Abstract
Methane, a potent greenhouse gas, has gained significant attention due to its environmental impact and economic potential. Chemical industries have focused on specialized catalytic systems, like zeolites, to convert methane into methanol. However, inherent limitations in selectivity, irreversibility, and pore blockages result in high costs and energy requirements, thus hindering their commercial viability and profitability. In contrast, biological methane conversion using methanotrophs has emerged as a promising alternative, offering higher conversion rates, self-renewability, improved selectivity, and economically feasible upstream processes. Nevertheless, biological methane oxidation encounters challenges including the difficulty in cultivating methanotrophs and their slow growth rates, which hinder large-scale bioprocessing. Another highlighted limitation is the limited mass transfer of methane into liquid in bioreactors. Practical strategies to enhance methane oxidation in biological systems, including optimizing reactor design to improve mass transfer, altering metal concentrations, genetic engineering of methane monooxygenases, enzyme encapsulation, and utilizing microbial consortia are discussed. By addressing the limitations of chemical approaches and highlighting the potential of biological methods, the review concluded that the utilization of genetically engineered methanotrophic biofilms on beads within a biotrickling reactor, along with enhanced aeration rates, will likely enhance methane oxidation and subsequent methane conversion rates.
1. Introduction
Methane plays a significant role as a greenhouse gas in our planet’s atmosphere, comprising around 18% of the overall greenhouse gas mixture [1]. The National Oceanic and Atmospheric Administration (NOAA) documented that the global level of methane has recently exceeded to 1925 ppb (as of December 2022) [2,3]. For a decade, the relentless increase in methane concentration in the atmosphere has been a severe worldwide issue. Methane possesses a remarkable potency in trapping heat in the atmosphere, exceeding carbon dioxide by over 25 times [4]. This significant disparity underscores the substantial environmental burden associated with methane emissions. The major anthropogenic sources of methane are landfill wastes, coal mines, and agricultural and human wastes (shown in Figure 1) [5,6]. Furthermore, the enormous use of fossil fuels due to industrialization and urbanization significantly contributes toward increasing the methane concentration in the environment [7]. However, methane exhibits a dual nature as both a potent greenhouse gas and a resource that can be produced using renewable methods. Biogas purification allows for the capture of methane from organic waste sources like agricultural residues, sewage, or landfill gases [8]. Additionally, advanced technologies like biological methanation enable carbon-negative methane production by combining CO2 and renewable hydrogen [9]. This process removes more CO2 from the atmosphere than it emits. Moreover, methane serves as a precursor to liquid fuels, providing an avenue to mitigate vented methane emissions [10,11]. Expanding the applications of methane, including C-C coupling reactions, similar to biomass processes, and oxidation to methanol, can unlock its potential as a versatile chemical building block across various industries, fostering a sustainable and circular economy [12]. However, the oxidation of methane is challenging in the field of catalytic conversion due to high C-H bond energy (104 kJ/mol), thus conceding only partial oxidation [13]. Additionally, the heterogenous reaction kinetics are slow due to the weak interaction of methane with the catalyst surface [14]. For the past few decades, microbially mediated methane oxidation has become a focus of research. Therefore, there is a constant search for the exploration of novel microbes that can convert methane into valuable products.
Among the microbial community, the methanotrophic and methylotrophic genera consist of fascinating enzymatic machinery—methane monooxygenase (MMO) that serves as a major sink for methane [15]. Methylotrophs are a diverse group of Gram-negative microorganisms that utilize C1 compounds such as methane, methanol, and other organic compounds as the source of carbon and energy [16]. Methanotrophs are a subset of methylotrophs that uses methane as the sole carbon and energy source [17]. Typically, aerobic methanotrophs can be categorized into three types—Type I (γ-proteobacteria), Type II (α-proteobacteria), and Type X (γ-proteobacteria)—on the basis of carbon assimilation pathways, membrane morphology, and phospholipid composition [18]. An out-group category, Verrucomicrobia also oxidizes methane but involves a far different mechanism from the above-stated types [19]. Therefore, the diverse methanotrophic community offers great potential for utilizing methanotrophs in the selective production of a wide range of value-added products. This approach agrees with the advantages of low cost, low energy requirements, and improved economic profitability.
Due to the fast-paced nature of the industry, projections remain uncertain, yet the 2020 McKinsey Global Institute report approximates that the global bioeconomy and has the potential to generate USD 2 to USD 4 trillion in direct annual economic impacts by 2030–2040 [20]. Despite challenges posed by low methane solubility and oxidation rates, methanotrophs have gained significant interest in recent years as they offer the potential for producing value-added products such as single-cell protein, biopolymers, and biofuels [21]. Solutions to address these challenges include optimizing bioreactor configuration, tuning physical parameters such as high pressure, and employing genetic engineering to enhance methane oxidation and create novel metabolic pathways [22,23,24]. Therefore, methanotrophic bioconversion has been an area of focus for several decades, given its potential to convert methane into useful products. The development of a bioeconomy based on methanotrophs has the potential to create significant economic impacts [25].
In this review, we discuss the challenges associated with methane oxidation using chemical and biological methods including the stability of the methane molecule, the availability of cofactors, and the diversity of methanotrophs and their metabolic pathways. We also discuss potential solutions such as the development of efficient bioreactor systems, the optimization of cultivation methods for methanotrophs, and the use of synthetic biology and genetic engineering. By addressing these challenges, methane oxidation can become a viable solution for mitigating the impact of methane emissions and contributing to a sustainable future.
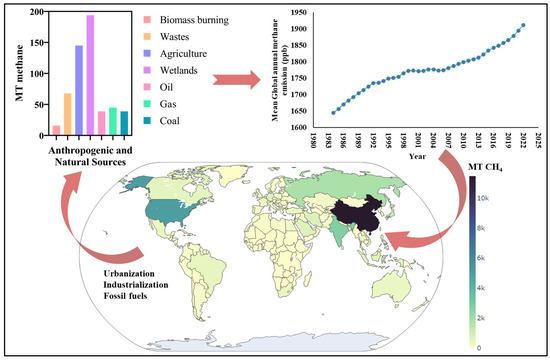
Figure 1.
Illustration of the production of methane in metric tons (MT) from both anthropogenic and natural sources, highlighting their collective impact on mean global annual methane emissions (in ppb) [26,27]. Additionally, a heat map is provided to depict the country-wise contributors to methane emissions [28].
2. Challenges in Methane Oxidation
According to the Global Methane Budget 2020 report, which estimated data from 2000–2017 using a bottom-up approach, methane emissions primarily originate from two categories: natural/ biogenic sources and anthropogenic sources [29]. Natural sources account for methane emissions of 367 Tg CH4 yr−1, while anthropogenic sources contribute 380 Tg CH4 yr−1. Among natural sources, wetlands are responsible for 145 Tg CH4 yr−1, while other natural sources including land sources and ocean sources contribute a combined 222 Tg CH4 yr−1. Within anthropogenic sources, agriculture and waste emissions account for 213 Tg CH4 yr−1, fossil fuel emissions contribute 135 Tg CH4 yr−1, and biomass and biofuel burning contribute 29 Tg CH4 yr−1 [29]. Terrestrial wetland habitats, characterized by waterlogged soils and permafrost, serve as significant carbon sinks. However, the warming climate poses a challenge as it causes wetland soils to warm or flood, resulting in the release of carbon into the atmosphere in the form of methane.
In the global transition towards a sustainable low-carbon economy, two key technologies, methane storage and methane capture, hold significant importance. Metal-organic framework (MOF) materials have demonstrated their potential in the field of gas adsorption storage due to their exceptional properties such as high surface area, excellent porosity, and an adjustable pore structure [30]. Regarding methane capture at atmospheric pressure, particular attention is given to CH4/N2 and CO2/CH4 separation, as well as methane capture technologies [30]. Notably, MOF material PCN-14 has achieved the highest reported methane capture at 298 K and 6.5 MPa, reaching an impressive volumetric uptake of 230 cm3 cm−3 [31]. Sadiq et al. developed a method to capture low-concentration methane emitted from landfills using aluminum fumarate and @MgFe2O4 magnetic framework composite [32]. This composite demonstrated methane uptake of 18.2 cm3 g−1 at 300 K and 0.1 MPa, particularly when the mass fraction of MgFe2O4 was only 1 wt%. In the oil and gas industry, methane recovery from storage tanks and well casinghead vent emissions can be achieved using a vapor recovery unit. Traditionally, the adsorbent material employed in these units has been porous carbon materials like zeolite 5A. Notably, at 303 K and 0.099 MPa, zeolite 5A has exhibited a methane uptake rate of 7.456 mg g−1 [33]. Indeed, the technologies discussed have demonstrated their effectiveness in capturing and storing methane for subsequent use as a precursor for liquid fuels. However, a crucial question arises: is the methane oxidation and conversion of methane into value-added products a challenging endeavor? The subsequent sections of this review comprehensively address this inquiry by highlighting the limitations encountered in methane conversion and presenting potential solutions.
Methane oxidation via chemical and biological approaches has unique challenges, that limits methane as a precursor for value-added products in commercial sectors. Table 1 presents a comparison between the chemical and biological methane oxidation with respect to operational conditions (e.g., temperature, pressure, and catalysts) and carbon conversion efficiency.

Table 1.
Comparison of operating parameters between chemical and biological methane oxidation.
Chemical methane oxidation is a promising approach for the conversion of methane into more useful chemicals, and the use of catalysts such as transition metal oxides has been explored as a strategy for facilitating the reaction [54]. However, the development of efficient and selective catalysts for methane oxidation remains a significant challenge. The high stability of the methane molecule presents a significant hurdle, making it difficult to activate and react with other molecules [55]. The C-H bond in methane is relatively unreactive and requires a high activation energy to break, making the process energy-intensive and adding to the cost of any industrial application [56]. To overcome this challenge, researchers have developed catalysts that can activate the C-H bond and promote the oxidation of methane [56,57]. These catalysts typically involve the use of transition metals, such as platinum, palladium, and rhodium, which can adsorb methane and facilitate the activation of the C-H bond but usually lead to the formation of carbonaceous deposits on the catalyst surface, leading to catalyst deactivation [58,59]. However, while these catalysts have shown promise in laboratory settings, developing efficient and selective chemical catalysts for methane oxidation for industrial purposes remains a significant challenge.
Biological methane oxidation offers several advantages over chemical methane oxidation, primarily due to its autobiocatalytic nature and tolerance towards impure methane. Unlike chemical catalysts that require periodic replacement, resulting in downtime and additional costs, a properly managed bioprocess operates continuously, with no additional operational expenses (OPEX) apart from hardware maintenance [60]. Additionally, the side and by-products generated during the process can be transformed into valuable additional products, supporting the goal of zero waste bioprocessing and fostering a more sustainable approach [61]. This is made possible by the self-replicating nature of the MMO in the bioprocess, highlighting the concept of autobiocatalysis. Furthermore, biological systems exhibit a notable tolerance to impurities present in methane-containing gas streams. Unlike chemical catalysts that are sensitive to impurities in methane or oxygen streams), biological catalysts can often withstand and effectively process impure methane sources [46]. This tolerance to impurities expands the range of methane feedstocks that can be utilized, including those derived from waste streams or unconventional sources. This versatility enhances the feasibility and flexibility of biological methane oxidation processes, offering opportunities for more diverse and sustainable methane utilization.
However, biological methane oxidation also faces several challenges that limit its potential for large-scale bioprocessing applications. One significant challenge is the requirement for copper and iron ions as cofactors in MMO enzymes, which are essential for methane oxidation [62]. These cofactors can be limiting factors in an uncontrolled natural environment, such as soils with low copper or iron content, and can affect the efficiency of the methane oxidation process [63,64]. However, in an applied process, one can easily address this challenge by supplementing the required cofactors, turning it into a mere triviality. Methanotrophs are often difficult to cultivate, and the slow growth rate can result in low productivity, making large-scale bioprocessing applications challenging [24,65]. Furthermore, the diversity of methanotrophs and their metabolic pathways can make it challenging to identify optimal strains for specific applications [66]. Methanotrophs, based on methane oxidation capacity, can be categorized as high-affinity or low-affinity based on their methane oxidation kinetics [67]. High-affinity methanotrophs are capable of oxidizing ambient methane concentrations (~2 ppm) [68], while low-affinity methanotrophs utilize methane at higher concentrations (>40 ppm) [69]. Also, different methanotrophs have different carbon assimilation pathways and require specific environmental conditions to maintain their activity, which can vary depending on the application [70]. Therefore, identifying the optimal strain for a specific application can be challenging and require extensive screening and characterization efforts. In addition, the methane oxidation process can lead to the formation of secondary byproducts such as formaldehyde, formic acid, and carbon monoxide [71]. These byproducts can reduce the net yield of the desirable product and limit the efficiency of the process. Researchers have explored different strategies to mitigate these side reactions such as the use of specific growth conditions and the development of new strains with improved selectivity.
