A Systematic Review of Machine Learning Models in Mental Health Analysis Based on Multi-Channel Multi-Modal Biometric Signals
Abstract
1. Introduction
2. Background and Prior Work
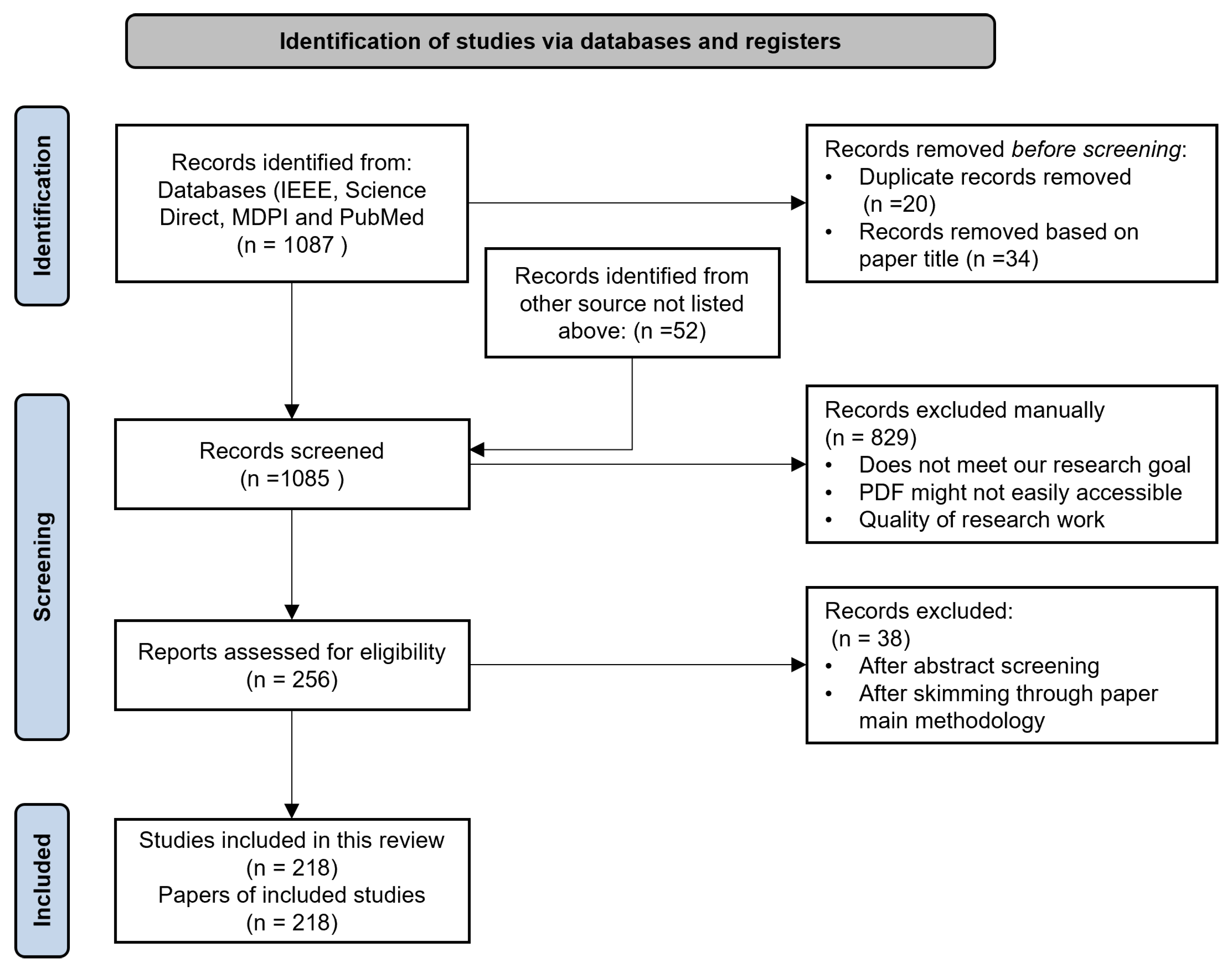
2.1. Literature Search Process
- Publications had to be released in 2017 or later.
- To deepen the understanding of the research questions, we also added 25 articles published between the years 2000 and 2016. This range was selected based on references from similar research.
- Articles had to have at least one or more of the keywords.
- Articles had to be published in recognized literature databases/websites.
- All selected papers had to be written in English.
- All papers were either studies, surveys, or reviews of the application of ML on PSG data and the classification of mental health issues using ML.
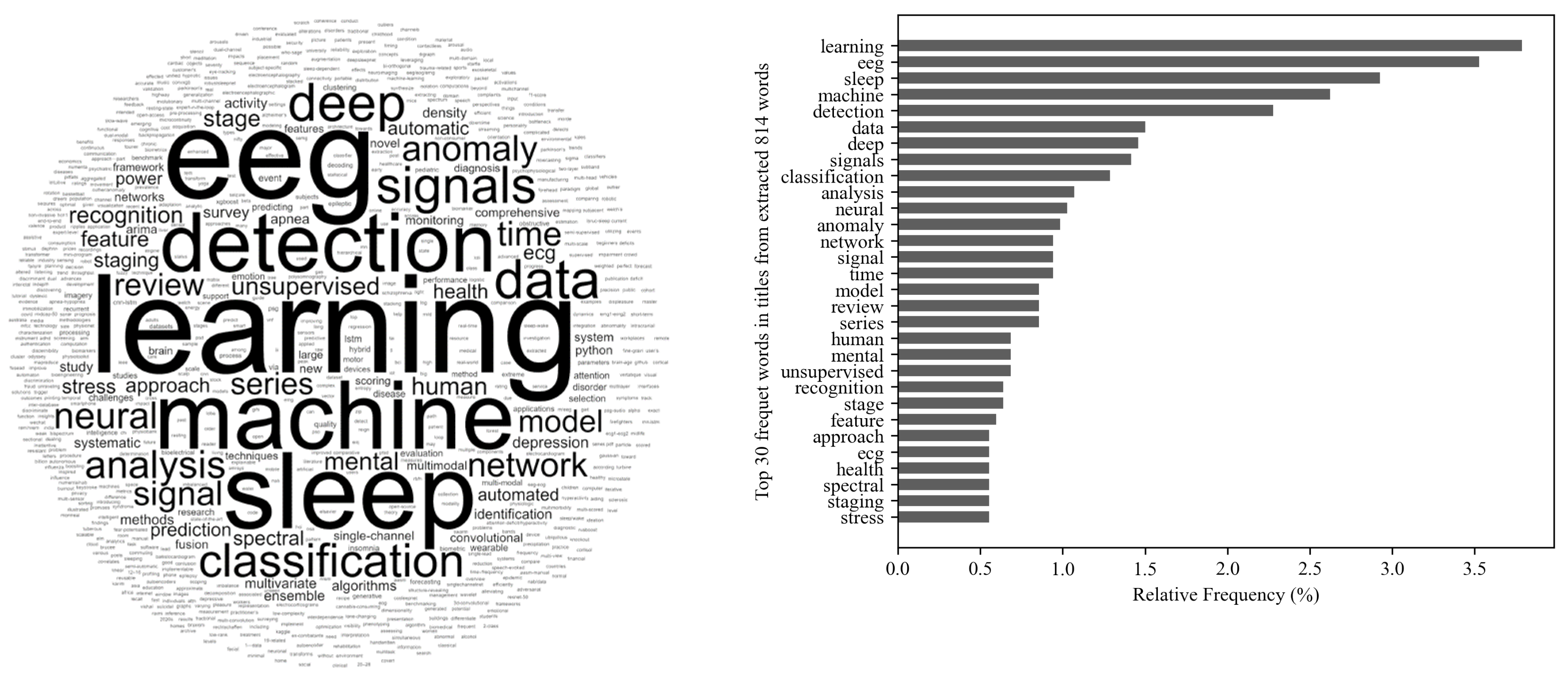
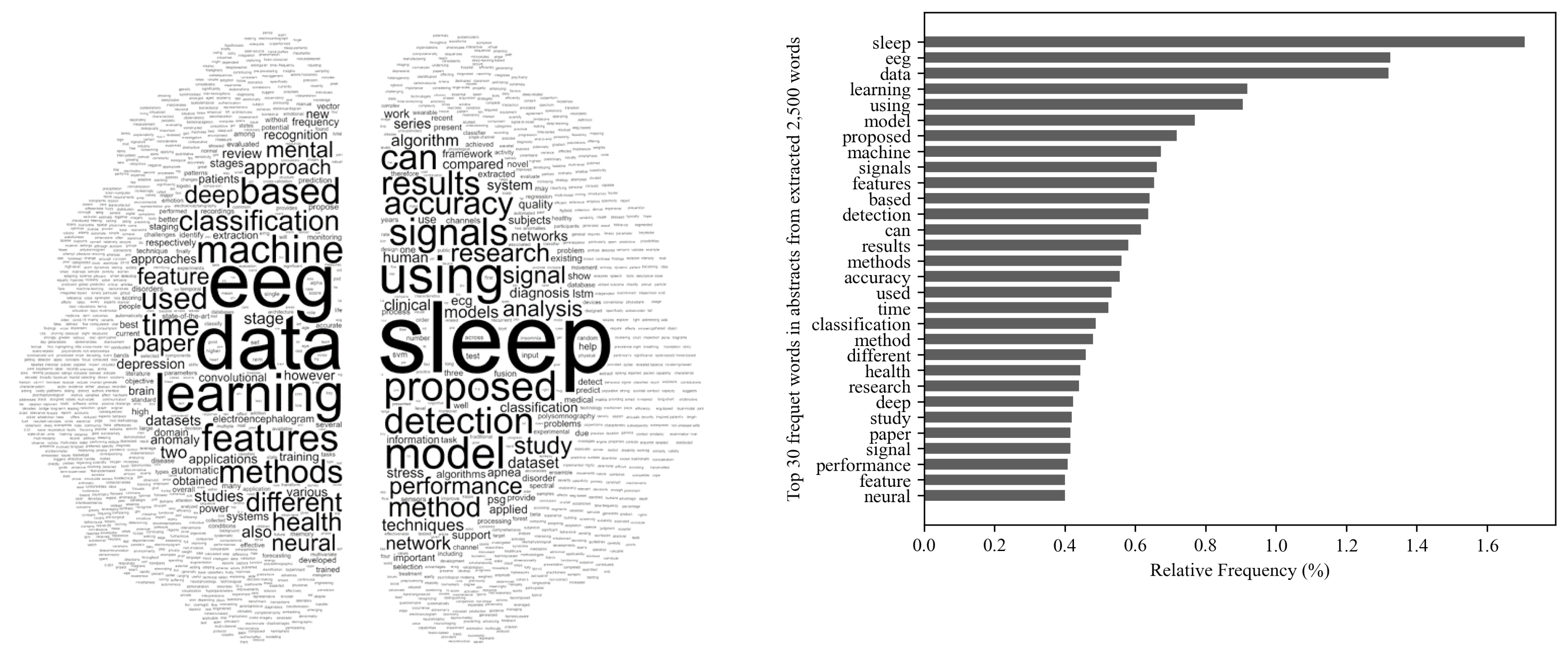
2.2. Word-Cloud Overview
2.3. Literature Distribution by Publication Year
2.4. Methods
2.4.1. Data Preparation
- An official website (https://sleepdata.org/; accessed on 21 February 2023) managed by The National Sleep Research Resource, provides open access for researchers and those interested in sleep studies, with large collections of physiological signals and clinical data elements. These datasets were collected from structured research cohorts and clinical trials. The Nationwide Children’s Hospital (NCH) Sleep Data Bank consisted of three folders: Sleep Data, Health Data, and the Sleep Data with annotated PSG data recordings [68].
- The Montreal Archive of Sleep Studies (MASS) (http://ceams-carsm.ca/en/MASS/; accessed on 21 February 2023) is an open-access and collaborative database of laboratory-based PSG recordings. It also comprised a cohort of subsets [25,69].
- PhysioNet (https://physionet.org/; accessed on 21 February 2023) is an open-access physiologic-signal data source that is managed by members of the MIT Laboratory for Computational Physiology [68,70,71].
