, C.; Shen, J. Community Structure of Phyllosphere Bacteria in Different Cultivars of Fingered Citron (Citrus medica ‘Fingered’) and Their Correlations With Fragrance. Front. Plant Sci. 2022, 13, 936252. [Google Scholar] [CrossRef]
- Yang, J.; Wang, Y.; Cui, X.; Xue, K.; Zhang, Y.; Yu, Z. Habitat filtering shapes the differential structure of microbial communities in the Xilingol grassland. Sci. Rep. 2019, 9, 19326. [Google Scholar] [CrossRef]
- Genitsaris, S.; Stefanidou, N.; Leontidou, K.; Matsi, T.; Karamanoli, K.; Mellidou, I. Bacterial Communities in the Rhizosphere and Phyllosphere of Halophytes and Drought-Tolerant Plants in Mediterranean Ecosystems. Microorganisms 2020, 8, 1708. [Google Scholar] [CrossRef] [PubMed]
- Rausch, C.; Daram, P.; Brunner, S.; Jansa, J.; Laloi, M.; Leggewie, G.; Amrhein, N.; Bucher, M. A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 2001, 414, 462–470. [Google Scholar] [CrossRef]
- Lopez-Velasco, G.; Welbaum, G.E.; Boyer, R.R.; Mane, S.P.; Ponder, M.A. Changes in spinach phylloepiphytic bacteria communities following minimal processing and refrigerated storage described using pyrosequencing of 16S rRNA amplicons. J. Appl. Microbiol. 2011, 110, 1203–1214. [Google Scholar] [CrossRef]
- Knief, C.; Delmotte, N.; Chaffron, S.; Stark, M.; Innerebner, G.; Wassmann, R.; von Mering, C.; Vorholt, A.J. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012, 6, 1378–1390. [Google Scholar] [CrossRef]
- Beattie, G.A.; Lindow, S.E. Bacterial colonization of leaves: A spectrum of strategies. Phytopathology 1999, 89, 353–359. [Google Scholar] [CrossRef]
- Innerebner, G.; Knief, C.; Vorholt, J.A. Protection of Arabidopsis thaliana against Leaf-Pathogenic Pseudomonas syringae by Sphingomonas Strains in a Controlled Model System. Appl. Environ. Microbiol. 2011, 77, 3202–3210. [Google Scholar] [CrossRef]
- Ottesen, A.R.; Gorham, S.; Pettengill, J.B.; Rideout, S.; Evans, P.; Brown, E. The impact of systemic and copper pesticide applications on the phyllosphere microflora of tomatoes. J. Sci. Food Agric. 2015, 95, 1116–1125. [Google Scholar] [CrossRef] [PubMed]
- Leducq, J.-B.; Sneddon, D.; Santos, M.; Condrain-Morel, D.; Bourret, G.; Martinez-Gomez, N.C.; Lee, J.A.; Foster, J.A.; Stolyar, S.; Shapiro, B.J.; et al. Comprehensive Phylogenomics of Methylobacterium Reveals Four Evolutionary Distinct Groups and Underappreciated Phyllosphere Diversity. Genome Biol. Evol. 2022, 14, evac123. [Google Scholar] [CrossRef] [PubMed]
- Roush, D.; Giraldo-Silva, A.; Garcia-Pichel, F. Cydrasil 3, a curated 16S rRNA gene reference package and web app for cyanobacterial phylogenetic placement. Sci. Data 2021, 8, 230. [Google Scholar] [CrossRef]
- Salzman, S.; Bustos-Díaz, E.D.; Whitaker, M.R.L.; Sierra, A.M.; Cibrián-Jaramillo, A.; Barona-Gómez, F.; Villarreal Aguilar, J.C. Chemical ecology of symbioses in cycads, an ancient plant lineage. New Phytol. 2025, 246, 1494–1504. [Google Scholar] [CrossRef] [PubMed]



| Season | Factors | Palo Seco | Site Santa Fe | El Copé |
|---|---|---|---|---|
| Rainy Season | Leaflet temperature (°C) | 22.5 ± 2.5 | 21.1 ± 1.8 | 23.4 ± 1.1 |
| Ambient temperature (°C) | 23.1 ± 3.6 | 23.8 ± 0.5 | 24.3 ± 1.2 | |
| Canopy cover (%) | 60.3 ± 11.0 | 57.2 ± 3.8 | 53.1 ± 5.5 | |
| Relative humidity (%) | 99.5 ± 12.1 | 98.7 ± 1.6 | 94.6 ± 1.5 | |
| Dry Season | Leaflet temperature (°C) | 18.8 ± 4.2 | 20.1 ± 1.6 | 24.3 ± 1.0 |
| Ambient temperature (°C) | 20.9 ± 2.6 | 25.3 ± 13.2 | 26.2 ± 1.5 | |
| Canopy cover (%) | 57.3 ± 5.6 | 58.2 ± 4.6 | 59.3 ± 4.1 | |