The bioreactor system can be employed for increased methane oxidation, but it has its own challenges. One major challenge in designing a gas-to-liquid bioreactor is the limited mass transfer of methane, which has low solubility in water at standard temperature and pressure (~22 mg/L at 20 °C and 1 atm) [72,73]. With respect to chemical methane oxidation, many reactor designs contain products and reactants in the liquid phase [74]. This is not feasible for a methanotrophic bioreactor for increasing mass transfer between gaseous methane and microorganisms [75]. The convective mass transfer coefficient kLa (h−1) is often used to measure the efficiency of gas–liquid mixing in bioreactors [76]. A greater kLa can be achieved through various reactor designs such as bubble columns, stirred tank reactors (STRs), or continuously stirred tank reactors (CSTRs) with gas spargers to produce additional bubbles [77]. However, these designs may require large reactor volumes that are beyond considerable. Additionally, there are some conventional techniques that can help improve mass transfer in bioreactors, such as operating at high pressure, reducing the size of gas bubbles, and increasing the number of bubbles [78]. Moreover, other designs involve immobilizing the biomass to increase protein density and overall productivity [79]. The use of 3D culture techniques can be beneficial for long-term experiments as flat surface cultures may not accurately reflect the system being studied [80]. Bioreactors can be filled with gel beads composed of alginate, silica, or hydrogels. Taylor et al. conducted an experiment in which they measured the methane uptake of Methylosinus trichosporium OB3b at different seeding densities in sodium alginate beads in batch and semi-steady state reactors [81]. Their objective was to inhibit methanol dehydrogenase using cyclopropane, which would result in the accumulation of methanol. However, the results showed that the use of alginate beads did not lead to any improvement in methanol production or methane uptake when compared to suspended biomass cultures [81].
Trickle bed reactors (TBRs) typically use ceramic beads or polyurethane foams to cultivate biofilms on the surface or within the pores of the foam [82]. TBRs offer advantages over STRs and bubble columns in terms of energy requirements, as the volumetric flow rate of the liquid component is primarily determined by gravity [83]. Encapsulated beads containing biofilm in a packed column can increase the surface area, which is crucial for gas-to-liquid mass transport [73]. To evaluate and compare the performance of various reactor types, including TBR, STR, and bubble columns, parameters such as syngas (CO and H2) conversion rates, as well as ethanol productivity, have been considered. This evaluation allows for a comprehensive comparison of TBR with other reactor types, highlighting its potential as an effective option for methane conversion processes. In a notable study conducted by Devarapalli et al., a TBR system utilizing Clostridium ragsdalei demonstrated a remarkable CO and H2 conversion rate of 90% along with a high ethanol productivity of 158 mg/L∙h at gas flow rates between 1.5 and 2.8 standard cubic centimeters per minute (sccm) [84]. These findings outperformed the ethanol productivities reported for C. carboxidivorans in hollow fiber membrane bioreactor (140 mg/L∙h), C. ljungdahlii in a CSTR without cell recycle (110 mg/L∙h), and Alkalibaculum bacchi in a CSTR (70 mg/L∙h) [85,86,87]. Furthermore, in a two-stage continuous syngas fermentation involving C. ljungdahlii, the CSTR exhibited CO and H2 conversion efficiencies of 46% and 49% at a gas flow rate of 23 sccm, while the bubble column achieved higher CO and H2 conversion efficiencies of 86% and 82% at a gas flow rate of 121 sccm [88]. Orgill et al. conducted a comparison of mass transfer coefficients for CO and H2 between TBR and STR [89]. Their findings revealed that a TBR with 6 mm bead size exhibited the highest total mass transfer coefficient, with a KTotA/L value of 421 h−1. In contrast, the STR demonstrated a lower KTotA/L of 114 h−1 under conditions of 400 sccm gas flow and 900 rpm stirring speed [89]. Considering the improved mass transfer coefficient for gas-to-liquid transfer, enhanced productivity, and higher conversion rates, it can be concluded that TBR holds promise as a viable option for methane conversion via methanotrophs. However, TBRs face challenges such as clogging due to the accumulation of biomass and unsteady operating conditions caused by an increase in pressure drop across the inlet and outlet [90]. These limitations restrict the use of TBRs in industrial settings for long-term operations. To conclude, chemical methane oxidation faces challenges such as the development of efficient and selective catalysts, high activation energy, and catalyst deactivation due to carbonaceous deposits. On the other hand, biological methane oxidation offers several advantages over chemical methane oxidation, primarily due to its autobiocatalytic nature and tolerance towards impure methane. However, biological methane oxidation faces its own set of challenges such as the requirement for copper and iron ions as cofactors, slow growth rates, optimal strain identification, byproduct formation, and limited mass transfer of methane. However, potential solutions exist to overcome these limitations in both chemical and biological methane oxidation, and in the later section we will discuss the promising small-scale increased biological methane oxidation strategies. Figure 2 shows the challenges via chemical and biological methane oxidation and potential biological solutions to overcome the limitations.
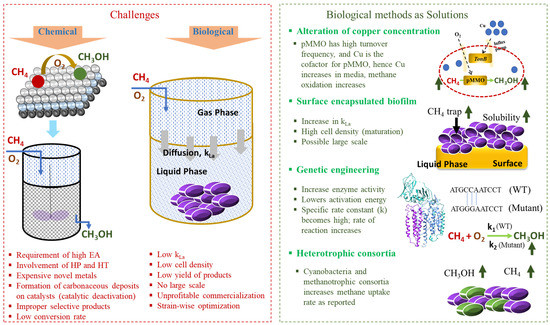
Figure 2.
The challenges of chemical and biological methane oxidation and the potential biological solutions to enhance methane oxidation. EA—activation energy; HP—high pressure; HT—high temperature; kLa—volumetric mass transfer coefficient; WT—wild type; k—specific rate constant.
3. Solution to Overcome the Challenges
3.1. Chemical Methods
3.1.1. Traditional Methods for Direct Atmospheric Methane Removal
Several chemical methods to mitigate atmospheric methane have been proposed and studied to help reduce methane emissions and limit its impact on the environment. Different chemical methods that are in practice to mitigate methane are listed in Table 2. One of the most promising chemical methods to mitigate atmospheric methane is the use of catalysts; the systems which can help oxidize methane into carbon dioxide [91]. The catalytic systems broadly use noble metals, transition metals, and metal oxides [92]. A study by Chen et al. reviewed that using a Ni-Au/Al2O3 catalyst could oxidize up to 84% of methane into methanol under a steam reforming process [93]. Inhibitors such as 3-nitrooxypropanol prevent methane-producing microbes from producing methane and are also reported to be promising to reduce methane production in cattle by up to 30% [94]. The first proposal towards direct atmospheric CH4 removal was laid by Boucher et al., with existing technologies such as zeolite minerals, cryogenic separation, molecular sieves, and adsorption filters [95]. Zeolites are the most popular chemical methane conversion catalyst in industries for the production of liquid fuels [96,97]. While zeolites offer several benefits, including reduced environmental corrosion, minimal waste generation, and easy continuous operation in fixed-bed reactors, they also come with drawbacks such as high sensitivity, irreversibility, and steric blockages [96]. Furthermore, structural tuning of zeolites into molecular sieve zeolites (e.g., Cobalt-ZSM-5) is beneficial to oxidize methane into methanol and formaldehyde and are effective in one-phase (gaseous) hydrocarbon separations [98]. However, the usage of these traditional materials or methods are limited to within chemical industries and are energetically and economically ineffective to treat large volume of atmospheric CH4 [99]. Ma et al. conducted a study for 63 days, where acidification levels (doses) ranging from 1.2 to 6.0 kg of concentrated sulfuric acid per cubic meter of slurry (pig excreta) were used across six distinct tanks [100]. They observed a substantial reduction in CH4 levels, with removal rates ranging from 46% (at 1.2 kg H2SO4) to 96% (at 6.0 kg H2SO4) and concluded that employing low-dose acidification proves to be a promising and viable strategy for mitigating CH4 emissions [100]. Highly interesting iron-salt aerosol method, which is a mimic of natural reactions related to mineral dust particles, is capable of direct atmospheric CH4 removal [101,102]. This method generates CH4 depleting chlorine atoms in the tropospheric atmosphere, thereby capable of removing the total atmospheric CH4 to some extent [103]. Furthermore, few speculative ideas are proposed by Lockley et al. such as CH4 ignition at point sources, lake sealing (impermeable cover/non-biodegradable foaming agents), and ducting CH4 bubble streams [104]. However, these methods may not be a complete solution to the problem of methane emissions, and further research is needed to determine their effectiveness in different environmental settings and to identify new and more effective methods for mitigating atmospheric methane.

Table 2.
Chemical methods for methane mitigation, their advantages, and disadvantages.
3.1.2. Chemical Methods and Reported Benchmark for Methane to Methanol Conversion
The multifaceted applications of methanol, coupled with its potential as a cleaner energy source, have firmly established it as a commercially valuable organic compound with a promising market outlook. Methanol has gained recognition as a promising alternative to fossil fuels in transportation and power generation due to its potential as a cleaner energy source [109,110,111,112]. Simultaneously, it plays a vital role as a feedstock for producing essential chemicals like formaldehyde, acetic acid, and dimethyl ether [113]. These chemicals find extensive application in the manufacturing of diverse industrial products including plastics, resins, and solvents [114]. Methanol’s versatile nature extends to other areas as well. It serves as an antifreeze agent in windshield washer fluid and natural gas pipelines, functions as an industrial solvent for inks, adhesives, and pharmaceutical ingredients, and also finds use in the production of building materials and high-tech devices [115,116,117,118]. The demand for methanol has been substantial, with the United States alone registering a demand of 19.7 million metric tons in 2022, valued at approximately USD 30 billion [119,120]. Furthermore, projections suggest a compound annual growth rate (CAGR) of 4.2%, which is expected to elevate the market value to USD 38 billion by 2028 [119].
Currently, the industrial process to synthesize methanol involves the use of syngas, which are usually prepared by methane reforming, but the cost of this process is high. Within the various chemical methods discussed earlier, lab and pilot scales have placed particular emphasis on utilizing specialized and optimized zeolitic catalytic systems to convert methane into methanol. However, there are several methods such as direct one-step oxidation, oxidation via MMO-mimicked catalyst, continuous methane–water–oxygen reaction with ZSM, and using non-thermal plasma.
The direct one-step oxidation of methane to methanol is more advantageous than the syngas route, and there are several reports that exist. In direct one-step oxidation noble metals (e.g., Au, Pd, and Rh) have been reported to exhibit excellent catalytic properties for the conversion of methane. Agarwal et al. achieved a high yield (92%) in the low-temperature (50 °C) selective oxidation of methane to methanol using colloidal gold-palladium nanoparticles as a catalyst in the presence of aqueous hydrogen peroxide (H2O2) [121]. Jin et al. developed a novel catalyst called a “molecular fence” by incorporating AuPd alloy nanoparticles into aluminosilicate zeolite crystals and modifying the external surface of the zeolite with organosilanes [122]. This catalyst demonstrated high efficiency in converting methane to methanol, achieving a production rate of 91.6 mmol gAuPd−1 h−1. Similarly, non-noble metals have also shown promise in this reaction. Taking inspiration from MMO biocatalysis, Grundner et al. developed a Cu-MOR catalyst through ion exchange, mimicking the active sites of MMO enzymes [23,123]. The catalyst, featuring single-site trinuclear copper oxygen clusters within mordenite zeolite, selectively converts methane to methanol by activating carbon–hydrogen bonds, resembling the enzymatic systems in terms of cluster rearrangements during the conversion process. The study achieved 80% methane conversion at 200 °C, with a methanol yield of approximately 160 µmol gcat–1 with Cu concentration of 500 µmol gcat–1 [123]. In another approach, Narsimhan et al. achieved continuous methane oxidation to methanol by introducing methane, oxygen, and water simultaneously into the reactor. Cu-Na-ZSM-5 and Cu-H-ZSM-5 catalysts demonstrated steady-state methanol production rates of 0.88 ± 0.02 μmol h–1 gcat–1 and 1.81 ± 0.01 μmol h–1 gcat–1, respectively [124]. Although the conversion rate was not particularly high in this study, it marked the first successful realization of continuous methane-to-methanol oxidation under continuous conditions. In one study, methanol synthesis from methane and water at room temperature and atmospheric pressure using non-thermal plasma was reported [125]. This approach prevents further decomposition and conversion of methanol by transferring it to the liquid phase. The addition of Ar or He gases, along with the use of plasma and semiconductor materials, significantly enhances methanol production rates and selectivity, with a maximum production rate of 56.7 mmol gcat−1 h−1 and 93% selectivity in the liquid phase, representing more than a five-fold increase compared to conditions without a catalyst and added gas [125].
