Exploring the Synergistic Impacts of Cover Crops and Fertilization on Soil Microbial Metabolic Diversity in Dryland Soybean Production Systems Using Biolog EcoPlates
Abstract
1. Introduction
- Soil microbial functional diversity and soil health characteristics were positively affected by cover crop mixtures and organic amendments in dryland soybean production systems.
- Cover crop mixture leads to higher microbial functional diversity compared to solo cover crop treatments.
2. Materials and Methods
2.1. Description of the Experimental Sites
2.2. Experimental Design and Field Methods
2.3. Soil Sampling and Analysis of Physicochemical Properties
2.4. BiologTM EcoPlates
2.5. Soil Microbial Communities Catabolic Profiling
2.5.1. Quantification of Average Well Color Development in Biolog EcoPlates Wells
2.5.2. Determination of Diversity Indices of Microbial Populations:
- (1)
- Shannon Diversity Index (H′):
- (2)
- Shannon Evenness Index (E):
- (3)
- Inverse Simpson Diversity Index (1/D):
- (4)
- McInthosh Index (U):
- (5)
- Substrate richness (SR):
- (6)
- Statistical analysis:
3. Results
3.1. Differences in AWCD over Time in Soils with Cover Crops and Fertilizer Source Treatment
3.2. Influence of Cover Cropping and Fertilizer Source Treatments on Microbial Metabolic Diversity Indices
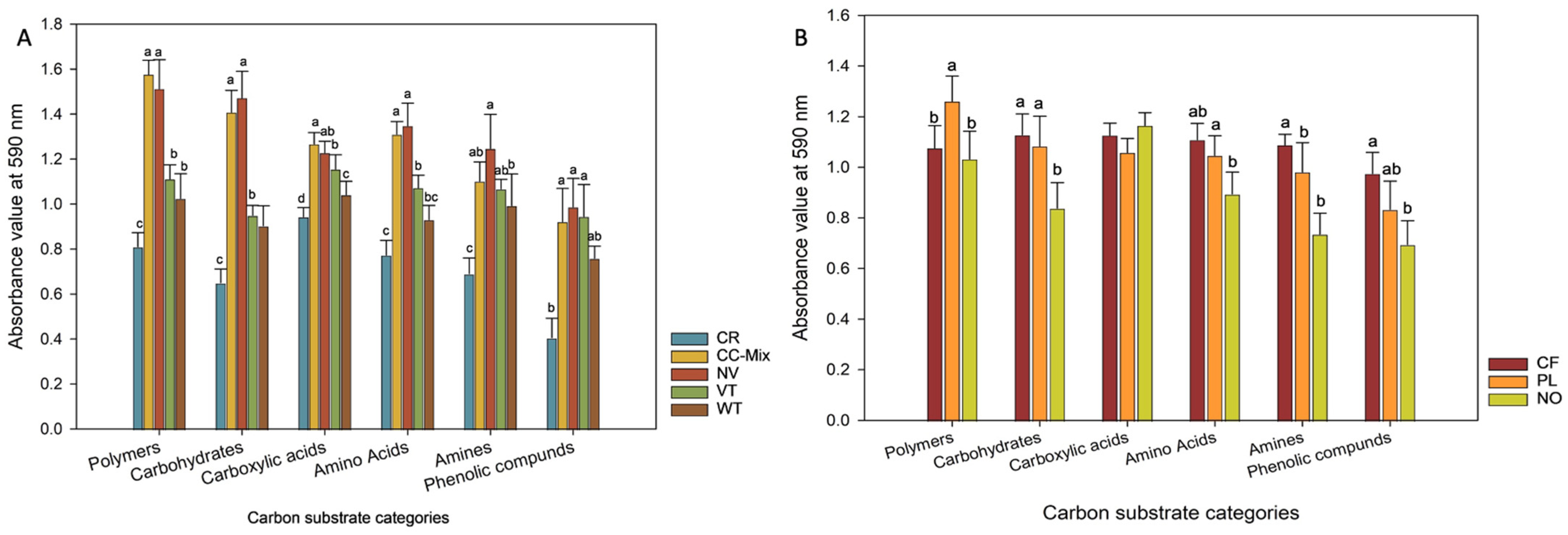
3.3. Classification of Carbon Substrate Utilization Categories in Biolog EcoPlate
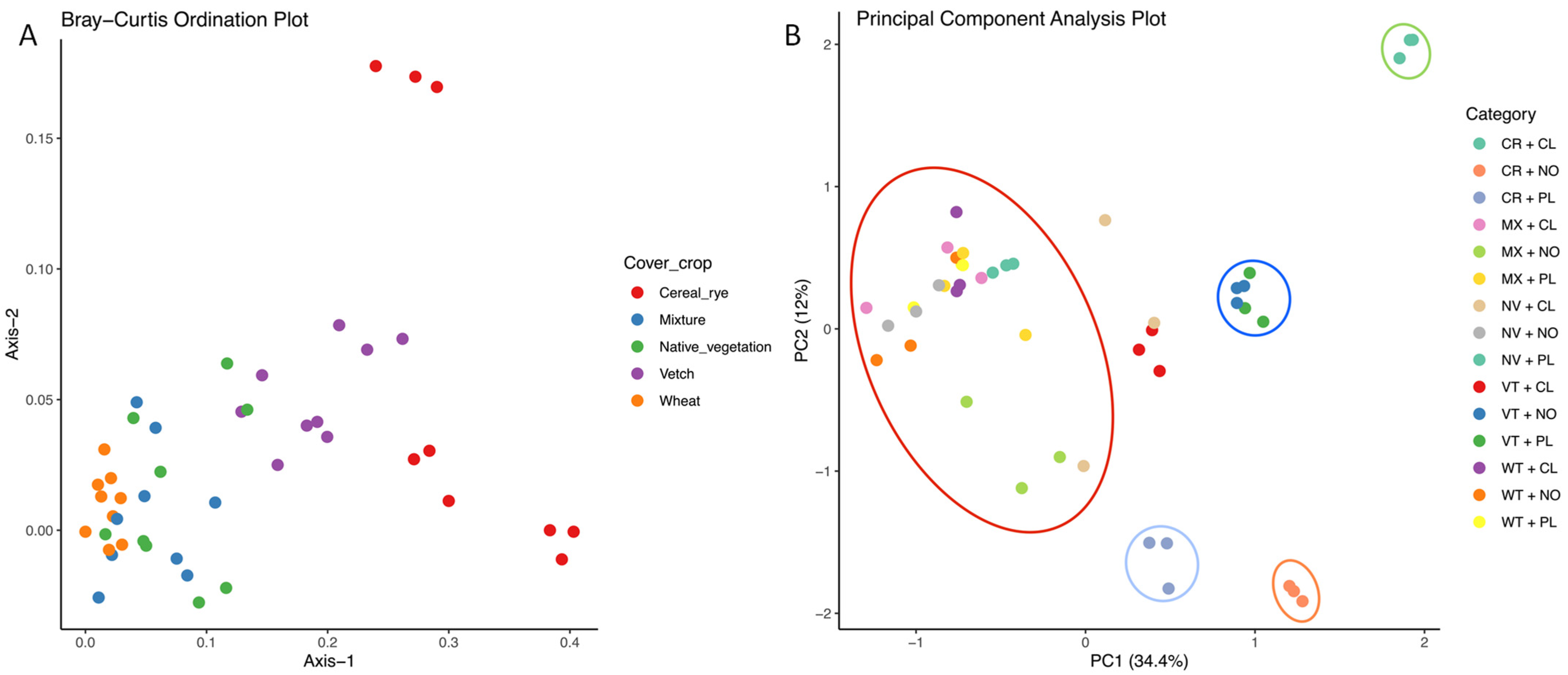
3.4. Analysis of Carbon Substrate Utilization by Soil Microbial Communities Using Principal Component Analysis (PCA)
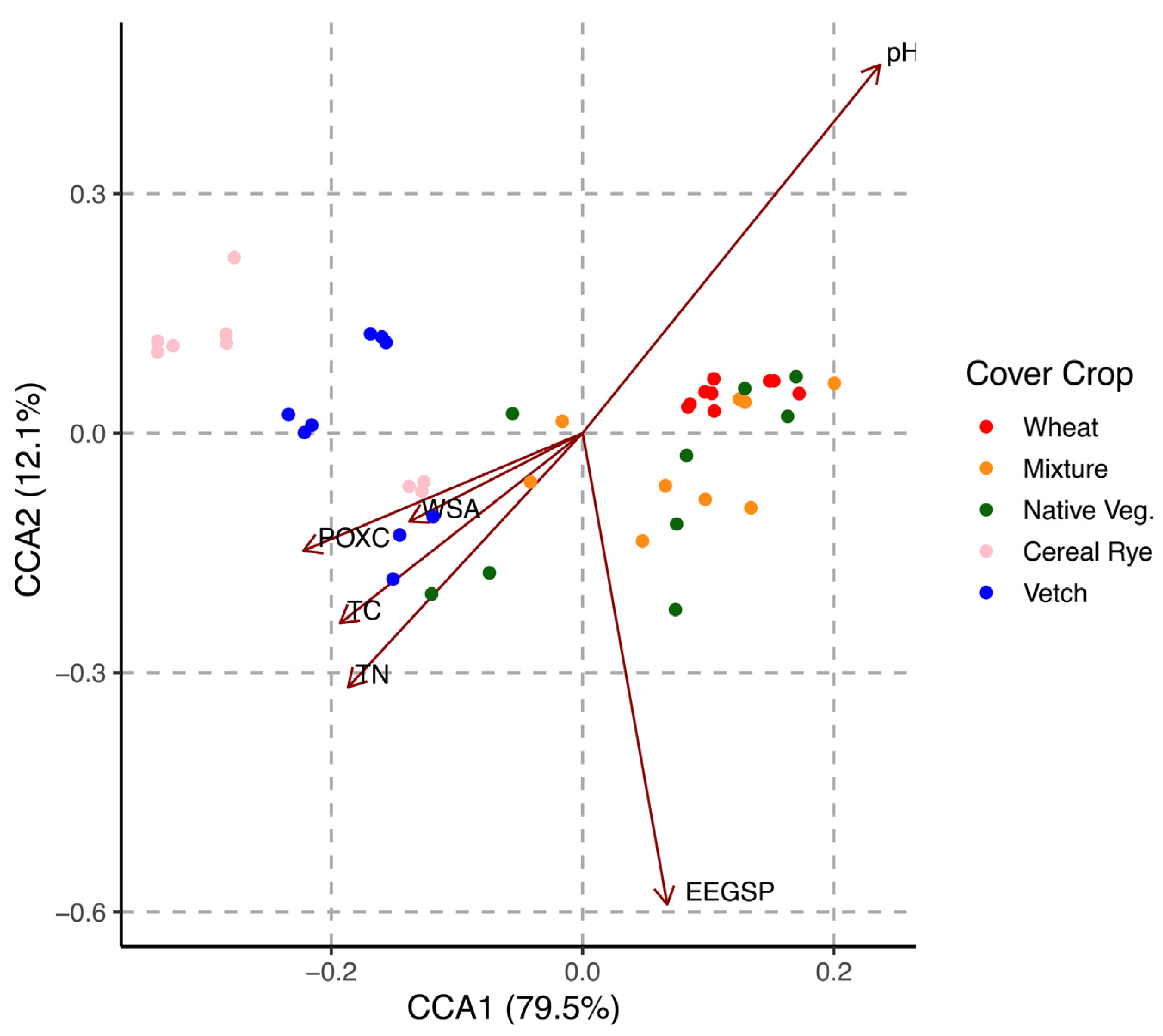
3.5. Association between Soil Microbial Metabolic Diversity and Soil Physicochemical Characteristics
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doran, J.W. Soil Health and Global Sustainability: Translating Science into Practice. Agric. Ecosyst. Environ. 2002, 88, 119–127. [Google Scholar] [CrossRef]
- Bogužas, V.; Skinulienė, L.; Butkevičienė, L.M.; Steponavičienė, V.; Petrauskas, E.; Maršalkienė, N. The Effect of Monoculture, Crop Rotation Combinations, and Continuous Bare Fallow on Soil CO2 Emissions, Earthworms, and Productivity of Winter Rye after a 50-Year Period. Plants 2022, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Norris, C.E.; Congreves, K.A. Alternative Management Practices Improve Soil Health Indices in Intensive Vegetable Cropping Systems: A Review. Front. Environ. Sci. 2018, 6, 50. [Google Scholar] [CrossRef]
- Friedel, J.K.; Gabel, D.; Stahr, K. Nitrogen Pools and Turnover in Arable Soils under Different Durations of Organic Farming: II: Source-and-Sink Function of the Soil Microbial Biomass or Competition with Growing Plants? J. Plant. Nutr. Soil Sci. 2001, 164, 421–429. [Google Scholar] [CrossRef]
- Lüneberg, K.; Schneider, D.; Siebe, C.; Daniel, R. Drylands Soil Bacterial Community Is Affected by Land Use Change and Different Irrigation Practices in the Mezquital Valley, Mexico. Sci. Rep. 2018, 8, 1413. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, K.; Makaju, S.; Ibrahim, R.; Missaoui, A. Current Progress in Nitrogen Fixing Plants and Microbiome Research. Plants 2020, 9, 97. [Google Scholar] [CrossRef]