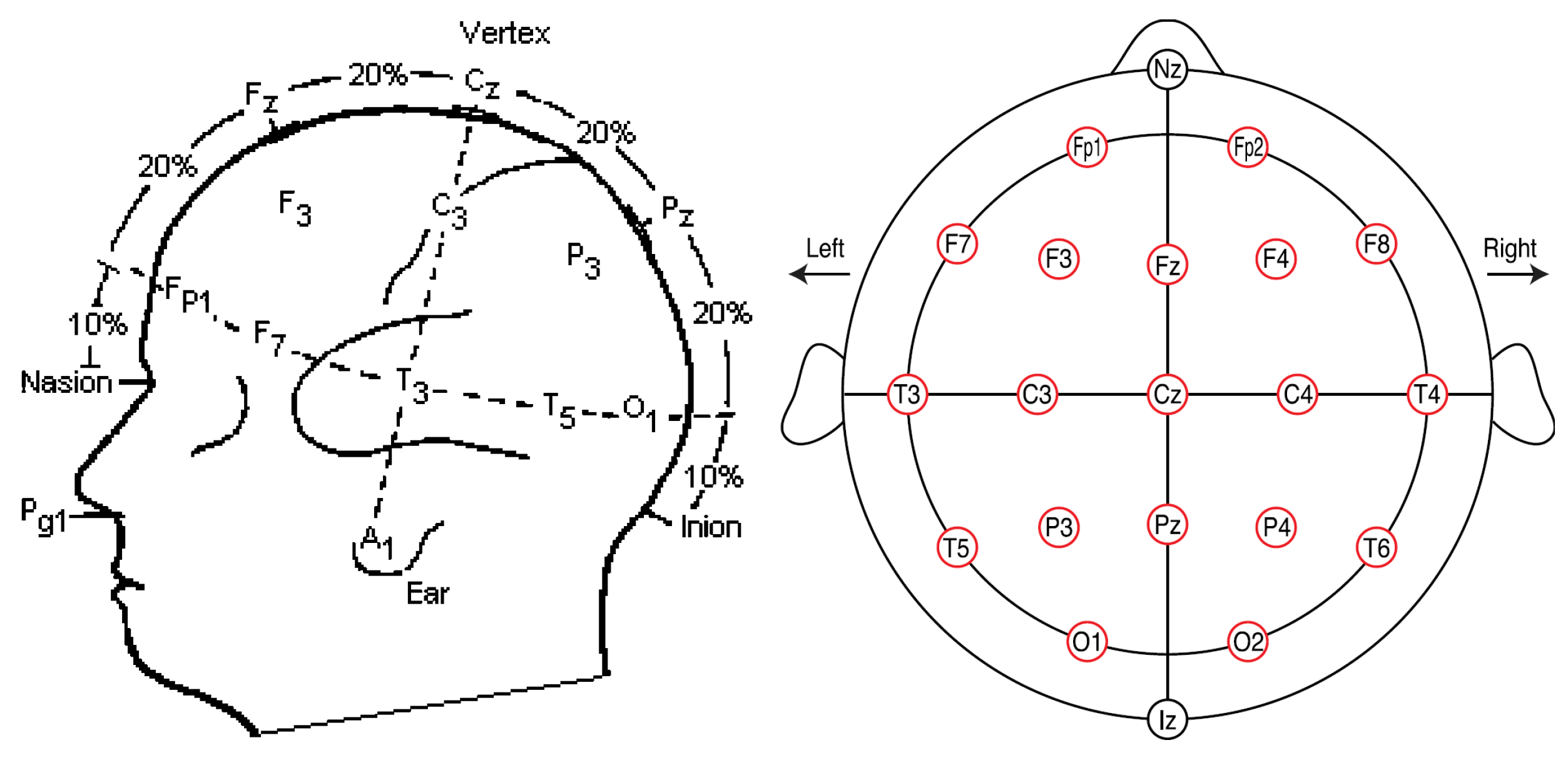
2.4.2. Data Acquisition
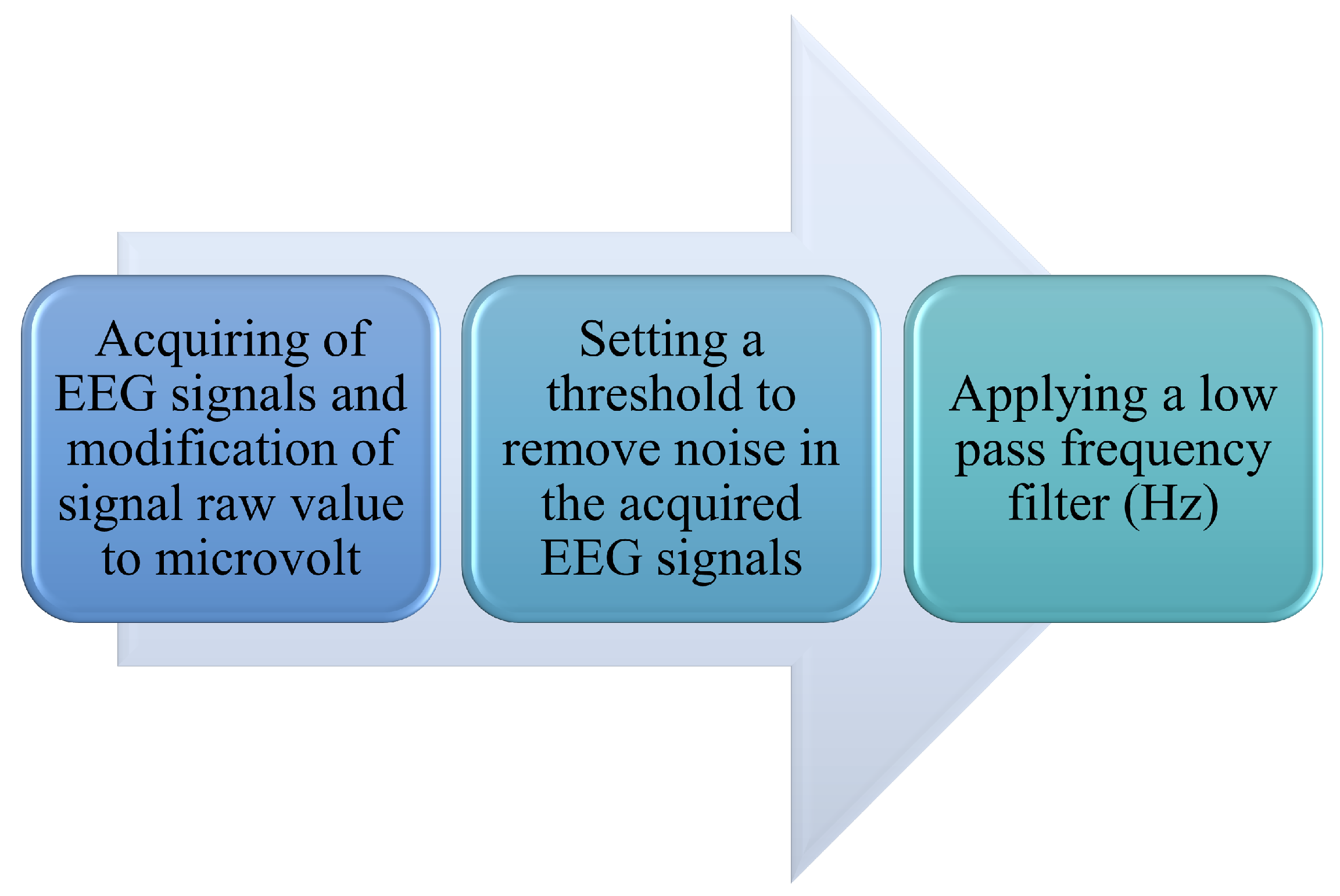
2.4.3. Pre-Processing of Signals
2.5. Feature Extraction
2.6. Balancing Datasets
2.7. Machine-Learning Modeling
2.8. Performance Evaluation
- Sensitivity: It is also known as recall. This measures the ratio of the number of samples correctly predicted to the total samples in the class. Sensitivity can be calculated based on true positive (TP) and false negative (FN) parameters [31,208,211]. Equation (1) shows a mathematical representation of the sensitivity computation.