Although there exist several effective catalytic approaches, the catalyzation approach have notable limitations, including inadequate selectivity, irreversibility, and pore blockages, which ultimately hinder the commercial viability and profitability of methanol production through methane oxidation. Hence, the pursuit for cost-effective and highly efficient CH4 conversion has motivated researchers to explore alternative biological approaches utilizing microbes. These methods are deemed environmentally friendly, sustainable, and economically viable, addressing the demand for more sustainable methane conversion processes.
3.2. Biological Methods
As previously described, methanotrophs utilize MMO, which exists in two forms, sMMO and pMMO, as their complex enzymatic machinery for biological methane oxidation. Both sMMO and pMMO have complex, multi-subunit scaffold, but with distinguishable structures, active sites, and reaction mechanisms [126]. Methanotrophs can express either both or one of these MMOs which is highly genus specific. The occurrence of both sMMO and pMMO in a particular methanotroph is rare and is limited to very few methanotrophic species (for example, M. trichosporium OB3b and M. capsulatus BATH) [127]. pMMO is present in almost all methanotrophs except for Methylocella and Methyloferula genera [70]. In methanotrophs where the genes for both forms exist, the expression of sMMO and pMMO is controlled by the availability of environmental copper, termed as “copper switch” [128]. Typically, high concentrations of copper (>0.85 µM) relative to biomass levels inhibit the activity of sMMO, whereas an excessive addition of iron has been observed to stimulate sMMO expression [129]. Specifically, when the copper-to-biomass ratio exceeds 0.86 µmol Cu.g−1 DW, methane oxidation occurs in the periplasmic membrane space mediated by pMMO, while at ratios below 0.86 µmol Cu.g−1 DW, methane oxidation is catalyzed by sMMO in the cytosol [127,130,131]. The involvement of metalloregulation in the expression of MMOs is due to the graved metal atoms within these enzymes, which serve as cofactors: sMMO has iron atoms confined to the non-heme diiron active site and pMMO has nucleated copper atoms [132]. Talking about metalloregulation, methanobactin (Mbn), a copper-chelating peptide, acts as chalkophores and copper scavenger, facilitating the transport and delivery of copper ions to MMO for its proper function while also playing a role in metalloregulation. Taking the example of M. trichosporium OB3b, the methanobactin operon consists of 11 genes organized as follows: import genes (MbnE periplasmic binding protein and MbnT Ton-B dependent transporter), export genes (MbnM MATE multidrug exporter, and regulatory sigma/anti-sigma factor pair MbnIR), and putative synthesis genes (MbnA precursor peptide, MbnC, and MbnN aminotransferase). Additionally, the methanobactin machinery in OB3b includes two hypothetical proteins, MbnP and MbnH, whose functions remain unknown. Figure 3 illustrates the intricate biological pathway of methane oxidation, highlighting the crucial role of methanobactin genes as regulators responsible for modulating the expression of methane monooxygenase (MMO). The reaction pertaining to sMMO and pMMO during the methane-to-methanol conversion is as follows:
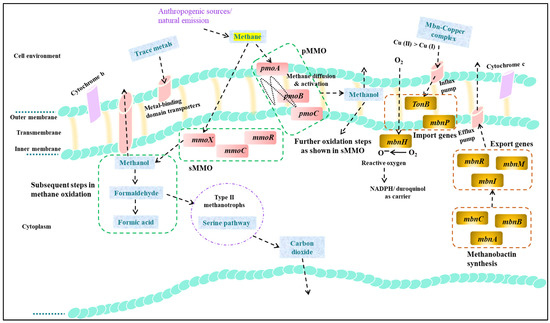
Figure 3.
The intricate biological pathway of methane oxidation in M. trichosporium OB3b is illustrated, highlighting the crucial role of methanobactin genes as regulators that govern the expression of MMO. In the first step of methane oxidation pathway, methane is oxidized by either sMMO (in cytoplasm, with main genes: mmoX, mmoC, and mmoB) or pMMO (in intracellular membrane, with main genes: pmoA, pmoB, and pmoC). The resulting product, methanol, is further oxidized to formaldehyde by methanol dehydrogenase. The Serine pathway (Type II; α-proteobacteria) is involved in the assimilation of formaldehyde, while the Ribulose monophosphate pathway is utilized in Type I/X (γ-proteobacteria). Eventually, formic acid and carbon dioxide (CO2) are generated. Notably, methanobactin, consisting of various genes involved in synthesis (mbn A, mbnB, and mbnC), export (mbnR, mbnM, and mbnI), import (mbnT, and MbnP), and reactive oxygen radical generation (mbnH) plays a vital role in facilitating pMMO or sMMO activity by supplying molecular oxygen.
pMMO: CH4 + O2 + 2e− + 2H+ → CH3OH + H2O
sMMO: CH4 + NAD(P)H2 + O2 → CH3OH + NAD(P+) + H2O
Therefore, to enhance the rates of methane oxidation in biological systems, various approaches can be employed. Firstly, mass transfer rates can be increased by employing different reactor configurations that facilitate efficient contact between the methanotrophs and methane substrate. Secondly, genetic modification of the MMO enzymes can be pursued to optimize their catalytic activity and improve methane oxidation efficiency. Finally, another strategy involves engineering the regulatory genes involved in the “copper switch” mechanism, which controls the expression of MMO enzymes based on the availability of environmental copper, thereby fine-tuning the methane oxidation process. By combining these approaches, it is possible to significantly enhance the efficiency of methane oxidation biologically. The later sub-sections describe the novel approaches that have been reported in lab-scale experiments to achieve higher methane oxidation. Table 3 presents an overview of different biological methods for methane mitigation, along with their advantages and disadvantages.

Table 3.
Biological methods for methane mitigation, their advantages, and disadvantages.
3.2.1. Bioreactor and Biofilter Based Method
Methane biofilters and bioreactors are crucial in mitigating the release of methane and relies on methanotrophs to convert methane into less harmful compounds [146]. In a bioreactor study involving iron-rich sediments from the Bothnian Sea, metagenomics was used to investigate shifts in microbial community composition under low oxygen concentrations [147]. The study identified marker genes for methane and iron cycling, as well as respiratory and fermentative metabolism, which provided insights into the metabolic potential of methanotrophs in coastal marine sediments. Novel Verrucomicrobia, Bacteroidetes, and Krumholzibacteria were recovered and found to have the potential for methane oxidation, organic matter degradation, and iron cycling, respectively [147]. The study suggests that the methane biofilter in coastal sediments may be more diverse than previously thought, with a variety of microorganisms contributing to methane oxidation. This diversity could be harnessed to improve the efficiency of bioreactors and biofilters for methane oxidation.
Moreover, biofilters with forced aeration have been investigated in experimental plants of different scales to understand their potential and limitations for methane oxidation [148]. In a study by Streese et al., a bench scale plant (total biofilter volume 60 L) and a pilot plant (4 m³) were used to evaluate the performance of biofilters for methane oxidation [148]. The forced aeration helps to maintain optimal conditions for the methane-oxidizing bacteria within the biofilter, ensuring sufficient oxygen supply and promoting efficient methane oxidation. In this study, the researchers observed that the methane oxidation rates were influenced by factors such as biofilter material, temperature, water content, and pH [148]. In another study by Farrokhzadeh et al., a multiple-level aeration biofilter design was proposed to improve conventional methane biofilter performance [49]. Laboratory flow-through column experiments were conducted to evaluate three actively aerated methane biofilter configurations, with air introduced at one, two, and three levels of the bed depth. The results indicated that the biofilter column with two aeration levels had the most even performance over time, maintaining 85% average oxidation efficiency over 95 days of experiments [49]. In a study by Jawad et al., a transient model was developed to predict methane concentration at various heights and times in a biofilter using composted sawdust as packing media [149]. The model was validated with experimental data and showed good agreement. The simulations suggested that under certain conditions, a removal efficiency of 95% might be achieved for a height of 2 m. One study analyzed the performance of different biofilter materials, including peat, compost, and a mixture of compost and lava rock [150]. The peat-filled biofilter did not show significant methane oxidation, while the other materials exhibited better performance after an adaptation period. This may limit the role of biofilter in methane oxidation under such conditions.
Biotrickling filters have been gaining attention for methane oxidation due to their potential for efficient gas treatment and lower energy consumption compared to other technologies [146]. Cáceres et al. conducted a study on the oxidation of methane in biotrickling filters inoculated with methanotrophic bacteria [151]. They found that the inoculation of the biotrickling filter with methanotrophic bacteria significantly increased the methane removal efficiency. The study also showed that the performance of the biotrickling filter was affected by factors such as the type of packing material, the presence of other gases (e.g., H2S and NH3), and environmental conditions. Further in the same study, Caceres et al. explored the potential of hydrophobic methanotrophs in two-liquid phase partitioning biotrickling filters for methane abatement [151]. The results indicated that the use of hydrophobic methanotrophs could enhance the methane removal efficiency and the overall performance of the biotrickling filter. In a study by La et al., a biotrickling filter packed with inert materials was investigated for methane treatment in the presence of a non-ionic surfactant [146]. The results showed that the presence of the surfactant enhanced the removal efficiency of methane, suggesting that the use of surfactants can improve the performance of biotrickling filters for methane oxidation [146]. In another study, Brandt et al. investigated the use of novel packing materials for improving methane oxidation in biofilters [152]. They found that the selection of appropriate packing materials could significantly affect the performance of the biofilter, with some materials showing better methane removal efficiency than others. Veillete et al. studied the effect of ammonium concentration on microbial population and performance of a biofilter treating air polluted with methane [153]. They found that higher ammonium concentrations led to a decrease in methane removal efficiency, highlighting the importance of controlling nutrient concentrations in biotrickling filters for optimal performance. Largely, biotrickling filters have shown promise for methane oxidation, with several studies demonstrating their potential for efficient methane removal. Factors such as packing material, the presence of surfactants, inoculation with methanotrophic bacteria, and nutrient concentrations can significantly influence the performance of biotrickling filters [75]. Further research on optimizing these factors, as well as understanding the interactions between methanotrophic bacteria and other microbial communities, can help improve the design and operation of biotrickling filters for methane mitigation.
Nevertheless, microbial fuel cells (MFCs) have gained significant attention in recent years for their potential towards direct conversion of methane to electricity via microorganisms as catalyst [154]. However, the direct conversion of methane to electricity in traditional fuel cells is challenging due to high-temperature operation requirements and catalytic instability [155]. In this context, microbial fuel cells offer a promising alternative by utilizing the biological conversion of methane to electricity through microbial electrochemical systems (MESs) [156]. MFCs operate at ambient temperatures and offer flexibility in scale, making them suitable for integration with catalytic processes to selectively produce desired chemical products [157]. However, achieving high efficiency in methane oxidation using MFCs has been a challenge due to the complexity of anaerobic oxidation of methane and the difficulty of microbial culturing.
Ren et. al. demonstrated an anaerobic reverse-methanogenesis process that directly converts methane into electricity using MFCs with high efficiency [154]. By carefully selecting a consortium of microorganisms, they were able to achieve efficient methane oxidation and current generation in the MFC system. Another study investigated enhancing methane oxidation in a bioelectrochemical membrane reactor using a soluble electron mediator [158]. The researchers focused on optimizing electrode materials to enhance power outputs in the MFC, addressing structural configurations and limitations of microbial fuel cells. Methane-driven MFCs have also been demonstrated to recover energy and mitigate dissolved methane emissions from anaerobic effluents [158]. This approach provides an effective solution for reducing methane emissions while simultaneously generating electricity from anaerobic wastewater treatment processes. Chen et al. conducted a study using two air–cathode single chamber MFCs containing anaerobic effluents and showed 85% dissolved methane removal and a maximum coulombic efficiency of 18% at 16 h hydraulic retention time. Further metagenomic 16S rRNA Illumina sequencing showed the presence of Geobacter sp. and methanotroph at the anode biofilm, which suggests the benefit of methane oxidation in electricity production [135]. Myung et al. used a two-staged MFC, whereas, in the first stage, methane-oxidizing bacteria was used to accumulate methanol in the high phosphate buffer (350 ± 42 mg/L), and in the second stage, the same accumulated methanol had been used to generate electricity and a maximum power density of 426 ± 17 mW/m2 was observed [159]. This implies that a multi-stage reactor can be used to address the issue of methanol accumulation in facultative methanotrophs. In the first stage, the growth of biomass is prioritized, while in the second stage, the activity of methanol dehydrogenase (MDH) is inhibited to allow methanol accumulation.
