Abstract
Curtobacterium species are increasingly recognized as plant pathogens and soil decomposers, but their prevalence and function as plant endophytes in aerial organs are less clear. In this study, we isolated six endophytic Curtobacterium species from the fruits, flower petals (previously unreported) and stem tissue of plants from diverse environments and examined their general characteristics. We found that all Curtobacterium endophytes belonging to three major Curtobacterium clusters—C. oceanosedimentum (a group not previously recognized as endophytic), C. luteum and C. flaccumfaciens—shared some common features. All or nearly all isolates tested were pigmented, displayed moderate salt tolerance and, surprisingly, were psychrotolerant, being able to grow at 6 °C. The exception was a fruit C. luteum isolate that appears to have evolved thermotolerance (up to 45 °C) instead as a likely adaptation to its environment. All isolates were able to metabolize starch and casein and solubilize inorganic phosphate, indicating conserved secreted hydrolase activity, but only isolates in the C. oceanosedimentum group were able to absorb and metabolize citrate. Finally, all endophytes tested were able to ferment the plant sugars sucrose and fructose, while they differed in their ability to use other sugars. Thus, this study documents common traits and adaptations in various Curtobacterium endophytes, and the presence of these isolates in floral and fruit organs implies the possible seed-borne inheritance of these isolates.
Keywords:
Curtobacterium; endophyte; fruit; flower; stem; cold tolerant; psychrotolerant; phylogenetic; pigmentation 1. Introduction
The genus Curtobacterium, meaning short rods [1], includes Gram-positive, obligate aerobic bacteria, belongs to the family Microbacteriaceae in the phylum Actinomycetota and is represented by over a dozen species [2]. Curtobacterium species are present in diverse environments including soil and plants, and the genus has been frequently described in the rhizosphere and also as endophytes in various parts of plants including roots, leaves, stems, fruit and seeds [2,3]. Curtobacterium was described as the predominant genus of the leaf litter community, likely decomposing plant debris [3].
The Curtobacterium species most commonly associated with plants is Curtobacterium flaccumfaciens, which includes pathogenic and non-pathogenic variants [4]. Several pathovars of C. flaccumfaciens are known to infect many species of plants including legumes, beet and flowering ornamentals, with significant economic losses; as a result, these pathovars are subject to strict quarantine in many countries [4]. Recently, C. allii (renamed C. flaccumfaciens pv. allii) was identified as a bulb rot pathogen in onion [5]. Pathogenic Curtobacterium spp. primarily colonize vascular tissues, resulting in wilt disease [4], and can also cause root rot [5]. Aside from C. flaccumfaciens though, most Curtobacterium species are not known to be pathogenic in plants; it is suggested that many species may perform more ecological roles and promote plant growth [6]. Other species of Curtobacterium include C. luteum, C. citreum, C. pusillum, C. herbarum and C. oceanosedimentum. The species C. plantarum, C. luteum and C. herbarum were reported as leaf endophytes in soybean [7], Citrus [8] and grass [9], respectively. C. luteum isolated from the sediment of seagrass meadow [10] and C. oceanosedimentum recovered from paddy soil [11] were both reported as plant growth promoters. C. citreum was isolated as a strawberry fruit endophyte [12], C. albidum (now considered C. citreum) was studied as a plant growth promoter in rice [13] and C. pusillum was found to be a human clinical specimen [14].
Compared to root endophytes, much less is known about endophytes in other parts of the plant, particularly reproductive organs. Pathogenic and commensal microbes that infect or colonize flowers and fruits can be vertically transmitted to offspring plants [15]. Such vertical transmission of commensals is also attractive from an evolutionary perspective as a model for plant–microbe coevolution [16]. Over one-third of the commensal bacteria in radish seeds were found to be derived from the buds, flowers and fruits of the parent plant [17]. Therefore, floral and fruit endophytes are likely undervalued in their contribution to plant health. Endophytes could stimulate plant growth through the synthesis of phytohormones and by enhancing the biotic and abiotic stress tolerance of plants. Endophytic Curtobacterium could be beneficial to plants, for example, through mitigating disease symptoms [18]. However, knowledge of endophytic Curtobacterium is restricted to rhizosphere and phyllosphere niches; there are a few reports of endophytic colonization in fruits such as strawberry and mulberry [12,19], but no flower endophytes have been reported. Additionally, Curtobacterium species have been predicted, based on genomic analysis, to be capable of digesting carbohydrates through glycosyl hydrolases [2], but functional evidence for such nutritional activities is limited. Furthermore, it is unclear if species other than C. flaccumfaciens are prominent endophytes in plants.
In this study, we comparatively characterized six endophytic Curtobacterium species isolated from fruits, stems and a previously unreported niche of Curtobacterium, flower petals. We found that all isolates share starch-degrading and phosphate-solubilizing capabilities as well as the ability to digest the plant sugars glucose, sucrose and fructose. Finally, we discovered that multiple isolates related to C. oceanosedimentum are plant endophytes. Surprisingly, we also found that nearly all tested Curtobacterium endophytes are psychrotolerant with some adaptive variants.
2. Methods
2.1. Bacterial Growth Media
Tryptic soy agar (TSA) was used for the general growth and propagation of bacteria, as well as to test growth at various temperatures. Milk agar plates were prepared by adding 3% skim milk to TSA. Starch agar, nutrient gelatin, Simmons citrate agar and urea broth were prepared according to the manufacturer’s instructions (Carolina Biological, Burlington, NC, USA), as was Pikovskaya’s agar (Himedia Inc., Thane, India). To test salt tolerance, TSA plates were prepared with 5%, 7.5% and 10% sodium chloride.
2.2. Isolation of Bacterial Endophytes
In an initial experiment, endophytic bacteria were isolated from eleven plant samples (one stem, two leaf, three flower petal and five fruit samples) (Supplementary Table S1). The six samples corresponding to the Curtobacterium isolates selected for this study were four fruits, one flower and one stem tissue (Table 1). The four fruits were two batches of store-bought rambutan in Springfield, Virginia, and Cleveland, Ohio, respectively, with the fruits most likely having Southeast Asian origin, steak tomato from a local store in Erie, Pennsylvania (PA), and a roughleaf dogwood wild berry fruit found on campus at Mercyhurst University (MU) in Erie, PA. Flower petals of a purple hydrangea at MU and the stem of Indian pipe, Monotropa uniflora, from Allegheny National Forest were the sources of the two remaining isolates. Endophytic bacteria were isolated by serial dilution plating of homogenized explants (0.5–1 g of tissue). The surface sterilization procedure was adapted from a previous study [20] and consisted of submersion in 95% ethanol for 15–20 s for thinner samples (petals, stems) followed by 3 thorough washes with sterile nanopure water. Bleach was omitted to minimize toxicity to endophytes in thin tissues, and imprints [21] of ethanol-sterilized Indian pipe stem and hydrangea petals showed no microbial growth on TSA plates. For larger fruit explants (tomato, rambutan, dogwood berry), fruits were completely submerged in 95% ethanol for up to a minute and rinsed thoroughly three times in sterile nanopure water. For Indian pipe stem, the exterior layers were shaved off using a sterile razor blade, and the rest of the tissue (likely cortex and vascular tissue) was weighed for processing. For fruits, surface-sterilized tissue was cut using a sterile razor blade, and using a separate blade, pulp was scooped to aseptically weigh 1 g for processing. All samples were homogenized in 9 mL of sterile water in a sterile mortar and pestle. Bacteria were mostly isolated in the first three dilutions (10−1 to 10−3) on tryptic soy agar (TSA) plates. Pure cultures of the bacteria were obtained by streaking for isolation. Bacteria identified as Curtobacterium species through 16S rRNA sequencing were further characterized. Bacteria were grown at 25 °C unless otherwise indicated.

Table 1.
Sequence identification of Curtobacterium species. The six isolates and their sources are listed. 16S rRNA contigs were subjected to NCBI Nucleotide search, and best matches are listed.