- Six, J.; Frey, S.D.; Thiet, R.K.; Batten, K.M. Bacterial and Fungal Contributions to Carbon Sequestration in Agroecosystems. Soil Sci. Soc. Am. J. 2006, 70, 555–569. [Google Scholar] [CrossRef]
- Enebe, M.C.; Babalola, O.O. The Impact of Microbes in the Orchestration of Plants’ Resistance to Biotic Stress: A Disease Management Approach. Appl. Microbiol. Biotechnol. 2019, 103, 9–25. [Google Scholar] [CrossRef]
- Selvakumar, G.; Panneerselvam, P.; Ganeshamurthy, A.N. Bacterial Mediated Alleviation of Abiotic Stress in Crops. In Bacteria in Agrobiology: Stress Management; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 205–224. ISBN 978-3-642-23465-1. [Google Scholar]
- Dubey, A.; Malla, M.A.; Khan, F.; Chowdhary, K.; Yadav, S.; Kumar, A.; Sharma, S.; Khare, P.K.; Khan, M.L. Soil Microbiome: A Key Player for Conservation of Soil Health under Changing Climate. Biodivers Conserv. 2019, 28, 2405–2429. [Google Scholar] [CrossRef]
- Tsiafouli, M.A.; Thébault, E.; Sgardelis, S.P.; de Ruiter, P.C.; van der Putten, W.H.; Birkhofer, K.; Hemerik, L.; de Vries, F.T.; Bardgett, R.D.; Brady, M.V.; et al. Intensive Agriculture Reduces Soil Biodiversity across Europe. Glob. Chang. Biol. 2015, 21, 973–985. [Google Scholar] [CrossRef]
- Fierer, N.; Strickland, M.S.; Liptzin, D.; Bradford, M.A.; Cleveland, C.C. Global Patterns in Belowground Communities. Ecol. Lett. 2009, 12, 1238–1249. [Google Scholar] [CrossRef]
- Wagg, C.; Bender, S.F.; Widmer, F.; van der Heijden, M.G.A. Soil Biodiversity and Soil Community Composition Determine Ecosystem Multifunctionality. Proc. Natl. Acad. Sci. USA 2014, 111, 5266–5270. [Google Scholar] [CrossRef]
- Baumann, K.; Marschner, P.; Smernik, R.J.; Baldock, J.A. Residue Chemistry and Microbial Community Structure during Decomposition of Eucalypt, Wheat and Vetch Residues. Soil Biol. Biochem. 2009, 41, 1966–1975. [Google Scholar] [CrossRef]
- Hättenschwiler, S.; Tiunov, A.V.; Scheu, S. Biodiversity and Litter Decomposition in Terrestrial Ecosystems. Annu. Rev. Ecol. Evol. Syst. 2005, 36, 191–218. [Google Scholar] [CrossRef]
- Kodadinne Narayana, N.; Kingery, W.L.; Shankle, M.W.; Ganapathi Shanmugam, S. Differential Response of Soil Microbial Diversity and Community Composition Influenced by Cover Crops and Fertilizer Treatments in a Dryland Soybean Production System. Agronomy 2022, 12, 618. [Google Scholar] [CrossRef]
- Fageria, N.K.; Baligar, V.C.; Bailey, B.A. Role of Cover Crops in Improving Soil and Row Crop Productivity. Commun. Soil Sci. Plant Anal. 2005, 36, 2733–2757. [Google Scholar] [CrossRef]
- Balota, E.; Calegari, A.; Nakatani, A.; Coyne, M. Benefits of Winter Cover Crops and No-Tillage for Microbial Parameters in a Brazilian Oxisol: A Long-Term Study. Agri. Ecosyst. Environ 2014, 197, 31–40. [Google Scholar] [CrossRef]
- Somenahally, A.; DuPont, J.I.; Brady, J.; McLawrence, J.; Northup, B.; Gowda, P. Microbial Communities in Soil Profile Are More Responsive to Legacy Effects of Wheat-Cover Crop Rotations than Tillage Systems. Soil Biol. Biochem. 2018, 123, 126–135. [Google Scholar] [CrossRef]