To conclude, the optimization of reactor design, including forced aeration and the selection of suitable packing materials, has shown promising results in enhancing methane oxidation rates. Biotrickling filters have emerged as an energy-efficient option for methane treatment, with the potential for enhanced removal efficiency through the use of specific packing materials, surfactants, and the inoculation of methanotrophic bacteria. Additionally, microbial fuel cells offer an alternative approach for the direct conversion of methane to electricity, with recent advancements demonstrating high efficiency and the potential for integration with catalytic processes. Further research is needed to optimize the factors influencing the performance of biotrickling filters and microbial fuel cells including nutrient concentrations, electrode materials, and microbial consortium selection.
3.2.2. Alteration in Metal Concentrations
Although there is no direct evidence from studies addressing the impact of altering metal concentrations on methane oxidation, observations from experiments involving different substrates such as trichloroethylene (TCE) suggest that manipulating metal concentrations, specifically copper, may have a positive effect on enhancing methane oxidation by methanotrophs. In a study utilizing methanotrophic consortia isolated from a landfill cover, it was observed that increasing copper (Cu2+) concentration from 0 to 15 μM increased the degradation rate of TCE from 74.41 nmol/(mgcell.h) at 15 μM Cu2+ to 654.99 nmol/(mgcell.h) at 0.03 μM Cu2+, with specific activity of sMMO ranging from 650 nmolnaphthol/(mgcell.h) at 0.03 μM to 100 nmolnaphthol/(mgcell.h) at 15 μM [160]. In another study, methanotrophic–heterotrophic communities enriched with Cu exhibited significant accumulation of polyhydroxybutyrate (PHB), with PHB content ranging from 43.2% to 45.9%, whereas cultures without Cu accumulated only small amounts of PHB (11.9% to 17.5%). Batch assays demonstrated that communities grown with Cu and higher oxygen levels synthesized more PHB, showing an extended optimal range of methane to oxygen ratio and achieving a high PHB content (48.7%) even in the presence of nitrogen [161]. In the study conducted by Lee et al., it was found that methanotrophs expressing the particulate form of pMMO had a competitive edge over cells expressing sMMO due to their higher growth rates. Furthermore, when exposed to excess Cu (100 µM), the pMMO-expressing cells exhibited enhanced growth and degraded the mixed chlorinated solvents at a faster rate within a shorter time frame [162]. Whole-cell assays on M. trichosporium OB3b expressing pMMO demonstrated that the affinity for TCE was enhanced with higher copper concentrations, as indicated by a decrease in Ks from 36 to 7.9 μM [163]. Therefore, the stimulation of methanotroph growth by higher copper concentrations highlights the importance of copper-mediated regulatory mechanisms in these organisms, suggesting a potential to enhance methane oxidation and increase the production of valuable bioproducts.
3.2.3. Synergetic Growth and Consortia
Consortia of methanotrophic bacteria can lead to enhanced methane oxidation rates due to their synergistic interactions and the ability to utilize excreted methane degradation metabolites [137]. One such example is the co-occurrence of type II methanotroph Methylocystis and the methylotroph Hyphomicrobium in heterotrophic-methanotrophic consortia. In this consortium, Methylocystis is responsible for oxidizing methane, while Hyphomicrobium can grow on the CH4-derived carbon intermediates produced by Methylocystis. This cross-feeding interaction improves both the methane-oxidation rate (9.2 ± 1.2 mg CH4 g−1 DWbiomass h−1) and biomass growth, leading to an efficient and stable methane oxidation system [138].
In this regard, aerobic methanotrophs, such as Methylobacter, have been found to form active methane-oxidizing communities even in anoxic environments. These methanotrophs can thrive in such conditions by closely associating with photosynthetic (micro) algae, which serves as an oxygen source. Light stimulation has been shown to enhance methane oxidation by gammaproteobacterial methanotrophs, including Methylobacter, in the anoxic water layers of oxygen-stratified lakes [139]. Aerobic methanotrophs may also rely on Sphagnum for molecular oxygen, enabling their proliferation in anoxic niches in peatland ecosystems. Alternatively, they can independently rely on incomplete denitrification [139]. Therefore, the interaction of aerobic methanotrophs with their biotic environment appears to be relevant in modulating community functioning and may help confer resilience during disturbances, prompting the use of alternative strategies and the formation of synergistic/antagonistic interactions.
3.2.4. Genetic Engineering of MMOs
As previously indicated, genetic studies offer an efficient approach to enhance MMO expression and methane oxidation for catalytic conversion of methane to methanol. To achieve this, it is crucial to explore the active sites of both sMMO and pMMO and identify the essential amino acid residues responsible for methane oxidation. While the genetic and mechanistic aspects of sMMO are well-established, there is a significant knowledge gap concerning the active site and metallic content of pMMO, presenting an opportunity for further investigation.
sMMO has three protein components (1) a hydroxylase (MMOH), (2) a reductase (MMOR), and (3) a regulatory (MMOB) component, which are collectively responsible for achieving its maximal catalytic activity. The MMOH which is an approximate 251 kDa α2β2γ2 homodimer and consists of three subunits, namely, α-subunit, β-subunit, and γ-subunit. The active site of sMMO is graved within the α-subunit of MMOH. On the other hand, pMMO catalyzes the oxidation of methane to methanol in the periplasmic space of the cell, and it is composed of three subunits— α-subunit (pmoA), β-subunit (pmoB), and γ-subunit (pmoC). The structure of pMMO is still not fully understood, but it is believed to be a heterotrimeric protein complex that contains copper ions coordinated by histidine residues. Furthermore, the active site within the pMMO is controversial and is, therefore, ambiguous between the three subunits.
By using site-directed mutagenesis to alter these crucial amino acid residues, researchers can enhance the methane oxidation rate in both sMMO and pMMO. Through computational methods, Samanta et al. identified five potential amino acid substitutions (B:Leu31Ser, B:Phe96Gly, B:Phe92Thr, B:Trp106Ala, and B:Tyr110Phe) in M. trichosporium OB3b that could potentially enhance methane oxidation rates mediated by pMMO [164]. Fei et al. in a review proposed that by implementing upstream strategies such as overexpression and protein engineering of methane monooxygenase, the rates of methane oxidation to methanol could be improved [21]. To date, the expression of sMMO in heterologous hosts has been challenging due to difficulties in efficiently expressing the sMMO hydroxylase component. This, along with the slow growth rate and low product yields of methanotrophic bacteria, hinders their industrial applications [165]. Developing a functional expression system for MMO genes in heterologous hosts that can achieve high cell density with additional carbon sources would greatly facilitate the advancement of Bio-GTL technologies [21]. Despite efforts to express sMMO in heterologous hosts such as Escherichia coli, Agrobacterium tumefaciens, Burkholderia cepacia, Pseudomonas mendocina, Pseudomonas putida, and Sinorhizobium meliloti, the sMMO activity remains relatively low and less robust compared to methanotrophic hosts like M. trichosporium OB3b, Methylomicrobium album, and Methylocystics parvus [127,166,167]. Nevertheless, Lloyd et al. successfully introduced sMMO genes from M. trichosporium OB3b and M. capsulatus BATH into Methylocystis parvus OBBP and Methylomicrobium album BG8, respectively, using conjugation with broad-host-range plasmids. The sMMO genes were expressed and produced active sMMO in these heterologous hosts, indicating that all the necessary genes for sMMO synthesis were present on the plasmids and effectively expressed [145]. The presence of conserved sMMO regulatory systems unique to methanotrophs further complicates heterologous expression [127,145]. In contrast, successful expression and regulation of pMMO, both homologously and heterologously, have been reported, although the construction of a robustly engineered methanotroph is yet to be achieved [142,144,168]. The ability to transfer the methanotrophic phenotype to various microorganisms would greatly impact the industrial utilization of this metabolic capability. However, until then, bioprocesses relying on the conversion of methane to value-added products primarily depend on natural methanotrophs.
These investigations underscore the promise of genetic engineering in advancing our comprehension of the active sites of methane monooxygenases and enabling their enhancement in catalytic activity and stability. Computational dynamics and site-directed mutagenesis have yielded valuable insights into the reaction mechanisms of sMMO and pMMO enzymes, elucidating the contributions of specific residues during the catalytic cycle. Through heterologous and homologous recombination, engineered variants of both MMOs have been developed, offering further opportunities to improve their activity and enhance methane oxidation.
3.2.5. Enzyme and Whole Cell Immobilization
Enzyme immobilization is a promising technique for improving enzyme stability and reusability, which can potentially increase methane oxidation in methanotrophs. This technique involves embedding gas-reacting enzymes within an organic, polymeric material, allowing for control over gas solubility, permeability, and surface area. While there is limited literature specifically discussing enzyme immobilization for methane oxidation in methanotrophs, there are relevant studies on methanol production. Mardina et al. utilized whole cell immobilized Methylocella tundra in alginate beads as biocatalysts and optimized production conditions for maximum methanol production [140]. They found that immobilized cells produced 5.18 mM of methanol, a significant improvement over free cells which produced only 0.66 mM, with 50% methane as a substrate [140]. Razumovsky et al. immobilized Methylosinus sporium B-2121 cells in polyvinyl alcohol cryogel and observed an improved methanol production of 62 ± 2 mg/L in 24 h, which was five times as compared to the free cells [141]. The improvement in methanol production is directly proportionate to the enhanced methane oxidation with accordance to methane oxidation. Blanchette et al. developed printed, mechanically robust, gas-permeable membranes with pMMO (from Methylococcus capsulatus BATH) based polymer material embedded within a silicon lattice to convert methane into methanol. They found that the printed cylinder with an area-to-volume ratio of 2.33 had an average pMMO activity of 128 ± 14 nmol MeOH min−1mg−1 per cylinder, which is reportedly the maximum activity of membrane-bound pMMO with NADH as a reducing agent [23]. This study established a direct proportionality between the area–volume ratio and methanol production [23]. The study resulted in the establishment of direct proportionality between the area–volume ratio and methanol production. Patel et al. encapsulated Methylosinus sporium cells in Na-alginate beads and silica gel and found maximum methanol production of 3.73 mM and 3.43 mM, respectively, while retaining 61.8% and 51.6% of the initial efficiency, which was far higher as compared to the free cells (11.5%) [169]. Overall, these studies suggest that enzyme immobilization techniques can significantly improve methanol production, and by extension, methane oxidation in methanotrophs.
4. Challenges in Upscaling of Biological Methods
Scaling up to commercial applications presents significant challenges that become increasingly pronounced at larger scales, requiring careful consideration. Though biotrickling filters show promise for removing methane, they have only been tested at small scales, from lab to pilot. The presence of other gases, such as hydrogen sulfide and ammonia, can negatively affect the performance of biotrickling filters for methane oxidation, as these gases can compete with methane for the active sites (e.g., pMMO) of methanotrophic bacteria [153]. Insufficient nutrient availability, such as nitrogen and phosphorus, can also limit the performance of biotrickling filters, as these nutrients are essential for the growth and activity of methanotrophic bacteria. These filters can be subject to periods of starvation or shock loads due to process variations or equipment malfunctions, leading to a decrease in viable bacteria numbers and negatively impacting performance. Furthermore, accurate evaluation of biotrickling filter performance can be challenging due to the complex interactions between microbial communities, packing materials, and environmental conditions. Addressing these challenges will be critical to realizing the full potential of biotrickling filters for large-scale methane removal.
In a pilot-scale study conducted by Karthikeyan et al., methane oxidation was investigated using methanotrophic consortia enriched from marine sediment [50,51]. They reported CH4 removal rates of 7–13% in a 10 L CSTR under high gas flow conditions of 2.5 L.min−1 (20–25% CH4). However, when the methanotrophic consortia were cultivated on plastic bio-balls, a significant improvement was observed, with CH4 removal reaching 95–97% in biofilters operating at a lower gas flow rate of 0.1 L.min−1 and with a CH4 concentration of 1%. Notably, the study highlighted that the presence of cyanobacteria, despite being a contamination, had positive effects on treating low-level CH4. The cyanobacteria provided additional oxygen for methane oxidation by the methanotrophic consortia, suggesting that co-cultivating MMC with cyanobacterial mats could be beneficial for CH4 remediation at an industrial scale without interfering with its performance [50]. Building upon the concept of a methanotroph–microalgae partnership, another study demonstrated the remarkable ability of methanotrophs and microalgae to jointly oxidize methane within bioflocs [170]. This particular study revealed that the microalgae within the bioflocs supplied the vital oxygen required for the methane oxidation process. Furthermore, an intriguing finding was that nearly all of the carbon derived from methane was assimilated into biomass by the partnership, without any significant release of carbon dioxide [170]. The study further supported the notion of a mutually beneficial relationship between methanotrophs and microalgae, highlighting their potential for efficient methane conversion at industrial sectors.