2.3. Visualization of Bacteria
Colony morphology analysis was performed by streaking for isolation and incubating at 25 °C for one week. Gram staining was performed as described previously [22] with 30 s sequential treatment of heat-fixed smears with crystal violet, Gram’s iodine, 95% ethanol and safranin.
2.4. Molecular Identification and Phylogenetic Analysis of Bacterial Isolates
To identify the bacteria, PCR was performed to amplify the 16S rRNA gene from the isolates. Without needing genomic DNA extraction from the bacteria, culture PCR was set up by adding 5 µL of overnight tryptic soy broth (TSB) cultures to 45 µL of master mix containing 1.25 µM primers—27F (5′-AGAGTTTGATYMTGGCTCAG-3′) and 1512R (5′-ACGGCTACCTTGTTACGACTT-3′) [23,24]—and DreamTaq Green PCR Master Mix (2X) (Thermo Scientific, Waltham, MA, USA). PCR was performed using the following conditions: 95 °C—2 min; 34 cycles of (94 °C—30 s, 57 °C—30 s, 72 °C—90 s); 72 °C—7 min. PCR amplification was confirmed through agarose gel (1.5%) electrophoresis and the PCR products were purified using the Invitrogen PureLink PCR Purification Kit (Thermo Scientific), and the purified PCR samples were submitted to Azenta Inc. (South Plainfield, NJ, USA) for Sanger sequencing. The forward and reverse sequences were combined to obtain a 1.4 kb contig (Supplementary Figure S1). All six 16S rRNA contig sequences have been submitted to Genbank, and the accession numbers are IPS11, PV019085; KB1, PV019086; PBH-A, PV019087; RMB2, PV019088; ST1.1, PV019089; WBB, PV019090. The 16S rRNA sequences were used for a Nucleotide BLAST search on NCBI. To construct a phylogenetic tree, M-Coffee [25] was first used to perform a multiple sequence alignment of the six contigs with reference Curtobacterium sequences obtained from the Genbank database. The alignment was then utilized to construct a maximum likelihood tree using the Tamura–Nei model of nucleotide substitutions [26] with 500 bootstrap replications on MEGA12 software [27], with Rathayibacter iranicus as the outgroup.
2.5. Detection of Bacterial Pigments
Carotenoid measurement was performed using a modification of a previous methanol-based extraction method [28]. Three replicate 3-milliliter aliquots of overnight TSB cultures for each isolate were centrifuged at 13,000 rpm to obtain pellets which were yellow or orange. The pellet was resuspended in 1 mL of 100% methanol, vortexed for 15–20 s to resuspend the pellet, incubated at 80 °C for 10 min (longer incubation reduced pigment yield) to extract the pigment and then centrifuged at 12,000 rpm for 1 min to pellet the cells. At this point, the pellet was discolored, and the supernatant acquired a yellow color with the extracted pigment. Aliquots of 200 µL of each replicate were loaded into a 96-well plate, and an absorption spectrum was generated and specific absorbance at 450 nm was measured using the Biotek Synergy H1M Microplate reader (Agilent Technologies, Santa Clara, CA, USA).
2.6. Metabolic Tests of Bacterial Isolates
The catalase test was performed by observing bubbling in 3% hydrogen peroxide. An oxidase test was conducted by adding one drop of oxidase reagent (BD oxidase reagent droppers) on a swab with bacteria and observing it for blue coloration as a positive result. Gelatinase was tested by looking for the liquefaction of nutrient gelatin (Difco) stabs. Citrate tests were positive if blue coloration was observed on Simmons citrate (Difco) slants or plates. To test amylase activity, bacteria were grown for 2 days on starch agar (Carolina Biological) and iodine was added to stain the starch in the plate. The presence of a halo around the bacterial growth indicated starch digestion and amylase activity. Caseinase activity was determined by growing bacteria on 3% skim milk agar plates in TSA. To quantify growth stimulation by milk, 3 replicate 20 µL drops (OD600 = 0.1) diluted from overnight cultures were plated on milk agar and allowed to grow for 5 days. All bacteria from each drop were suspended in 10 mL of sterile water and absorbance was read at 600 nm to quantify growth. Phosphatase activity was determined by swabbing or inoculating 20 µL drops (OD600 = 0.3) onto Pikovskaya’s agar [29] containing insoluble calcium phosphate, and a halo around bacterial growth was indicative of secreted phosphatase activity. Laccase activity was detected using LB agar plates supplemented with 0.35 mM copper sulfate and 0.01% 2,6-dimethoxyphenol, a chromogenic substrate that produces a red color when metabolized by a laccase enzyme [30,31].
2.7. Stress Tolerance of Bacterial Isolates
Mild, moderate and high salt tolerance was determined by growth on 5%, 7.5% and 10% sodium chloride, respectively, as described previously [32]. Growth at various temperatures was tested by swabbing bacteria onto TSA plates and incubating them at various temperatures (2 °C, 6 °C, 25 °C, 37 °C, 42 °C). To quantify growth at a higher temperature, isolates were inoculated in TSB cultures and allowed to sit at 25 °C, 42 °C and 45 °C for 2 days. Absorbance at 600 nm was recorded for cultures at day 0 and day 2 to quantify growth. To quantify growth in cold conditions, overnight TSB cultures of bacteria, normalized to OD600 = 0.1, were used to plate three replicate 20 µL drops on TSA plates and incubated at 25 °C and 6 °C for 5 days. Bacteria from each replicate were homogenized in 10 mL of sterile water and absorbance at 600 nm was observed to quantify growth.
2.8. Statistical Tests
Experiments were performed with three replicates, and each experiment was repeated 2–3 times. Results are presented as mean ± standard deviation. The significance of the results among the treatments was determined using Student t-tests (p < 0.05) and one-way ANOVA followed by Tukey’s post hoc test (https://astatsa.com/OneWay_Anova_with_TukeyHSD/, accessed on 20 January 2025).
3. Results
3.1. Curtobacterium Species Isolated from Various Sources
Twenty-three endophytic bacteria were isolated using serial dilution plating from eleven wild and store-bought stem, flower and fruit samples as part of a class project and identified using 16S rRNA sequencing (Supplementary Table S1). Since Curtobacterium was strikingly the most common genus from diverse explants and since knowledge of endophytic Curtobacterium is relatively scarce, six Curtobacterium species isolates were selected for comparative analysis (Table 1). Four of the isolates were from fruit pulp, one from flower petals, and one from the stem of the parasitic plant, Monotropa uniflora, commonly referred to as Indian pipe (Table 1). A 1.4 kb amplicon of the 16S rRNA gene was amplified from each of the isolates and sequenced to assemble ~1.4 kb contigs. A BLAST search of the contigs and phylogenetic analysis of the six endophytic isolates revealed best matches with three species: C. oceanosedimentum (IPS11, KB1, PBH-A), C. luteum (RMB2) and C. flaccumfaciens (ST1.1, WBB) (Table 1, Figure 1). Colony morphology was generally similar among the isolates (Table 2). Colonies were flat or raised, distinctly glossy and mostly circular with a tendency to fuse with neighboring colonies, resulting in ovoid colonies in a paint splatter pattern (Figure 2). All isolates were visualized as short Gram-positive rods in Gram staining, catalase-positive and oxidase-negative, as expected for Curtobacterium species (Figure 2; Table 3).
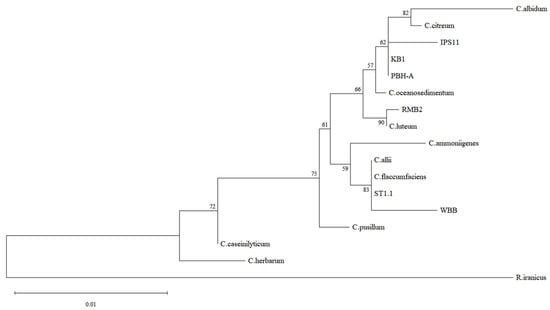
Figure 1.
