Bioinformatics Analysis of Unique High-Density Lipoprotein-MicroRNAs Cargo Reveals Its Neurodegenerative Disease Potential
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition and Pre-Processing
2.2. MiRNA Target Gene Prediction
2.3. GO and KEGG Pathway Enrichment Analysis
2.4. Integration of the Protein–Protein Interaction (PPI) Network
2.5. Data Visualization
3. Results
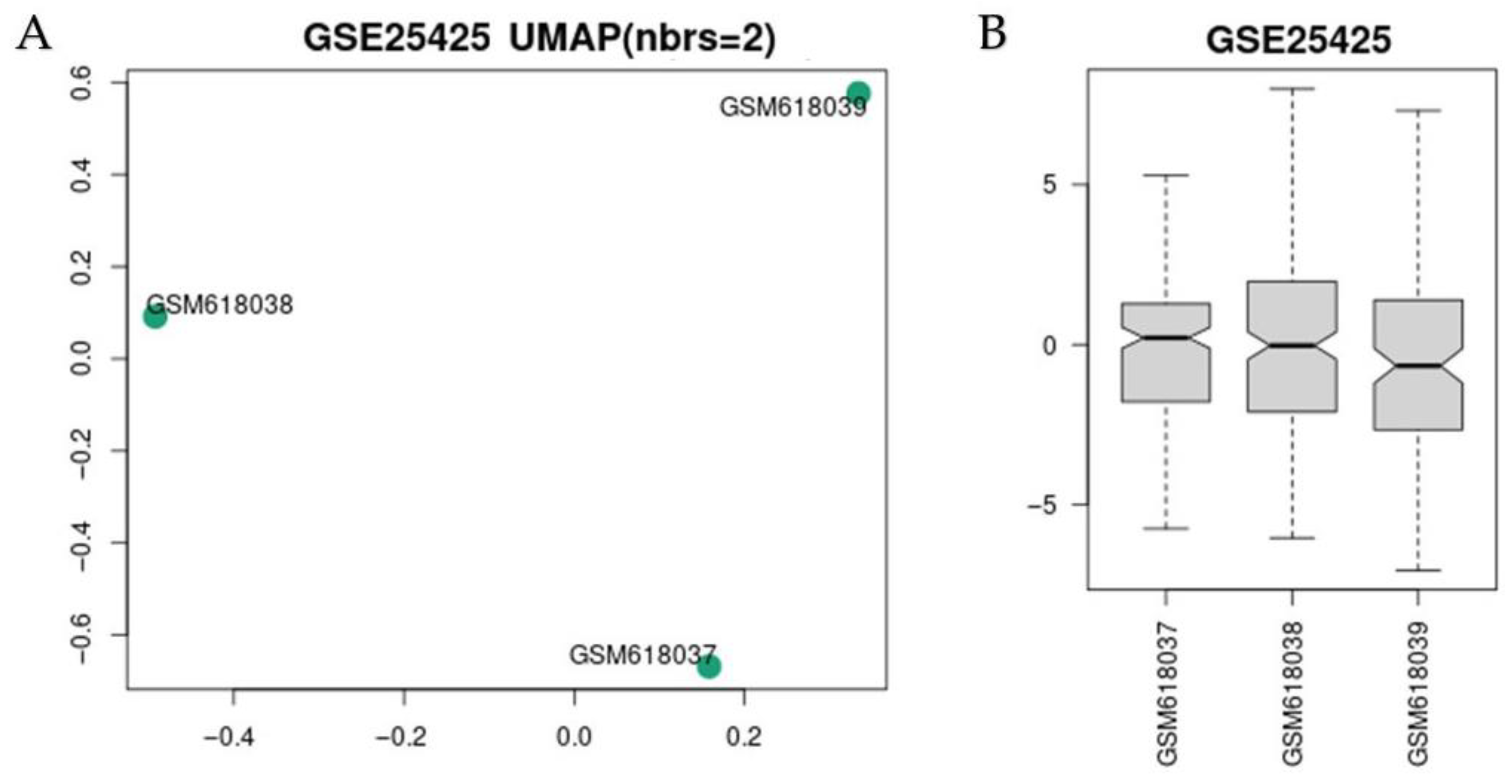
3.1. GEO Dataset Information
3.2. Analysis of Exosome, LDL, and HDL Dataset
3.3. Identification of Unique HDL-miRNA Cargo and KEGG Pathways Analysis
3.4. Protein–Protein Interactions
4. Discussion
5. Future Perspectives
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AKT1 | Protein kinase B (AKT1 is a specific isoform) |
| APP | Amyloid Precursor Protein |
| APOE | Apolipoprotein E |
| BACE | Beta-Site Amyloid Precursor Protein Cleaving Enzyme |
| BCL2 | B-cell lymphoma 2 (a family of regulator proteins) |
| BDNF | Brain-Derived Neurotrophic Factor |
| BP | Biological Processes |
| CALM3 | Calmodulin 3 (calcium-binding messenger protein) |
| CC | Cellular Components |
| EV | Extracellular Vesicle |
| FPLC | Fast Protein Liquid Chromatography |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| GPL | Gene Expression Omnibus Platform |
| GSK3B | Glycogen Synthase Kinase 3 Beta |
| HDL | High-Density Lipoprotein |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KRAS | Kirsten rat sarcoma viral oncogene homolog (gene encoding a small GTPase protein) |
| MCODE | Molecular Complex Detection |
| MF | Molecular Functions |
| miRNA | MicroRNA |
| mRNA | Messenger RNA |
| mTOR | Mechanistic Target of Rapamycin |
| PI3K | Phosphoinositide 3-Kinase |
| PPI | Protein–Protein Interaction |
| RNA | Ribonucleic Acid |
| RPS | Ribosomal Protein Subunit (RPS27A in the document refers to Ribosomal Protein S27a) |
| TORC1 | Target of Rapamycin Complex 1 |
| UBB | Ubiquitin B |
| VCP | Valosin-Containing Protein |
References