The National Renewable Energy Laboratory (NREL) isolated a strain called Methylomicrobium alcaliphilum 20Z that is capable of consuming a mixed stream of commercial biogas (consisting of methane, carbon dioxide, and hydrogen sulfide) [171,172]. NREL concluded that CO2 does not have a significant impact on methanotroph growth and that biogas with varying compositions can be used for culturing. Two critical factors in this regard are low operating costs and scalability. Chemical plants are unable to economically purify waste streams selectively, and the process of purification and separation often requires large volumes of reactants to be profitable. Using biological processes can circumvent these issues, as it requires no high-tech facilities to remove unwanted substances. A novel approach to address this is the use of a bioreactor that utilizes gas–solid mass transfer of methane gas through a solid hydrogel. The Lawrence Livermore National Lab (LLNL) immobilizes methanotrophs in a hydrogel that is supported by a cylindrical scaffold, aiding in structural integrity and increasing mass transfer between the gas and solid (immobilized methanotrophs) [73].
Therefore, experimental studies at National Laboratories have demonstrated the potential for future enhancements in methane conversion to value-added products at higher rates and with improved economic feasibility. Strategies such as co-cultivating methanotrophs with cyanobacteria or microalgae and utilizing innovative bioreactor designs like gas–solid mass transfer systems offer promising avenues for achieving efficient methane conversion on an industrial scale. These advancements pave the way for realizing the potential of methane as a valuable resource in various sectors.
5. Conclusions
In conclusion, while challenges exist in both chemical and biological methane conversion methods, laboratory-scale studies have demonstrated that biological approaches offer more potential for enhancing methane oxidation rates. One effective solution that may show potential is the amalgamation of enzyme immobilization in biotrickling reactors. Co-cultivating methanotrophs with cyanobacteria or microalgae in biotrickling reactors and utilizing aeration systems can enhance gas–solid mass transfer, offering a promising avenue for achieving efficient methane conversion on an industrial scale. Encapsulated beads containing methanotrophic culture biofilm in a packed column can significantly increase the surface area, which is crucial for efficient gas-to-liquid mass transport. Pilot-scale studies have reported impressive results, with approximately 97% methane conversion achieved using plastic balls as the medium. This demonstrates the feasibility of scaling up these technologies and achieving high conversion rates at larger operational scales. In addition to process optimization, genetic engineering approaches hold promise for enhancing enzymatic activity and thus improving methane oxidation rates. By employing genetic engineering techniques to fine-tune the activity of MMOs, it is possible to enhance the catalytic efficiency of these enzymes. This approach opens up new avenues for improving the overall methane conversion efficiency. However, it is important to note that there are still several challenges that need to be addressed to achieve widespread implementation of these technologies. Factors such as reactor design, catalyst selection, nutrient supply, and process control must be carefully optimized to maximize the efficiency of gas-to-liquid mass transfer and methane oxidation. Additionally, the economic viability and scalability of these approaches need to be thoroughly evaluated to ensure their practical applicability on an industrial scale. Therefore, continued research, development, and collaboration among researchers, engineers, and industry stakeholders are crucial for further advancements and the successful implementation of these technologies in real-world applications. With sustained efforts, it is possible to overcome the challenges and contribute to a more sustainable and environmentally friendly approach to commercially produce methane-based bioproducts.
Author Contributions
Conceptualization, D.S. and R.K.S.; methodology, D.S.; software, D.S.; validation, D.S. and R.K.S.; formal analysis, D.S.; investigation, D.S. and R.K.S.; resources, R.K.S.; data curation, D.S.; writing—original draft preparation, D.S.; writing—review and editing, D.S. and R.K.S.; visualization, D.S.; supervision, R.K.S.; project administration, R.K.S.; funding acquisition, R.K.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Foundation in the form of the BuG ReMeDEE initiative (Award #1736255) and other NSF TII T2 (#1849206, and #1920954).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors acknowledge the support by the Department of Chemical and Biological Engineering at the South Dakota School of Mines and Technology.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Skea, J.; Shukla, P.; Kılkış, Ş. Climate Change 2022: Mitigation of Climate Change; Cambridge University Press: Cambridge, MA, USA, 2022. [Google Scholar]
- Wuebbles, D.J.; Jain, A.K. Concerns About Climate Change and the Role of Fossil Fuel Use. Fuel Process. Technol. 2001, 71, 99–119. [Google Scholar] [CrossRef]
- Administration, National Oceanic and Atmospheric. Trends in Methane Emission. Available online: https://gml.noaa.gov/ccgg/trends_ch4/ (accessed on 18 February 2023).
- Agency, United States Environmental Protection. Global Methane Initiative. Environmental Protection Agency. Available online: https://www.epa.gov/gmi/importance-methane#:~:text=Methane%20is%20more%20than%2025,due%20to%20human%2Drelated%20activities (accessed on 1 May 2023).
- Halford, J.O.; Miller, G.A. Standard Heat Capacities of Gaseous Methanol, Ethanol, Methane and Ethane at 279 K. By Thermal Conductivity. J. Phys. Chem. 1957, 61, 1536–1539. [Google Scholar] [CrossRef]
- Karakurt, I.; Aydin, G.; Aydiner, K. Sources and mitigation of methane emissions by sectors: A critical review. Renew. Energy 2012, 39, 40–48. [Google Scholar] [CrossRef]
- Raihan, A.; Muhtasim, D.A.; Farhana, S.; Pavel, M.I.; Faruk, O.; Rahman, M.; Mahmood, A. Nexus between carbon emissions, economic growth, renewable energy use, urbanization, industrialization, technological innovation, and forest area towards achieving environmental sustainability in Bangladesh. Energy Clim. Chang. 2022, 3, 100080. [Google Scholar] [CrossRef]
- Chandra, R.; Takeuchi, H.; Hasegawa, T. Methane production from lignocellulosic agricultural crop wastes: A review in context to second generation of biofuel production. Renew. Sustain. Energy Rev. 2012, 16, 1462–1476. [Google Scholar] [CrossRef]
- Meylan, F.D.; Piguet, F.-P.; Erkman, S. Power-to-gas through CO2 methanation: Assessment of the carbon balance regarding EU directives. J. Energy Storage 2017, 11, 16–24. [Google Scholar] [CrossRef]
- Samanta, D.; Govil, T.; Salem, D.R.; Krumholz, L.R.; Gerlach, R.; Gadhamshetty, V.; Sani, R.K. Methane Monooxygenases: Their Regulations and Applications in Biofuel Production. In Microbes for Sustainable Development and Bioremediation; CRC Press: Boca Raton, FL, USA, 2019; pp. 187–206. [Google Scholar]
- Muh, E.; Tabet, F.; Amara, S. Biomass Conversion to Fuels and Value-Added Chemicals: A Comprehensive Review of the Thermochemical Processes. Curr. Altern. Energy 2021, 4, 3–25. [Google Scholar] [CrossRef]
- Soulivong, D.; Norsic, S.; Taoufik, M.; Coperet, C.; Thivolle-Cazat, J.; Chakka, S.; Basset, J.-M. Non-Oxidative Coupling Reaction of Methane to Ethane and Hydrogen Catalyzed by the Silica-Supported Tantalum Hydride: (SiO)2Ta−H. J. Am. Chem. Soc. 2008, 130, 5044–5045. [Google Scholar] [CrossRef]
- Ravi, M.; Sushkevich, V.L.; Knorpp, A.J.; Newton, M.A.; Palagin, D.; Pinar, A.B.; Ranocchiari, M.; van Bokhoven, J.A. Misconceptions and challenges in methane-to-methanol over transition-metal-exchanged zeolites. Nat. Catal. 2019, 2, 485–494. [Google Scholar] [CrossRef]
- Horn, R.; Schlögl, R. Methane activation by heterogeneous catalysis. Catal. Lett. 2015, 145, 23–39. [Google Scholar] [CrossRef]
- Samanta, D.; Govil, T.; Salem, D.R.; Krumholz, L.R.; Gerlach, R.; Gadhamshetty, V.; Sani, R.K. 12 Methane Monooxygenases. In Microbes for Sustainable Development and Bioremediation, 1st ed.; CRC Press: Boca Raton, FL, USA, 2019; p. 187. [Google Scholar] [CrossRef]
- Lidstrom, M.E. Aerobic methylotrophic prokaryotes. Prokaryotes 2006, 2, 618. [Google Scholar]
- AlSayed, A.; Fergala, A.; Eldyasti, A. Sustainable biogas mitigation and value-added resources recovery using methanotrophs intergrated into wastewater treatment plants. Rev. Environ. Sci. Bio/Technol. 2018, 17, 351–393. [Google Scholar] [CrossRef]
- Murrell, J.C.; McDonald, I.R.; Bourne, D.G. Molecular methods for the study of methanotroph ecology. FEMS Microbiol. Ecol. 1998, 27, 103–114. [Google Scholar] [CrossRef]
- Khadem, A.F. Methanotrophy under Extreme Conditions: Biochemistry and Physiology of Methylacidiphilum Fumariolicum SolV. Ph.D Thesis, Radboud University, Nijmegen, The Netherlands, 2012. [Google Scholar]
- McKinsey Global Institute. The BioRevolution. In Innovations Transforming Economies, Societies, and Our Lives; McKinsey Global Institute: New York, NY, USA, 2020; pp. 1–200. [Google Scholar]
- Fei, Q.; Guarnieri, M.T.; Tao, L.; Laurens, L.M.; Dowe, N.; Pienkos, P.T. Bioconversion of natural gas to liquid fuel: Opportunities and challenges. Biotechnol. Adv. 2014, 32, 596–614. [Google Scholar] [CrossRef]
- Comesaña-Gándara, B.; García-Depraect, O.; Santos-Beneit, F.; Bordel, S.; Lebrero, R.; Muñoz, R. Recent trends and advances in biogas upgrading and methanotrophs-based valorization. Chem. Eng. J. Adv. 2022, 11, 100325. [Google Scholar] [CrossRef]
- Blanchette, C.D.; Knipe, J.M.; Stolaroff, J.K.; DeOtte, J.R.; Oakdale, J.S.; Maiti, A.; Lenhardt, J.M.; Sirajuddin, S.; Rosenzweig, A.C.; Baker, S.E. Printable enzyme-embedded materials for methane to methanol conversion. Nat. Commun. 2016, 7, 11900. [Google Scholar] [CrossRef]
- Sahoo, K.K.; Goswami, G.; Das, D. Biotransformation of methane and carbon dioxide into high-value products by methanotrophs: Current state of art and future prospects. Front. Microbiol. 2021, 12, 636486. [Google Scholar] [CrossRef]
- Menon, A.; Lyng, J.G. Circular bioeconomy solutions: Driving anaerobic digestion of waste streams towards production of high value medium chain fatty acids. Rev. Environ. Sci. Bio/Technol. 2021, 20, 189–208. [Google Scholar] [CrossRef]
- (IEA). International Energy Agency. Sources of Methane Emissions. Available online: https://www.iea.org/data-and-statistics/charts/sources-of-methane-emissions-2 (accessed on 1 May 2023).
- (NOAA). National Oceanic and Atmospheric Administration. Trends in Atmospheric Methane (Global CH4 Monthly Means). Available online: https://gml.noaa.gov/ccgg/trends_ch4/ (accessed on 10 May 2023).
- (IEA). International Energy Agency. Total Methane Emissions and Methane Intensity of Production in Selected Oil and Gas Producers. 2021. Available online: https://www.iea.org/data-and-statistics/charts/total-methane-emissions-and-methane-intensity-of-production-in-selected-oil-and-gas-producers-2021 (accessed on 1 May 2023).