Phylogenetic analysis of Curtobacterium species. 16S rRNA sequences of the six Curtobacterium isolates and representative Curtobacterium species were subjected to multiple sequence alignment using M-Coffee. The alignment was used in MEGA12 software to prepare a maximum likelihood tree using the Tamura–Nei model of nucleotide substitutions with 500 bootstrap replications. Genbank accession numbers of reference sequences: C. allii—OK275102; C. albidum—NR_026156.1; C. ammoniigenes—AB266597; C. caseinilyticum—OR143695; C. citreum—X77436; C. flaccumfaciens—AJ312209; C. herbarum—AJ310413; C. luteum—X77437; C. pusillum—AJ784400; Rathayibacter iranicus—NR_042575.1 (Microbacteriaceae relative outgroup).

Table 2.
Colony morphology analysis of Curtobacterium isolates. Five-day-old colonies streaked for isolation on TSA at 25 °C were assessed for the morphological features listed in the table.
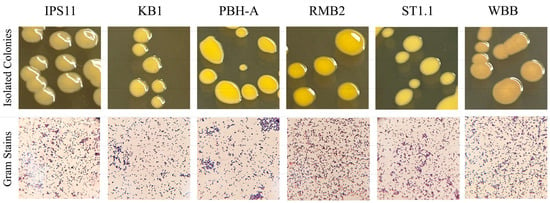
Figure 2.
Colony morphology and Gram staining of Curtobacterium isolates. Upper panel, Four-to-five-day-old colonies on TSA grown at 25 °C were used to document morphology. Lower panel, Gram staining was performed on 2-day-old colonies, and Gram-stained cells were documented at 100× magnification.

Table 3.
Enzyme tests of Curtobacterium isolates. Amylase, caseinase and phosphatase activities were tested by growing the isolates on 1% starch agar, 3% milk agar and Pikovskaya’s agar, respectively, for 4–5 days at 25 °C. Citrate utilization was tested on Simmons citrate agar slants as well as plates for 3–4 days. Urea hydrolysis test was performed in urea broth for 3–5 days. Laccase activity was tested on LB medium containing 2,6-dimethoxyphenol for 4–5 days. nd—not determined due to no growth.
3.2. Pigmentation in Curtobacterium Species
All Curtobacterium isolates, except IPS11, which generally produced pale yellow colonies, and WBB, which produced orange colonies, were characterized by vibrant yellow colonies (Figure 2 and Figure 3). A spectral scan of the extract of a representative isolate, RMB2, revealed a peak with an absorption maximum at 450 nm, which was absent in the non-pigmented control Bacillus species (Figure 3). Relatively high absorption at 450 nm was observed in all Curtobacterium isolates, suggestive of a potential carotenoid peak that could contribute to the yellow/orange coloration of the isolates.
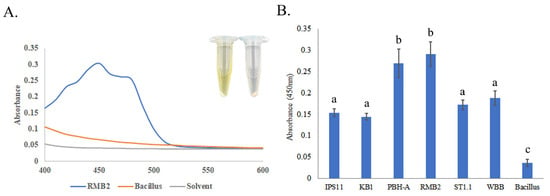
Figure 3.
Pigmentation in Curtobacterium isolates. Overnight TSB cultures of bacteria were used for pigment extraction in 100% methanol. (A) A spectral scan was performed with a methanol extract of RMB2 as well as the non-pigmented Bacillus subtilis. (B) Absorbance at 450 nm of all Curtobacterium isolates to quantify pigmentation. Error bars reflect standard deviation across three replicates. Statistical significance was determined using one-way ANOVA, and the letters above the bars indicate statistical grouping following Tukey’s post hoc test (p < 0.01).
3.3. All Curtobacterium Isolates Could Digest Starch, Casein and Insoluble Phosphate, but Differed in Their Ability to Utilize Citrate
All Curtobacterium species in this study appear to have certain conserved secreted enzyme activities. All isolates displayed amylase activity in digesting starch on starch agar, caseinase activity in digesting milk protein in milk agar and phosphatase activity in their ability to solubilize inorganic phosphate on Pikovskaya’s agar (Figure 4A,B). Furthermore, the growth of nearly all Curtobacterium species is remarkably stimulated by skim milk, suggesting that milk protein, sugars and/or minerals could enhance the growth of these isolates (Figure 4A). Interestingly, all isolates displayed an exceptionally mucoid and watery phenotype on Pikovskaya’s agar plates (Supplementary Figure S2). Two of the six isolates corresponding to the C. oceanosedimentum group, IPS11 and KB1, were able to utilize citrate and grow better on Simmons citrate agar, unlike the other isolates (Figure 4C). Interestingly, the isolate WBB did not grow on Simmons citrate agar, suggesting possible inhibition by bromothymol blue or other ingredients of the medium (Figure 4C). None of the isolates appeared to display gelatinase, urease or laccase enzyme activity (Table 3).
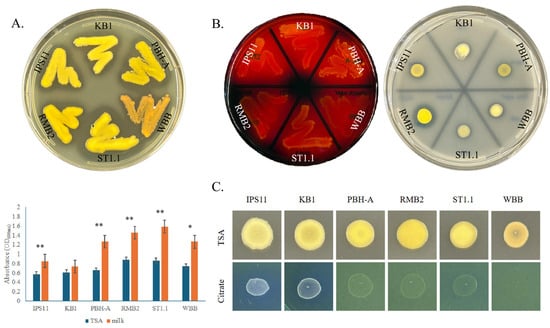
Figure 4.
Metabolic capabilities of Curtobacterium isolates. (A) Upper panel: Bacteria were swabbed onto 3% milk agar and grown for 2 days at 25 °C. Halo indicates positive casein hydrolysis. Lower panel: Graph indicating stimulation of growth of isolates on milk agar; 20 µL drops (OD600 = 0.1) of each isolate grown on TSA or milk agar for 5 days and bacteria quantified with absorbance at 600 nm. Error bars represent standard deviation, and statistical significance was confirmed with t-test. ns—no significant difference; *, p < 0.05, **, p < 0.01. (B) Amylase and phosphatase activity visualized by growth on starch agar and Pikovskaya’s agar, respectively, for 2 days. Clear zones around bacteria after adding iodine indicate amylase activity. Clear zones on Pikovskaya’s agar indicate inorganic phosphate solubilization. (C) Growth on TSA (control) and Simmons citrate agar after 5 days; 20 µL of overnight cultures (OD600 = 0.1) spotted in triplicate.
3.4. All Curtobacterium Isolates Could Ferment Fructose, Sucrose and Glucose, but Some Isolates Developed Specialized Sugar Fermentation Capacity
The ability of the Curtobacterium isolates to ferment ten different sugars was tested in phenol red broth with sugar supplements. All the isolates were able to ferment fructose, sucrose, glucose, galactose and arabinose, based on their ability to acidify the medium through sugar fermentation and turn it yellow (Table 4). However, only IPS11 was able to ferment mannitol, and only IPS11 and RMB2 were able to break down maltose. None of the Curtobacterium isolates were able to ferment lactose, sorbitol or trehalose.

Table 4.
Sugar tests of Curtobacterium isolates. Sugar fermentation was tested using phenol red broth supplemented with various sugars (1% final concentration). Yellow color observed in 2–5 dpi was indicative of sugar fermentation (+). Ara (Arabinose); Fru (Fructose); Gal (Galactose); Glu (Glucose); Lac (Lactose); Mal (Maltose); Man (Mannitol); Sor (Sorbitol); Suc (Sucrose); Tre (Trehalose).
3.5. All Curtobacterium Isolates Are Psychrotolerant, with the Exception of One Isolate That Is Thermotolerant
All Curtobacterium isolates, except RMB2, demonstrated cold tolerance, being able to grow at 6 °C, with one isolate, ST1.1, displaying superior cold tolerance (Figure 5A). Interestingly, RMB2 was the only isolate that could grow at high temperatures up to 45 °C (Figure 5B). Besides temperature tolerance, all Curtobacterium bacterium species displayed moderate salt tolerance based on their ability to grow on 7.5% sodium chloride (Table 5).
