Abstract
The branched-chain amino acids (BCAAs) leucine, isoleucine and valine are synthesized via a common biosynthetic pathway. Ketol-acid reductoisomerase (KARI) is the second enzyme in this pathway. In addition to its role in BCAA biosynthesis, KARI catalyzes two rate-limiting steps that are key components of a cell-free biofuel biosynthesis route. For industrial applications, reaction temperature and enzyme stability are key factors that affect process robustness and product yield. Here, we have solved the cryo-EM structure (2.94 Å resolution) of a homododecameric Class I KARI (from Campylobacter jejuni) and demonstrated how a triad of amino acid side chains plays a crucial role in promoting the oligomerization of this enzyme. Importantly, both its thermal and solvent stability are greatly enhanced in the dodecameric state when compared to its dimeric counterpart (apparent melting temperatures (Tm) of 83.1 °C and 51.5 °C, respectively). We also employed protein design (PROSS) for a tetrameric Class II KARI (from Escherichia coli) to generate a variant with improved thermal and solvent stabilities. In total, 34 mutations were introduced, which did not affect the oligomeric state of this enzyme but resulted in a fully functional catalyst with a significantly elevated Tm (58.5 °C vs. 47.9 °C for the native version).
1. Introduction
The branched-chain amino acids’ (BCAAs) pathway for the biosynthesis of leucine, isoleucine and valine is present in bacteria, fungi and plants, but not in animals [1]. Consequently, this biosynthesis route has long been recognized as a target for herbicides and antimicrobial agents [2,3,4,5,6]. More recently, enzymes from this pathway attracted increasing interest as components of designed enzymatic cascades to convert renewable raw material into high-value products [7]. For example, in Escherichia coli, the BCAA pathway was re-engineered to produce higher alcohols, including isobutanol, 1-butanol, 2-methyl-1-butanol, 3-methyl-1-butanol and 2-phenylethanol from glucose [7]. These products are alternatives to current fossil fuels because they exhibit a higher energy density, lower hygroscopicity, lower vapor pressure and are compatible with existing fossil fuel infrastructures [8].
Recently, ab initio designed, cell-free biosynthesis routes were reported that facilitate the conversion of renewable sugar streams into the biofuel building blocks ethanol or isobutanol [9]. This approach is partly reliant on the use of the BCAA pathway enzymes, acetohydroxyacid synthase (AHAS), ketol-acid reductoisomerase (KARI) and dihydroxyacid dehydratase (DHAD) [10,11,12,13,14,15]. Compared to conventional fermentation approaches, cell-free strategies have several advantages. The cell-free production route enhances the resilience of the process towards accumulating reaction products, which are toxic to cellular production systems. Furthermore, reactions can be performed at higher temperatures, which can improve the rate and efficiency of production leading to higher product yields.
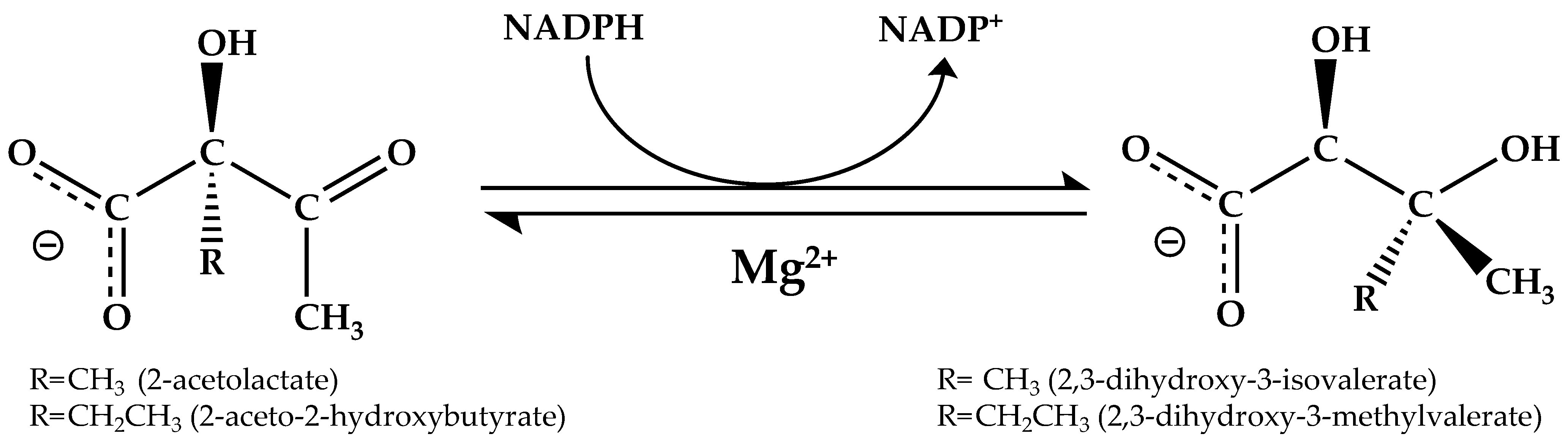
KARI is a bifunctional enzyme that converts 2-acetolactate into 2,3-dihydroxy-3-isovalerate (precursor of valine and leucine) or 2-aceto-2-hydroxybutyrate into 2,3-dihydroxy-3-methylvalerate (precursor of isoleucine) via a two-step reaction within a single active site [16,17,18,19]. The initial Mg2+-dependent alkyl group rearrangement is followed by a NAD(P)H-dependent reduction (Figure 1) [16,18].
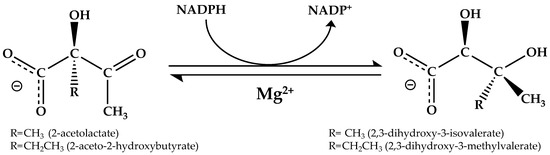
Figure 1.
Reaction catalyzed by KARI. NADPH is the commonly used cofactor, but some KARIs can use NADH as the preferred reductant. KARI catalyzes the concerted isomerization and reduction in the native substrate, 2-acetolactate.
KARIs can be divided into two classes according to the length of the polypeptide. Class I KARIs are ~330 amino acids in length, containing an N-terminal domain with a Rossmann-fold and a C-terminal domain that is largely α-helical [19]. Class I KARIs require the C-terminal domain from an adjacent subunit to complete the active site, forming a dimer (e.g., KARI from Mycobacterium tuberculosis (MtKARI) [20] or Staphylococcus aureus (SaKARI) [21]) or dodecamer (e.g., Pseudomonas aeruginosa KARI (PaKARI) [22] and Sulfolobus solfataricus KARI (SsKARI) [23]); the dodecamer assembly requires six interlocking dimeric units [22,24]; see below. Class II KARIs, exemplified by enzymes from E. coli (EcKARI) [5] and plants [25], are ~500 amino acids in length and contain an additional C-terminal domain that appears to have arisen as the result of a gene duplication event. With this extension, Class II KARIs can form a complete active site within a single subunit, a feature that could be favorable for cell-free chemical production strategies, as this may simplify attachment of the enzyme to a solid support.
The advantages of using enzymes as catalysts include their extremely high efficiency, exquisite substrate specificity, stereo-selectivity and potentially low environmental impact. Such properties have been widely exploited in applications that include biosensors, bioreactors and in enzymatic fuel cells [26,27,28,29]. However, compared with chemical catalysts, a major drawback of using enzymes in industry is their short half-life and low thermal stability [30]. Another major limitation inherent to KARIs is their low turnover number, which is the lowest amongst the enzymes in the BCAA pathway [9]. However, previous studies demonstrated that KARIs are amenable to modifications which may enhance their potential for industrial applications. For instance, the cofactor preference of Slackia exigua KARI was switched from NADPH to NADH (a more stable hydride donor) through targeted mutations [31]. More recently, we employed ancestral sequence reconstruction (ASR) to design an ancient KARI variant that both increased thermal stability and enhanced reactivity towards the substrate, 2-acetolactate [10].
In this study, we focused our attention on (i) investigating factors that contribute to the thermal stability of KARI and (ii) designing a variant with enhanced stability using the protein design algorithm, Protein Repair One Stop Shop (PROSS) [32]. Specifically, we demonstrate that the emergence of higher oligomeric forms of KARI may be a response for the need to have enhanced thermal stability. The cumulative data provide detailed molecular insight into factors that are essential to engineer KARI variants suitable for industrial applications.