- Saunois, M.; Stavert, A.R.; Poulter, B.; Bousquet, P.; Canadell, J.G.; Jackson, R.B.; Raymond, P.A.; Dlugokencky, E.J.; Houweling, S.; Patra, P.K. The global methane budget 2000–2017. Earth Syst. Sci. Data 2020, 12, 1561–1623. [Google Scholar] [CrossRef]
- Li, D.-z.; Chen, L.; Liu, G.; Yuan, Z.-y.; Li, B.-f.; Zhang, X.; Wei, J.-q. Porous metal–organic frameworks for methane storage and capture: Status and challenges. New Carbon Mater. 2021, 36, 468–496. [Google Scholar] [CrossRef]
- Peng, Y.; Krungleviciute, V.; Eryazici, I.; Hupp, J.T.; Farha, O.K.; Yildirim, T. Methane storage in metal–organic frameworks: Current records, surprise findings, and challenges. J. Am. Chem. Soc. 2013, 135, 11887–11894. [Google Scholar] [PubMed]
- Sadiq, M.M.; Rubio-Martinez, M.; Zadehahmadi, F.; Suzuki, K.; Hill, M.R. Magnetic framework composites for low concentration methane capture. Ind. Eng. Chem. Res. 2018, 57, 6040–6047. [Google Scholar] [CrossRef]
- Bakhtyari, A.; Mofarahi, M. Pure and binary adsorption equilibria of methane and nitrogen on zeolite 5A. J. Chem. Eng. Data 2014, 59, 626–639. [Google Scholar] [CrossRef]
- Goodman, E.D.; Dai, S.; Yang, A.-C.; Wrasman, C.J.; Gallo, A.; Bare, S.R.; Hoffman, A.S.; Jaramillo, T.F.; Graham, G.W.; Pan, X. Uniform Pt/Pd bimetallic nanocrystals demonstrate platinum effect on palladium methane combustion activity and stability. ACS Catal. 2017, 7, 4372–4380. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, S.; Shan, J.-j.; Nguyen, L.; Zhan, S.; Gu, X.; Tao, F. In situ surface chemistries and catalytic performances of ceria doped with palladium, platinum, and rhodium in methane partial oxidation for the production of syngas. Acs Catal. 2013, 3, 2627–2639. [Google Scholar]
- Ross, M.O.; Rosenzweig, A.C. A tale of two methane monooxygenases. JBIC J. Biol. Inorg. Chem. 2017, 22, 307–319. [Google Scholar] [PubMed]
- Ross, M.O.; MacMillan, F.; Wang, J.; Nisthal, A.; Lawton, T.J.; Olafson, B.D.; Mayo, S.L.; Rosenzweig, A.C.; Hoffman, B.M. Particulate methane monooxygenase contains only mononuclear copper centers. Science 2019, 364, 566–570. [Google Scholar] [PubMed]
- Guerrero-Caballero, J.; Kane, T.; Haidar, N.; Jalowiecki-Duhamel, L.; Löfberg, A. Ni, Co, Fe supported on Ceria and Zr doped Ceria as oxygen carriers for chemical looping dry reforming of methane. Catal. Today 2019, 333, 251–258. [Google Scholar]
- Visvanathan, C.; Pokhrel, D.; Cheimchaisri, W.; Hettiaratchi, J.; Wu, J.S. Methanotrophic activities in tropical landfill cover soils: Effects of temperature, moisture content and methane concentration. Waste Manag. Res. 1999, 17, 313–323. [Google Scholar] [CrossRef]
- Nie, H.; Howe, J.Y.; Lachkov, P.T.; Chin, Y.-H.C. Chemical and structural dynamics of nanostructures in bimetallic Pt–Pd catalysts, their inhomogeneity, and their roles in methane oxidation. Acs Catal. 2019, 9, 5445–5461. [Google Scholar] [CrossRef]
- Bharadwaj, S.; Schmidt, L.D. Catalytic partial oxidation of natural gas to syngas. Fuel Process. Technol. 1995, 42, 109–127. [Google Scholar]
- Alrashed, W.; Chandra, R.; Abbott, T.; Lee, H.-S. Nitrite reduction using a membrane biofilm reactor (MBfR) in a hypoxic environment with dilute methane under low pressures. Sci. Total Environ. 2022, 841, 156757. [Google Scholar] [PubMed]
- Claridge, J.B.; York, A.P.; Brungs, A.J.; Marquez-Alvarez, C.; Sloan, J.; Tsang, S.C.; Green, M.L. New catalysts for the conversion of methane to synthesis gas: Molybdenum and tungsten carbide. J. Catal. 1998, 180, 85–100. [Google Scholar]
- Scheller, S.; Ermler, U.; Shima, S. Catabolic pathways and enzymes involved in anaerobic methane oxidation. Anaerob. Util. Hydrocarb. Oils Lipids 2020, 31–59. [Google Scholar] [CrossRef]
- Cai, X.; Hu, Y.H. Advances in catalytic conversion of methane and carbon dioxide to highly valuable products. Energy Sci. Eng. 2019, 7, 4–29. [Google Scholar]
- de Klerk, A. Engineering evaluation of direct methane to methanol conversion. Energy Sci. Eng. 2015, 3, 60–70. [Google Scholar] [CrossRef]
- Lawton, T.J.; Rosenzweig, A.C. Methane-oxidizing enzymes: An upstream problem in biological gas-to-liquids conversion. J. Am. Chem. Soc. 2016, 138, 9327–9340. [Google Scholar]
- Dummer, N.F.; Willock, D.J.; He, Q.; Howard, M.J.; Lewis, R.J.; Qi, G.; Taylor, S.H.; Xu, J.; Bethell, D.; Kiely, C.J.; et al. Methane Oxidation to Methanol. Chem. Rev. 2022, 123, 6359–6411. [Google Scholar] [CrossRef]
- Farrokhzadeh, H.; Hettiaratchi, J.P.A.; Jayasinghe, P.; Kumar, S. Aerated biofilters with multiple-level air injection configurations to enhance biological treatment of methane emissions. Bioresour. Technol. 2017, 239, 219–225. [Google Scholar] [CrossRef]
- Karthikeyan, O.P.; Saravanan, N.; Cirés, S.; Alvarez-Roa, C.; Razaghi, A.; Chidambarampadmavathy, K.; Velu, C.; Subashchandrabose, G.; Heimann, K. Culture scale-up and immobilisation of a mixed methanotrophic consortium for methane remediation in pilot-scale bio-filters. Environ. Technol. 2017, 38, 474–482. [Google Scholar] [CrossRef]
- Patricia, R.-R.; Gómez-Borraz, T.L.; Revah, S.; Morales, M. Methanotroph-Microalgae Co-Culture for Greenhouse Gas Mitigation: Effect of Initial Biomass Ratio and Methane Concentration. Chemosphere 2020, 259, 127418. [Google Scholar] [CrossRef]
- Lee, S.G.; Goo, J.H.; Kim, H.G.; Oh, J.-I.; Kim, Y.M.; Kim, S.W. Optimization of methanol biosynthesis from methane using Methylosinus trichosporium OB3b. Biotechnol. Lett. 2004, 26, 947–950. [Google Scholar] [CrossRef] [PubMed]
- Xin, J.-y.; Cui, J.-r.; Niu, J.-z.; Hua, S.-f.; Xia, C.-g.; Li, S.-b.; Zhu, L.-m. Production of methanol from methane by methanotrophic bacteria. Biocatal. Biotransform. 2004, 22, 225–229. [Google Scholar] [CrossRef]
- Ravi, M.; Ranocchiari, M.; van Bokhoven, J.A. The direct catalytic oxidation of methane to methanol—A critical assessment. Angew. Chem. Int. Ed. 2017, 56, 16464–16483. [Google Scholar]
- Alvarez-Galvan, M.; Mota, N.; Ojeda, M.; Rojas, S.; Navarro, R.; Fierro, J. Direct methane conversion routes to chemicals and fuels. Catal. Today 2011, 171, 15–23. [Google Scholar] [CrossRef]
- Labinger, J.A.; Bercaw, J.E. Understanding and exploiting C–H bond activation. Nature 2002, 417, 507–514. [Google Scholar]
- Lunsford, J.H. The catalytic oxidative coupling of methane. Angew. Chem. Int. Ed. Engl. 1995, 34, 970–980. [Google Scholar] [CrossRef]
- Zaera, F. Selectivity in hydrocarbon catalytic reforming: A surface chemistry perspective. Appl. Catal. A Gen. 2002, 229, 75–91. [Google Scholar]
- Puliyalil, H.; Jurković, D.L.; Dasireddy, V.D.; Likozar, B. A review of plasma-assisted catalytic conversion of gaseous carbon dioxide and methane into value-added platform chemicals and fuels. RSC Adv. 2018, 8, 27481–27508. [Google Scholar]
- Viswanathan, M.B.; Raman, D.R.; Rosentrater, K.A.; Shanks, B.H. A technoeconomic platform for early-stage process design and cost estimation of Joint fermentative catalytic bioprocessing. Processes 2020, 8, 229. [Google Scholar] [CrossRef]
- Nair, L.G.; Agrawal, K.; Verma, P. An insight into the principles of lignocellulosic biomass-based zero-waste biorefineries: A green leap towards imperishable energy-based future. Biotechnol. Genet. Eng. Rev. 2022, 38, 288–338. [Google Scholar] [CrossRef] [PubMed]
- Koo, C.W.; Rosenzweig, A.C. Biochemistry of aerobic biological methane oxidation. Chem. Soc. Rev. 2021, 50, 3424–3436. [Google Scholar] [CrossRef] [PubMed]
- Glass, J.B.; Orphan, V.J. Trace metal requirements for microbial enzymes involved in the production and consumption of methane and nitrous oxide. Front. Microbiol. 2012, 3, 61. [Google Scholar] [CrossRef]
- Mancinelli, R.L. The regulation of methane oxidation in soil. Annu. Rev. Microbiol. 1995, 49, 581–605. [Google Scholar] [CrossRef]
- Kapoore, R.V.; Padmaperuma, G.; Maneein, S.; Vaidyanathan, S. Co-culturing microbial consortia: Approaches for applications in biomanufacturing and bioprocessing. Crit. Rev. Biotechnol. 2022, 42, 46–72. [Google Scholar] [CrossRef] [PubMed]
- Che, S.; Men, Y. Synthetic microbial consortia for biosynthesis and biodegradation: Promises and challenges. J. Ind. Microbiol. Biotechnol. 2019, 46, 1343–1358. [Google Scholar] [CrossRef] [PubMed]
- Reay, D.S.; Nedwell, D.B. Methane oxidation in temperate soils: Effects of inorganic N. Soil Biol. Biochem. 2004, 36, 2059–2065. [Google Scholar] [CrossRef]
- Smith, T.; Murrell, J. Methanotrophy/methane oxidation. Encycl. Microbiol. 2009, 3, 293–298. [Google Scholar]
- Tikhonova, E.N.; Kravchenko, I.K. Activity and diversity of aerobic methanotrophs in thermal springs of the russian far east. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–30. [Google Scholar]
- Nazaries, L.; Murrell, J.C.; Millard, P.; Baggs, L.; Singh, B.K. Methane, microbes and models: Fundamental understanding of the soil methane cycle for future predictions. Environ. Microbiol. 2013, 15, 2395–2417. [Google Scholar] [CrossRef]
- Larkin, D.; Caldwell, T.; Lobban, L.; Mallinson, R.G. Oxygen pathways and carbon dioxide utilization in methane partial oxidation in ambient temperature electric discharges. Energy Fuels 1998, 12, 740–744. [Google Scholar] [CrossRef]
- Meraz, J.L.; Dubrawski, K.L.; El Abbadi, S.H.; Choo, K.-H.; Criddle, C.S. Membrane and fluid contactors for safe and efficient methane delivery in methanotrophic bioreactors. J. Environ. Eng. 2020, 146, 03120006. [Google Scholar] [CrossRef]
- Hwee, N. Methane Remediation Using Biocatalysts in Batch Gas-Solid Mass Transfer System Challenges and Prospects. 2022. Available online: https://www.osti.gov/servlets/purl/1860693 (accessed on 1 May 2023).