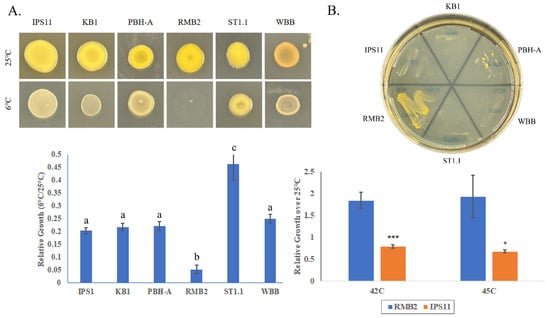
Figure 5.
Temperature tolerance of Curtobacterium isolates. (A) Upper panel: overnight TSB cultures diluted to OD600 of 0.1 were spotted in triplicate and incubated at 6 °C or 25 °C for 5 days. Lower panel: Growth was quantified by suspending each replicate in 10 mL of sterile water and determining absorbance at 600 nm. Error bars are standard deviation. One-way ANOVA was performed followed by Tukey’s post hoc test. Letters indicate significance groupings (p < 0.01). (B) Upper panel: growth on TSA after 4 days at 42 °C. Lower panel: Absorbance (OD600) of cultures after incubating for 2 days at 25 °C, 42 °C or 45 °C. Error bars represent standard deviation. t-test was performed, and significant differences are as follows: * p ≤ 0.05, *** p ≤ 0.001.

Table 5.
Temperature and salt tolerance of Curtobacterium isolates. Salt tolerance was examined by swabbing the isolates on TSA containing 5%, 7.5% and 10% sodium chloride and incubating at 25 °C for 5 days. Cold and heat tolerance was tested by growing the isolates on TSA for up to a week at various temperatures; plates were incubated for 2 weeks at 2 °C.
4. Discussion
In this study, we compared six endophytic isolates of Curtobacterium species isolated from various plant sources, specifically fruit, flower and stem tissue. The isolates appeared to be mostly related to C. flaccumfaciens, C. luteum and C. oceanosedimentum species based on a BLAST search and phylogenetic analysis of 16S rRNA sequences. All tested isolates were yellow- or orange-pigmented, short or curt Gram-positive rods (hence the name Curtobacterium), mostly psychrotolerant and capable of starch, casein, and insoluble phosphate hydrolysis, able to digest common plant sugars and differentially utilized citrate and other sugars. We also found a thermotolerant Curtobacterium isolate, suggesting novel adaptations.
4.1. Curtobacterium Species as Plant Endophytes
Four of the six Curtobacterium isolates in this study were isolated from fruits—KB1, RMB2, ST1.2 and WBB (Table 1). Consistent with our observations, Curtobacterium species have previously been found on the surface or interior of a number of fruits, suggesting that Curtobacterium is a common pomophyte, inhabiting fruits. Specifically, Curtobacterium has been isolated as an epiphyte from blueberry [33], wild cranberry fruit [34], withered grapes [35] and nectarine [36], and as an endophyte from the fruits of coffee berry [37], mulberry [19], and strawberry [12]. Typically, C. flaccumfaciens and C. citreum have been reported in fruits, which makes our observation of C. luteum (RMB2) and C. oceanosedimentum (KB1) in fruits novel. One of the Curtobacterium strains was isolated from purple hydrangea flower petals (PBH-A) and is the first characterization of Curtobacterium from flower petals, since only one other study has reported the floral isolation of Curtobacterium, specifically from apple flower stigmas [38]. Curtobacterium has also been commonly found on or inside the stem of dry bean (Phaseolus sp.) [39] Eucalyptus [40], sugarcane [41], tea chrysanthemum [42], tomato [43], willow tree [44] and yerba mate (Ilex sp.) [45]. Our isolation of Curtobacterium from the stem of the parasitic plant Monotropa uniflora (IPS11) in this study adds to the knowledge that Curtobacterium is a stem endophyte in a diversity of plants. C. flaccumfaciens isolates are mostly reported as pathogenic strains from diseased plants. Rarely, C. flaccumfaciens have also been isolated from healthy tissue as endophytes [46], so non-pathogenic strains or opportunistic pathogens of C. flaccumfaciens could exist. Indeed, non-pathogenic variants of pathogens such as Pseudomonas syringae have been reported [47], with the variants missing key enzymes and transcriptional regulators corresponding to virulence based on genome analysis. Since all strains in this study were isolated from apparently healthy tissue, it is possible that these isolates are commensals that are supported by the host tissue and protect the host from potential pathogens or could themselves be opportunistic pathogens.
4.2. Morphological Features of Curtobacterium Species
Curtobacterium has primarily been described as a yellow-pigmented genus; some C. flaccumfaciens isolates have been noted to be orange or pink [48]. Consistently, nearly all our isolates were yellow, with the exception of the C. flaccumfaciens isolate WBB, which displayed an orange color (Figure 2 and Figure 3). There are two reported lineages of C. flaccumfaciens—yellow and orange colony variants [4]—and our two C. flaccumfaciens isolates, ST1.1 (yellow) and WBB (orange), could correspond to these lineages. Interestingly, all isolates, including WBB, showed a similar absorption spectrum with a maximal absorption at 450 nm (Figure 3), suggesting the presence of a potential carotenoid pigment producing the yellow color [49], akin to Pantoea stewarti, which is also yellow with an absorption maximum of 450 nm due to a carotenoid pigment [50]. Since the orange-colored WBB had a similar absorption spectrum to the others, the orange color may be a reflection of a different internal environment (perhaps pH) in WBB that may allow the same pigment to display a different color. Carotenoids are pigments that can serve as blue light filters by absorbing at 450 nm and thus protect from damage by excess light. They could also protect cells from reactive oxygen species arising from light exposure or other sources. The apparent presence of carotenoids or other pigments in all our isolates (even endophytes that may have limited light exposure) as well as every Curtobacterium reported in the literature suggests that the pigments could be intimately conserved in Curtobacterium species as antioxidant guardians. Carotenoids are also known to possess antimicrobial properties [51,52], and the carotenoid extract of the endophytic Citricoccus displayed antibacterial characteristics [52]. Bacteria can secrete carotenoids using extracellular vesicles [53], and thus, it is possible that the pigmentation in endophytic Curtobacterium species may also correspond to a bioactive plant protective function.
Colonies of all six Curtobacterium isolates on tryptic soy agar plates were circular to ovoid and generally flat or raised (Figure 2), which may be reflective of the obligate aerobic nature of Curtobacterium species [54], which is interesting for endophytes that appear to be living in oxygen-limiting conditions inside plant tissues. Colonies of some of the isolates were mucoid as has been reported for C. pusillum [14], which may reflect the ability of some of the isolates to synthesize water-retaining extracellular polysaccharides (EPSs), possibly using sugars from the medium.
4.3. Nutritional Preferences of Curtobacterium Species
All six isolates tested were capable of degrading the plant carbohydrate starch and the milk protein casein as well as solubilizing phosphate based on plate assays (Figure 4, Table 3). Since all strains were isolated as endophytes from plants (fruit, flower, stem), this may be reflective of their reliance on the starch in their environment for nutrition. Casein hydrolysis, mediated by the exoenzyme caseinase, is confirmed by the presence of a clear zone around the bacteria on a milk agar plate, and all isolates in this study were caseinase-positive. Caseinase is not only found in bacteria associated with milk and dairy products but also appears to be a virulence factor in digesting host proteins that are structurally similar to casein [55]. All six diverse Curtobacterium isolates in our study were caseinase-positive, and the enzyme may perhaps digest casein-like proteins in the host or remodel its own secreted proteins or extracellular matrix with protease activity. Indeed, every reference that tested casein hydrolysis reported that the Curtobacterium isolates were caseinase-positive [43,56,57,58,59], suggesting that casein hydrolysis by Curtobacterium species may be universally conserved. Furthermore, casein hydrolysis is one of the confirmatory markers used to identify pathogenic C. flaccumfaciens, as recommended by the US National Seed Health System in the Be 4.3 Selective Media Assay (University of Idaho). It is interesting to note that milk supplementation enhanced the growth of many of the Curtobacterium isolates (Figure 4), likely due to enrichment with milk protein, sugars, minerals and vitamins. This suggests that Curtobacterium growth in culture could be enhanced by adding skim milk.