- Accuracy: This is the fraction of samples that were correctly classified. Accuracy can be expressed as the ratio of the summation of true-positive (TP) and true-negative (TN) parameters to the total sample size, which includes true positive (TP), false positive (FP), false negative (FN), and true negative (TN) [31,137,208,211]. Equation (2) shows a mathematical representation of accuracy.
- Precision: It is the ratio of the samples correctly predicted to the total predicted positive samples. Equation (3) shows a mathematical representation of the precision computation.
- Specificity: It measures how many healthy (negative) samples were identified as healthy (negative) samples by a model. Equation (4) shows a mathematical representation of the specificity computation.
- F1-score: It is a function of precision and sensitivity (recall). It is represented as the harmonic mean of sensitivity and precision. Equation (5) shows a mathematical representation of F1-score computation. F1-scores range from 0 to 1, with 1 being a perfect precision sensitivity (recall) and 0 being the lowest precision sensitivity. Equation (5) shows a mathematical representation of the F1-score computation.
3. Summary and Discussion
3.1. Challenges of Using ML on Multi-Channel and Multi-Modal PSG Datasets
3.2. Research Gaps
4. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Nomenclatures
| AASM | American Academy of Sleep Medicine |
| ADHD | Attention-Deficit Hyperactivity Disorder |
| AI | Artificial Intelligence |
| ANN | Artificial Neural Network |
| CNN | Convolutional Neural Networks |
| ECG | Electrocardiograph |
| EEG | Electroencephalogram |
| ELM | Extreme Learning Machine |
| EMG | Electromyogram |
| EOG | Electro-oculogram |
| FFT | Fast Fourier Transformation |
| HRV | Heart Rate Variability |
| LR | Logistic Regression |
| LSTM | Long-Short Term Memory |
| ML | Machine Learning |
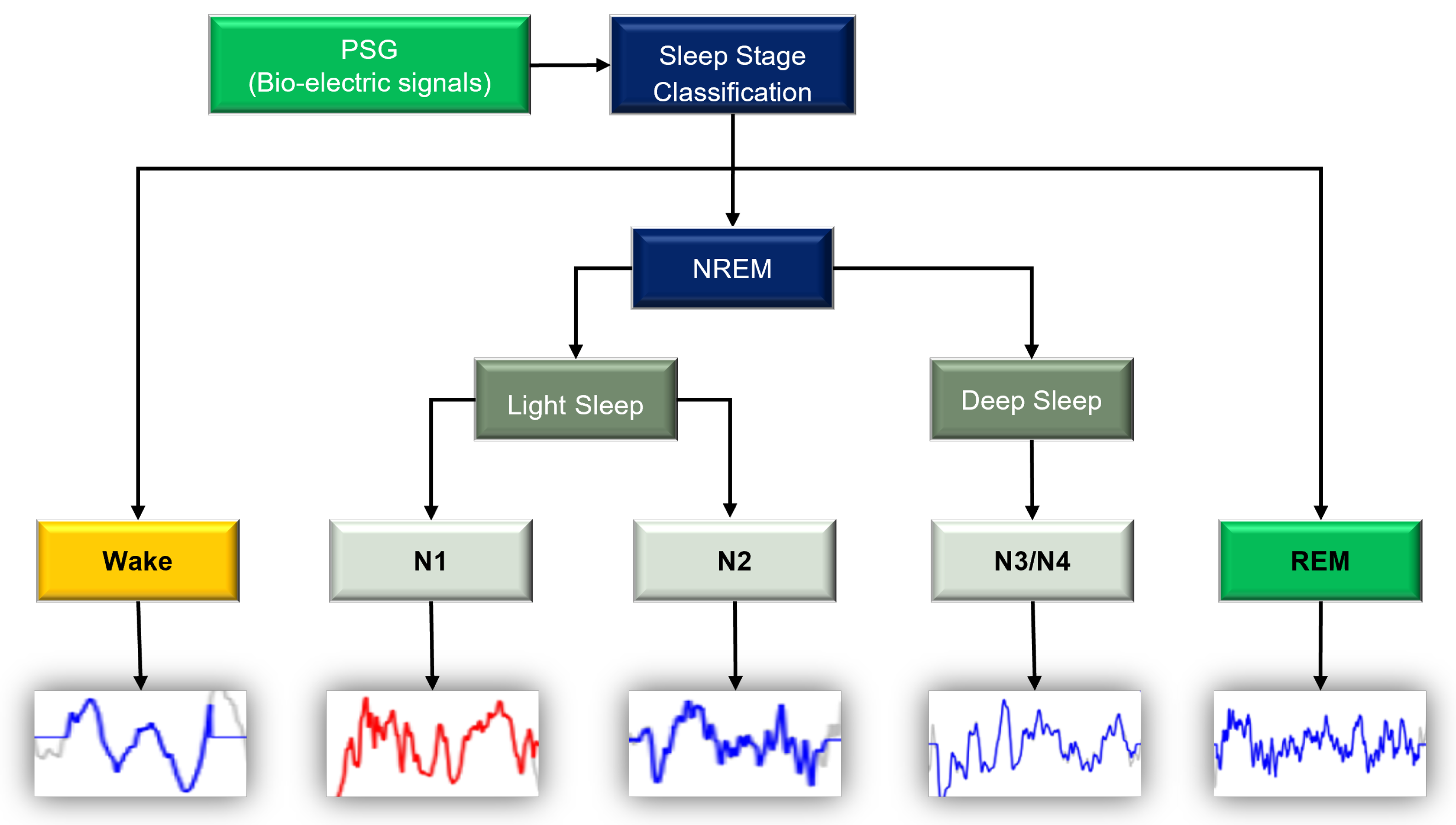
| NREM | Non-Rapid Eye Movement |
| REM | Rapid Eye Movement |
| RF | Random Forest |
| RNN | Recurrent Neural Networks |
| RK | Rechtschaffen and Kales |
| PCA | Principal Component Analysis |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| PSD | Power Spectral Density |
| SNN | Spike Neural Network |
| PSG | Polysomnography |
| SPO2 | Saturation of Peripheral Oxygen |
| SVM | Support Vector Machine |
References
- Goetz, C.; Bavaresco, R.; Kunst, R.; Barbosa, J. Industrial intelligence in the care of workers’ mental health: A review of status and challenges. Int. J. Ind. Ergon. 2022, 87, 103234. [Google Scholar] [CrossRef]
- Ramos-Lima, L.F.; Waikamp, V.; Antonelli-Salgado, T.; Passos, I.C.; Freitas, L.H.M. The use of machine learning techniques in trauma-related disorders: A systematic review. J. Psychiatr. Res. 2020, 121, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, N.; Zhang, L.; Liu, Y.; Zhang, T.; Li, D.; Bai, D.; Liu, X.; Li, L. Predicting PTSD symptoms in firefighters using a fear-potentiated startle paradigm and machine learning. J. Affect. Disord. 2022, 319, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Sumathi, M.S.; Joshi, C.S.; Thomas, R.R.; Reethu, G. Analysis and Performance of Machine Learning Algorithms on Disease Diagnosis. In Proceedings of the 2020 3rd International Conference on Energy, Power and Environment: Towards Clean Energy Technologies, Shanghai, China, 12–13 July 2021; IEEE: Shillong, India, 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Rejaibi, E. MFCC-based Recurrent Neural Network for automatic clinical depression recognition and assessment from speech. Biomed. Signal Process. Control. 2022, 11. [Google Scholar] [CrossRef]
- Gore, E.; Rathi, S. Types of Data with Algorithms for Assessing Mental Health Conditions. In Proceedings of the 2019 5th International Conference on Computing, Communication, Control and Automation (ICCUBEA), Pune, India, 19–21 September 2019; IEEE: Pune, India, 2019; pp. 1–8. [Google Scholar] [CrossRef]
- Lu, W.; Li, J.; Li, Y.; Sun, A.; Wang, J. A CNN-LSTM-Based Model to Forecast Stock Prices. Complexity 2020, 2020, 6622927. [Google Scholar] [CrossRef]
- Zhang, C.; Song, D.; Chen, Y.; Feng, X.; Lumezanu, C.; Cheng, W.; Ni, J.; Zong, B.; Chen, H.; Chawla, N.V. A Deep Neural Network for Unsupervised Anomaly Detection and Diagnosis in Multivariate Time Series Data. Proc. AAAI Conf. Artif. Intell. 2019, 33, 1409–1416. [Google Scholar] [CrossRef]
- Li, J.; Izakian, H.; Pedrycz, W.; Jamal, I. Clustering-based anomaly detection in multi-variate time-series data. Appl. Soft Comput. 2021, 100, 106919. [Google Scholar] [CrossRef]
- Manjunath, S.; Nathaniel, A.; Druce, J.; German, S. Improving the Performance of Fine-Grain Image Classifiers via Generative Data Augmentation. arXiv 2020. [Google Scholar] [CrossRef]