- Krishna, R.; Sie, S. Strategies for multiphase reactor selection. Chem. Eng. Sci. 1994, 49, 4029–4065. [Google Scholar] [CrossRef]
- Estrada, J.M.; Lebrero, R.; Quijano, G.; Pérez, R.; Figueroa-González, I.; García-Encina, P.A.; Muñoz, R. Methane abatement in a gas-recycling biotrickling filter: Evaluating innovative operational strategies to overcome mass transfer limitations. Chem. Eng. J. 2014, 253, 385–393. [Google Scholar] [CrossRef]
- Amaral, P.F.; Freire, M.G.; Rocha-Leão, M.H.M.; Marrucho, I.M.; Coutinho, J.A.; Coelho, M.A.Z. Optimization of oxygen mass transfer in a multiphase bioreactor with perfluorodecalin as a second liquid phase. Biotechnol. Bioeng. 2008, 99, 588–598. [Google Scholar] [CrossRef]
- Stone, K.A.; Hilliard, M.V.; He, Q.P.; Wang, J. A mini review on bioreactor configurations and gas transfer enhancements for biochemical methane conversion. Biochem. Eng. J. 2017, 128, 83–92. [Google Scholar] [CrossRef]
- Ramezani, M.; Legg, M.J.; Haghighat, A.; Li, Z.; Vigil, R.D.; Olsen, M.G. Experimental investigation of the effect of ethyl alcohol surfactant on oxygen mass transfer and bubble size distribution in an air-water multiphase Taylor-Couette vortex bioreactor. Chem. Eng. J. 2017, 319, 288–296. [Google Scholar] [CrossRef]
- Dong, T.; Knoshaug, E.P.; Pienkos, P.T.; Laurens, L.M. Lipid recovery from wet oleaginous microbial biomass for biofuel production: A critical review. Appl. Energy 2016, 177, 879–895. [Google Scholar] [CrossRef]
- Antoni, D.; Burckel, H.; Josset, E.; Noel, G. Three-dimensional cell culture: A breakthrough in vivo. Int. J. Mol. Sci. 2015, 16, 5517–5527. [Google Scholar] [CrossRef]
- Taylor, A.; Molzahn, P.; Bushnell, T.; Cheney, C.; LaJeunesse, M.; Azizian, M.; Semprini, L. Immobilization of Methylosinus trichosporium OB3b for methanol production. J. Ind. Microbiol. Biotechnol. 2018, 45, 201–211. [Google Scholar] [CrossRef]
- Tiwari, H.; Sonwani, R.K.; Singh, R.S. Bioremediation of dyes: A brief review of bioreactor performance. Environ. Technol. Rev. 2023, 12, 83–128. [Google Scholar] [CrossRef]
- Datsevich, L.B.; Datsevich, L.B. Analysis of Conventional Industrial Processes. Conv. Three-Phase Fixed-Bed Technol. Anal. Crit. 2012, 7, 33–62. [Google Scholar]
- Devarapalli, M.; Lewis, R.S.; Atiyeh, H.K. Continuous ethanol production from synthesis gas by Clostridium ragsdalei in a trickle-bed reactor. Fermentation 2017, 3, 23. [Google Scholar] [CrossRef]
- Liu, K.; Atiyeh, H.K.; Stevenson, B.S.; Tanner, R.S.; Wilkins, M.R.; Huhnke, R.L. Continuous syngas fermentation for the production of ethanol, n-propanol and n-butanol. Bioresour. Technol. 2014, 151, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Datar, R.P.; Shenkman, R.M.; Cateni, B.G.; Huhnke, R.L.; Lewis, R.S. Fermentation of biomass-generated producer gas to ethanol. Biotechnol. Bioeng. 2004, 86, 587–594. [Google Scholar] [CrossRef]
- Shen, Y.; Brown, R.; Wen, Z. Enhancing mass transfer and ethanol production in syngas fermentation of Clostridium carboxidivorans P7 through a monolithic biofilm reactor. Appl. Energy 2014, 136, 68–76. [Google Scholar] [CrossRef]
- Martin, M.E.; Richter, H.; Saha, S.; Angenent, L.T. Traits of selected Clostridium strains for syngas fermentation to ethanol. Biotechnol. Bioeng. 2016, 113, 531–539. [Google Scholar] [CrossRef]
- Orgill, J.J.; Atiyeh, H.K.; Devarapalli, M.; Phillips, J.R.; Lewis, R.S.; Huhnke, R.L. A comparison of mass transfer coefficients between trickle-bed, hollow fiber membrane and stirred tank reactors. Bioresour. Technol. 2013, 133, 340–346. [Google Scholar] [CrossRef]
- Gunes, B. A critical review on biofilm-based reactor systems for enhanced syngas fermentation processes. Renew. Sustain. Energy Rev. 2021, 143, 110950. [Google Scholar] [CrossRef]
- Havran, V.; Dudukovic, M.P.; Lo, C.S. Conversion of methane and carbon dioxide to higher value products. Ind. Eng. Chem. Res. 2011, 50, 7089–7100. [Google Scholar] [CrossRef]
- Liotta, L. Catalytic oxidation of volatile organic compounds on supported noble metals. Appl. Catal. B Environ. 2010, 100, 403–412. [Google Scholar] [CrossRef]
- Chen, Y.; Mu, X.; Luo, X.; Shi, K.; Yang, G.; Wu, T. Catalytic conversion of methane at low temperatures: A critical review. Energy Technol. 2020, 8, 1900750. [Google Scholar] [CrossRef]
- Hristov, A.N.; Oh, J.; Giallongo, F.; Frederick, T.W.; Harper, M.T.; Weeks, H.L.; Branco, A.F.; Moate, P.J.; Deighton, M.H.; Williams, S.R.O. An inhibitor persistently decreased enteric methane emission from dairy cows with no negative effect on milk production. Proc. Natl. Acad. Sci. USA 2015, 112, 10663–10668. [Google Scholar] [CrossRef] [PubMed]
- Boucher, O.; Folberth, G.A. New directions: Atmospheric methane removal as a way to mitigate climate change? Atmos. Environ. 2010, 44, 3343–3345. [Google Scholar] [CrossRef]
- Perot, G.; Guisnet, M. Advantages and disadvantages of zeolites as catalysts in organic chemistry. J. Mol. Catal. 1990, 61, 173–196. [Google Scholar] [CrossRef]
- Wang, P.; Sun, Q.; Zhang, Y.; Cao, J. Effective removal of methane using nano-sized zeolite 4A synthesized from kaolin. Inorg. Chem. Commun. 2020, 111, 107639. [Google Scholar] [CrossRef]
- Beznis, N.V.; van Laak, A.N.C.; Weckhuysen, B.M.; Bitter, J.H. Oxidation of methane to methanol and formaldehyde over Co–ZSM-5 molecular sieves: Tuning the reactivity and selectivity by alkaline and acid treatments of the zeolite ZSM-5 agglomerates. Microporous Mesoporous Mater. 2011, 138, 176–183. [Google Scholar] [CrossRef]
- Ming, T.; Li, W.; Yuan, Q.; Davies, P.; De Richter, R.; Peng, C.; Deng, Q.; Yuan, Y.; Caillol, S.; Zhou, N. Perspectives on removal of atmospheric methane. Adv. Appl. Energy 2022, 5, 100085. [Google Scholar] [CrossRef]
- Ma, C.; Dalby, F.R.; Feilberg, A.; Jacobsen, B.H.; Petersen, S.O. Low-Dose Acidification as a Methane Mitigation Strategy for Manure Management. ACS Agric. Sci. Technol. 2022, 2, 437–442. [Google Scholar] [CrossRef]
- Sturtz, T.M.; Jenkins, P.T.; de Richter, R. Environmental Impact Modeling for a Small-Scale Field Test of Methane Removal by Iron Salt Aerosols. Sustainability 2022, 14, 14060. [Google Scholar] [CrossRef]
- Oeste, F.D.; de Richter, R.; Ming, T.; Caillol, S. Climate engineering by mimicking natural dust climate control: The iron salt aerosol method. Earth Syst. Dyn. 2017, 8, 1–54. [Google Scholar] [CrossRef]
- Ming, T.; de Richter, R.; Oeste, F.D.; Tulip, R.; Caillol, S. A nature-based negative emissions technology able to remove atmospheric methane and other greenhouse gases. Atmos. Pollut. Res. 2021, 12, 101035. [Google Scholar] [CrossRef]
- Stolaroff, J.K.; Bhattacharyya, S.; Smith, C.A.; Bourcier, W.L.; Cameron-Smith, P.J.; Aines, R.D. Review of methane mitigation technologies with application to rapid release of methane from the Arctic. Environ. Sci. Technol. 2012, 46, 6455–6469. [Google Scholar] [CrossRef]
- Lackner, K.S. Practical constraints on atmospheric methane removal. Nat. Sustain. 2020, 3, 357. [Google Scholar] [CrossRef]
- Post, M.F. Diffusion in zeolite molecular sieves. In Studies in Surface Science and Catalysis; Elsevier: Amsterdam, The Netherlands, 1991; Volume 58, pp. 391–443. [Google Scholar]
- Jiang, L.; Liu, W.; Wang, R.; Gonzalez-Diaz, A.; Rojas-Michaga, M.; Michailos, S.; Pourkashanian, M.; Zhang, X.; Font-Palma, C. Sorption direct air capture with CO2 utilization. Prog. Energy Combust. Sci. 2023, 95, 101069. [Google Scholar] [CrossRef]
- Su, S.; Beath, A.; Guo, H.; Mallett, C. An assessment of mine methane mitigation and utilisation technologies. Prog. Energy Combust. Sci. 2005, 31, 123–170. [Google Scholar] [CrossRef]
- Shih, C.F.; Zhang, T.; Li, J.; Bai, C. Powering the future with liquid sunshine. Joule 2018, 2, 1925–1949. [Google Scholar] [CrossRef]
- Chauhan, M.; Vanshika; Kumar, A.; Chauhan, D.; Jain, A.K. Renewable Feedstocks for Biofuels. Biofuel Extr. Technol. 2023, 151–176. [Google Scholar] [CrossRef]
- Srivastava, R.K. Biological Approaches for Advanced Fuels Development from Biological/Plant Residues or Wastes. In Biotechnology for Waste Biomass Utilization; Apple Academic Press: Cambridge, UK, 2022; pp. 129–148. [Google Scholar]
- Bicer, Y.; Khalid, F. Life cycle environmental impact comparison of solid oxide fuel cells fueled by natural gas, hydrogen, ammonia and methanol for combined heat and power generation. Int. J. Hydrogen Energy 2020, 45, 3670–3685. [Google Scholar] [CrossRef]
- Mohammadi, R.; Harandi, M.H. Methanol to Olefin (MTO) Value Chain Management. New Appl. Stud. Manag. Econ. Account. 2023, 6, 7–17. [Google Scholar]
- Galusnyak, S.C.; Petrescu, L.; Chisalita, D.A.; Cormos, C.-C. Life cycle assessment of methanol production and conversion into various chemical intermediates and products. Energy 2022, 259, 124784. [Google Scholar] [CrossRef]
- Sarangi, P.K.; Nanda, S.; Vo, D.-V.N. Technological advancements in the production and application of biomethanol. Biorefinery Altern. Resour. Target. Green Fuels Platf. Chem. 2020, 127–139. [Google Scholar] [CrossRef]
- Oklu, N.K.; Matsinha, L.C.; Makhubela, B.C. Bio-solvents: Synthesis, industrial production and applications. Solvents Ion. Liq. Solvent Eff. 2019, 1–24. [Google Scholar] [CrossRef]
- Han, D.; Karmakar, T.; Bjelobrk, Z.; Gong, J.; Parrinello, M. Solvent-mediated morphology selection of the active pharmaceutical ingredient isoniazid: Experimental and simulation studies. Chem. Eng. Sci. 2019, 204, 320–328. [Google Scholar] [CrossRef]
- Liu, K.; Wang, X.; Yan, Y. Network analysis of industrial symbiosis in chemical industrial parks: A case study of Nanjing Jiangbei new materials high-tech park. Sustainability 2022, 14, 1381. [Google Scholar] [CrossRef]
- Markets and Markets. Methanol Market. In Methanol Market by Feedstock; Markets and Markets Publisher: Maharashtra, India, 2023. [Google Scholar]
- Newswire, C.P.R. Global Methanol Industry. In Global Methanol Market to Reach 99.8 Million Metric Tons by 2030; Global Industry Analysts Publisher: San Jose, CA, USA, 2023; p. 523. [Google Scholar]
- Agarwal, N.; Freakley, S.J.; McVicker, R.U.; Althahban, S.M.; Dimitratos, N.; He, Q.; Morgan, D.J.; Jenkins, R.L.; Willock, D.J.; Taylor, S.H. Aqueous Au-Pd colloids catalyze selective CH4 oxidation to CH3OH with O2 under mild conditions. Science 2017, 358, 223–227. [Google Scholar] [CrossRef]
- Jin, Z.; Wang, L.; Zuidema, E.; Mondal, K.; Zhang, M.; Zhang, J.; Wang, C.; Meng, X.; Yang, H.; Mesters, C. Hydrophobic zeolite modification for in situ peroxide formation in methane oxidation to methanol. Science 2020, 367, 193–197. [Google Scholar] [CrossRef]
- Grundner, S.; Markovits, M.A.; Li, G.; Tromp, M.; Pidko, E.A.; Hensen, E.J.; Jentys, A.; Sanchez-Sanchez, M.; Lercher, J.A. Single-site trinuclear copper oxygen clusters in mordenite for selective conversion of methane to methanol. Nat. Commun. 2015, 6, 7546. [Google Scholar] [CrossRef]
- Narsimhan, K.; Iyoki, K.; Dinh, K.; Román-Leshkov, Y. Catalytic oxidation of methane into methanol over copper-exchanged zeolites with oxygen at low temperature. ACS Cent. Sci. 2016, 2, 424–429. [Google Scholar] [CrossRef]
- Bi, W.; Tang, Y.; Li, X.; Dai, C.; Song, C.; Guo, X.; Ma, X. One-step direct conversion of methane to methanol with water in non-thermal plasma. Commun. Chem. 2022, 5, 124. [Google Scholar] [CrossRef]