Phosphate is abundantly present in soil, but most of it is insoluble and inaccessible, making many plants rely on microbes that solubilize phosphate by secreting extracellular phosphatases [60]. All six isolates in this study could solubilize inorganic phosphate based on a halo on Pikovskaya’s agar, which contains insoluble calcium phosphate (Figure 4). This may be indicative of their ability to hydrolyze insoluble phosphate while in soil or within the plant’s internal tissue (fruit, flower, stem) while they are endophytes. Perhaps the endophytes could utilize structural phosphate present around plant cells or stored in fruits and other tissues [61]. Curtobacterium has been broadly reported as a phosphate-solubilizing bacterium [10,62,63,64,65,66], supporting our observation and indicating that the secreted phosphatase activity may be widely conserved. Remarkably, all Curtobacterium isolates exhibited a highly mucoid and runny phenotype only on phosphate-containing Pikovskaya’s, agar suggesting that phosphate could promote mucoidal growth, perhaps by stimulating the production of water-retaining extracellular polysaccharides or phosphorylated polysaccharides. Indeed, phosphate appeared to be required for EPS production in Enterobacter sp. [67]
Citrate utilization as a carbon and energy source by Curtobacterium species has rarely been published, and the reported species were unable to absorb or metabolize citrate [10,68]. Some of the isolates in our study—specifically, those belonging to the C. oceanosedimentum group—were able to utilize citrate as observed on Simmons citrate agar, while others were not (Figure 4). This may suggest local nutritional adaptations of these strains based on their environment. Fruits and other plant organs are rich in nutrients such as minerals and sugars, particularly fructose, sucrose and glucose [69]. Not surprisingly, all six isolates in this study were able to ferment these three sugars in addition to galactose and arabinose (Table 4), and the fruit/plant sugars could be supporting the endophytic growth of these Curtobacterium species. Indeed, many of the strains previously reported appeared to be capable of fermenting these sugars [14,58,70,71].
4.4. Temperature Adaptations of Curtobacterium Species
All Curtobacterium isolates displayed moderate levels of salt stress tolerance, based on their growth on 7.5% sodium chloride (Table 5), suggesting that osmotic stress tolerance may be conserved. The ability to grow at various temperatures was surprising. Microbes that are able to grow at temperatures below 7 °C are considered psychrotolerant [72,73]. It appears from evidence in the literature that Curtobacterium species are generally mesophilic bacteria with an optimum growth temperature of 25–30 °C [14,58], with only an isolated report of a strain being able to grow at low temperatures (5 °C) [74]. Surprisingly, we found five out of six isolates to be cold-tolerant, being able to grow comfortably at 6 °C (Table 5, Figure 5). This suggests that psychrotolerance may be conserved in two major clusters of Curtobacterium: C. oceanosedimentum (IPS11, KB1, PBH-A) and C. flaccumfacciens (ST1.1, WBB). Cold adaptation may be necessary for the survival of these Curtobacterium isolates in their endophytic environments, especially with the higher water contents in fruits that raise the risk of lethality by icing and freezing.
Psychrotolerance in endophytes could have implications for the health of the host plant. Cold-tolerant endophytes have been shown to enhance cold tolerance in their hosts. For instance, Microbacterium testaceum isolated from seeds of a cold-tolerant rice variety could confer cold tolerance to a cold-sensitive rice cultivar [75]. Similarly, psychrotolerant bacteria isolated from the Antarctic plant Colobanthus stimulated the synthesis of secondary metabolites like salicylic acid, implicated in cold tolerance [76]. Since cold-tolerant bacteria secrete antifreeze proteins to minimize freezing and increase their survival [77], the possible secretion of such proteins by endophytic Curtobacterium spp. could also protect water-rich fruit and stem endophytic compartments and thus protect the host plant from freezing. It has been suggested that carotenoids in the membrane could increase membrane flexibility under low temperatures, so it is possible that the pigmentation in Curtobacterium species also promotes cold tolerance.
Temperature at or over 45 °C is considered the beginning of the thermotolerance range for bacteria [78], and growth at 42 °C in some cases is considered a transition from a mesophile habit to thermotolerance [79]. Only one of the isolates, RMB2 (C. luteum), isolated from tropical rambutan fruit, lacked the ability to grow at low temperatures in this study. Intriguingly, RMB2 showed relatively exceptional heat tolerance in contrast to the other five isolates, being able to grow at temperatures as high as 45 °C (Figure 5). This observation aligns with a previous study demonstrating the ability of C. luteum isolated from seagrass meadow to grow at up to 45 °C [10] and suggests that C. luteum isolates of Asian origin may have evolved heat adaptations. Heat-tolerant strains of Curtobacterium could widen their applications in areas such as plant growth promotion in the face of climate change and in commercial enzyme production. The Curtobacterium strains in this study were isolated from diverse sources—the Indian pipe, dogwood berry and hydrangea samples were wild samples from Western Pennsylvania, the tomato fruits were locally grown in Erie, PA, and the rambutan fruits, purchased from markets, are most likely of Southeast Asian origin. It is unclear how the differences in the isolates correlate with their natural environments, but it is striking that one of the two rambutan isolates, KB1, was similar to the other isolates in most aspects, while the other Rambutan isolate (RMB2) appeared to have developed distinct temperature tolerance capabilities, possibly due to unknown local environmental constraints.
5. Conclusions
In this study, we found two new species of Curtobacterium—C. luteum and C. oceanosedimentum—to be endophytes in plants, specifically in understudied niches, floral and fruit tissues. Surprisingly, we found that nearly all endophytic isolates were psychrotolerant with potential benefits to the host during cold stress, and this capacity of Curtobacterium appears to be underevaluated. Only one of the isolates (C. luteum) was distinctly cold-sensitive and, exceptionally, turned out to be heat-tolerant, suggesting the differential evolution of stress tolerance in Curtobacterium species and also implying that Curtobacterium species could be a source of cold-active and thermostable enzymes. All the plant isolates in this study, consistent with their environment, were able to digest plant carbohydrates—starch, sucrose and fructose—in addition to inorganic phosphate and casein. The growth stimulation of these isolates by milk suggests that Curtobacterium growth in culture could be enhanced through milk supplementation. The endophytic nature of these bacteria not only suggests the potential for plant protection or plant growth promotion but also implies possible vertical seed-borne transmission to the next generation, and these prospects could be tested in future studies. In summary, the study of endophytic Curtobacterium species revealed many conserved traits with variations that may imply local adaptations, and the presence of Curtobacterium isolates in flowers and fruits suggests that vertical transmission is possible to perpetuate the endophytic habit through generations.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/bacteria4020025/s1, Figure S1: Contigs of 16S rRNA amplified from Curtobacterium isolates. Figure S2: Growth of Curtobacterium isolates on Pikovskayas agar. Table S1: Endophytes isolated in this study and selection of Curtobacterium species.
Author Contributions
Conceptualization, R.M.; methodology, A.A. and R.M.; software, I.F., S.W. and R.M.; validation, A.A. and R.M.; formal analysis, A.A., S.W. and R.M.; investigation, A.A., S.W., K.B., K.C., K.S. and R.M.; resources, R.M.; data curation, A.A. and R.M.; writing—original draft preparation, A.A., S.W., K.B., K.C., K.S. and R.M.; writing—review and editing, S.W. and R.M.; visualization, I.F., A.A. and R.M.; supervision, R.M.; project administration, R.M.; funding acquisition, R.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Darbaker Prize in Microscopical Biology awarded to R.M. and undergraduate research grants awarded by the Pennsylvania Academy of Sciences (2024).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All sequence data has been deposited in the Genbank database as explained in the methods.