- Goldstein, M.; Uchida, S. A Comparative Evaluation of Unsupervised Anomaly Detection Algorithms for Multivariate Data. PLoS ONE 2016, 11, e0152173. [Google Scholar] [CrossRef]
- Sabry, F.; Eltaras, T.; Labda, W.; Alzoubi, K.; Malluhi, Q. Machine Learning for Healthcare Wearable Devices: The Big Picture. J. Healthc. Eng. 2022, 2022, 4653923. [Google Scholar] [CrossRef]
- Mehdiyev, N.; Lahann, J.; Emrich, A.; Enke, D.; Fettke, P.; Loos, P. Time Series Classification using Deep Learning for Process Planning: A Case from the Process Industry. Procedia Comput. Sci. 2017, 114, 242–249. [Google Scholar] [CrossRef]
- Li, J. Multi-modal bio-electrical signal fusion analysis based on different acquisition devices and scene settings: Overview, challenges, and novel orientation. Inf. Fusion 2022, 79, 229–247. [Google Scholar] [CrossRef]
- Saeidi, M.; Karwowski, W.; Farahani, F.V.; Fiok, K.; Taiar, R.; Hancock, P.A.; Al-Juaid, A. Neural Decoding of EEG Signals with Machine Learning: A Systematic Review. Brain Sci. 2021, 11, 1525. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.; Yoon, H.; Park, K. A Novel Wearable Forehead EOG Measurement System for Human Computer Interfaces. Sensors 2017, 17, 1485. [Google Scholar] [CrossRef] [PubMed]
- Hallett, M.; DelRosso, L.M.; Elble, R.; Ferri, R.; Horak, F.B.; Lehericy, S.; Mancini, M.; Matsuhashi, M.; Matsumoto, R.; Muthuraman, M.; et al. Evaluation of movement and brain activity. Clin. Neurophysiol. 2021, 132, 2608–2638. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wei, X.; Guo, J.; Zheng, Y.; Li, J.; Du, S. Research Progress of Rehabilitation Exoskeletal Robot and Evaluation Methodologies Based on Bio-electrical Signals. In Proceedings of the 2019 IEEE 9th Annual International Conference on CYBER Technology in Automation, Control, and Intelligent Systems (CYBER), Suzhou, China, 29 July–2 August 2019; IEEE: Suzhou, China, 2019; pp. 826–831. [Google Scholar] [CrossRef]
- Bleakley, L.E.; Keenan, R.J.; Graven, R.D.; Metha, J.A.; Ma, S.; Daykin, H.; Cornthwaite-Duncan, L.; Hoyer, D.; Reid, C.A.; Jacobson, L.H. Altered EEG power spectrum, but not sleep-wake architecture, in HCN1 knockout mice. Behav. Brain Res. 2023, 437, 114105. [Google Scholar] [CrossRef] [PubMed]
- Bi, X.; Wang, H. Early Alzheimer’s disease diagnosis based on EEG spectral images using deep learning. Neural Netw. 2019, 114, 119–135. [Google Scholar] [CrossRef]
- Martinek, R.; Ladrova, M.; Sidikova, M.; Jaros, R.; Behbehani, K.; Kahankova, R.; Kawala-Sterniuk, A. Advanced Bio-electrical Signal Processing Methods: Past, Present and Future Approach—Part II: Brain Signals. Sensors 2021, 21, 6343. [Google Scholar] [CrossRef]
- Kaplan, K.A.; Hardas, P.P.; Redline, S.; Zeitzer, J.M. Correlates of sleep quality in midlife and beyond: A machine learning analysis. Sleep Med. 2017, 34, 162–167. [Google Scholar] [CrossRef]
- Gerla, V.; Kremen, V.; Macas, M.; Dudysova, D.; Mladek, A.; Sos, P.; Lhotska, L. Iterative expert-in-the-loop classification of sleep PSG recordings using a hierarchical clustering. J. Neurosci. Methods 2019, 317, 61–70. [Google Scholar] [CrossRef]
- Biswal, S.; Sun, H.; Goparaju, B.; Westover, M.B.; Sun, J.; Bianchi, M.T. Expert-level sleep scoring with deep neural networks. J. Am. Med. Inform. Assoc. 2018, 25, 1643–1650. [Google Scholar] [CrossRef]
- Chambon, S.; Galtier, M.N.; Arnal, P.J.; Wainrib, G.; Gramfort, A. A Deep Learning Architecture for Temporal Sleep Stage Classification Using Multivariate and Multimodal Time Series. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Patel, V.; Acharya, U.R. Automated identification of insomnia using optimal bi-orthogonal wavelet transform technique with single-channel EEG signals. Knowl.-Based Syst. 2021, 224, 107078. [Google Scholar] [CrossRef]
- Zaffaroni, A.; Coffey, S.; Dodd, S.; Kilroy, H.; Lyon, G.; O’Rourke, D.; Lederer, K.; Fietze, I.; Penzel, T. Sleep Staging Monitoring Based on Sonar Smartphone Technology. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; IEEE: Berlin, Germany, 2019; pp. 2230–2233. [Google Scholar] [CrossRef]
- Ravan, M. A machine learning approach using EEG signals to measure sleep quality. AIMS Electron. Electr. Eng. 2019, 3, 347–358. [Google Scholar] [CrossRef]
- Wang, H.; Lu, C.; Zhang, Q.; Hu, Z.; Yuan, X.; Zhang, P.; Liu, W. A novel sleep staging network based on multi-scale dual attention. Biomed. Signal Process. Control 2022, 74, 103486. [Google Scholar] [CrossRef]
- Ramachandran, A.; Karuppiah, A. A Survey on Recent Advances in Machine Learning Based Sleep Apnea Detection Systems. Healthcare 2021, 9, 914. [Google Scholar] [CrossRef] [PubMed]