- Kaspar, T.C.; Jaynes, D.B.; Parkin, T.B.; Moorman, T.B.; Singer, J.W. Effectiveness of Oat and Rye Cover Crops in Reducing Nitrate Losses in Drainage Water. Agric. Water Manag. 2012, 110, 25–33. [Google Scholar] [CrossRef]
- Haramoto, E.R.; Gallandt, E.R. Brassica Cover Cropping for Weed Management: A Review. Renew. Agric. Food Syst. 2004, 19, 187–198. [Google Scholar] [CrossRef]
- Kruger, D.H.M.; Fourie, J.C.; Malan, A.P. Cover Crops with Biofumigation Properties for the Suppression of Plant-Parasitic Nematodes: A Review. S. Afr. J. Enol. Vi. 2013, 34, 287–295. [Google Scholar] [CrossRef]
- Xu, W.; Ge, Z.; Poudel, D.R. Application and Optimization of Biolog EcoPlates in Functional Diversity Studies of Soil Microbial Communities. MATEC Web Conf. 2015, 22, 04015. [Google Scholar] [CrossRef]
- Lundquist, E.J.; Jackson, L.E.; Scow, K.M.; Hsu, C. Changes in Microbial Biomass and Community Composition, and Soil Carbon and Nitrogen Pools after Incorporation of Rye into Three California Agricultural Soils. Soil Biol. Biochem. 1999, 31, 221–236. [Google Scholar] [CrossRef]
- Abdul-Baki, A.A.; Teasdale, J.R.; Korcak, R.; Chitwood, D.J.; Huettel, R.N. Fresh-Market Tomato Production in a Low-Input Alternative System Using Cover-Crop Mulch. HortScience 1996, 31, 65–69. [Google Scholar] [CrossRef]
- Carrera, L.M.; Buyer, J.S.; Vinyard, B.; Abdul-Baki, A.A.; Sikora, L.J.; Teasdale, J.R. Effects of Cover Crops, Compost, and Manure Amendments on Soil Microbial Community Structure in Tomato Production Systems. Appl. Soil Ecol. 2007, 37, 247–255. [Google Scholar] [CrossRef]
- Buyer, J.S.; Teasdale, J.R.; Roberts, D.P.; Zasada, I.A.; Maul, J.E. Factors Affecting Soil Microbial Community Structure in Tomato Cropping Systems. Soil Biol. Biochem. 2010, 42, 831–841. [Google Scholar] [CrossRef]
- Nair, A.; Ngouajio, M. Soil Microbial Biomass, Functional Microbial Diversity, and Nematode Community Structure as Affected by Cover Crops and Compost in an Organic Vegetable Production System. Appl. Soil Ecol. 2012, 58, 45–55. [Google Scholar] [CrossRef]
- Chu, M.; Jagadamma, S.; Walker, F.R.; Eash, N.S.; Buschermohle, M.J.; Duncan, L.A. Effect of Multispecies Cover Crop Mixture on Soil Properties and Crop Yield. Agric. Environ. Lett. 2017, 2, 170030. [Google Scholar] [CrossRef]
- Sytske, M.D.; Rutgers, M.; Wouterse, M.; Wietse de Boer, P.; Bodelier, L.E. Decomposition of Mixtures of Cover Crop Residues Increases Microbial Functional Diversity. Geoderma 2020, 361, 114060. [Google Scholar] [CrossRef]
- Redin, M.; Recous, S.; Aita, C.; Dietrich, G.; Skolaude, A.C.; Ludke, W.H.; Schmatz, R.; Giacomini, S.J. How the Chemical Composition and Heterogeneity of Crop Residue Mixtures Decomposing at the Soil Surface Affects C and N Mineralization. Soil Biol. Biochem. 2014, 78, 65–75. [Google Scholar] [CrossRef]
- Chavarría, D.N.; Verdenelli, R.A.; Serri, D.L.; Restovich, S.B.; Andriulo, A.E.; Meriles, J.M.; Vargas-Gil, S. Effect of Cover Crops on Microbial Community Structure and Related Enzyme Activities and Macronutrient Availability. Eur. J. Soil Biol. 2016, 76, 74–82. [Google Scholar] [CrossRef]