2. Materials and Methods
2.1. Expression and Purification of the Enzymes and Their Mutants
Here, CjKARI is defined as the wild-type KARI from Campylobacter jejuni, and CjKARI_Dm is the K290A/L294A double mutant. The CjKARI and CjKARI_Dm genes were synthesized by Epoch Life Science Inc. The DNA fragments were inserted between the NdeI and XhoI sites and cloned into a pET-21a(+) vector. The final constructs contain the gene sequences of the enzyme and a -LEHHHHHH- sequence at the C-terminus for purification. The plasmids containing the enzyme were then transformed into E. coli BL21 (DE3) cells. These cells were then grown in LB medium in the presence of 100 mg/L ampicillin at 37 °C until the OD600 reached ~0.6. The expression of protein was induced by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG), followed by growth for 10 hours at 20 °C. Cells were centrifuged and the pellet resuspended in ice-cold lysis buffer (20 mM Tris-HCl pH 8.0, 20 mM imidazole, 250 mM MgCl2, 500 mM NaCl and 10% glycerol). The cells were homogenized by ultrasonication, followed by further centrifugation. The supernatant was collected for protein purification using a QIAGEN Ni-NTA-agarose resin column and by gel filtration chromatography with a Superdex 200 Hiload 26/60 column (GE Healthcare) in gel filtration buffer (20 mM Tris-HCl, pH 8.0, 250 mM MgCl2, 500 mM NaCl and 10% glycerol). The protein concentration was determined by measurement of A280. The molar extinction coefficient, 20,400 cm−1 M−1, was calculated using the method of Gill and von Hippel [33] by considering the theoretical molecular weight for the monomer and adding the two additional residues from the construct (-LE-) and the hexa-histidine tag (38032 Da for CjKARI and 37,933 Da for CjKARI_Dm). Purity was assessed by 12% SDS-PAGE. For long-term storage, the enzyme was kept at −70 °C in gel filtration buffer. The preparation of MtKARI and EcKARI (including its mutant) followed previously described protocols [5,20].
2.2. Preparation of the CjKARI for Cryo-EM Studies
Crosslinking was performed with the enzyme in 50 mM HEPES, pH 8.0, by incubating with 0.05% glutaraldehyde for 15 min at 25 °C. The reaction was terminated by adding ~65 mM Tris-HCl, pH 8.2. The crosslinked enzyme was repurified at 4 °C by gel filtration using a Superose 6 Increase 10/300 GL column (GE Healthcare). The buffer consisted of 50 mM NaCl and 20 mM Tris-HCl, pH 8.0. The concentration of the enzyme was ~1.1 mg/mL.
2.3. Cryo-Electron Microscopy Studies
The enzyme samples were applied to ultrathin carbon film supported by a holey carbon film on Quantifoil R1.2/1.3 300 mesh copper grids. Prior to sample application, the grids were glow-discharged using a Gatan Solarus 950 Advanced Plasma cleaning system (Gatan, Inc., Pleasanton, CA, USA). Dithiothreitol (DTT; 5 mM) was added to the protein, and 3 μL of the prepared sample were applied onto the grids. The grids were blotted for 3.5 s and then plunge-frozen in liquid ethane cooled with liquid nitrogen using a FEI Vitrobot Mark IV (FEI company, Hillsboro, OR, USA) operated at 10 °C and 100% humidity. A Titan Krios transmission electron microscope was used to image the grids under 300 kV, while maintaining the specimen at liquid nitrogen temperatures. Automated data acquisition was performed using SerialEM. Images for these samples were recorded using a Gatan, Inc. K3 direct detection camera at 29,000-fold magnification operated in super-resolution counting mode with a physical pixel size of 0.41 Å and binned to a pixel size of 0.82 Å. The defocus range was −1.2–−1.8 μm, and each exposure had an accumulated dose of ~50 e−/Å2 and a total of 40 frames per micrograph.
2.4. Cryo-EM Image Processing, Model Building, Refinement and Validation
A number of micrograph movies were collected, initial and final particles picked, and all the information related to data collection and refinement is summarized in Table 1. All processing steps were performed using CryoSPARC [34]. All the original images were aligned, and local motion correction was conducted by patching motion correction. Suitable particles were automatically selected through reference-free 2D classification and used for ab initio 3D reconstruction to generate 3D initial models as a reference for heterogeneous refinement. After heterogeneous refinement, final particles were selected for non-uniform refinement (Nu-refinement) to generate the final cryo-EM map. The final resolution was estimated according to a global-standard Fourier shell correlation cutoff of 0.5 [35]. Local resolution maps were also generated by CryoSPARC. The cryo-EM maps of CjKARI were used for model building. The maps were segmented into submaps where each contains a dimer (i.e., two monomers) using UCSF Chimera [36]. Then, all the residues in the dimer model were manually fitted into the 3D map density using Coot [37]. The fitted structure was further refined with real space refinement in PHENIX [38] to avoid over-fitting. All the figures were generated with UCSF Chimera and Pymol.

Table 1.
Cryo-EM data statistics.
2.5. Oligomeric States Analysis
Size-exclusion chromatography-multi-angle laser light scattering (SEC-MALS) was performed using an inline Superdex Increase 200 5/150 GL SEC column (GE Healthcare) combined with a Dawn Heleos II 18-angle light-scattering detector coupled with an Optilab TrEX refractive index detector (Wyatt Technology, Santa Barbara, CA, USA). A total of 100 μg of purified CjKARI and CjKARI_Dm were eluted at 0.25 ml/min in 10 mM Tris-HCl, pH 8.0, 250 mM MgCl2 and 500 mM NaCl at room temperature. The molecular mass calculations were performed using Astra6.1 software (Wyatt Technology). The input of the refractive increment (dn/dc values) was fixed at 0.186 mL/g, with an assumption that dn/dc is invariable for unmodified proteins. The molecular mass was determined across the protein elution peak.
2.6. Enzyme Kinetics and Thermal Stability Studies
To remove glycerol from the stored enzyme samples, the enzymes were loaded onto a PD-10 column using a buffer containing 20 mM Tris-HCl, pH 8.0, 250 mM MgCl2 and 500 mM NaCl. The enzyme concentrations were measured using Direct Detect. The activity of the enzymes was monitored by measuring the conversion of NADPH to NADP+ at 340 nm using 2-acetolactate or 3-hydroxypyruvate as the substrate. In a standard assay, the enzyme activity was measured at 25 °C 100 mM Tris-HCl pH 8.0, 4 mM MgCl2 and 0.2 mM of NADPH. The catalytic parameters (kcat, Km) were calculated using the Michaelis–Menten equation in Prism 6.0. In order to determine the thermal stability of the KARI variants, samples were aliquoted into 100 μLs volumes and placed into PCR tubes at a final concentration of 1 mg/mL. The samples were incubated on a heating block at temperatures ranging from 30 °C to 95 °C for 10 min. The samples were then assayed under standard conditions (see above). The activity of the samples incubated at 30 °C for 10 min was considered to represent 100% activity. The thermal stability profile of the enzyme was plotted using Prism 6.0. In order to assess the effect of isobutanol on KARI activity, 10% isobutanol (v/v) was added to a protein sample, which was then incubated at 25 °C. The activity was assayed under standard conditions at various time points.
2.7. PROSS Design of EcKARI
The PROSS algorithm was applied as previously described [32] with the sequence of EcKARI as the query for the sequence homologue search and its structure (PDB entry: 1YRL) [19] as the template structure. The final model used for expression was selected by manual examination of each mutation position of the design models. The redesigned EcKARI is defined as EcKARI_De.
3. Results and Discussion
Recombinantly expressed KARIs from C. jejuni (CjKARI) and E. coli (EcKARI) were selected as representatives for a dodecameric Class I and a tetrameric Class II KARI, respectively, to probe and optimize their thermal stability.
3.1. Overall Structure of the Class I CjKARI and the Relevance of the Oligomeric State on Enzyme Stability
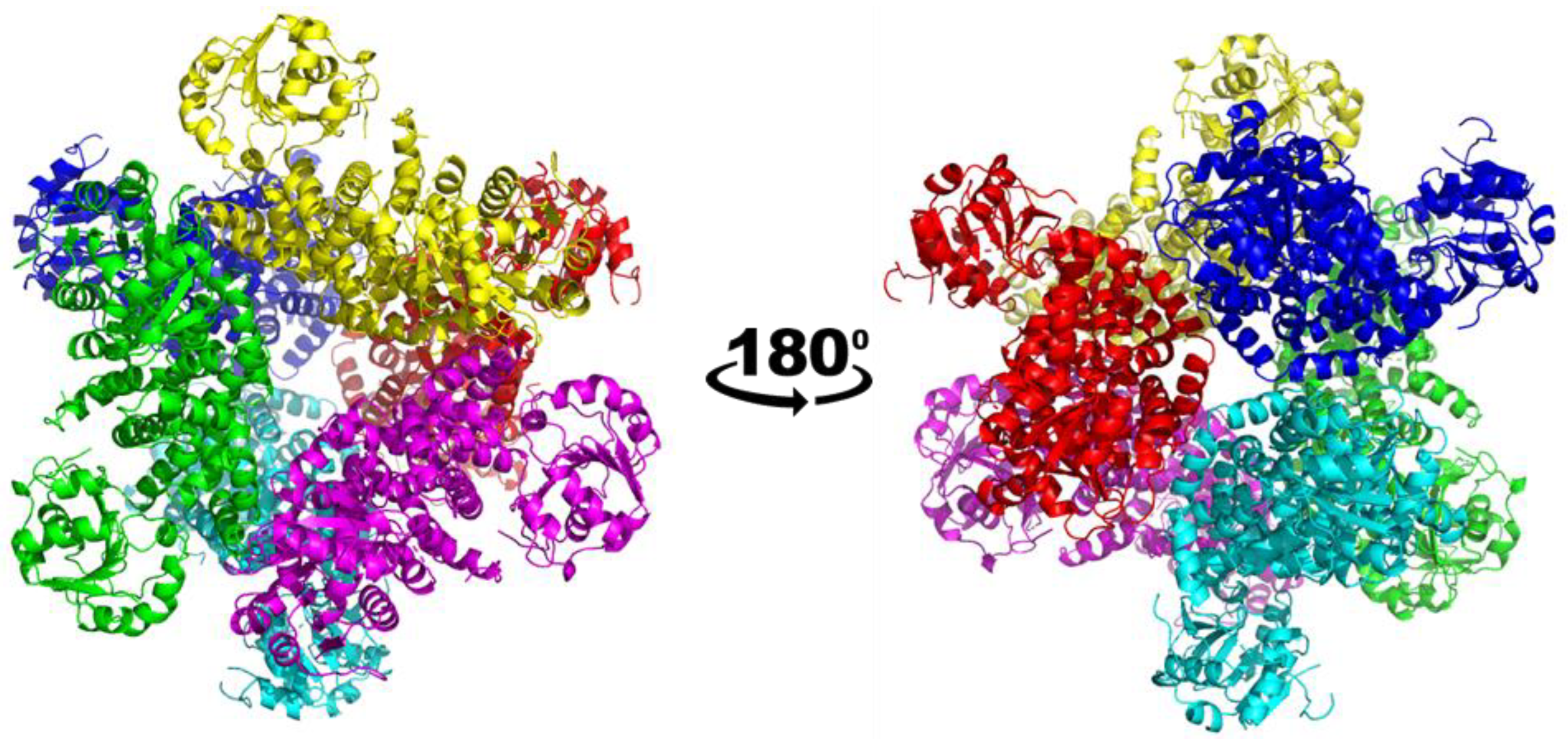
The structure of the apo form of CjKARI was solved by cryo-electron microscopy (cryo-EM) at 2.94 Å resolution, showing the homododecameric structure of the enzyme (Figure 2; Table 1). The backbone of the polypeptide could be traced throughout except for two regions, 50–59 and 142–147. The former represents the region where the adenine of NADPH is expected to bind. Based on other KARI structures, this region is known to be flexible [20,21,22,23,24,25]. The latter is a loop in the C-terminal domain and is located on the surface of the protein.