- Aboalayon, K.; Faezipour, M.; Almuhammadi, W.; Moslehpour, S. Sleep Stage Classification Using EEG Signal Analysis: A Comprehensive Survey and New Investigation. Entropy 2016, 18, 272. [Google Scholar] [CrossRef]
- Xu, S.; Faust, O.; Seoni, S.; Chakraborty, S.; Barua, P.D.; Loh, H.W.; Elphick, H.; Molinari, F.; Acharya, U.R. A review of automated sleep disorder detection. Comput. Biol. Med. 2022, 150, 106100. [Google Scholar] [CrossRef]
- Acharya, U.R.; Oh, S.L.; Hagiwara, Y.; Tan, J.H.; Adeli, H.; Subha, D.P. Automated EEG-based screening of depression using deep convolutional neural network. Comput. Methods Programs Biomed. 2018, 161, 103–113. [Google Scholar] [CrossRef]
- Liu, S.; Shen, J.; Li, Y.; Wang, J.; Wang, J.; Xu, J.; Wang, Q.; Chen, R. EEG Power Spectral Analysis of Abnormal Cortical Activations During REM/NREM Sleep in Obstructive Sleep Apnea. Front. Neurol. 2021, 12, 643855. [Google Scholar] [CrossRef]
- Ay, B.; Yildirim, O.; Talo, M.; Baloglu, U.B.; Aydin, G.; Puthankattil, S.D.; Acharya, U.R. Automated Depression Detection Using Deep Representation and Sequence Learning with EEG Signals. J. Med. Syst. 2019, 43, 205. [Google Scholar] [CrossRef]
- Zoubek, L.; Charbonnier, S.; Lesecq, S.; Buguet, A.; Chapotot, F. Feature selection for sleep/wake stages classification using data driven methods. Biomed. Signal Process. Control 2007, 2, 171–179. [Google Scholar] [CrossRef]
- Rezaei, M.; Mohammadi, H.; Khazaie, H. EEG/EOG/EMG data from a cross sectional study on psychophysiological insomnia and normal sleep subjects. Data Brief 2017, 15, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Satapathy, S.K.; Bhoi, A.K.; Loganathan, D.; Khandelwal, B.; Barsocchi, P. Machine learning with ensemble stacking model for automated sleep staging using dual-channel EEG signal. Biomed. Signal Process. Control 2021, 69, 102898. [Google Scholar] [CrossRef]
- Gao, W.; Xu, Y.; Li, S.; Fu, Y.; Zheng, D.; She, Y. Obstructive sleep apnea syndrome detection based on ballistocardiogram via machine learning approach. Math. Biosci. Eng. 2019, 16, 5672–5686. [Google Scholar] [CrossRef]
- Salama, E.S. A 3D-convolutional neural network framework with ensemble learning techniques for multi-modal emotion recognition. Egypt. Inform. J. 2021, 22, 167–176. [Google Scholar] [CrossRef]
- Langer, N.; Plomecka, M.B.; Tröndle, M.; Negi, A.; Popov, T.; Milham, M.; Haufe, S. A benchmark for prediction of psychiatric multimorbidity from resting EEG data in a large pediatric sample. NeuroImage 2022, 258, 119348. [Google Scholar] [CrossRef]
- Uwaechia, A.N.; Ramli, D.A. A Comprehensive Survey on ECG Signals as New Biometric Modality for Human Authentication: Recent Advances and Future Challenges. IEEE Access 2021, 9, 97760–97802. [Google Scholar] [CrossRef]
- Rakshith, V.; Apoorv, V.; Akarsh, N.K.; Arjun, K.; Krupa, B.N.; Pratima, M.; Vedamurthachar, A. A novel approach for the identification of chronic alcohol users from ECG signals. In Proceedings of the TENCON 201–2017 IEEE Region 10 Conference, Penang, Malaysia, 5–8 November 2017; IEEE: Penang, Malaysia, 2017; pp. 1321–1326. [Google Scholar] [CrossRef]
- Vuksanovic, B. Analysis of Human Electrocardiogram for Biometric Recognition Using Analytic and AR Modeling Extracted Parameters. Int. J. Inf. Electron. Eng. 2014, 4. [Google Scholar] [CrossRef]
- Said, S.; Nait-ali, A. Machine-Learning based Wearable Multi-Channel sEMG Biometrics Modality for User’s Identification. In Proceedings of the 2021 4th International Conference on Bio-Engineering for Smart Technologies (BioSMART), Paris/Créteil, France, 8–10 December 2021; p. 4. [Google Scholar]
- Pigoni, A.; Delvecchio, G.; Madonna, D.; Bressi, C.; Soares, J.; Brambilla, P. Can Machine Learning help us in dealing with treatment resistant depression? A review. J. Affect. Disord. 2019, 259, 21–26. [Google Scholar] [CrossRef]
- Combrisson, E.; Vallat, R.; Eichenlaub, J.B.; O’Reilly, C.; Lajnef, T.; Guillot, A.; Ruby, P.M.; Jerbi, K. Sleep: An Open-Source Python Software for Visualization, Analysis, and Staging of Sleep Data. Front. Neuroinform. 2017, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Supratak, A.; Dong, H.; Wu, C.; Guo, Y. DeepSleepNet: A Model for Automatic Sleep Stage Scoring Based on Raw Single-Channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1998–2008. [Google Scholar] [CrossRef] [PubMed]
- Moser, D.; Anderer, P.; Gruber, G.; Parapatics, S.; Loretz, E.; Boeck, M.; Kloesch, G.; Heller, E.; Schmidt, A.; Danker-Hopfe, H.; et al. Sleep Classification According to AASM and Rechtschaffen & Kales: Effects on Sleep Scoring Parameters. Sleep 2009, 32, 139–149. [Google Scholar]
- The AASM-Manual for Scoring Sleep and Associated Event. Available online: https://aasm.org/clinical-resources/scoring-manual/ (accessed on 21 February 2023).