- Housman, M.; Tallman, S.; Jones, C.; Miller, P.; Zabinski, C. Soil Biological Response to Multi-Species Cover Crops in the Northern Great Plains. Agric. Ecosyst. Environ. 2021, 313, 107373. [Google Scholar] [CrossRef]
- Bowles, T.M.; Acosta-Martínez, V.; Calderón, F.; Jackson, L.E. Soil Enzyme Activities, Microbial Communities, and Carbon and Nitrogen Availability in Organic Agroecosystems across an Intensively-Managed Agricultural Landscape. Soil Biol. Biochem. 2014, 68, 252–262. [Google Scholar] [CrossRef]
- Bundy, J.G.; Davey, M.P.; Viant, M.R. Environmental Metabolomics: A Critical Review and Future Perspectives. Metabolomics 2009, 5, 3–21. [Google Scholar] [CrossRef]
- Prosser, J.I. Molecular and Functional Diversity in Soil Micro-Organisms. In Diversity and Integration in Mycorrhizas; Smith, S.E., Smith, F.A., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 9–17. ISBN 978-90-481-5933-8. [Google Scholar]
- Garland, J.L.; Mills, A.L. Classification and Characterization of Heterotrophic Microbial Communities on the Basis of Patterns of Community-Level Sole-Carbon-Source Utilization. Appl. Environ. Microbiol. 1991, 57, 2351–2359. [Google Scholar] [CrossRef]
- Miura, T.; Makoto, K.; Niwa, S.; Kaneko, N.; Sakamoto, K. Comparison of Fatty Acid Methyl Ester Methods for Characterization of Microbial Communities in Forest and Arable Soil: Phospholipid Fraction (PLFA) versus Total Ester Linked Fatty Acids (EL-FAME). Pedobiologia 2017, 63, 14–18. [Google Scholar] [CrossRef]
- Kim, J.-W.; Rehmann, L.; Ray, M.B. Development of Microalgal Bioassay Based on the Community Level Physiological Profiling (CLPP). Algal Res. 2017, 25, 47–53. [Google Scholar] [CrossRef]
- Ge, Z.; Du, H.; Gao, Y.; Qiu, W. Analysis on Metabolic Functions of Stored Rice Microbial Communities by BIOLOG ECO Microplates. Front. Microbiol. 2018, 9, 1375. [Google Scholar] [CrossRef]
- Sofo, A.; Ricciuti, P. A Standardized Method for Estimating the Functional Diversity of Soil Bacterial Community by Biolog® EcoPlatesTM Assay—The Case Study of a Sustainable Olive Orchard. Appl. Sci. 2019, 9, 4035. [Google Scholar] [CrossRef]
- Preston-Mafham, J.; Boddy, L.; Randerson, P.F. Analysis of Microbial Community Functional Diversity Using Sole-Carbon-Source Utilisation Profiles—A Critique. FEMS Microbiol. Ecol. 2002, 42, 1–14. [Google Scholar] [CrossRef]
- Gresshoff, P.M.; Hayashi, S.; Biswas, B.; Mirzaei, S.; Indrasumunar, A.; Reid, D.; Samuel, S.; Tollenaere, A.; van Hameren, B.; Hastwell, A.; et al. The Value of Biodiversity in Legume Symbiotic Nitrogen Fixation and Nodulation for Biofuel and Food Production. J. Plant Physiol. 2015, 172, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Crop Rotations and Nitrogen Fertilization to Manage Soil Organic Carbon Dynamics—Studdert—2000—Soil Science Society of America Journal—Wiley Online Library. Available online: https://acsess.onlinelibrary.wiley.com/doi/full/10.2136/sssaj2000.6441496x?casa_token=1K9nk6ndXnMAAAAA%3Ah-zsB8QZf6qSdNskQBnm21ylLKSzeoTDor686VzO4EwZS6cHQX78UbjNq95WtJli_P927aW69lnldo (accessed on 24 November 2022).