- Ben-Aicha, S.; Escate, R.; Casaní, L.; Padró, T.; Peña, E.; Arderiu, G.; Mendieta, G.; Badimón, L.; Vilahur, G. High-density lipoprotein remodelled in hypercholesterolaemic blood induce epigenetically driven down-regulation of endothelial HIF-1α expression in a preclinical animal model. Cardiovasc. Res. 2020, 116, 1288–1299. [Google Scholar] [CrossRef] [PubMed]
- Peronace, C.; Cione, E.; Abrego-Guandique, D.M.; Fazio, M.D.; Panduri, G.; Caroleo, M.C.; Cannataro, R.; Minchella, P. FAM19A4 and hsa-miR124-2 Double Methylation as Screening for ASC-H- and CIN1 HPV-Positive Women. Pathogens 2024, 13, 312. [Google Scholar] [CrossRef] [PubMed]
- Michell, D.L.; Vickers, K.C. Lipoprotein carriers of microRNAs. Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 2016, 1861, 2069–2074. [Google Scholar] [CrossRef]
- Baldán, Á.; de Aguiar Vallim, T.Q. miRNAs and High-Density Lipoprotein metabolism. Biochim. Biophys. Acta—Mol. Cell Biol. Lipids 2016, 1861, 2053–2061. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Tussy, P.; Ruz-Maldonado, I.; Fernández-Hernando, C. MicroRNAs and Circular RNAs in Lipoprotein Metabolism. Curr. Atheroscler. Rep. 2021, 23, 33. [Google Scholar] [CrossRef]
- Canfrán-Duque, A.; Lin, C.S.; Goedeke, L.; Suárez, Y.; Fernández-Hernando, C. Micro-RNAs and high-density lipoprotein metabolism. Arterioscler. Thromb. Vasc. Biol. 2016, 36, 1076–1084. [Google Scholar] [CrossRef]
- Rossi-Herring, G.; Belmonte, T.; Rivas-Urbina, A.; Benítez, S.; Rotllan, N.; Crespo, J.; Llorente-Cortés, V.; Sánchez-Quesada, J.L.; de Gonzalo-Calvo, D. Circulating lipoprotein-carried miRNome analysis reveals novel VLDL-enriched microRNAs that strongly correlate with the HDL-microRNA profile. Biomed. Pharmacother. 2023, 162, 114623. [Google Scholar] [CrossRef]
- Graham, A. Modulation of the Cellular microRNA Landscape: Contribution to the Protective Effects of High-Density Lipoproteins (HDL). Biology 2023, 12, 1232. [Google Scholar] [CrossRef]
- Hussain, S.M.; Robb, C.; Tonkin, A.M.; Lacaze, P.; Chong, T.T.-J.; Beilin, L.J.; Yu, C.; Watts, G.F.; Ryan, J.; Ernst, M.E.; et al. Association of plasma high-density lipoprotein cholesterol level with risk of incident dementia: A cohort study of healthy older adults. Lancet Reg. Health—West. Pacific 2024, 43, 100963. [Google Scholar] [CrossRef]