- Huang, C.S.; Lin, C.L.; Ko, L.W.; Liu, S.Y.; Sua, T.P.; Lin, C.T. A hierarchical classification system for sleep stage scoring via forehead EEG signals. In Proceedings of the 2013 IEEE Symposium on Computational Intelligence, Cognitive Algorithms, Mind, and Brain (CCMB), Singapore, 16–19 April 2013; IEEE: Singapore, 2013; pp. 1–5. [Google Scholar] [CrossRef]
- Ghaderyan, P.; Moghaddam, F.; Khoshnoud, S.; Shamsi, M. New interdependence feature of EEG signals as a biomarker of timing deficits evaluated in Attention-Deficit/Hyperactivity Disorder detection. Measurement 2022, 199, 111468. [Google Scholar] [CrossRef]
- Lee, W.; Kim, G.; Yu, J.; Kim, Y. Model Interpretation Considering Both Time and Frequency Axes Given Time Series Data. Appl. Sci. 2022, 12, 12807. [Google Scholar] [CrossRef]
- Dev, A.; Roy, N.; Islam, M.K.; Biswas, C.; Ahmed, H.U.; Amin, M.A.; Sarker, F.; Vaidyanathan, R.; Mamun, K.A. Exploration of EEG-Based Depression Biomarkers Identification Techniques and Their Applications: A Systematic Review. IEEE Access 2022, 10, 16756–16781. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Sharma, L.D.; Bohat, V.K.; Habib, M.; Al-Zoubi, A.M.; Faris, H.; Aljarah, I. Evolutionary inspired approach for mental stress detection using EEG signal. Expert Syst. Appl. 2022, 197, 116634. [Google Scholar] [CrossRef]
- Motwani, A.; Shukla, P.K.; Pawar, M. Ubiquitous and smart healthcare monitoring frameworks based on machine learning: A comprehensive review. Artif. Intell. Med. 2022, 134, 102431. [Google Scholar] [CrossRef]
- Barros, C.; Silva, C.A.; Pinheiro, A.P. Advanced EEG-based learning approaches to predict schizophrenia: Promises and pitfalls. Artif. Intell. Med. 2021, 114, 102039. [Google Scholar] [CrossRef]
- Mirchi, N.; Warsi, N.M.; Zhang, F.; Wong, S.M.; Suresh, H.; Mithani, K.; Erdman, L.; Ibrahim, G.M. Decoding Intracranial EEG With Machine Learning: A Systematic Review. Front. Hum. Neurosci. 2022, 16, 913777. [Google Scholar] [CrossRef] [PubMed]
- Thieme, A.; Belgrave, D.; Doherty, G. Machine Learning in Mental Health: A Systematic Review of the HCI Literature to Support the Development of Effective and Implementable ML Systems. ACM Trans. Comput.-Hum. Interact. 2020, 27, 1–53. [Google Scholar] [CrossRef]
- Boukobza, A.; Burgun, A.; Roudier, B.; Tsopra, R. Deep Neural Networks for Simultaneously Capturing Public Topics and Sentiments During a Pandemic: Application on a COVID-19 Tweet Data Set. JMIR Med. Inform. 2022, 10, e34306. [Google Scholar] [CrossRef] [PubMed]
- Gramfort, A.; Luessi, M.; Larson, E.; Engemann, D.A.; Strohmeier, D.; Brodbeck, C.; Goj, R.; Jas, M.; Brooks, T.; Parkkonen, L.; et al. MEG and EEG Data Analysis with MNE-Python. Front. Neurosci. 2013, 7, 1–13. [Google Scholar] [CrossRef]
- Sekkal, R.N.; Bereksi-Reguig, F.; Ruiz-Fernandez, D.; Dib, N.; Sekkal, S. Automatic sleep-stage classification: From classical machine learning methods to deep learning. Biomed. Signal Process. Control 2022, 77, 103751. [Google Scholar] [CrossRef]
- Pepi, C.; Mercier, M.; Carfì Pavia, G.; de Benedictis, A.; Vigevano, F.; Rossi-Espagnet, M.C.; Falcicchio, G.; Marras, C.E.; Specchio, N.; de Palma, L. Can Presurgical Interhemispheric EEG Connectivity Predict Outcome in Hemispheric Surgery? A Brain Machine Learning Approach. Brain Sci. 2023, 13, 71. [Google Scholar] [CrossRef]
- ElMoaqet, H.; Eid, M.; Ryalat, M.; Penzel, T. A Deep Transfer Learning Framework for Sleep Stage Classification with Single-Channel EEG Signals. Sensors 2022, 22, 8826. [Google Scholar] [CrossRef]
- Ehiabhi, J.; Wang, H. An Unsupervised Anomaly Detection Model for Multivariate Time Series Data. In Proceedings of the IISE ANNUAL Conference, Seattle, DC, USA, 21 May 2022; p. 7. [Google Scholar]
- Ameera, A.; Saidatul, A.; Ibrahim, Z. Analysis of EEG Spectrum Bands Using Power Spectral Density for Pleasure and Displeasure State. IOP Conf. Ser. Mater. Sci. Eng. 2019, 557, 012030. [Google Scholar] [CrossRef]
- Lee, H.; Li, B.; DeForte, S.; Splaingard, M.; Huang, Y.; Chi, Y.; Linwood, S.L. A Large Collection of Real-world Pediatric Sleep Studies. Sci. Data 2022, 9, 421. [Google Scholar] [CrossRef]
- O’Reilly, C.; Gosselin, N.; Carrier, J.; Nielsen, T. Montreal Archive of Sleep Studies: An open-access resource for instrument benchmarking and exploratory research. J. Sleep Res. 2014, 23, 628–635. [Google Scholar] [CrossRef]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a New Research Resource for Complex Physiologic Signals. Circulation 2000, 101. [Google Scholar] [CrossRef] [PubMed]
- Memis, G.; Sert, M. Multimodal Classification of Obstructive Sleep Apnea Using Feature Level Fusion. In Proceedings of the 2017 IEEE 11th International Conference on Semantic Computing (ICSC), San Diego, CA, USA, 30 January–1 February 2017; IEEE: San Diego, CA, USA, 2017; pp. 85–88. [Google Scholar] [CrossRef]
- Khosla, A.; Khandnor, P.; Chand, T. Automated diagnosis of depression from EEG signals using traditional and deep-learning approaches: A comparative analysis. Biocybern. Biomed. Eng. 2022, 42, 108–142. [Google Scholar] [CrossRef]
- Engemann, D.A.; Mellot, A.; Höchenberger, R.; Banville, H.; Sabbagh, D.; Gemein, L.; Ball, T.; Gramfort, A. A reusable benchmark of brain-age prediction from M/EEG resting-state signals. NeuroImage 2022, 262, 119521. [Google Scholar] [CrossRef] [PubMed]