- Fernandez-Gnecco, G.; Smalla, K.; Maccario, L.; Sørensen, S.J.; Barbieri, P.; Consolo, V.F.; Covacevich, F.; Babin, D. Microbial Community Analysis of Soils under Different Soybean Cropping Regimes in the Argentinean South-Eastern Humid Pampas. FEMS Microb. Ecol. 2021, 97, fiab007. [Google Scholar] [CrossRef] [PubMed]
- McDaniel, M.D.; Grandy, A.S. Soil Microbial Biomass and Function Are Altered by 12 Years of Crop Rotation. SOIL 2016, 2, 583–599. [Google Scholar] [CrossRef]
- Venter, Z.S.; Jacobs, K.; Hawkins, H.-J. The Impact of Crop Rotation on Soil Microbial Diversity: A Meta-Analysis. Pedobiologia 2016, 59, 215–223. [Google Scholar] [CrossRef]
- Tamburini, G.; Bommarco, R.; Wanger, T.C.; Kremen, C.; van der Heijden, M.G.A.; Liebman, M.; Hallin, S. Agricultural Diversification Promotes Multiple Ecosystem Services without Compromising Yield. Sci. Adv. 2020, 6, eaba1715. [Google Scholar] [CrossRef] [PubMed]
- IPS—Record of Climatological Observations—Select Month|IPS|National Climatic Data Center (NCDC). Available online: https://www.ncdc.noaa.gov/IPS/coop/coop.html?_page=2&state=MS&foreign=false&stationID=227111&_target3=Next+%3E (accessed on 18 April 2023).
- Pokhrel, S.; Kingery, W.L.; Cox, M.S.; Shankle, M.W.; Shanmugam, S.G. Impact of Cover Crops and Poultry Litter on Selected Soil Properties and Yield in Dryland Soybean Production. Agronomy 2021, 11, 119. [Google Scholar] [CrossRef]
- Zak, J.C.; Willig, M.R.; Moorhead, D.L.; Wildman, H.G. Functional Diversity of Microbial Communities: A Quantitative Approach. Soil Biol. Biochem. 1994, 26, 1101–1108. [Google Scholar] [CrossRef]
- Simpson’s Diversity Index. Available online: https://geographyfieldwork.com/Simpson%27sDiversityIndex.htm (accessed on 25 November 2022).
- Lan, X.; Du, H.; Peng, W.; Liu, Y.; Fang, Z.; Song, T. Functional Diversity of the Soil Culturable Microbial Community in Eucalyptus Plantations of Different Ages in Guangxi, South China. Forests 2019, 10, 1083. [Google Scholar] [CrossRef]
- Zhen, T.; Fan, W.; Wang, H.; Cao, X.; Xu, X. Monitoring Soil Microorganisms with Community-Level Physiological Profiles Using Biolog EcoPlatesTM in Chaohu Lakeside Wetland, East China. Eurasian Soil Sc. 2020, 53, 1142–1153. [Google Scholar] [CrossRef]
- YaNan, T.; HongQi, W. Application of biolog to study of environmental microbial function diversity. Environ. Sci. Technol. 2011, 34, 50–57. [Google Scholar]
- Németh, I.; Molnár, S.; Vaszita, E.; Molnár, M. The Biolog EcoPlateTM Technique for Assessing the Effect of Metal Oxide Nanoparticles on Freshwater Microbial Communities. Nanomaterials 2021, 11, 1777. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis, 2nd ed; Use R! Springer: Cham, Switzerland, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Anderson, M.J.; Walsh, D.C.I. PERMANOVA, ANOSIM, and the Mantel Test in the Face of Heterogeneous Dispersions: What Null Hypothesis Are You Testing? Ecol. Monogr. 2013, 83, 557–574. [Google Scholar] [CrossRef]
- Ter Braak, C.J.F. The Analysis of Vegetation-Environment Relationships by Canonical Correspondence Analysis. Vegetatio 1987, 69, 69–77. [Google Scholar] [CrossRef]
- Oksanen, J. Vegan: Community Ecology Package. 2010. Available online: http://CRAN.R-project.org/package=vegan (accessed on 6 June 2023).