- Saraceno, G.F.; Abrego-Guandique, D.M.; Cannataro, R.; Caroleo, M.C.; Cione, E. Machine Learning Approach to Identify Case-Control Studies on ApoE Gene Mutations Linked to Alzheimer’s Disease in Italy. BioMedInformatics 2024, 4, 600–622. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef]
- Dorrity, M.W.; Saunders, L.M.; Queitsch, C.; Fields, S.; Trapnell, C. Dimensionality reduction by UMAP to visualize physical and genetic interactions. Nat. Commun. 2020, 11, 1537. [Google Scholar] [CrossRef]
- Ru, Y.; Kechris, K.J.; Tabakoff, B.; Hoffman, P.; Radcliffe, R.A.; Bowler, R.; Mahaffey, S.; Rossi, S.; Calin, G.A.; Bemis, L.; et al. The multiMiR R package and database: Integration of microRNA–target interactions along with their disease and drug associations. Nucleic Acids Res. 2014, 42, e133. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003, 4, 2. [Google Scholar] [CrossRef]
- Nasab, A.S.; Noorani, F.; Paeizi, Z.; Khani, L.; Banaei, S.; Sadeghi, M.; Shafeghat, M.; Shafie, M.; Mayeli, M. A Comprehensive Investigation of the Potential Role of Lipoproteins and Metabolite Profile as Biomarkers of Alzheimer’s Disease Compared to the Known CSF Biomarkers. Int. J. Alzheimers. Dis. 2023, 2023, 3540020. [Google Scholar] [CrossRef]
- Ruiz, M.; Frej, C.; Holmér, A.; Guo, L.J.; Tran, S.; Dahlbäck, B. High-Density Lipoprotein–Associated Apolipoprotein M Limits Endothelial Inflammation by Delivering Sphingosine-1-Phosphate to the Sphingosine-1-Phosphate Receptor 1. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 118–129. [Google Scholar] [CrossRef]
- Wagner, J.; Riwanto, M.; Besler, C.; Knau, A.; Fichtlscherer, S.; Röxe, T.; Zeiher, A.M.; Landmesser, U.; Dimmeler, S. Characterization of levels and cellular transfer of circulating lipoprotein-bound microRNAs. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1392–1400. [Google Scholar] [CrossRef]