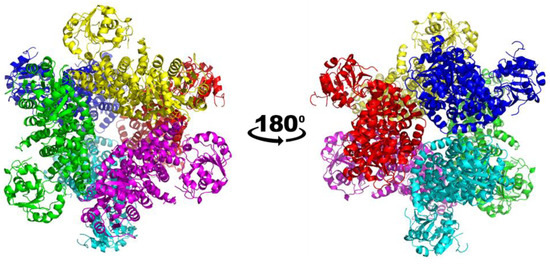
Figure 2.
Overall structure of CjKARI in two orientations. The cryo-EM structure of CjKARI was solved to a resolution of 2.94 Å and illustrates how the dodecamer is composed of six dimeric units.
The cryo-EM map resolved the location of the majority of the side chains including Y258, L294 and K290 (Figure 3). Similar to PaKARI and SsKARI, the dodecamer of CjKARI represents an assembly of six identical dimers with the active sites located between an N-domain and two interlocking C-domains. The C-domain is deeply knotted [39] and harbors the catalytic pocket that accommodates two essential metal ions (Mg2+ in most naturally occurring KARIs). The interactions that hold the CjKARI dodecamer together involve conserved residues from the C-domains (Figure 3).
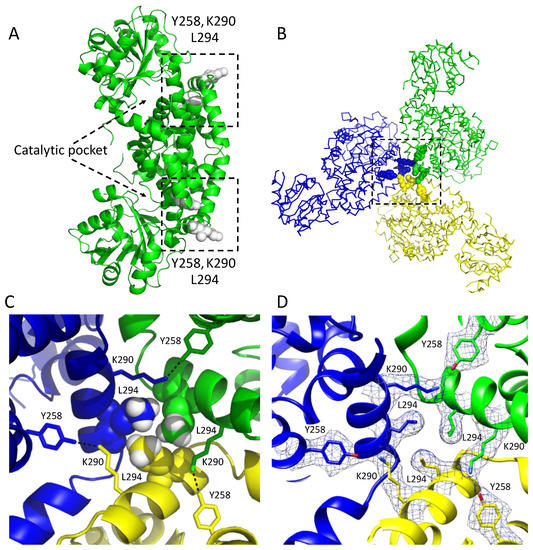
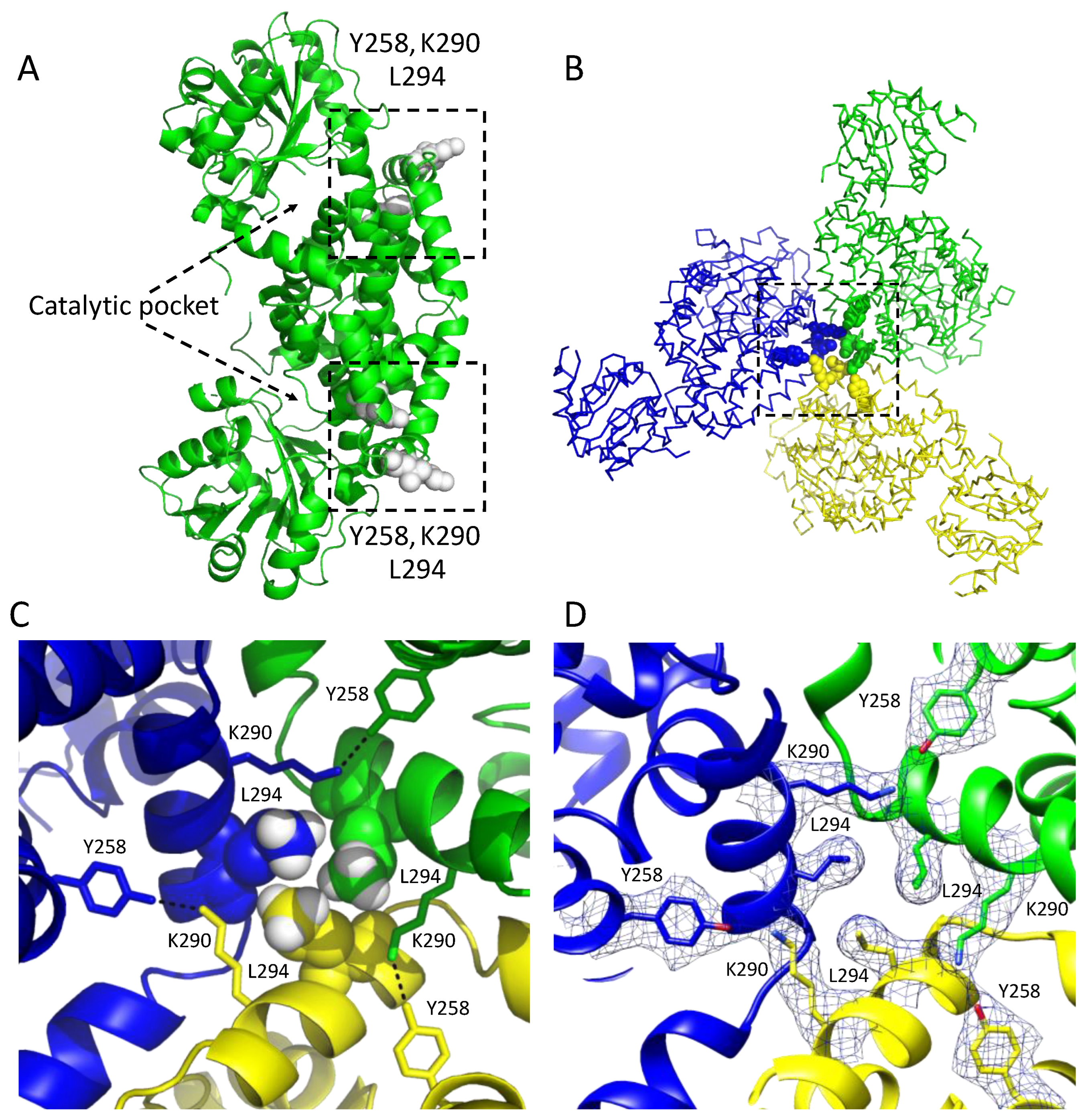
Figure 3.
Subunit interactions in a dodecameric KARI. (A) A dimeric unit showing the location of the regions (boxed) involved in the formation of the dodecamer in CjKARI. Residues colored white in the box are the contact residues that are key to stabilization of the three dimers in this image. (B) The assembly of three of the six dimers in the dodecamer. An equivalent arrangement occurs with the other three dimers of the dodecamer. (C) Zoom showing the interactions of three contact residues Y258, K290 and L294. The hydrogen atoms (attached as riding models) of the distal carbon that make contact with the neighboring subunits are shown as white CPK models. Y258 and K290 from the different subunits form hydrogen bonds (3.0 Å). (D) Cryo-EM map of the residues Y258, K290 and L294.
In particular, the side chains of Y258, K290 and L294 (Figure 3A) appear to be critically important for interactions that maintain the dodecameric structure. These residues are highly conserved among the dodecameric KARIs. Specifically, Y258 from one subunit forms a hydrogen bond with K290 from a neighboring subunit, while the L294 residues from three subunits form a hydrophobic cluster (Figure 3B–D). We, therefore, predicted that mutations of K290 and L294 to alanine would be sufficient to alter the oligomeric state from a dodecamer to a dimer. Hence, we generated the K290A/L294A double mutant of CjKARI (CjKARI_Dm) by site-directed mutagenesis and expressed and purified this enzyme using the same protocol as for its wild-type counterpart.