- Choi, K.-H.; Dobbs, F.C. Comparison of Two Kinds of Biolog Microplates (GN and ECO) in Their Ability to Distinguish among Aquatic Microbial Communities. J. Microbiol. Methods 1999, 36, 203–213. [Google Scholar] [CrossRef]
- Wang, Y.; Ouyang, Z.; Zheng, H.; Wang, X.; Chen, F.; Zeng, J. Carbon Metabolism of Soil Microbial Communities of Restored Forests in Southern China. J. Soils Sediments 2011, 11, 789–799. [Google Scholar] [CrossRef]
- Wortman, S.E.; Francis, C.A.; Bernards, M.L.; Drijber, R.A.; Lindquist, J.L. Optimizing Cover Crop Benefits with Diverse Mixtures and an Alternative Termination Method. Agron. J. 2012, 104, 1425–1435. [Google Scholar] [CrossRef]
- Garbeva, P.; van Veen, J.A.; van Elsas, J.D. Microbial Diversity in Soil: Selection Microbial Populations by Plant and Soil Type and Implications for Disease Suppressiveness. Annu. Rev. Phytopathol. 2004, 42, 243–270. [Google Scholar] [CrossRef]
- Perucci, P. Enzyme Activity and Microbial Biomass in a Field Soil Amended with Municipal Refuse. Biol. Fertil. Soils 1992, 14, 54–60. [Google Scholar] [CrossRef]
- Marinari, S.; Masciandaro, G.; Ceccanti, B.; Grego, S. Influence of Organic and Mineral Fertilisers on Soil Biological and Physical Properties. Bioresour. Technol. 2000, 72, 9–17. [Google Scholar] [CrossRef]
- Bulluck, L.R.; Brosius, M.; Evanylo, G.K.; Ristaino, J.B. Organic and Synthetic Fertility Amendments Influence Soil Microbial, Physical and Chemical Properties on Organic and Conventional Farms. Appl. Soil Ecol. 2002, 19, 147–160. [Google Scholar] [CrossRef]
- Naidu, R. Influence of Lime, Fertilizer and Manure Applications on Soil Organic Matter Content and Soil Physical Conditions: A Review. Nutr. Cycl. Agroecosyst. 1998, 51, 123–137. [Google Scholar]
- Melero, S.; Porras, J.C.R.; Herencia, J.F.; Madejon, E. Chemical and Biochemical Properties in a Silty Loam Soil under Conventional and Organic Management. Soil Tillage Res. 2006, 90, 162–170. [Google Scholar] [CrossRef]
- Marschner, H. Ion Uptake Mechanisms of Individual Cells and Roots. In Mineral Nutrition of Higher Plants; Elsevier: Amsterdam, The Netherlands, 1995; pp. 6–78. ISBN 978-0-12-473542-2. [Google Scholar]
- Nivelle, E.; Verzeaux, J.; Habbib, H.; Kuzyakov, Y.; Decocq, G.; Roger, D.; Lacoux, J.; Duclercq, J.; Spicher, F.; Nava-Saucedo, J.-E.; et al. Functional Response of Soil Microbial Communities to Tillage, Cover Crops and Nitrogen Fertilization. Appl. Soil Ecol. 2016, 108, 147–155. [Google Scholar] [CrossRef]
- Wu, Y.; Lyu, T.; Yue, B.; Tonoli, E.; Verderio, E.A.M.; Ma, Y.; Pan, G. Enhancement of Tomato Plant Growth and Productivity in Organic Farming by Agri-Nanotechnology Using Nanobubble Oxygation. J. Agric. Food Chem. 2019, 67, 10823–10831. [Google Scholar] [CrossRef] [PubMed]
- Nunan, N.; Lerch, T.Z.; Pouteau, V.; Mora, P.; Changey, F.; Kätterer, T.; Giusti-Miller, S.; Herrmann, A.M. Metabolising Old Soil Carbon: Simply a Matter of Simple Organic Matter? Soil Biol. Biochem. 2015, 88, 128–136. [Google Scholar] [CrossRef]
- Grandy, A.S.; Neff, J.C. Molecular C Dynamics Downstream: The Biochemical Decomposition Sequence and Its Impact on Soil Organic Matter Structure and Function. Sci. Total Environ. 2008, 404, 297–307. [Google Scholar] [CrossRef]
- Chen, Y.-H.; Liu, X.-W.; Huang, J.-L.; Baloch, S.; Xu, X.; Pei, X.-F. Microbial Diversity and Chemical Analysis of Shuidouchi, Traditional Chinese Fermented Soybean. Food Res. Int. 2019, 116, 1289–1297. [Google Scholar] [CrossRef]
- Eller, M.H.; Warner, A.L.; Knap, H.T. Genomic Organization and Expression Analyses of Putrescine Pathway Genes in Soybean. Plant Physiol. Biochem. 2006, 44, 49–57. [Google Scholar] [CrossRef]
- Guanghua, W.; Junjie, L.; Xiaoning, Q.; Jian, J.; Yang, W.; Xiaobing, L. Effects of Fertilization on Bacterial Community Structure and Function in a Black Soil of Dehui Region Estimated by Biolog and PCR-DGGE Methods. Acta Ecol. Sin. 2008, 28, 220–226. [Google Scholar] [CrossRef]