- Niculescu, L.S.; Simionescu, N.; Sanda, G.M.; Carnuta, M.G.; Stancu, C.S.; Popescu, A.C.; Popescu, M.R.; Vlad, A.; Dimulescu, D.R.; Simionescu, M. MiR-486 and miR-92a identified in circulating HDL discriminate between stable and vulnerable coronary artery disease patients. PLoS ONE 2015, 10, e0140958. [Google Scholar] [CrossRef] [PubMed]
- Kawahata, I.; Fukunaga, K. Protein Kinases and Neurodegenerative Diseases. Int. J. Mol. Sci. 2023, 24, 5574. [Google Scholar] [CrossRef] [PubMed]
- Etchegaray, J.I.; Elguero, E.J.; Tran, J.A.; Sinatra, V.; Feany, M.B.; McCall, K. Defective Phagocytic Corpse Processing Results in Neurodegeneration and Can Be Rescued by TORC1 Activation. J. Neurosci. 2016, 36, 3170. [Google Scholar] [CrossRef]
- Schmidt, M.F.; Gan, Z.Y.; Komander, D.; Dewson, G. Ubiquitin signalling in neurodegeneration: Mechanisms and therapeutic opportunities. Cell Death Differ. 2021, 28, 570–590. [Google Scholar] [CrossRef] [PubMed]
- Heras-Sandoval, D.; Pérez-Rojas, J.M.; Hernández-Damián, J.; Pedraza-Chaverri, J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cell. Signal. 2014, 26, 2694–2701. [Google Scholar] [CrossRef] [PubMed]
- Morrison, K.R.; Solly, E.L.; Shemesh, T.; Psaltis, P.J.; Nicholls, S.J.; Brown, A.; Bursill, C.A.; Tan, J.T.M. Elevated HDL-bound miR-181c-5p level is associated with diabetic vascular complications in Australian Aboriginal people. Diabetologia 2021, 64, 1402–1411. [Google Scholar] [CrossRef]
- Jääskeläinen, O.; Solje, E.; Hall, A.; Katisko, K.; Korhonen, V.; Tiainen, M.; Kangas, A.J.; Helisalmi, S.; Pikkarainen, M.; Koivisto, A.; et al. Low Serum High-Density Lipoprotein Cholesterol Levels Associate with the C9orf72 Repeat Expansion in Frontotemporal Lobar Degeneration Patients. J. Alzheimers Dis. 2019, 72, 127–137. [Google Scholar] [CrossRef]
- Wu, Q.; Sheng, Q.; Michell, D.; Ramirez-Solano, M.; Posey, O.; Phothisane, A.; Shaik, S.; Vickers, K.C.; Ormseth, M.J. Anti-Inflammatory Effect of High-Density Lipoprotein Blunted by Delivery of Altered MicroRNA Cargo in Patients With Rheumatoid Arthritis. Arthritis Rheumatol. 2024, 76, 684–695. [Google Scholar] [CrossRef]