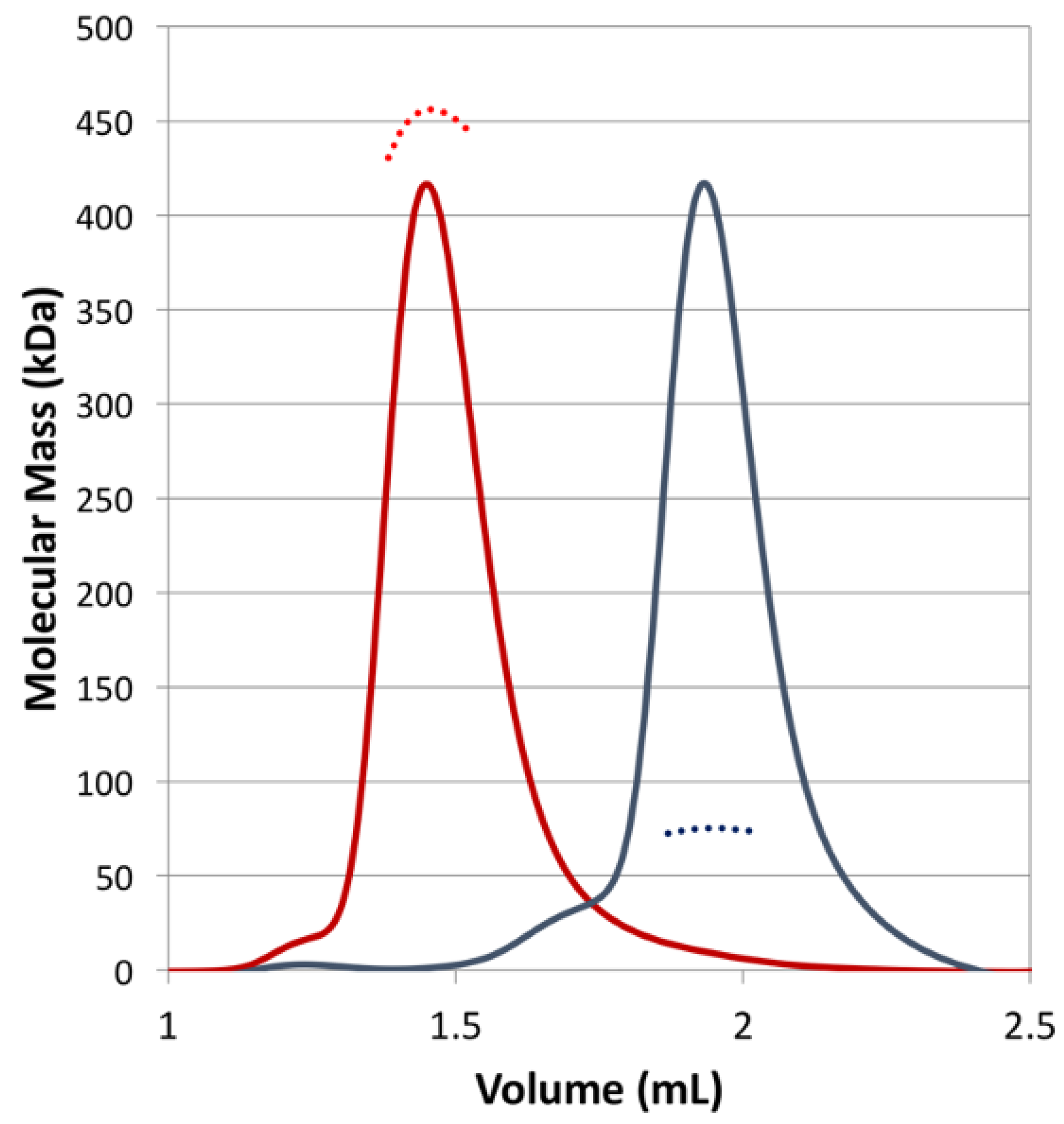
Using size-exclusion chromatography (SEC), coupled with multi-angle laser light scattering (MALS), the oligomerization status of CjKARI and CjKARI_Dm was investigated. As expected, CjKARI has a molecular mass of ~450 kDa consistent with a dodecamer (the theoretical molecular mass of the dodecamer is 456 kDa). In contrast, CjKARI_Dm elutes later and has a refractive index corresponding to a molecular mass of ~74.5 kDa, consistent with the formation of a dimer (the theoretical molecular mass of a dimer is 75.8 kDa). Thus, the two mutations, as predicted, disrupt formation of the dodecamer (Figure 4).
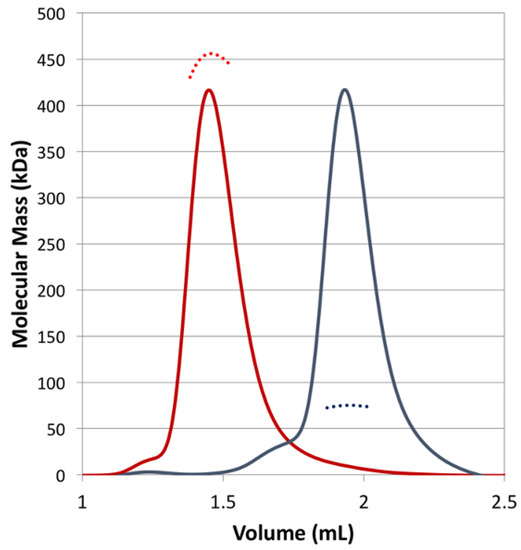
Figure 4.
SEC-MALS of CjKARI and CjKARI_Dm. The chromatogram (refractive index) and the measured molecular mass for CjKARI are represented by a red solid line and a red dashed line, respectively. For CjKARI_Dm, these are represented by grey lines. CjKARI elutes with a MW of ~450 kDa, consistent with it being a dodecamer. CjKARI_Dm elutes with a MW of 74.5 kDa, consistent with it being a dimer.
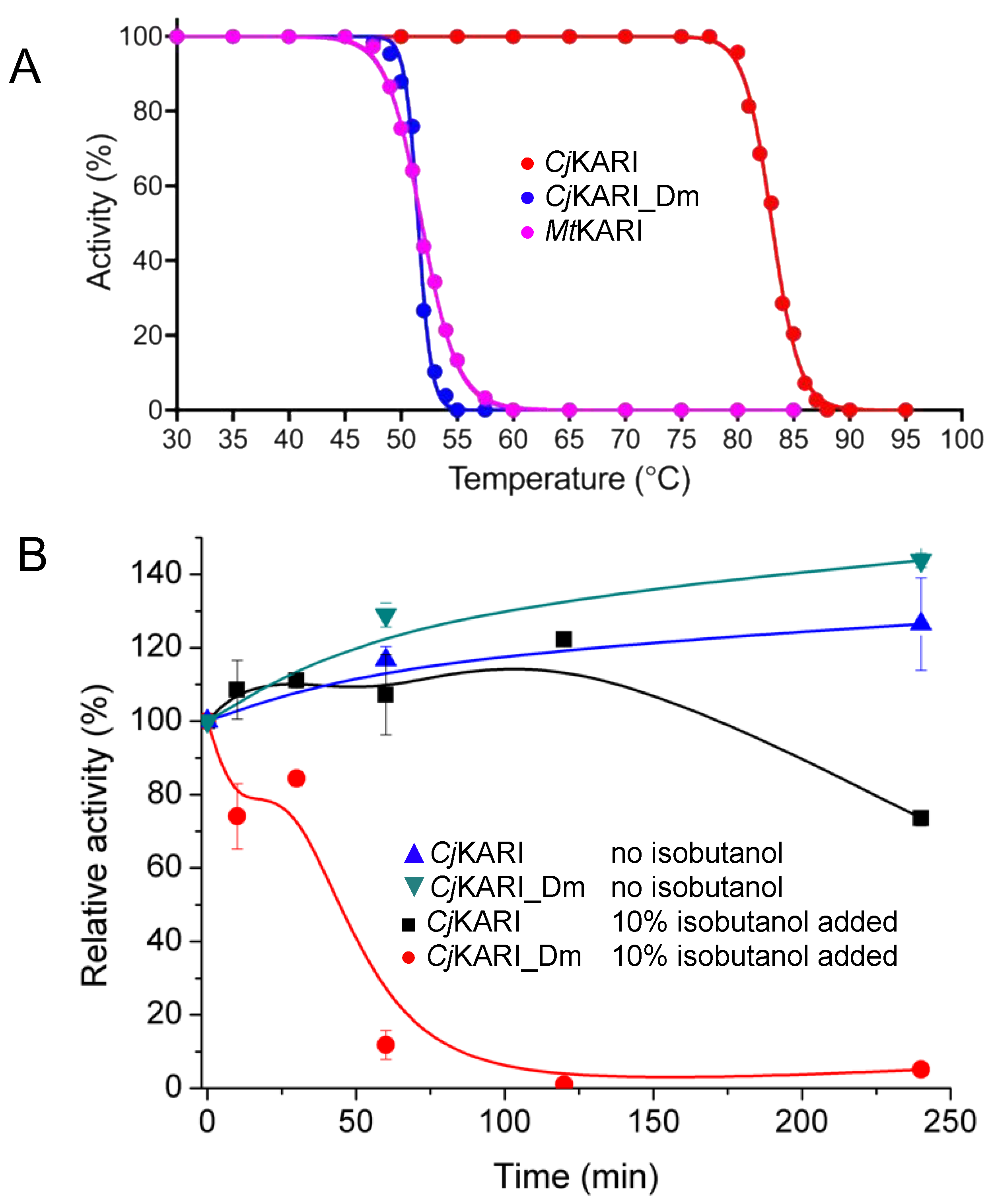
Kinetic assays show that CjKARI_Dm has a kcat of 0.47 ± 0.02 s−1 (Km: 1.98 ± 0.19 mM) for the reaction with the native substrate, 2-acetolactate, and a kcat of 3.30 ± 0.15 s−1 (Km: 4.35 ± 0.40 mM) when 3-hydroxypyruvate (Figure 1) is used as substrate. In comparison, the wild-type enzyme has kcat values of 0.85 ± 0.05 s−1 (Km: 881 ± 50 µM) and 2.20 ± 0.05 s−1 (Km: 1.70 ± 0.10 mM) for 2-acetolactate and 3-hydroxypyruvate, respectively. Thus, the catalytic efficiency (i.e., kcat/Km) towards the two substrates is four- and two-fold greater, respectively, for the native enzyme. However, while the effect of the mutations on the catalytic parameters is relatively modest, their effect on the apparent melting temperature (Tm) is more dramatic (83.1 °C vs. 51.5 °C for CjKARI vs. CjKARI_Dm; Figure 5A). MtKARI, which is dimeric in its native form [20], has a similar thermal stability as CjKARI_Dm with a Tm of 51.8 °C.