- Gomez, E.; Ferreras, L.; Toresani, S. Soil Bacterial Functional Diversity as Influenced by Organic Amendment Application. Bioresour. Technol. 2006, 97, 1484–1489. [Google Scholar] [CrossRef]
- Bucher, A.E.; Lanyon, L.E. Evaluating Soil Management with Microbial Community-Level Physiological Profiles. Appl. Soil Ecol. 2005, 29, 59–71. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The Rhizosphere Microbiome and Plant Health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Kanasugi, M.; Sarkodee-Addo, E.; Ansong Omari, R.; Mohammad Golam Dastogeer, K.; Fujii, Y.; Oppong Abebrese, S.; Bam, R.; Asuming-Brempong, S.; Okazaki, S. Exploring Rice Root Microbiome; The Variation, Specialization and Interaction of Bacteria and Fungi In Six Tropic Savanna Regions in Ghana. Sustainability 2020, 12, 5835. [Google Scholar] [CrossRef]
- Tawaraya, K.; Horie, R.; Shinano, T.; Wagatsuma, T.; Saito, K.; Oikawa, A. Metabolite Profiling of Soybean Root Exudates under Phosphorus Deficiency. Soil Sci. Plant Nutr. 2014, 60, 679–694. [Google Scholar] [CrossRef]
- Timotiwu, P.B.; Sakurai, N. Identification of Mono-, Oligo-, and Polysaccharides Secreted from Soybean Roots. J. Plant Res. 2002, 115, 0077–0085. [Google Scholar] [CrossRef] [PubMed]

| AWCD1 | H’ | E | U | R | 1/D | |
| Cover Crop | ||||||
| Cereal rye | 0.76 (0.04) d | 3.14 (0.02) d | 0.91 (0.006) d | 5.15 (0.27) d | 16.55 (1.00) d | 67.79 (3.60) |
| CC-mix 2 | 1.32 (0.04) a | 3.31 (0.009) a | 0.96 (0.002) a | 8.03 (0.26) a | 25.6 (0.37) a | 71.88 (3.70) |
| Native vegetation | 1.34 (0.07) a | 3.29 (0.016) a | 0.96 (0.004) a | 8.25 (0.40) a | 25.33 (0.78) a | 66.30 (2.72) |
| Vetch | 1.05 (0.03) b | 3.24 (0.004) b | 0.94 (0.001) b | 6.77 (0.19) b | 21.44 (0.92) b | 84.41 (7.46) |
| Wheat | 0.95 (0.07) c | 3.20 (0.01) c | 0.93 (0.003) c | 6.23 (0.44) c | 19.88 (1.67) c | 84.00 (8.83) |
| CC*FT 3 | s* | s* | s* | s* | s* | ns |
| Fertilizer Source | ||||||
| CL | 1.11 (0.054) a | 3.21 (0.019) b | 0.93 (0.005) b | 7.13 (0.26) a | 22.46 (0.58) a | 72.02 (2.12) |
| None | 0.99 (0.07) b | 3.22 (0.024) b | 0.94 (0.007) b | 6.32 (0.39) b | 20.46 (1.54) b | 77.67 (6.10) |
| PL | 1.15 (0.08) a | 3.27 (0.011) a | 0.95 (0.003) a | 7.21 (0.47) a | 22.4 (1.21) a | 74.97 (5.53) |
| Treatment 2 | AWCD1 | H’ | E | U | R |
|---|---|---|---|---|---|
| MX +PL | 1.57 (0.036) a | 3.33 (0.018) a | 0.97 (0.0032) a | 9.47 (0.28) a | 27 (0.83) a |
| MX + CL | 1.47 (0.038) ab | 3.32 (0.012) a | 0.967 (0.0032) a | 8.87 (0.29) ab | 26.33 (1.03) ab |
| NV + NO | 1.34 (0.047) bc | 3.32 (0.0134) a | 0.968 (0.0023) a | 8.09 (0.28) bc | 26 (0.93) ab |
| NV + PL | 1.32 (0.074) bc | 3.31 (0.0324) ab | 0.964 (0.0093) ab | 8.05 (0.23) bc | 26.33 (1.22) ab |
| WT + PL | 1.23 (0.048) cd | 3.26 (0.0284) bcd | 0.95 (0.0042) bcd | 7.76 (0.31) c | 25 (0.73) abc |
| MX + NO | 1.17 (0.083) cd | 3.30 (0.0173) abc | 0.963 (0.0024) abc | 7.17 (0.32) cd | 24.67 (0.69) abc |
| NV + CL | 1.12 (0.037) de | 3.23 (0.0182) d | 0.943 (0.0043) d | 7.18 (0.35) cd | 22.67 (0.73) cde |
| VT + NO | 1.12 (0.028) de | 3.24 (0.0284) cd | 0.943 (0.0032) cd | 7.15 (0.29) cd | 23.67 (0.83) bcd |
| VT + CL | 1.10 (0.047) de | 3.24 (0.0482) cd | 0.944 (0.0024) cd | 7.12 (0.23) cd | 22.67 (0.93) cde |
| WT + CL | 0.94 (0.058) ef | 3.17 (0.0284) ef | 0.923 (0.0032) ef | 6.26 (0.32) de | 21 (1.02) de |
| VT + PL | 0.93 (0.057) ef | 3.24 (0.0138) cd | 0.944 (0.0023) cd | 6.04 (0.29) e | 18 (1.10) fg |
| CR + CL | 0.91 (0.075) fg | 3.11 (0.0324) fg | 0.905 (0.0024) fg | 6.22 (0.28) de | 20 (1.12) ef |
| CR + PL | 0.73 (0.036) gh | 3.23 (0.0231) de | 0.940 (0.043) de | 4.76 (0.23) f | 16.33 (1.11) gh |
| WT + NO | 0.70 (0.037) h | 3.19 (0.2842) de | 0.931 (0.0024) de | 4.69 (0.24) f | 13.67 (1.03) hi |
| CR + NO | 0.63 (0.048) h | 3.07 (0.0124) g | 0.894 (0.0036) g | 4.47 (0.25) f | 13.33 (1.20) i |
| Carbon Substrate | PC1 | PC2 |
|---|---|---|
| Polymers | ||
| Tween 40 | 0.96179268 | −0.03904354 |
| Tween 80 | 0.34211498 | 0.03092762 |
| Alpha-Cyclodextrin | −0.83462371 | −0.41750008 |