- Rosenson, R.S.; Brewer, H.B.; Ansell, B.J.; Barter, P.; Chapman, M.J.; Heinecke, J.W.; Kontush, A.; Tall, A.R.; Webb, N.R. Dysfunctional HDL and atherosclerotic cardiovascular disease. Nat. Rev. Cardiol. 2016, 13, 48–60. [Google Scholar] [CrossRef]
- Watts, M.E.; Williams, S.M.; Nithianantharajah, J.; Claudianos, C. Hypoxia-Induced MicroRNA-210 Targets Neurodegenerative Pathways. Non-Coding RNA 2018, 4, 10. [Google Scholar] [CrossRef]
- Sauer, M.; Was, N.; Ziegenhals, T.; Wang, X.; Hafner, M.; Becker, M.; Fischer, U. The miR-26 family regulates neural differentiation-associated microRNAs and mRNAs by directly targeting REST. J. Cell Sci. 2021, 134, jcs257535. [Google Scholar] [CrossRef] [PubMed]
- Barros-Viegas, A.T.; Carmona, V.; Ferreiro, E.; Guedes, J.; Cardoso, A.M.; Cunha, P.; Pereira de Almeida, L.; Resende de Oliveira, C.; Pedro de Magalhães, J.; Peça, J.; et al. miRNA-31 Improves Cognition and Abolishes Amyloid-β; Pathology by Targeting APP and BACE1 in an Animal Model of Alzheimer’s Disease. Mol. Ther. Nucleic Acids 2020, 19, 1219–1236. [Google Scholar] [CrossRef] [PubMed]
- Wallach, T.; Mossmann, Z.J.; Szczepek, M.; Wetzel, M.; Machado, R.; Raden, M.; Miladi, M.; Kleinau, G.; Krüger, C.; Dembny, P.; et al. MicroRNA-100-5p and microRNA-298-5p released from apoptotic cortical neurons are endogenous Toll-like receptor 7/8 ligands that contribute to neurodegeneration. Mol. Neurodegener. 2021, 16, 80. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.B.; Mufson, E.J.; Counts, S.E. Evidence for a neuroprotective microRNA pathway in amnestic mild cognitive impairment. Front. Neurosci. 2015, 9, 430. [Google Scholar] [CrossRef]
- Peña-Bautista, C.; Álvarez-Sánchez, L.; Cañada-Martínez, A.J.; Baquero, M.; Cháfer-Pericás, C. Epigenomics and Lipidomics Integration in Alzheimer Disease: Pathways Involved in Early Stages. Biomedicines 2021, 9, 1812. [Google Scholar] [CrossRef]