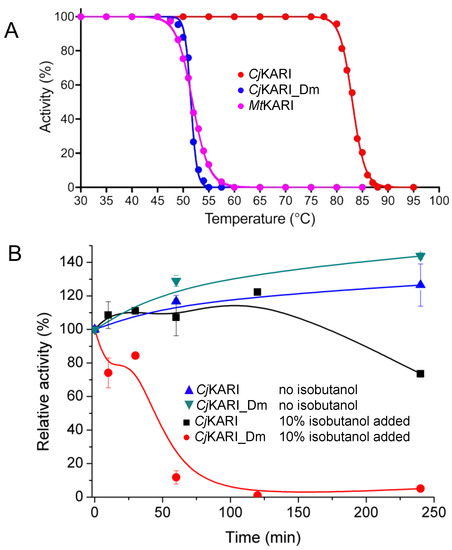
Figure 5.
Activities of dimeric and dodecameric KARIs. (A) The normalized activities of CjKARI, CjKARI_Dm and MtKARI after 10 min incubation at temperatures ranging from 30 °C to 95 °C. (B) Stability of CjKARI and CjKARI_Dm in the presence or absence of 10% isobutanol.
Cell-based isobutanol production systems have a maximal total volumetric yield as low as 1–2% before the growth of the cell culture is affected, due to end-product toxicity effects [7,40,41]. We thus tested the activities of wild-type CjKARI and CjKARI_Dm in the presence of isobutanol at a concentration of 10% (v/v) (Figure 5B). After four hours of incubation at 25 °C, the dodecamer retained 80% of its activity, whereas the dimeric enzyme was inactive. Thus, not only does the dodecameric enzyme have a higher apparent thermal stability compared to its dimeric counterpart, it also is less susceptible to loss of activity in the presence of isobutanol.
Studies to modify the quaternary structures of dimeric Class II KARIs (i.e., plant KARIs) have been reported [42]. In an attempt to correlate the oligomeric state of such enzymes with their function, a loop that crosses over from one subunit to the other was removed via mutagenesis. This led to the dissociation of the dimer into monomers, with a concomitant 3.8-fold reduction in the specific activity at 30 °C. Furthermore, the denaturation of the monomeric enzyme occurred at 35 °C, whereas the wild-type enzyme was still fully active at temperatures > 50 °C [42]. Thus, the higher oligomeric states of both classes of KARI appear to be critical for maintaining stability and activity.
In addition to CjKARI, the structures of a number of dodecameric class I KARIs have been determined. These include PaKARI, Alicyclobacillus acidocaldarius KARI (AaKARI), Azotobacter vinelandii KARI (AvKARI) and SsKARI [22,23,24,43]. Note that SsKARI from the thermophile S. solfataricus is thermostable but catalytically inefficient [9] for industrial applications as it catalyzes reactions two to three orders of magnitude slower than CjKARI. A comparison of these dodecameric structures reveals similar dimeric unit assemblies as CjKARI (Figure 3). Thus, these dimer-dimer interactions can be predicted by sequence analyses, which can then be used as a guide to distinguish potentially highly thermostable Class I dodecameric KARIs from their dimeric counterparts. As the dimeric units that assemble in dimeric class I KARIs are also found in the dodecamer, new dodecameric enzymes can be assembled from dimers using a protein-protein interface design.
3.2. Improving Enzyme Stability through Molecular Design
Above, we demonstrated how changes to the oligomeric structure contribute to enhanced stability of a Class I KARI. We also explored the possibility to increase the thermal stability of a Class II KARI using a computational analysis with the program Protein Repair One Stop Shop (PROSS) [32], which combines phylogenetic analysis with Rosetta atomistic design calculations to increase thermal stability without impairing activity. This computational approach effectively bypasses laborious iterative design/experiment cycles typically used in directed evolution. We used PROSS to predict a set of mutations in EcKARI, which were further qualified through a detailed evolutionary and structural analysis. We collected 634 full-length reviewed proteins of the gene ilvC in UniProt, aligned them using MAFFT [44] and generated a phylogenetic tree using RAxML [45] (where the root was placed to represent the hypothesized internal domain duplication event, separating Class I from Class II KARIs). We inferred the most probable amino acid states at all ancestral branch points to recover the joint substitutions that best explain the composition of existent KARI sequences. The crystallographic coordinates for chain A of EcKARI (PDB code: 1YRL) were used as a guide structure, and seven structural models were generated using Rosetta. PROSS predicted a model EcKARI_De with 34 changes. These changes were examined by (a) matching each target amino acid to inferred substitutions between the Class II ancestral branch point and EcKARI, (b) mapping and rationalizing each mutation relative to chain A of EcKARI and (c) experimentally characterizing EcKARI_De.
From the 34 predicted substitutions in EcKARI_De, 24 matched changes were suggested by our evolutionary analysis as the most likely to have occurred sometime between the class II ancestor and EcKARI [10]. This indicates that a majority of (predicted) thermal stability-enhancing mutations are also recovered by ASR (and hence not reliant on access to a structure). Only eight substitutions corresponded to changes that are assigned near-zero probability to have occurred in nature (K69P/EQ where E or Q are the most likely ancestral states, not P; A73D/AK; D124S/KQ; K184R/QHK; M188W/L; S314K/RS; E384P/E; G456N/Q). We note that these eight mutations suggested by PROSS are available broadly in the KARI family alignment [10] but not favored within the EcKARI branch or across close homologs. Exceptional cases are E384P, where P is associated almost exclusively with class I KARI, and G456N where N is generally rare across the KARI family.