- Koo, C.W. Structural Studies of Particulate Methane Monooxygenase in a Native Lipid Bilayer. Ph.D. Thesis, Northwestern University, Evanston, IL, USA, 2022. [Google Scholar]
- Murrell, J.C.; Gilbert, B.; McDonald, I.R. Molecular biology and regulation of methane monooxygenase. Arch. Microbiol. 2000, 173, 325–332. [Google Scholar] [CrossRef]
- Semrau, J.D.; DiSpirito, A.A.; Gu, W.; Yoon, S. Metals and methanotrophy. Appl. Environ. Microbiol. 2018, 84, e02289-17. [Google Scholar] [CrossRef] [PubMed]
- Begonja, A.; Hrsak, D. Effect of growth conditions on the expression of soluble methane monooxygenase. Food Technol. Biotechnol. 2001, 39, 29–36. [Google Scholar]
- Chan, S.I.; Chen, K.H.-C.; Yu, S.S.-F.; Chen, C.-L.; Kuo, S.S.-J. Toward delineating the structure and function of the particulate methane monooxygenase from methanotrophic bacteria. Biochemistry 2004, 43, 4421–4430. [Google Scholar] [CrossRef]
- Nielsen, A.K.; Gerdes, K.; Murrell, J.C. Copper-dependent reciprocal transcriptional regulation of methane monooxygenase genes in Methylococcus capsulatus and Methylosinus trichosporium. Mol. Microbiol. 1997, 25, 399–409. [Google Scholar] [CrossRef]
- Srinivas, V.; Banerjee, R.; Lebrette, H.; Jones, J.C.; Aurelius, O.; Kim, I.-S.; Pham, C.C.; Gul, S.; Sutherlin, K.D.; Bhowmick, A. High-resolution XFEL structure of the soluble methane monooxygenase hydroxylase complex with its regulatory component at ambient temperature in two oxidation states. J. Am. Chem. Soc. 2020, 142, 14249–14266. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.; Carey, J.N.; Semrau, J.D. Feasibility of atmospheric methane removal using methanotrophic biotrickling filters. Appl. Microbiol. Biotechnol. 2009, 83, 949–956. [Google Scholar] [CrossRef]
- Sun, M.-T.; Zhao, Y.-Z.; Yang, Z.-M.; Shi, X.-S.; Wang, L.; Dai, M.; Wang, F.; Guo, R.-B. Methane Elimination Using Biofiltration Packed With Fly Ash Ceramsite as Support Material. Front. Bioeng. Biotechnol. 2020, 8, 351. [Google Scholar] [CrossRef]
- Chen, S.; Smith, A.L. Methane-driven microbial fuel cells recover energy and mitigate dissolved methane emissions from anaerobic effluents. Environ. Sci. Water Res. Technol. 2018, 4, 67–79. [Google Scholar] [CrossRef]
- Jawaharraj, K.; Dhiman, S.S.; Bedwell, S.; Vemuri, B.; Islam, J.; Sani, R.K.; Gadhamshetty, V. Electricity from methane by Methylococcus capsulatus (Bath) and Methylosinus trichosporium OB3b. Bioresour. Technol. 2021, 321, 124398. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Kim, T. Development of a novel methanotrophic process with the helper micro-organism Hyphomicrobium sp. NM3. J. Appl. Microbiol. 2019, 126, 534–544. [Google Scholar] [CrossRef]
- Chidambarampadmavathy, K.; Karthikeyan, O.P.; Huerlimann, R.; Maes, G.E.; Heimann, K. Response of mixed methanotrophic consortia to different methane to oxygen ratios. Waste Manag. 2017, 61, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Cruz, S.; Vaksmaa, A.; Horn, M.A.; Niemann, H.; Pijuan, M.; Ho, A. Methanotrophs: Discoveries, environmental relevance, and a perspective on current and future applications. Front. Microbiol. 2021, 12, 678057. [Google Scholar] [CrossRef] [PubMed]
- Mardina, P.; Li, J.; Patel, S.K.; Kim, I.-W.; Lee, J.-K.; Selvaraj, C. Potential of immobilized whole-cell Methylocella tundrae as a biocatalyst for methanol production from methane. J. Microbiol. Biotechnol. 2016, 26, 1234–1241. [Google Scholar] [CrossRef]
- Razumovsky, S.; Efremenko, E.; Makhlis, T.; Senko, O.; Bikhovsky, M.Y.; Podmaster’Ev, V.; Varfolomeev, S. Effect of immobilization on the main dynamic characteristics of the enzymatic oxidation of methane to methanol by bacteria Methylosinus sporium B-2121. Russ. Chem. Bull. 2008, 57, 1633–1636. [Google Scholar] [CrossRef]
- Chistoserdova, L. Modularity of methylotrophy, revisited. Environ. Microbiol. 2011, 13, 2603–2622. [Google Scholar] [CrossRef]
- Smith, T.; Dalton, H. Biocatalysis by methane monooxygenase and its implications for the petroleum industry. Stud. Surf. Sci. Catal. 2004, 151, 177–192. [Google Scholar]
- Smith, S.M.; Balasubramanian, R.; Rosenzweig, A.C. Metal reconstitution of particulate methane monooxygenase and heterologous expression of the pmoB subunit. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2011; Volume 495, pp. 195–210. [Google Scholar]
- Lloyd, J.S.; De Marco, P.; Dalton, H.; Murrell, J.C. Heterologous expression of soluble methane monooxygenase genes in methanotrophs containing only particulate methane monooxygenase. Arch. Microbiol. 1999, 171, 364–370. [Google Scholar] [CrossRef]
- La, H.; Hettiaratchi, J.P.A.; Achari, G.; Dunfield, P.F. Biofiltration of methane. Bioresour. Technol. 2018, 268, 759–772. [Google Scholar] [CrossRef]
- Dalcin Martins, P.; de Jong, A.; Lenstra, W.K.; van Helmond, N.A.; Slomp, C.P.; Jetten, M.S.; Welte, C.U.; Rasigraf, O. Enrichment of novel Verrucomicrobia, Bacteroidetes, and Krumholzibacteria in an oxygen-limited methane-and iron-fed bioreactor inoculated with Bothnian Sea sediments. MicrobiologyOpen 2021, 10, e1175. [Google Scholar] [CrossRef]
- Streese, J.; Stegmann, R. Potentials and limitations of biofilters for methane oxidation. In Proceedings of the Sardinia ‘05, International Waste Management and Landfill Symposium, Cagliari, Italy, 3–7 October 2005. [Google Scholar]
- Jawad, J.; Khalil, M.J.; Sengar, A.K.; Zaidi, S.J. Experimental analysis and modeling of the methane degradation in a three stage biofilter using composted sawdust as packing media. J. Environ. Manag. 2021, 286, 112214. [Google Scholar] [CrossRef]
- Streese, J.; Stegmann, R. Design of biofilters for methane oxidation. In Proceedings of the Sardinia 2003, Ninth International Waste Management and Landfill Symposium, Cagliari, Italy, 6–10 October 2003; pp. 6–10. [Google Scholar]
- Cáceres, M.; Dorado, A.D.; Gentina, J.C.; Aroca, G. Oxidation of methane in biotrickling filters inoculated with methanotrophic bacteria. Environ. Sci. Pollut. Res. 2017, 24, 25702–25712. [Google Scholar] [CrossRef] [PubMed]
- Brandt, E.M.F.; Duarte, F.V.; Vieira, J.P.R.; Melo, V.M.; Souza, C.L.; Araújo, J.C.; Chernicharo, C.A.L. The use of novel packing material for improving methane oxidation in biofilters. J. Environ. Manag. 2016, 182, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Veillette, M.; Viens, P.; Ramirez, A.A.; Brzezinski, R.; Heitz, M. Effect of ammonium concentration on microbial population and performance of a biofilter treating air polluted with methane. Chem. Eng. J. 2011, 171, 1114–1123. [Google Scholar] [CrossRef]
- Ren, Z.J. Microbial fuel cells: Running on gas. Nat. Energy 2017, 2, 17093. [Google Scholar] [CrossRef]
- Song, C. Fuel processing for low-temperature and high-temperature fuel cells: Challenges, and opportunities for sustainable development in the 21st century. Catal. Today 2002, 77, 17–49. [Google Scholar] [CrossRef]
- Pandit, S.; Savla, N.; Sonawane, J.M.; Sani, A.M.d.; Gupta, P.K.; Mathuriya, A.S.; Rai, A.K.; Jadhav, D.A.; Jung, S.P.; Prasad, R. Agricultural waste and wastewater as feedstock for bioelectricity generation using microbial fuel cells: Recent advances. Fermentation 2021, 7, 169. [Google Scholar] [CrossRef]
- Li, W.-W.; Yu, H.-Q.; He, Z. Towards sustainable wastewater treatment by using microbial fuel cells-centered technologies. Energy Environ. Sci. 2014, 7, 911–924. [Google Scholar] [CrossRef]
- Zhang, X.; Rabiee, H.; Frank, J.; Cai, C.; Stark, T.; Virdis, B.; Yuan, Z.; Hu, S. Enhancing methane oxidation in a bioelectrochemical membrane reactor using a soluble electron mediator. Biotechnol. Biofuels 2020, 13, 173. [Google Scholar] [CrossRef]
- Myung, J.; Saikaly, P.E.; Logan, B.E. A two-staged system to generate electricity in microbial fuel cells using methane. Chem. Eng. J. 2018, 352, 262–267. [Google Scholar] [CrossRef]
- Xing, Z.; Zhao, T.; Zhang, L.; Gao, Y.; Liu, S.; Yang, X. Effects of copper on expression of methane monooxygenases, trichloroethylene degradation, and community structure in methanotrophic consortia. Eng. Life Sci. 2018, 18, 236–243. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, X.; Zhou, J.; Zhang, Y. Enrichments of methanotrophic–heterotrophic cultures with high poly-β-hydroxybutyrate (PHB) accumulation capacities. J. Environ. Sci. 2018, 65, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-W.; Keeney, D.R.; Lim, D.-H.; Dispirito, A.A.; Semrau, J.D. Mixed pollutant degradation by Methylosinus trichosporium OB3b expressing either soluble or particulate methane monooxygenase: Can the tortoise beat the hare? Appl. Environ. Microbiol. 2006, 72, 7503–7509. [Google Scholar] [CrossRef] [PubMed]
- Lontoh, S.; Semrau, J.D. Methane and trichloroethylene degradation by Methylosinus trichosporium OB3b expressing particulate methane monooxygenase. Appl. Environ. Microbiol. 1998, 64, 1106–1114. [Google Scholar] [CrossRef] [PubMed]
- Samanta, D.; Govil, T.; Saxena, P.; Gadhamshetty, V.; Krumholz, L.R.; Salem, D.R.; Sani, R.K. Enhancement of Methane Catalysis Rates in Methylosinus trichosporium OB3b. Biomolecules 2022, 12, 560. [Google Scholar] [CrossRef]
- Yu, S.S.-F.; Chen, K.H.-C.; Tseng, M.Y.-H.; Wang, Y.-S.; Tseng, C.-F.; Chen, Y.-J.; Huang, D.-S.; Chan, S.I. Production of high-quality particulate methane monooxygenase in high yields from Methylococcus capsulatus (Bath) with a hollow-fiber membrane bioreactor. J. Bacteriol. 2003, 185, 5915–5924. [Google Scholar] [CrossRef]
- Jahng, D.; Wood, T.K. Trichloroethylene and chloroform degradation by a recombinant pseudomonad expressing soluble methane monooxygenase from Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 1994, 60, 2473–2482. [Google Scholar] [CrossRef]
- Jahng, D.; Kim, C.S.; Hanson, R.S.; Wood, T.K. Optimization of trichloroethylene degradation using soluble methane monooxygenase of Methylosinus trichosporium OB3b expressed in recombinant bacteria. Biotechnol. Bioeng. 1996, 51, 349–359. [Google Scholar] [CrossRef]
- Gou, Z.; Xing, X.-H.; Luo, M.; Jiang, H.; Han, B.; Wu, H.; Wang, L.; Zhang, F. Functional expression of the particulate methane mono-oxygenase gene in recombinant Rhodococcus erythropolis. FEMS Microbiol. Lett. 2006, 263, 136–141. [Google Scholar] [CrossRef]
- Patel, S.K.; Jeong, J.-H.; Mehariya, S.; Otari, S.V.; Madan, B.; Haw, J.R.; Lee, J.-K.; Zhang, L.; Kim, I.-W. Production of methanol from methane by encapsulated Methylosinus sporium. J. Microbiol. Biotechnol. 2016, 26, 2098–2105. [Google Scholar] [CrossRef]
- Van der Ha, D.; Bundervoet, B.; Verstraete, W.; Boon, N. A sustainable, carbon neutral methane oxidation by a partnership of methane oxidizing communities and microalgae. Water Res. 2011, 45, 2845–2854. [Google Scholar] [CrossRef]
- Tays, C. Growth Characterization and Transcriptomics of Methanotrophic Bacteria as Effected by Carbon and Nitrogen Sources. Ph.D. Thesis, University of Alberta, Edmonton, AB, Canada, 2019. [Google Scholar]
- Akberdin, I.R.; Thompson, M.; Hamilton, R.; Desai, N.; Alexander, D.; Henard, C.A.; Guarnieri, M.T.; Kalyuzhnaya, M.G. Methane utilization in Methylomicrobium alcaliphilum 20ZR: A systems approach. Sci. Rep. 2018, 8, 2512. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).