- Krüger, D.M.; Pena-Centeno, T.; Liu, S.; Park, T.; Kaurani, L.; Pradhan, R.; Huang, Y.; Risacher, S.L.; Burkhardt, S.; Schütz, A.; et al. The plasma miRNAome in ADNI: Signatures to aid the detection of at-risk individuals. Alzheimers Dement. 2024, 20, 7479–7494. [Google Scholar] [CrossRef]
- Liu, S.; Park, T.; Krüger, D.M.; Pena-Centeno, T.; Burkhardt, S.; Schutz, A.; Huang, Y.; Rosewood, T.; Chaudhuri, S.; Cho, M.; et al. Plasma miRNAs across the Alzheimer’s disease continuum: Relationship to central biomarkers. Alzheimers Dement. 2024, 20, 7698–7714. [Google Scholar] [CrossRef]
- Abrego-Guandique, D.M.; Cione, E.; Caroleo, M.C.; Bonilla, D.A.; Cannataro, R. Ketogenic diet and microRNAs: Focus on cognitive function. Front. Nutr. 2025, 12, 1545832. [Google Scholar] [CrossRef]
- Abrego Guandique, D.M.; Radbakhsh, S.; Bacchetti, T.; Aghaee-Bakhtiari, S.H.; Mahmoudi, A.; Akhonpour Manteghi, A.; Bazyari, M.J.; Cione, E.; Ferretti, G.; Sahebkar, A. Trehalose effect in patients with Alzheimer’s disease: Focus on circulating microRNAs assessed by direct hybridization. FEBS Open Bio 2024, 14, 2–91. [Google Scholar] [CrossRef]
- Abrego-Guandique, D.M.; Saraceno, G.F.; Cannataro, R.; de Burnside, M.M.; Caroleo, M.C.; Cione, E. Apolipoprotein E and Alzheimer’s Disease in Italian Population: Systematic Review and Meta-Analysis. Brain Sci. 2024, 14, 908. [Google Scholar] [CrossRef] [PubMed]
- Fortea, J.; Pegueroles, J.; Alcolea, D.; Belbin, O.; Dols-Icardo, O.; Vaqué-Alcázar, L.; Videla, L.; Gispert, J.D.; Suárez-Calvet, M.; Johnson, S.C.; et al. APOE4 homozygozity represents a distinct genetic form of Alzheimer’s disease. Nat. Med. 2024, 30, 1284–1291. [Google Scholar] [CrossRef] [PubMed]
- Hong, B.V.; Zheng, J.; Agus, J.K.; Tang, X.; Lebrilla, C.B.; Jin, L.-W.; Maezawa, I.; Erickson, K.; Harvey, D.J.; DeCarli, C.S.; et al. High-Density Lipoprotein Changes in Alzheimer’s Disease Are APOE Genotype-Specific. Biomedicines 2022, 10, 1495. [Google Scholar] [CrossRef] [PubMed]
- Tabet, F.; Vickers, K.C.; Cuesta Torres, L.F.; Wiese, C.B.; Shoucri, B.M.; Lambert, G.; Catherinet, C.; Prado-Lourenco, L.; Levin, M.G.; Thacker, S.; et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat. Commun. 2014, 5, 3292. [Google Scholar] [CrossRef]