| Relative humidity (%) | 99.9 ± 10.8 | 99.9 ± 0.9 | 87.1 ± 14.8 |
| 16S rRNA | nifH | |||||
|---|---|---|---|---|---|---|
| Pairwise Comparisons | R2 | p-Value | p-Adjusted | R2 | p-Value | p-Adjusted |
| El Copé dry vs. Palo Seco dry | 0.128 | 0.002 | 0.030 | 0.150 | 0.003 | 0.045 |
| El Copé dry vs. Santa Fe dry | 0.077 | 0.031 | 0.465 | 0.075 | 0.196 | 1.000 |
| El Copé dry vs. El Copé rainy | 0.090 | 0.015 | 0.225 | 0.060 | 0.289 | 1.000 |
| El Copé dry vs. Palo Seco rainy | 0.132 | 0.001 | 0.015 | 0.106 | 0.004 | 0.060 |
| El Copé dry vs. Santa Fe rainy | 0.098 | 0.004 | 0.060 | 0.073 | 0.142 | 1.000 |
| Palo Seco dry vs. Santa Fe dry | 0.118 | 0.002 | 0.030 | 0.109 | 0.077 | 1.000 |
| Palo Seco dry vs. El Copé rainy | 0.141 | 0.001 | 0.015 | 0.100 | 0.039 | 0.585 |
| Palo Seco dry vs. Palo Seco rainy | 0.005 | 0.258 | 1.000 | 0.075 | 0.321 | 1.000 |
| Palo Seco dry vs. Santa Fe rainy | 0.099 | 0.008 | 0.120 | 0.145 | 0.009 | 0.135 |
| Santa Fe dry vs. El Copé rainy | 0.084 | 0.012 | 0.180 | 0.074 | 0.173 | 1.000 |
| Santa Fe dry vs. Palo Seco rainy | 0.129 | 0.001 | 0.015 | 0.090 | 0.085 | 1.000 |
| Santa Fe dry vs. Santa Fe rainy | 0.056 | 0.321 | 1.000 | 0.060 | 0.583 | 1.000 |
| El Copé rainy vs. Palo Seco rainy | 0.154 | 0.001 | 0.015 | 0.068 | 0.181 | 1.000 |
| El Copé rainy vs. Santa Fe rainy | 0.103 | 0.002 | 0.030 | 0.075 | 0.090 | 1.000 |
| Palo Seco rainy vs. Santa Fe rainy | 0.118 | 0.001 | 0.015 | 0.122 | 0.007 | 0.105 |
| Phylum | Family | 16S rRNA Gene | nifH Gene |
|---|---|---|---|
| Cyanobacteria | Tolypothrichaceae | - | ✓ |
| Dermocarpellaceae | - | ✓ | |
| Leptolyngbyaceae | - | ✓ | |
| Rivulariaceae | - | ✓ | |
| Scytonemataceae | - | ✓ | |
| Stigonemataceae | - | ✓ | |
| Nostocaceae | ✓ | ✓ | |
| Bacillota | Clostridiaceae | - | ✓ |
| Desulfitobacteriaceae | - | ✓ | |
| Eubacteriales Family XIII Incertae Sedis | - | ✓ | |
| Thermodesulfobacteriota | Desulfobulbaceae | - | ✓ |
| Bacteroidota | Prolixibacteraceae | - | ✓ |
| Firmicutes | Paenibacillaceae | ✓ | ✓ |
| Proteobacteria | Boseaceae | - | ✓ |
| Burkholderiales incertae sedis | - | ✓ | |
| Chromatiaceae | - | ✓ | |
| Methylocystaceae | - | ✓ | |
| Nitrobacteraceae | - | ✓ | |
| Beijerinckiaceae | ✓ | ✓ | |
| Enterobacteriaceae | ✓ | ✓ | |
| Erwiniaceae | ✓ | ✓ | |
| Hyphomicrobiaceae | ✓ | ✓ | |
| Pseudomonadaceae | ✓ | ✓ | |
| Xanthomonadaceae | ✓ | ✓ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Castro, L.; Sierra, A.M.; Villarreal Aguilar, J.C.; Saltonstall, K. Spatial and Seasonal Analysis of Phyllosphere Bacterial Communities of the Epiphytic Gymnosperm Zamia pseudoparasitica. Appl. Biosci. 2025, 4, 35. https://doi.org/10.3390/applbiosci4030035
Rodríguez-Castro L, Sierra AM, Villarreal Aguilar JC, Saltonstall K. Spatial and Seasonal Analysis of Phyllosphere Bacterial Communities of the Epiphytic Gymnosperm Zamia pseudoparasitica. Applied Biosciences. 2025; 4(3):35. https://doi.org/10.3390/applbiosci4030035
Chicago/Turabian StyleRodríguez-Castro, Lilisbeth, Adriel M. Sierra, Juan Carlos Villarreal Aguilar, and Kristin Saltonstall. 2025. "Spatial and Seasonal Analysis of Phyllosphere Bacterial Communities of the Epiphytic Gymnosperm Zamia pseudoparasitica" Applied Biosciences 4, no. 3: 35. https://doi.org/10.3390/applbiosci4030035
APA StyleRodríguez-Castro, L., Sierra, A. M., Villarreal Aguilar, J. C., & Saltonstall, K. (2025). Spatial and Seasonal Analysis of Phyllosphere Bacterial Communities of the Epiphytic Gymnosperm Zamia pseudoparasitica. Applied Biosciences, 4(3), 35. https://doi.org/10.3390/applbiosci4030035