| Glycogen | −0.63170406 | −0.23706666 |
| Carbohydrates | ||
| Glucose-1-Phosphate | −0.25453698 | 0.58790260 |
| D-L-Alpha-Glycerol Phosphate | 0.19979821 | −0.74993661 |
| D-Cellubiose | −0.45806275 | 0.15852001 |
| Alpha-D-Lactose | −0.50881668 | 0.77378681 |
| Beta-Methyl-D-Glucoside | −0.70499935 | 0.02685358 |
| D-Xylose | −0.74269362 | 0.35992147 |
| I-Erythritol | −0.56968692 | 0.59688337 |
| D-Mannitol | 0.85824297 | −0.38592539 |
| N-Acetyl-D-Glucosamine | 0.58653954 | −0.02068943 |
| Carboxylic acids | ||
| Pyruvic Acid Methyl Ester | 0.42378445 | −0.26569367 |
| D-Glucosaminic acid | 0.52295150 | −0.39975288 |
| D-Galactonic Acid-Gamma-Lactone | 0.63099447 | −0.06814916 |
| D-Galacturonic acid | 0.92169910 | −0.40273613 |
| Gamma-Amino-Butyric Acid | 0.75517974 | −0.05258175 |
| Itaconic Acid | −0.23717004 | −0.45819712 |
| Beta-Keto Butyric Acid | −0.65735782 | −0.34116388 |
| D-Malic Acid | 0.93858412 | −0.04106865 |
| Amino acids | ||
| L-Arginine | 0.60169520 | 0.39097528 |
| L-Asparagine | 1.00149965 | 0.01168636 |
| L-Phenylalanine | −0.58543371 | −0.02388333 |
| L-Serine | 0.08844583 | 0.36381245 |
| L-Threonine | −0.62958879 | −0.42939563 |
| Glycyl-L-Glutamic Acid | −0.78420635 | −0.36112807 |
| Amines | ||
| Phenythyl-Amine | −0.52359729 | 0.15857889 |
| Putrescine | 0.84357073 | 0.29691745 |
| Phenolic acids | ||
| 2-Hydroxy Benzonic Acid | −0.18237857 | −0.16476193 |
| 4-Hydroxy Benzonic Acid | 0.23406065 | 0.47218892 |
| TC 1 (%) | TN (%) | WSA (%) | EEGSP (mg/kg) | POXC (mg/kg) | pH | |
|---|---|---|---|---|---|---|
| Cover crop | ||||||
| Cereal rye | 1.66 (0.104) | 0.160 (0.006) ab | 52.11 (2.67) | 82.55 (2.86) | 550.77 (21.39) | 5.64 (0.12) ab |
| CC-mix 2 | 1.70 (0.07) | 0.173 (0.009) ab | 49.11 (2.38) | 89.88 (3.42) | 569.33 (28.61) | 5.56 (0.08) ab |
| Native vegetation | 1.58 (0.08) | 0.156 (0.006) ab | 56.77 (2.89) | 85.77 (2.43) | 521.11 (18.94) | 5.75 (0.09) a |
| Vetch | 1.72 (0.10) | 0.178 (0.01) a | 55 (3.08) | 83.11 (3.34) | 561 (40.02) | 5.48 (0.07) b |
| Wheat | 1.52 (0.07) | 0.153 (0.005) b | 46.44 (2.63) | 79.88 (0.44) | 503.22 (1.67) | 5.76 (0.06) a |
| CC*FT 3 | ns | ns | ns | ns | ns | ns |
| Fertilizer source | ||||||
| Mineral | 1.58 (0.06) | 0.15 (0.005) | 49.73 (2.12) | 81.2 (2.57) | 537.13 (22.53) | 5.48 (0.063) b |
| None | 1.60 (0.06) | 0.16 (0.006) | 52.2 (2.84) | 82.4 (2.52) | 530 (21.60) | 5.766 (0.08) a |
| Poultry litter | 1.72 (0.07) | 0.17 (0.007) | 53.73 (1.66) | 89.13 (2.20) | 555.46 (21.34) | 5.68 (0.04) a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chinthalapudi, D.P.M.; Pokhrel, S.; Kingery, W.L.; Shankle, M.W.; Ganapathi Shanmugam, S. Exploring the Synergistic Impacts of Cover Crops and Fertilization on Soil Microbial Metabolic Diversity in Dryland Soybean Production Systems Using Biolog EcoPlates. Appl. Biosci. 2023, 2, 328-346. https://doi.org/10.3390/applbiosci2030022
Chinthalapudi DPM, Pokhrel S, Kingery WL, Shankle MW, Ganapathi Shanmugam S. Exploring the Synergistic Impacts of Cover Crops and Fertilization on Soil Microbial Metabolic Diversity in Dryland Soybean Production Systems Using Biolog EcoPlates. Applied Biosciences. 2023; 2(3):328-346. https://doi.org/10.3390/applbiosci2030022
Chicago/Turabian StyleChinthalapudi, Durga P. M., Sapna Pokhrel, William L. Kingery, Mark W. Shankle, and Shankar Ganapathi Shanmugam. 2023. "Exploring the Synergistic Impacts of Cover Crops and Fertilization on Soil Microbial Metabolic Diversity in Dryland Soybean Production Systems Using Biolog EcoPlates" Applied Biosciences 2, no. 3: 328-346. https://doi.org/10.3390/applbiosci2030022
APA StyleChinthalapudi, D. P. M., Pokhrel, S., Kingery, W. L., Shankle, M. W., & Ganapathi Shanmugam, S. (2023). Exploring the Synergistic Impacts of Cover Crops and Fertilization on Soil Microbial Metabolic Diversity in Dryland Soybean Production Systems Using Biolog EcoPlates. Applied Biosciences, 2(3), 328-346. https://doi.org/10.3390/applbiosci2030022