- Cione, E.; Michelini, S.; Abrego-Guandique, D.M.; Vaia, N.; Michelini, S.; Puleo, V.; Bertelli, M.; Caroleo, M.C.; Cannataro, R. Identification of Specific microRNAs in Adipose Tissue Affected by Lipedema. Curr. Issues Mol. Biol. 2024, 46, 11957–11974. [Google Scholar] [CrossRef]



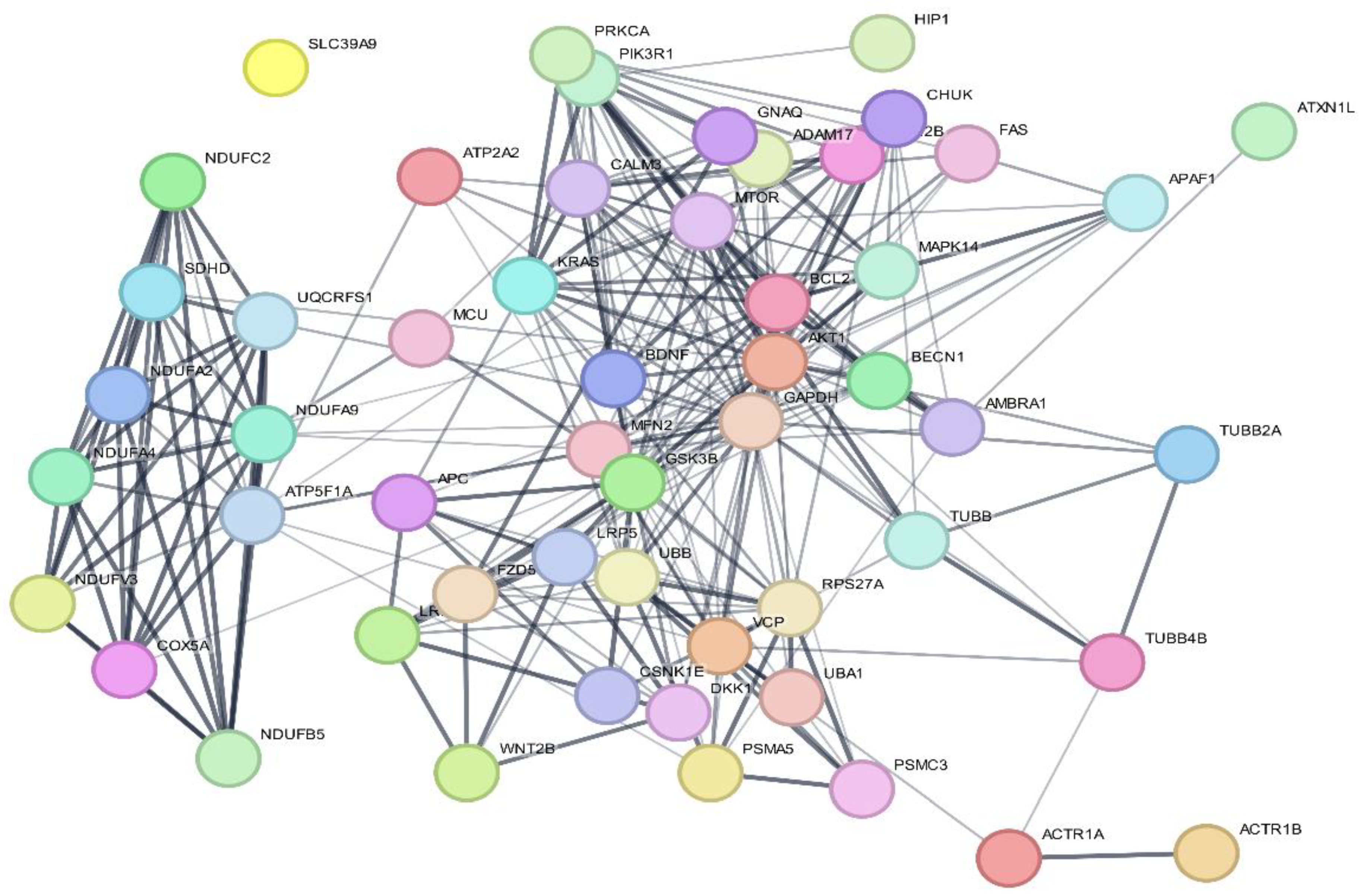
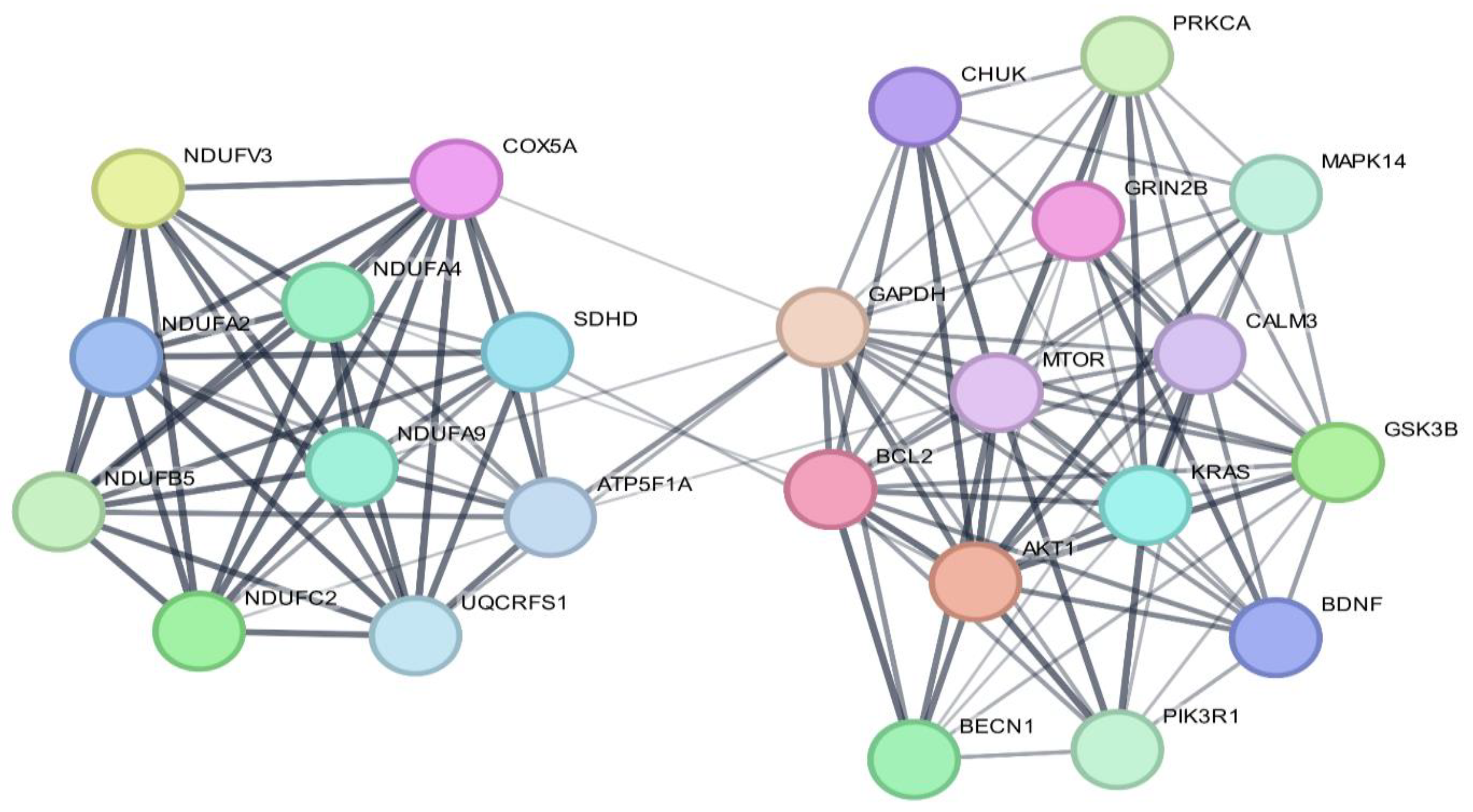
| Nodes | Node Degree | Betweenness | Closeness |
|---|---|---|---|
| GAPDH | 34 | 0.2515 | 0.7285 |
| AKT1 | 29 | 0.1513 | 0.6799 |
| GSK3B | 26 | 0.0826 | 0.6219 |
| MTOR | 19 | 0.0311 | 0.6071 |
| BCL2 | 19 | 0.0474 | 0.5930 |
| CALM3 | 19 | 0.0364 | 0.5730 |
| KRAS | 17 | 0.0133 | 0.5483 |
| VCP | 17 | 0.0870 | 0.5862 |
| RPS27A | 17 | 0.0251 | 0.5483 |
| UBB | 15 | 0.0173 | 0.5312 |
| BDNF | 9 | 0.0013 | 0.5609 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abrego-Guandique, D.M.; Caroleo, M.C.; Luciani, F.; Cione, E. Bioinformatics Analysis of Unique High-Density Lipoprotein-MicroRNAs Cargo Reveals Its Neurodegenerative Disease Potential. Appl. Biosci. 2025, 4, 34. https://doi.org/10.3390/applbiosci4030034
Abrego-Guandique DM, Caroleo MC, Luciani F, Cione E. Bioinformatics Analysis of Unique High-Density Lipoprotein-MicroRNAs Cargo Reveals Its Neurodegenerative Disease Potential. Applied Biosciences. 2025; 4(3):34. https://doi.org/10.3390/applbiosci4030034
Chicago/Turabian StyleAbrego-Guandique, Diana Marisol, Maria Cristina Caroleo, Filippo Luciani, and Erika Cione. 2025. "Bioinformatics Analysis of Unique High-Density Lipoprotein-MicroRNAs Cargo Reveals Its Neurodegenerative Disease Potential" Applied Biosciences 4, no. 3: 34. https://doi.org/10.3390/applbiosci4030034
APA StyleAbrego-Guandique, D. M., Caroleo, M. C., Luciani, F., & Cione, E. (2025). Bioinformatics Analysis of Unique High-Density Lipoprotein-MicroRNAs Cargo Reveals Its Neurodegenerative Disease Potential. Applied Biosciences, 4(3), 34. https://doi.org/10.3390/applbiosci4030034