Acknowledgments
We would like to thank Justin Baker and Allison Cortina for assistance with the isolation of strains.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
| EPS | Extracellular Polysaccharide |
| IPS11 | Indian Pipe Stem 1 |
| OD | Optical Density |
| PCR | Polymerase Chain Reaction |
| TSA(B) | Tryptic Soy Agar (Broth) |
References
- Saddler, G.S.; Guimarāes, P.M. Curtobacterium. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons, Ltd.: Chichester, UK, 2015; pp. 1–14. [Google Scholar] [CrossRef]
- Evseev, P.; Lukianova, A.; Tarakanov, R.; Tokmakova, A.; Shneider, M.; Ignatov, A.; Miroshnikov, K. Curtobacterium spp. and Curtobacterium flaccumfaciens: Phylogeny, Genomics-Based Taxonomy, Pathogenicity, and Diagnostics. Curr. Issues Mol. Biol. 2022, 44, 889–927. [Google Scholar] [CrossRef]
- Chase, A.B.; Arevalo, P.; Polz, M.F.; Berlemont, R.; Martiny, J.B. Evidence for Ecological Flexibility in the Cosmopolitan Genus Curtobacterium. Front. Microbiol. 2016, 7, 1874. [Google Scholar] [CrossRef] [PubMed]
- Osdaghi, E.; Taghavi, S.M.; Calamai, S.; Biancalani, C.; Cerboneschi, M.; Tegli, S.; Harveson, R.M. Phenotypic and Molecular-Phylogenetic Analysis Provide Novel Insights into the Diversity of Curtobacterium flaccumfaciens. Phytopathology 2018, 108, 1154–1164. [Google Scholar] [CrossRef] [PubMed]
- Khanal, M.; Bhatta, B.P.; Malla, S. Isolation and Characterization of Bacteria Associated with Onion and First Report of Onion Diseases Caused by Five Bacterial Pathogens in Texas, U.S.A. Plant Dis. 2023, 107, 1721–1729. [Google Scholar] [CrossRef] [PubMed]
- Sturz, A.V.; Christie, B.R.; Matheson, B.G.; Nowak, J. Biodiversity of endophytic bacteria which colonize red clover nodules, roots, stems and foliage and their influence on host growth. Biol. Fertil. Soils 1997, 25, 13–19. [Google Scholar] [CrossRef]
- DUNLEAVY, J.M. Curtobacterium plantarum sp. nov. Is Ubiquitous in Plant Leaves and Is Seed Transmitted in Soybean and Corn†. Int. J. Syst. Evol. Microbiol. 1989, 39, 240–249. [Google Scholar] [CrossRef]
- Munir, S.; Li, Y.; He, P.; Huang, M.; He, P.; He, P.; Cui, W.; Wu, Y.; He, Y. Core endophyte communities of different citrus varieties from citrus growing regions in China. Sci. Rep. 2020, 10, 3648. [Google Scholar] [CrossRef]
- Behrendt, U.; Ulrich, A.; Schumann, P.; Naumann, D.; Suzuki, K.I. Diversity of grass-associated Microbacteriaceae isolated from the phyllosphere and litter layer after mulching the sward; polyphasic characterization of Subtercola pratensis sp. nov., Curtobacterium herbarum sp. nov. and Plantibacter flavus gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 1441–1454. [Google Scholar] [CrossRef]
- Saranya, K.; Sundaramanickam, A.; Manupoori, S.; Kanth, S.V. Screening of multi-faceted phosphate-solubilising bacterium from seagrass meadow and their plant growth promotion under saline stress condition. Microbiol. Res. 2022, 261, 127080. [Google Scholar] [CrossRef]
- Patel, M.; Patel, K.; Al-Keridis, L.A.; Alshammari, N.; Badraoui, R.; Elasbali, A.M.; Al-Soud, W.A.; Hassan, M.I.; Yadav, D.K.; Adnan, M. Cadmium-Tolerant Plant Growth-Promoting Bacteria Curtobacterium oceanosedimentum Improves Growth Attributes and Strengthens Antioxidant System in Chili (Capsicum frutescens). Sustainability 2022, 14, 4335. [Google Scholar] [CrossRef]
- de Melo Pereira, G.V.; Magalhaes, K.T.; Lorenzetii, E.R.; Souza, T.P.; Schwan, R.F. A multiphasic approach for the identification of endophytic bacterial in strawberry fruit and their potential for plant growth promotion. Microb. Ecol. 2012, 63, 405–417. [Google Scholar] [CrossRef]
- Vimal, S.R.; Patel, V.K.; Singh, J.S. Plant growth promoting Curtobacterium albidum strain SRV4: An agriculturally important microbe to alleviate salinity stress in paddy plants. Ecol. Indic. 2019, 105, 553–562. [Google Scholar] [CrossRef]
- Funke, G.; Aravena-Roman, M.; Frodl, R. First description of Curtobacterium spp. isolated from human clinical specimens. J. Clin. Microbiol. 2005, 43, 1032–1036. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Zhao, Y.; Shen, W.; Han, D.; Yang, M. Seed-to-Seed: Plant Core Vertically Transmitted Microbiota. J. Agric. Food Chem. 2023, 71, 19255–19264. [Google Scholar] [CrossRef] [PubMed]
- Fagorzi, C.; Mengoni, A. Endophytes: Improving Plant Performance. Microorganisms 2022, 10, 1777. [Google Scholar] [CrossRef]
- Chesneau, G.; Torres-Cortes, G.; Briand, M.; Darrasse, A.; Preveaux, A.; Marais, C.; Jacques, M.-A.; Shade, A.; Barret, M. Temporal dynamics of bacterial communities during seed development and maturation. FEMS Microbiol. Ecol. 2020, 96, fiaa190. [Google Scholar] [CrossRef]
- Bulgari, D.; Minio, A.; Casati, P.; Quaglino, F.; Delledonne, M.; Bianco, P.A. Curtobacterium sp. Genome Sequencing Underlines Plant Growth Promotion-Related Traits. Genome Announc. 2014, 2, e00592-14. [Google Scholar] [CrossRef]
- Xu, Z.Y.; Li, X.P.; Du, Y.; Xu, B.T.; Zhang, X.L. Responses of soil microbial communities in mulberry rhizophere to intercropping and nitrogen application. Ying Yong Sheng Tai Xue Bao J. Appl. Ecol. 2019, 30, 1983–1992. [Google Scholar] [CrossRef]
- Burgdorf, R.J.; Laing, M.D.; Morris, C.D.; Jamal-Ally, S.F. A procedure to evaluate the efficiency of surface sterilization methods in culture-independent fungal endophyte studies. Braz. J. Microbiol. Publ. Braz. Soc. Microbiol. 2014, 45, 977–983. [Google Scholar] [CrossRef]
- Qin, S.; Li, J.; Chen, H.H.; Zhao, G.Z.; Zhu, W.Y.; Jiang, C.L.; Xu, L.H.; Li, W.J. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl. Environ. Microbiol. 2009, 75, 6176–6186. [Google Scholar] [CrossRef]
- Moyes, R.B.; Reynolds, J.; Breakwell, D.P. Differential Staining of Bacteria: Gram Stain. Curr. Protoc. Microbiol. 2009, 15, A.3C.1–A.3C.8. [Google Scholar] [CrossRef]
- Suzuki, M.T.; Giovannoni, S.J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 1996, 62, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Kane, M.D.; Poulsen, L.K.; Stahl, D.A. Monitoring the enrichment and isolation of sulfate-reducing bacteria by using oligonucleotide hybridization probes designed from environmentally derived 16S rRNA sequences. Appl. Environ. Microbiol. 1993, 59, 682–686. [Google Scholar] [CrossRef] [PubMed]