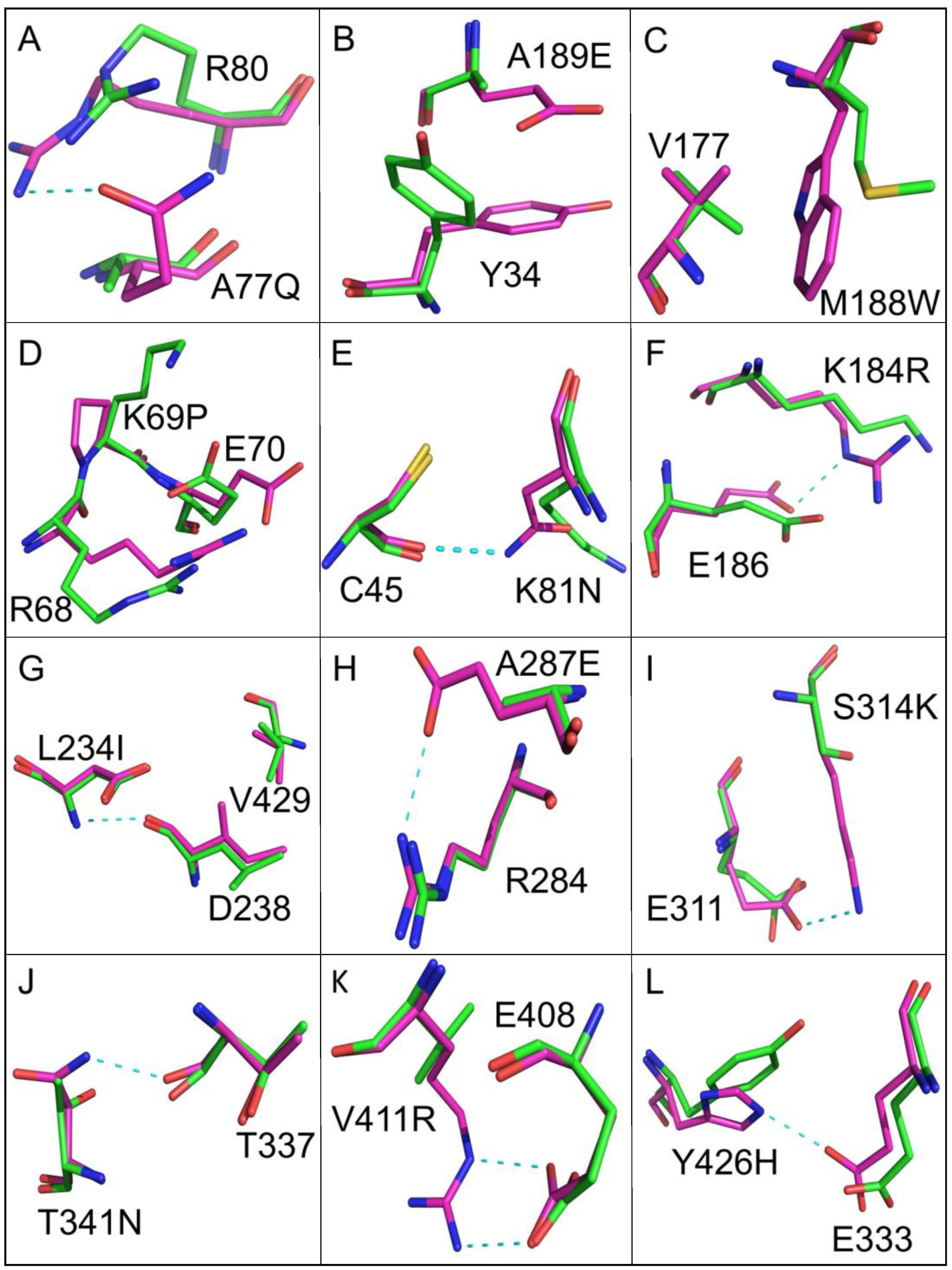
A comparison of the crystal structure of wild-type EcKARI and the Ec_KARI_De Rosetta/PROSS model indicates that the protein backbones are similar (RMSD of 0.276 Å after superposition of all Cα-atoms). However, the orientation of the side chains of several mutated residues varies to create new interactions (Table 2). These can be categorized into four types of change that increase structural stability by (i) introducing polarity to the surface of the enzyme, (ii) contributing to the core packing, (iii) strengthening turn structures by inclusion of a proline side-chain and (iv) adding new hydrogen bonds or salt bridges. Examples of substitutions are illustrated in Figure 6. To enhance the polarity of the surface, thirteen changes were made to the enzyme. For eight examples, this could be achieved by replacing a small hydrophobic residue (e.g., glycine or alanine) with a bulkier polar or charged residue (Figure 6A,B; Table 2). For the A189E change, it was predicted that a change in rotamer conformation of the nearby Y34 could occur, creating additional hydrophobic interactions to stabilize the enzyme (Figure 6B). To improve the core packing, three changes were made. In Figure 6C, the effect of the M188W mutation is depicted, which, as well as introducing a bulkier hydrophobic group, induces a conformational change in the nearby valine side chain to enhance interactions further. Three changes were also made to facilitate the introduction of stabilizing proline side chains. In two cases, charged amino acids were substituted by proline, and in the third example an alanine was replaced by proline. The K69P mutation is illustrated in Figure 6D; mutations to Pro may lower the entropy loss upon folding, thereby improving folding free energy [46]. Conformational changes are also predicted to occur to the side chains of R68 and E70. Nine changes were made that are predicted to result in an increase in the number of hydrogen bonds or salt bridges in the structure. These are illustrated in Figure 6E–L. Three other changes appeared to show no apparent effect that may lead to improved stabilization, but similarly these changes also did not seem to be detrimental to stability.

Table 2.
Differences in amino acid composition between wild-type EcKARI and redesigned EcKARI_De.
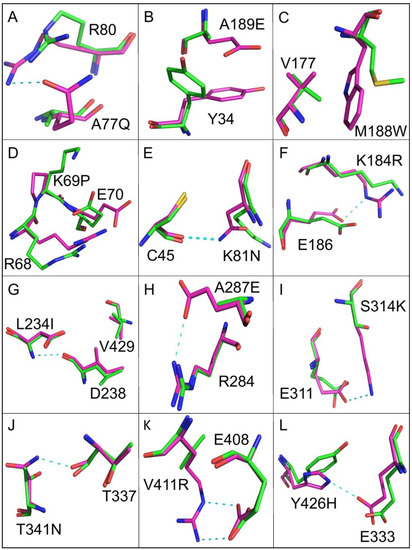
Figure 6.
The redesign of EcKARI. Superposition of the crystal structure of EcKARI (shown in green) and the PROSS-Rosetta model of EcKARI_De (shown in magenta). The carbon atoms of the residues of the wild-type enzyme are shown in green, and carbon atoms of the designed enzyme are shown in magenta. Panels (A–L) show the individual site mutations when EcKARI and EcKARI_De are compared. Dashed cyan lines represent the new hydrogen bond interactions formed as a result of the change. In B and C new hydrophobic contacts are formed. In D there are multiple conformational changes due to the K69P change.
EcKARI and EcKARI_De were expressed and purified to homogeneity. The catalytic efficiencies of wild-type EcKARI and EcKARI_De are similar despite the introduction of 34 mutations in the latter. With 2-acetolactate as the substrate, EcKARI_De has a kcat of 0.75 ± 0.01 s−1 (Km of 490 ± 25 µM), while for the reaction with 3-hydroxypyruvate a kcat of 8.1 ± 0.2 s−1 (Km of 3.4 ± 0.2 mM) was determined. The corresponding values for EcKARI are: kcat = 2.15 ± 0.02 s−1 and Km = 230 ± 8 µM (for 2-acetolactate), and kcat = 7.9 ± 0.2 s−1 and Km = 3.5 ± 0.2 mM (for 3-hydroxypyruvate). Thus, while the catalytic efficiency for the full reaction is approximately six-fold greater in the wild-type enzyme, for the reduction-only reaction, the two enzymes are virtually identical. The structure of EcKARI was determined in the apo-form [19] and with both cofactors (i.e., Mg2+ and NADPH) [5]. The major difference in the two complexes is that the active site is closed in the apo-form but is open when the cofactors are bound. Therefore, there are several ligand-induced conformational changes that occur to prepare the enzyme for substrate binding, during catalysis and upon release of the products [5]. This inherent flexibility is therefore of prime importance to allow catalysis to commence. As none of the mutations in the designed mutant is close to the active- or the NADPH-binding site, the reduction in catalytic efficiency may be due to the fact that the enzyme has lost some structural flexibility important for preparing the active site for catalysis.
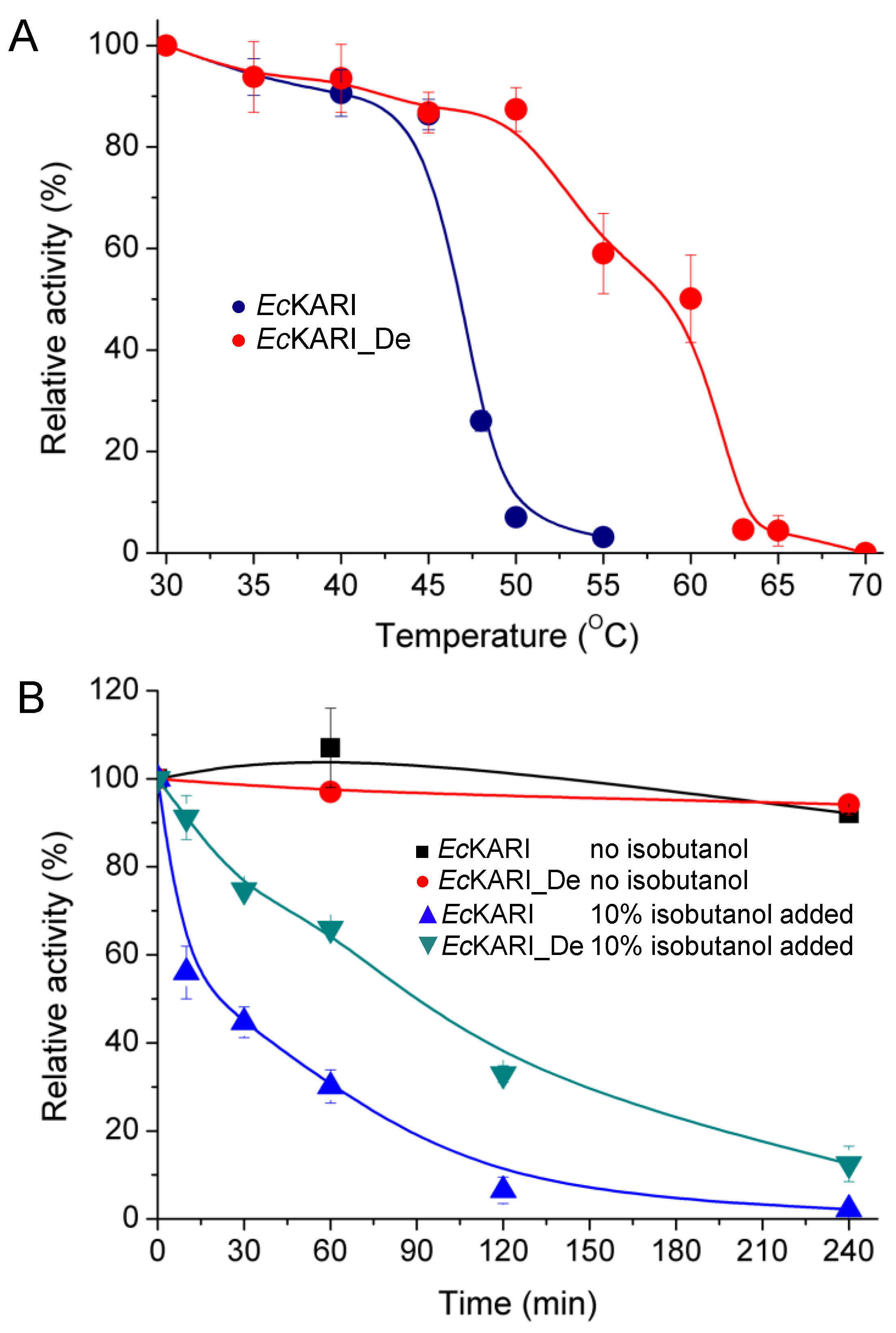
The gel-filtration elution time of EcKARI_De was the same as for the wild-type protein, showing that the tetrameric state of EcKARI_De is maintained, but the Tm of EcKARI_De increased by over 10 °C from 47.9 °C to 58.5 °C (Figure 7A). Next, we tested how a prolonged incubation with 10% (v/v) isobutanol affects the activity of both EcKARI and EcKARI_De (Figure 7B). While not as impressive as wild-type CjKARI (Figure 5B), the rate of activity loss was significantly reduced for the designed protein when compared to EcKARI. EcKARI_De retained ~75% activity after 1 hour of incubation with 10% isobutanol, while the wild-type enzyme had less than 40% of its initial value. Even after four hours of incubation, the mutant retained nearly 20% of its initial activity, while EcKARI was inactive.