- Wallace, I.M.; O’Sullivan, O.; Higgins, D.G.; Notredame, C. M-Coffee: Combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006, 34, 1692–1699. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Sanderford, M.; Sharma, S.; Tamura, K. MEGA12: Molecular Evolutionary Genetic Analysis Version 12 for Adaptive and Green Computing. Mol. Biol. Evol. 2024, 41, msae263. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.R.; Mahawar, H.; Bajpai, A.; Dubey, G.; Parmar, R.; Atoliya, N.; Devi, M.H.; Singh, A.B.; Jain, D.; Patra, A.; et al. Methylotroph bacteria and cellular metabolite carotenoid alleviate ultraviolet radiation-driven abiotic stress in plants. Front. Microbiol. 2022, 13, 899268. [Google Scholar] [CrossRef]
- Pikovskaya, R.I. Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Microbiology 1948, 17, 362–370. [Google Scholar]
- Wan, Y.; Du, Y.; Miyakoshi, T. Enzymatic catalysis of 2,6-dimethoxyphenol by laccases and products characterization in organic solutions. Sci. China Ser. B Chem. 2008, 51, 669–676. [Google Scholar] [CrossRef]
- Neifar, M.; Chouchane, H.; Mahjoubi, M.; Jaouani, A.; Cherif, A. Pseudomonas extremorientalis BU118: A new salt-tolerant laccase-secreting bacterium with biotechnological potential in textile azo dye decolourization. 3 Biotech 2016, 6, 107. [Google Scholar] [CrossRef]
- Schillaci, M.; Raio, A. Pseudomonas and Curtobacterium Strains from Olive Rhizosphere Characterized and Evaluated for Plant Growth Promoting Traits. Plants 2022, 11, 2245. [Google Scholar] [CrossRef] [PubMed]
- Chacon, F.I.; Sineli, P.E.; Mansilla, F.I.; Pereyra, M.M.; Diaz, M.A.; Volentini, S.I.; Poehlein, A.; Meinhardt, F.; Daniel, R.; Dib, J.R. Native Cultivable Bacteria from the Blueberry Microbiome as Novel Potential Biocontrol Agents. Microorganisms 2022, 10, 969. [Google Scholar] [CrossRef]
- Kooner, A.; Soby, S. Draft Genome Sequence of Curtobacterium sp. Strain MWU13-2055, Isolated from a Wild Cranberry Fruit Surface in Massachusetts, USA. Microbiol. Resour. Announc. 2022, 11, e0056522. [Google Scholar] [CrossRef]
- Lorenzini, M.; Zapparoli, G. Epiphytic bacteria from withered grapes and their antagonistic effects on grape-rotting fungi. Int. J. Food Microbiol. 2020, 319, 108505. [Google Scholar] [CrossRef] [PubMed]
- Janisiewicz, W.J.; Buyer, J.S. Culturable bacterial microflora associated with nectarine fruit and their potential for control of brown rot. Can. J. Microbiol. 2010, 56, 480–486. [Google Scholar] [CrossRef]
- Vega, F.E.; Pava-Ripoll, M.; Posada, F.; Buyer, J.S. Endophytic bacteria in Coffea arabica L. J. Basic Microbiol. 2005, 45, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Steven, B.; Zeng, Q. Complete Genome Sequences of Curtobacterium, Pantoea, Erwinia, and Two Pseudomonas sp. Strains, Isolated from Apple Flower Stigmas from Connecticut, USA. Microbiol. Resour. Announc. 2021, 10, e00154-21. [Google Scholar] [CrossRef]
- Harveson, R.M.; Schwartz, H.F.; Urrea, C.A.; Yonts, C.D. Bacterial Wilt of Dry-Edible Beans in the Central High Plains of the U.S.: Past, Present, and Future. Plant Dis. 2015, 99, 1665–1677. [Google Scholar] [CrossRef]
- Procopio, R.E.; Araujo, W.L.; Maccheroni, W., Jr.; Azevedo, J.L. Characterization of an endophytic bacterial community associated with Eucalyptus spp. Genet. Mol. Res. GMR 2009, 8, 1408–1422. [Google Scholar] [CrossRef]
- Magnani, G.S.; Didonet, C.M.; Cruz, L.M.; Picheth, C.F.; Pedrosa, F.O.; Souza, E.M. Diversity of endophytic bacteria in Brazilian sugarcane. Genet. Mol. Res. GMR 2010, 9, 250–258. [Google Scholar] [CrossRef]
- Sun, T.; Yang, Y.; Duan, K.; Liao, Y.; Zhang, Z.; Guan, Z.; Chen, S.; Fang, W.; Chen, F.; Zhao, S. Biodiversity of Endophytic Microbes in Diverse Tea Chrysanthemum Cultivars and Their Potential Promoting Effects on Plant Growth and Quality. Biology 2023, 12, 986. [Google Scholar] [CrossRef] [PubMed]
- Kizheva, Y.; Pandova, M.; Dimitrova, M.; Gladicheva, Y.; Garkova, M.; Pirnareva, D.; Donchev, D.; Moncheva, P.; Hristova, P. First Report of Curtobacterium flaccumfaciens in Bulgaria. Pathogens 2024, 13, 483. [Google Scholar] [CrossRef]
- Freeman, J.; Firrincieli, A.; Baker, D.; Doty, S. Curtobacterium salicis sp. nov., isolated from willow tree stems in Washington state. Antonie Leeuwenhoek 2024, 117, 62. [Google Scholar] [CrossRef]
- Perez, M.L.; Collavino, M.M.; Sansberro, P.A.; Mroginski, L.A.; Galdeano, E. Diversity of endophytic fungal and bacterial communities in Ilex paraguariensis grown under field conditions. World J. Microbiol. Biotechnol. 2016, 32, 61. [Google Scholar] [CrossRef] [PubMed]
- Gagne-Bourgue, F.; Aliferis, K.A.; Seguin, P.; Rani, M.; Samson, R.; Jabaji, S. Isolation and characterization of indigenous endophytic bacteria associated with leaves of switchgrass (Panicum virgatum L.) cultivars. J. Appl. Microbiol. 2013, 114, 836–853. [Google Scholar] [CrossRef] [PubMed]
- Passera, A.; Compant, S.; Casati, P.; Maturo, M.G.; Battelli, G.; Quaglino, F.; Antonielli, L.; Salerno, D.; Brasca, M.; Toffolatti, S.L.; et al. Not Just a Pathogen? Description of a Plant-Beneficial Pseudomonas syringae Strain. Front. Microbiol. 2019, 10, 1409. [Google Scholar] [CrossRef]
- Osdaghi, E.; Young, A.J.; Harveson, R.M. Bacterial wilt of dry beans caused by Curtobacterium flaccumfaciens pv. flaccumfaciens: A new threat from an old enemy. Mol. Plant Pathol. 2020, 21, 605–621. [Google Scholar] [CrossRef]
- Stahl, W.; Sies, H. Antioxidant activity of carotenoids. Mol. Asp. Med. 2003, 24, 345–351. [Google Scholar] [CrossRef]
- Mohammadi, M.; Burbank, L.; Roper, M.C. Biological role of pigment production for the bacterial phytopathogen Pantoea stewartii subsp. stewartii. Appl. Environ. Microbiol. 2012, 78, 6859–6865. [Google Scholar] [CrossRef]
- Ungureanu, C.; Ferdeș, M. Evaluation of Antioxidant and Antimicrobial Activities of Torularhodin. Adv. Sci. Lett. 2012, 18, 50–53. [Google Scholar] [CrossRef]
- Karpiński, T.M.; Ożarowski, M. What Do We Know about Antimicrobial Activity of Astaxanthin and Fucoxanthin? Mar. Drugs 2021, 20, 36. [Google Scholar] [CrossRef] [PubMed]
- Biller, S.J.; Lundeen, R.A. Prochlorococcus extracellular vesicles: Molecular composition and adsorption to diverse microbes. Environ. Microbiol. 2022, 24, 420–435. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, J.C.; Lang, J.M.; Darling, A.E.; Eisen, J.A.; Coil, D.A. Draft Genome Sequence of Curtobacterium flaccumfaciens Strain UCD-AKU (Phylum Actinobacteria). Genome Announc. 2013, 1, e00244-13. [Google Scholar] [CrossRef] [PubMed]
- Preda, M.; Mihai, M.M. Phenotypic and genotypic virulence features of staphylococcal strains isolated from difficult-to-treat skin and soft tissue infections. PLoS ONE 2021, 16, e0246478. [Google Scholar] [CrossRef]
- Feng, G.-D.; Li, J.; Yang, S.; Zhang, J.; Zhu, H. Curtobacterium caseinilyticum sp. nov., Curtobacterium subtropicum sp. nov. and Curtobacterium citri sp. nov., isolated from citrus phyllosphere. Int. J. Syst. Evol. Microbiol. 2023, 73, 006152. [Google Scholar] [CrossRef]
- Krimi, Z.; Ziouche, C.; Tafifet, L.; Djellout, H.; Mohamed-Mahmoud, F.; Raio, A. Euphorbia helioscopia a Putative Plant Reservoir of Pathogenic Curtobacterium flaccumfaciens. Curr. Microbiol. 2023, 80, 154. [Google Scholar] [CrossRef]
- Chen, Y.F.; Yin, Y.N.; Zhang, X.M.; Guo, J.H. Curtobacterium flaccumfaciens pv. beticola, A New Pathovar of Pathogens in Sugar Beet. Plant Dis. 2007, 91, 677–684. [Google Scholar] [CrossRef]
- Gonzalez, A.J.; Tello, J.C.; Rodicio, M.R. Bacterial Wilt of Beans (Phaseolus vulgaris) Caused by Curtobacterium flaccumfaciens in Southeastern Spain. Plant Dis. 2005, 89, 1361. [Google Scholar] [CrossRef]
- Rodríguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Weigend, M.; Mustafa, A.; Ensikat, H.J. Calcium phosphate in plant trichomes: The overlooked biomineral. Planta 2018, 247, 277–285. [Google Scholar] [CrossRef]
- Pereira, S.I.; Castro, P.M. Diversity and characterization of culturable bacterial endophytes from Zea mays and their potential as plant growth-promoting agents in metal-degraded soils. Environ. Sci. Pollut. Res. Int. 2014, 21, 14110–14123. [Google Scholar] [CrossRef] [PubMed]
- Diez-Mendez, A.; Rivas, R. Improvement of saffron production using Curtobacterium herbarum as a bioinoculant under greenhouse conditions. AIMS Microbiol. 2017, 3, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Kirui, C.K.; Njeru, E.M.; Runo, S. Diversity and Phosphate Solubilization Efficiency of Phosphate Solubilizing Bacteria Isolated from Semi-Arid Agroecosystems of Eastern Kenya. Microbiol. Insights 2022, 15, 11786361221088991. [Google Scholar] [CrossRef]
- Kandel, S.L.; Firrincieli, A.; Joubert, P.M.; Okubara, P.A.; Leston, N.D.; McGeorge, K.M.; Mugnozza, G.S.; Harfouche, A.; Kim, S.H.; Doty, S.L. An In vitro Study of Bio-Control and Plant Growth Promotion Potential of Salicaceae Endophytes. Front. Microbiol. 2017, 8, 386. [Google Scholar] [CrossRef]
- Verma, S.K.; White, J.F. Indigenous endophytic seed bacteria promote seedling development and defend against fungal disease in browntop millet (Urochloa ramosa L.). J. Appl. Microbiol. 2018, 124, 764–778. [Google Scholar] [CrossRef] [PubMed]
- Concórdio-Reis, P.; Pereira, J.R.; Torres, C.A.V.; Sevrin, C.; Grandfils, C.; Freitas, F. Effect of mono- and dipotassium phosphate concentration on extracellular polysaccharide production by the bacterium Enterobacter A47. Process Biochem. 2018, 75, 16–21. [Google Scholar] [CrossRef]
- Kim, M.K.; Kim, Y.J.; Kim, H.B.; Kim, S.Y.; Yi, T.H.; Yang, D.C. Curtobacterium ginsengisoli sp. nov., isolated from soil of a ginseng field. Int. J. Syst. Evol. Microbiol. 2008, 58, 2393–2397. [Google Scholar] [CrossRef]
- Vincente, A.R.; Manganaris, G.A.; Ortiz, C.M.; Sozzi, G.O.; Crisosto, C.H. Chapter 5—Nutritional Quality of Fruits and Vegetables. In Postharvest Handling, 3rd ed.; Florkowski, W.J., Shewfelt, R.L., Brueckner, B., Prussia, S.E., Eds.; Academic Press: San Diego, CA, USA, 2014; pp. 69–122. [Google Scholar]
- Aizawa, T.; Ve, N.B.; Kimoto, K.I.; Iwabuchi, N.; Sumida, H.; Hasegawa, I.; Sasaki, S.; Tamura, T.; Kudo, T.; Suzuki, K.I.; et al. Curtobacterium ammoniigenes sp. nov., an ammonia-producing bacterium isolated from plants inhabiting acidic swamps in actual acid sulfate soil areas of Vietnam. Int. J. Syst. Evol. Microbiol. 2007, 57, 1447–1452. [Google Scholar] [CrossRef]
- Tokmakova, A.D.; Tarakanov, R.I.; Lukianova, A.A.; Evseev, P.V.; Dorofeeva, L.V.; Ignatov, A.N.; Dzhalilov, F.S.; Subbotin, S.A.; Miroshnikov, K.A. Phytopathogenic Curtobacterium flaccumfaciens Strains Circulating on Leguminous Plants, Alternative Hosts and Weeds in Russia. Plants 2024, 13, 667. [Google Scholar] [CrossRef]
- Marshall, R.J. Food and nutritional analysis|Dairy Products. In Encyclopedia of Analytical Science, 2nd ed.; Worsfold, P., Townshend, A., Poole, C., Eds.; Elsevier: Oxford, UK, 2005; pp. 312–319. [Google Scholar]
- Cortez, M.A.S.; Duarte, M.C.K.H.; de Melo, L.R.B. Chapter 1—Relevant factors for raw milk quality for dairy foods manufacture. In Dairy Foods; Cruz, A.G.d., Ranadheera, C.S., Nazzaro, F., Mortazavian, A.M., Eds.; Woodhead Publishing: Sawston, UK, 2022; pp. 1–17. [Google Scholar]
- Kuddus, M.; Ramteke, P.W. A cold-active extracellular metalloprotease from Curtobacterium luteum (MTCC 7529): Enzyme production and characterization. J. Gen. Appl. Microbiol. 2008, 54, 385–392. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, X.; Hou, L.; Xu, G.; Guan, F.; Zhang, W.; Luo, H.; Wu, N.; Yao, B.; Zhang, C.; et al. The seed endophytic microbe Microbacterium testaceum M15 enhances the cold tolerance and growth of rice (Oryza sativa L.). Microbiol. Res. 2024, 289, 127908. [Google Scholar] [CrossRef] [PubMed]
- Licciardello, G.; Doppler, M.; Sicher, C.; Bueschl, C.; Ruso, D.; Schuhmacher, R.; Perazzolli, M. Metabolic changes in tomato plants caused by psychrotolerant Antarctic endophytic bacteria might be implicated in cold stress mitigation. Physiol. Plant. 2024, 176, e14352. [Google Scholar] [CrossRef]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef]
- Hurtado-Bautista, E.; Pérez Sánchez, L.F.; Islas-Robles, A.; Santoyo, G.; Olmedo-Alvarez, G. Phenotypic plasticity and evolution of thermal tolerance in bacteria from temperate and hot spring environments. PeerJ 2021, 9, e11734. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Gong, H.; Xu, Y.; Cai, C.; Hua, Y.; Li, L.; Dai, L.; Dai, X. The transition temperature (42 °C) from mesophilic to thermophilic micro-organisms enhances biomethane potential of corn stover. Sci. Total Environ. 2021, 759, 143549. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).