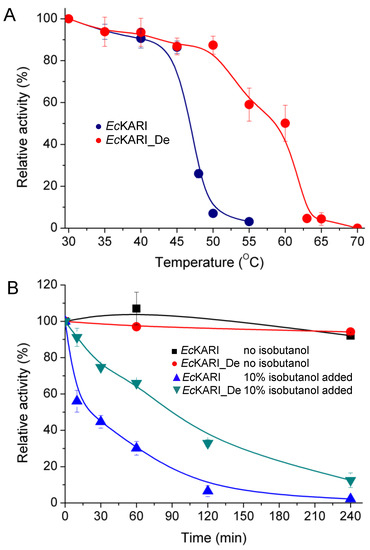
Figure 7.
Activity of EcKARI and EcKARI_De. (A) The normalized activity of EcKARI and EcKARI_De after 10 min incubation at temperatures ranging from 30 °C to 70 °C. (B) Stability of EcKARI and EcKARI_De in the presence or absence of 10% isobutanol.
4. Conclusions
KARI is an established target for biocidal agents (herbicides and antimicrobials) [2,3,6,21,47,48,49,50], but this enzyme also plays a central role in synthetic biomanufacturing cascades [7,9,51]. This study demonstrated that the oligomeric status is critical to the thermostability of KARI. Here, we demonstrated that while CjKARI can function as a dimer, its apparent Tm value increases by ~30 °C upon dodecamer formation. Furthermore, we could provide evidence that the EcKARI thermostability and resistance to the high energy biofuel isobutanol can be enhanced by rational protein design. Enzyme engineering has been shown to benefit from guidance from both evolution and structure; here, we rationalize a range of changes suggested by PROSS by reference to substitutions that trace back to ancestral variants and/or by reference to structure. This is the first PROSS application that has enhanced the thermostability of an E. coli protein, impressively in the presence of a denaturant (i.e., isobutanol). Thus, an integrated approach of structure and phylogenetic analysis holds great promise for enhancing the properties of enzymes in a wide range of applications.
Author Contributions
Conceptualization, Y.L., V.S., S.J.F., G.S. and L.W.G.; methodology, Y.L., S.Z., F.L., Y.G., A.K., X.Y., S.W. and B.W.; software, A.G., M.B. and S.J.F.; formal analysis, Y.L., S.Z. and B.W.; investigation, Y.L., S.Z., A.G., F.L., Y.G., X.Y., A.K., S.W. and B.W.; data curation, A.G., M.J.L., M.B., Z.R., S.J.F. and L.W.G.; writing—original draft preparation, Y.L., S.Z., G.S. and L.W.G.; writing—review and editing, T.B.B., M.A.S., M.J.L., V.S., M.B., G.S. and L.W.G.; visualization, S..Z., F.L., Y.G. and M.J.L.; supervision, R.P.M., B.K., M.A.S., Z.R., S.J.F.,M.J.L., G.S. and L.W.G.; project administration, G.S. and L.W.G.; funding acquisition, R.P.M., V.S., G.S. and L.W.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by a Discovery Grant from the Australian Research Council (ARC; DP210101802) to G.S., L.W.G. and V.S. Research in the Fleishman laboratory was funded by a Starter’s Grant from the European Research Council (335439) and by the Israel Science Foundation through its Center for Research Excellence in Structural Cell Biology (1775/12). V.S. and G.S. would like to thank the Federal Ministry of Education and Research (BMBF), Germany for funding. T.B. gratefully acknowledges funding by the Werner Siemens foundation to establish the new field of Synthetic Biotechnology at the Technical University of Munich, Germany. B.K. is an ARC Laureate Fellow (FL180100109).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The coordinates for the structure of CjKARI have been deposited to the Protein Databank (PDB) and the Electron Microscopy Data Bank (EMDB).
Acknowledgments
The authors are grateful to Thierry Lonhienne for his careful proof-reading of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Duggleby, R.G.; Pang, S.S. Acetohydroxyacid synthase. J. Biochem. Mol. Biol. 2000, 33, 1–36. [Google Scholar]
- Grandoni, J.A.; Marta, P.T.; Schloss, J.V. Inhibitors of branched-chain amino acid biosynthesis as potential antituberculosis agents. J. Antimicrob. Chemother. 1998, 4, 475–482. [Google Scholar] [CrossRef] [Green Version]
- Schulz, A.; Sponemann, P.; Kocher, H.; Wengenmayer, F. The herbicidally active experimental compound Hoe 704 is a potent inhibitor of the enzyme acetolactate reductoisomerase. FEBS Lett. 1988, 238, 375–378. [Google Scholar] [CrossRef] [Green Version]
- Epelbaum, S.; LaRossa, R.A.; VanDyk, T.K.; Elkayam, T.; Chipman, D.M.; Barak, Z. Branched-chain amino acid biosynthesis in Salmonella typhimurium: A quantitative analysis. J. Bacteriol. 1998, 180, 4056–4067. [Google Scholar] [CrossRef] [Green Version]
- Wong, S.H.; Lonhienne, T.G.; Winzor, D.J.; Schenk, G.; Guddat, L.W. Bacterial and plant ketol-acid reductoisomerases have different mechanisms of induced fit during the catalytic cycle. J. Mol. Biol. 2012, 424, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Pue, N.; Guddat, L.W. Acetohydroxyacid synthase: A target for antimicrobial drug discovery. Curr. Pharm. Des. 2014, 20, 740–753. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, S.; Hanai, T.; Liao, J.C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 2008, 451, 86–89. [Google Scholar] [CrossRef]
- Wang, B.W.; Shi, A.Q.; Tu, R.; Zhang, X.L.; Wang, Q.H.; Bai, F.W. Branched-chain higher alcohols. Adv. Biochem. Eng. Biotechnol. 2012, 128, 101–118. [Google Scholar]
- Guterl, J.K.; Garbe, D.; Carsten, J.; Steffler, F.; Sommer, B.; Reisse, S.; Philipp, A.; Haack, M.; Ruhmann, B.; Koltermann, A.; et al. Cell-free metabolic engineering: Production of chemicals by minimized reaction cascades. ChemSusChem 2012, 5, 2165–2172. [Google Scholar] [CrossRef]
- Gumulya, Y.; Baek, J.-M.; Wun, S.-J.; Thomson, R.E.S.; Harris, K.L.; Hunter, D.J.B.; Behrendorff, J.B.Y.H.; Kulig, J.; Zheng, S.; Wu, X.; et al. Engineering highly functional thermostable proteins using ancestral sequence reconstruction. Nature Catal. 2018, 1, 878–888. [Google Scholar] [CrossRef]
- Lonhienne, T.; Low, Y.S.; Garcia, M.D.; Croll, T.; Gao, Y.; Wang, Q.; Brillault, L.; Williams, C.M.; Fraser, J.A.; McGeary, R.P.; et al. Structures of fungal and plant acetohydroxyacid synthases. Nature 2020, 586, 317–321. [Google Scholar] [CrossRef]
- Flint, D.H.; Emptage, M.H.; Finnegan, M.G.; Fu, W.; Johnson, M.K. The role and properties of the iron-sulfur cluster in Escherichia coli dihydroxy-acid dehydratase. J. Biol. Chem. 1993, 268, 14732–14742. [Google Scholar] [CrossRef]
- Sutiono, S.; Teshima, M.; Beer, B.; Schenk, G.; Sieber, V. Enabling the direct enzymatic dehydration of D-glycerate to pyruvate as the key step in synthetic enzyme cascades used in the cell-free production of fine chemicals. ACS Catal. 2020, 10, 3110–3118. [Google Scholar] [CrossRef]
- Melse, O.; Sutiono, S.; Haslbeck, M.; Schenk, G.; Antes, I.; Sieber, V. Structure-guided modulation of the catalytic properties of dihydroxyacid dehydratases. ChemBioChem 2022, 23, e202200088. [Google Scholar] [CrossRef]
- Bayaraa, T.; Gaete, J.; Sutiono, S.; Kurz, J.; Lonhienne, T.; Harmer, J.R.; Bernhardt, P.V.; Sieber, V.; Guddat, L.W.; Schenk, G. Dihydroxy-acid dehydratases from pathogenic bacteria: Emerging drug targets to combat antibiotic resistance. Chem. Eur. J. 2022, 28, e202200927. [Google Scholar] [CrossRef] [PubMed]
- Tadrowski, S.; Pedroso, M.M.; Sieber, V.; Larrabee, J.A.; Guddat, L.W.; Schenk, G. Metal ions play an essential catalytic role in the mechanism of ketol-acid reductoisomerase. Chemistry 2016, 22, 7427–7436. [Google Scholar] [CrossRef] [PubMed]
- Dumas, R.; Biou, V.; Halgand, F.; Douce, R.; Duggleby, R.G. Enzymology, structure, and dynamics of acetohydroxy acid isomeroreductase. Acc. Chem. Res. 2001, 34, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, R.; Lee, Y.T.; Guddat, L.W.; Duggleby, R.G. Probing the mechanism of the bifunctional enzyme ketol-acid reductoisomerase by site-directed mutagenesis of the active site. FEBS J. 2005, 272, 593–602. [Google Scholar] [CrossRef]
- Tyagi, R.; Duquerroy, S.; Navaza, J.; Guddat, L.W.; Duggleby, R.G. The crystal structure of a bacterial class II ketol-acid reductolsomerase: Domain conservation and evolution. Protein Sci. 2005, 14, 3089–3100. [Google Scholar] [CrossRef] [Green Version]
- Lv, Y.; Kandale, A.; Wun, S.J.; McGeary, R.P.; Williams, S.J.; Kobe, B.; Sieber, V.; Schembri, M.A.; Schenk, G.; Guddat, L.W. Crystal structure of Mycobacterium tuberculosis ketol-acid reductoisomerase at 1.0 Å resolution-a potential target for anti-tuberculosis drug discovery. FEBS J. 2016, 283, 1184–1196. [Google Scholar] [CrossRef]
- Patel, K.; Teran, D.; Zheng, S.; Kandale, A.; Garcia, M.; Lv, Y.; Schembri, M.A.; McGeary, R.P.; Schenk, G.; Guddat, L.W. Crystal structures of Staphylococcus aureus ketol-acid reductoisomerase in complex with two transition state analogs that have biocidal activity. Chem. Eur. J. 2017, 23, 18289–18295. [Google Scholar] [CrossRef] [PubMed]
- Ahn, H.J.; Eom, S.J.; Yoon, H.J.; Lee, B.I.; Cho, H.J.; Suh, S.W. Crystal structure of class I acetohydroxy acid isomeroreductase from Pseudomonas aeruginosa. J. Mol. Biol. 2003, 328, 505–515. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chang, Y.C.; Lin, B.L.; Lin, K.F.; Huang, C.H.; Hsieh, D.L.; Ko, T.P.; Tsai, M.D. Use of Cryo-EM to uncover structural bases of pH effect and cofactor bispecificity of ketol-acid reductoisomerase. J. Am. Chem. Soc. 2019, 141, 6136–6140. [Google Scholar] [CrossRef] [PubMed]
- Cahn, J.K.; Brinkmann-Chen, S.; Spatzal, T.; Wiig, J.A.; Buller, A.R.; Einsle, O.; Hu, Y.; Ribbe, M.W.; Arnold, F.H. Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases. Biochem. J. 2015, 468, 475–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, E.W.W.; Guddat, L.W. Conformational changes in a plant ketol-acid reductoisomerase upon Mg2+ and NADPH binding as revealed by two crystal structures. J. Mol. Biol. 2009, 389, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Kirk, O.; Borchert, T.V.; Fuglsang, C.C. Industrial enzyme applications. Curr. Opin. Biotechnol. 2002, 13, 345–351. [Google Scholar] [CrossRef]
- Van Beilen, J.B.; Li, Z. Enzyme technology: An overview. Curr. Opin. Biotechnol. 2002, 13, 338–344. [Google Scholar] [CrossRef]
- Ahuja, S.K.; Ferreira, G.M.; Moreira, A.R. Utilization of enzymes for environmental applications. Crit. Rev. Biotechnol. 2004, 24, 125–154. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A. Engineering a more sustainable world through catalysis and green chemistry. J. R. Soc. Interface 2016, 13. [Google Scholar] [CrossRef] [Green Version]
- Bornscheuer, U.T. Trends and challenges in enzyme technology. Adv. Biochem. Eng. Biotechnol. 2005, 100, 181–203. [Google Scholar]
- Brinkmann-Chen, S.; Flock, T.; Cahn, J.K.; Snow, C.D.; Brustad, E.M.; McIntosh, J.A.; Meinhold, P.; Zhang, L.; Arnold, F.H. General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH. Proc. Natl. Acad. Sci. USA 2013, 110, 10946–10951. [Google Scholar] [CrossRef] [Green Version]
- Goldenzweig, A.; Goldsmith, M.; Hill, S.E.; Gertman, O.; Laurino, P.; Ashani, Y.; Dym, O.; Unger, T.; Albeck, S.; Prilusky, J.; et al. Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Mol. Cell 2016, 63, 337–346. [Google Scholar] [CrossRef] [Green Version]
- Gill, S.C.; von Hippel, P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989, 182, 319–326. [Google Scholar] [CrossRef]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Grigorieff, N. Frealign: An exploratory tool for single-particle cryo-EM. Methods Enzym. 2016, 579, 191–226. [Google Scholar]
- Pintilie, G.D.; Zhang, J.; Goddard, T.D.; Chiu, W.; Gossard, D.C. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 2010, 170, 427–438. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Taylor, W.R. A deeply knotted protein structure and how it might fold. Nature 2000, 406, 916–919. [Google Scholar] [CrossRef]
- Atsumi, S.; Wu, T.Y.; Machado, I.M.; Huang, W.C.; Chen, P.Y.; Pellegrini, M.; Liao, J.C. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol. Syst. Biol. 2010, 6, 449. [Google Scholar] [CrossRef]
- Brynildsen, M.P.; Liao, J.C. An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol. Syst. Biol. 2009, 5, 277. [Google Scholar] [CrossRef] [PubMed]
- Wessel, P.M.; Biou, V.; Douce, R.; Dumas, R. A loop deletion in the plant acetohydroxy acid isomeroreductase homodimer generates an active monomer with reduced stability and altered magnesium affinity. Biochemistry 1998, 37, 12753–12760. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann-Chen, S.; Cahn, J.K.B.; Arnold, F.H. Uncovering rare NADH-preferring ketol-acid reductoisomerases. Metab. Eng. 2014, 26, 17–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, K.D.; Tomii, K.; Katoh, K. Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics 2016, 32, 3246–3251. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.W.; Nicholson, H.; Becktel, W.J. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc. Natl. Acad. Sci. USA 1987, 84, 6663–6667. [Google Scholar] [CrossRef] [Green Version]
- Bayaraa, T.; Kurz, J.L.; Patel, K.M.; Hussein, W.M.; West, N.P.; Schenk, G.; McGeary, R.P.; Guddat, L.W. Discovery, synthesis and evaluation of a novel ketol-acid reductoisomerase inhibitor. Chem. Eur. J. 2020, 26, 8958–8968. [Google Scholar] [CrossRef]
- Wun, S.J.; Johnson, L.A.; McGeary, R.P.; Schenk, G.; Guddat, L.W. Inhibition studies of ketol-acid reductoisomerases from pathogenic microorganisms. Arch. Biochem. Biophys. 2020, 692, 108516. [Google Scholar] [CrossRef]
- Lin, X.; Kurz, J.L.; Patel, K.M.; Wun, S.J.; Hussein, W.M.; Lonhienne, T.; West, N.P.; McGeary, R.P.; Schenk, G.; Guddat, L.W. Discovery of a pyrimidine-dione derivative with potent inhibitory activity against Mycobacterium tuberculosis ketol-acid reductoisomerase. Chem. Eur. J. 2020, 27, 3130–3141. [Google Scholar] [CrossRef]
- Kandale, A.; Patel, K.; Waleed, H.; Wun, S.J.; Zheng, S.; Tan, L.; West, N.; Schenk, G.; Guddat, L.W.; McGeary, R.P. Analogs of the herbicide, N-hydroxy-N-isopropyloxamate (IpOHA), inhibit Mycobacterium tuberculosis ketol-acid reductoisomerase and their prodrugs are promising anti-TB drug leads. J. Med. Chem. 2021, 64, 1670–1684. [Google Scholar] [CrossRef]
- Valera, A.; Wang, S.; Carr, R.; Trembleau, L.; Deng, H. Characterization of a class II ketol-acid reductoisomerase from Mycobacterium tuberculosis. RSC Adv. 2022, 12, 10540–10544. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).