- Khalighi, S.; Sousa, T.; Santos, J.M.; Nunes, U. ISRUC-Sleep: A comprehensive public dataset for sleep researchers. Comput. Methods Programs Biomed. 2016, 124, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, R.; Sahana, B.C. Automatic Detection of Mental Health Status using Alpha Subband of EEG Data. In Proceedings of the 2022 IEEE International Symposium on Medical Measurements and Applications (MeMeA), Messina, Italy, 2–24 June 2022; IEEE: Messina, Italy, 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Jain, R.; Ganesan, R.A. Reliable sleep staging of unseen subjects with fusion of multiple EEG features and RUSBoost. Biomed. Signal Process. Control 2021, 70, 103061. [Google Scholar] [CrossRef]
- Supakar, R.; Satvaya, P.; Chakrabarti, P. A deep learning based model using RNN-LSTM for the Detection of Schizophrenia from EEG data. Comput. Biol. Med. 2022, 151, 106225. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, K.; Qadir, J.; O’Brien, T.J.; Kuhlmann, L.; Razi, A. A Generative Model to Synthesize EEG Data for Epileptic Seizure Prediction. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 2322–2332. [Google Scholar] [CrossRef]
- Chatterjee, R.; Maitra, T.; Hafizul Islam, S.; Hassan, M.M.; Alamri, A.; Fortino, G. A novel machine learning based feature selection for motor imagery EEG signal classification in Internet of medical things environment. Future Gener. Comput. Syst. 2019, 98, 419–434. [Google Scholar] [CrossRef]
- Alam, M.N.; Ibrahimy, M.I.; Motakabber, S.M.A. Feature Extraction of EEG Signal by Power Spectral Density for Motor Imagery Based BCI. In Proceedings of the 2021 8th International Conference on Computer and Communication Engineering (ICCCE), Kuala Lumpur, Malaysia, 9–10 April 2021; IEEE: Kuala Lumpur, Malaysia, 2021; pp. 234–237. [Google Scholar] [CrossRef]
- Tang, M.; Zhang, Z.; He, Z.; Li, W.; Mou, X.; Du, L.; Wang, P.; Zhao, Z.; Chen, X.; Li, X.; et al. Deep adaptation network for subject-specific sleep stage classification based on a single-lead ECG. Biomed. Signal Process. Control 2022, 75, 103548. [Google Scholar] [CrossRef]
- Alvarez-Estevez, D.; Rijsman, R.M. Inter-database validation of a deep-learning approach for automatic sleep scoring. PLoS ONE 2021, 16, e0256111. [Google Scholar] [CrossRef]
- Palotti, J.; Mall, R.; Aupetit, M.; Rueschman, M.; Singh, M.; Sathyanarayana, A.; Taheri, S.; Fernandez-Luque, L. Benchmark on a large cohort for sleep-wake classification with machine learning techniques. NPJ Digit. Med. 2019, 2, 50. [Google Scholar] [CrossRef]
- Zarei, A.; Beheshti, H.; Asl, B.M. Detection of sleep apnea using deep neural networks and single-lead ECG signals. Biomed. Signal Process. Control 2022, 71, 103125. [Google Scholar] [CrossRef]
- Zhao, R.; Xia, Y.; Wang, Q. Dual-modal and multi-scale deep neural networks for sleep staging using EEG and ECG signals. Biomed. Signal Process. Control 2021, 66, 102455. [Google Scholar] [CrossRef]
- Pisipati, M.; Nandy, A. Human Emotion Recognition using EEG Signal in Music Listening. In Proceedings of the 2021 IEEE 18th India Council International Conference (INDICON), Guwahati, India, 19–21 December 2021; IEEE: Guwahati, India, 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Galvão, F.; Alarcão, S.M.; Fonseca, M.J. Predicting Exact Valence and Arousal Values from EEG. Sensors 2021, 21, 3414. [Google Scholar] [CrossRef]
- Qu, W.; Kao, C.H.; Hong, H.; Chi, Z.; Grunstein, R.; Gordon, C.; Wang, Z. Single-channel EEG based insomnia detection with domain adaptation. Comput. Biol. Med. 2021, 139, 104989. [Google Scholar] [CrossRef]
- Abdelhameed, A.M.; Bayoumi, M. Semi-Supervised EEG Signals Classification System for Epileptic Seizure Detection. IEEE Signal Process. Lett. 2019, 26, 1922–1926. [Google Scholar] [CrossRef]
- Pourmohammadi, S.; Maleki, A. Stress detection using ECG and EMG signals: A comprehensive study. Comput. Methods Programs Biomed. 2020, 193, 105482. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Qi, J.; Hu, J.; Hao, S. A new approach for product evaluation based on integration of EEG and eye-tracking. Adv. Eng. Inform. 2022, 52, 101601. [Google Scholar] [CrossRef]
- Sridhar, N.; Shoeb, A.; Stephens, P.; Kharbouch, A.; Shimol, D.B.; Burkart, J.; Ghoreyshi, A.; Myers, L. Deep learning for automated sleep staging using instantaneous heart rate. NPJ Digit. Med. 2020, 3, 106. [Google Scholar] [CrossRef] [PubMed]
- Petroff, O.A.; Spencer, D.D.; Goncharova, I.I.; Zaveri, H.P. A comparison of the power spectral density of scalp EEG and subjacent electrocorticograms. Clin. Neurophysiol. 2016, 127, 1108–1112. [Google Scholar] [CrossRef] [PubMed]
- Unde, S.A.; Shriram, R. Coherence Analysis of EEG Signal Using Power Spectral Density. In Proceedings of the 2014 Fourth International Conference on Communication Systems and Network Technologies, Bhopal, India, 7–9 April 2014; IEEE: Bhopal, India, 2014; pp. 871–874. [Google Scholar] [CrossRef]
- Nagar, P.; Sethia, D. Brain Mapping Based Stress Identification Using Portable EEG Based Device. In Proceedings of the 2019 11th International Conference on Communication Systems & Networks (COMSNETS), Bengaluru, India, 7–11 January 2019; IEEE: Bengaluru, India, 2019; pp. 601–606. [Google Scholar] [CrossRef]
- Weon, H.W.; Byun, Y.E.; Lim, H.J. Quantitative EEG (QEEG) Analysis of Emotional Interaction between Abusers and Victims in Intimate Partner Violence: A Pilot Study. Brain Sci. 2021, 11, 570. [Google Scholar] [CrossRef] [PubMed]
- El-Sappagh, S.; Alonso, J.M.; Islam, S.M.R.; Sultan, A.M.; Kwak, K.S. A multilayer multi-modal detection and prediction model based on explainable artificial intelligence for Alzheimer’s disease. Sci. Rep. 2021, 11, 2660. [Google Scholar] [CrossRef] [PubMed]
- Vidhya, R.B.; Jerritta, S. Pre-processing ECG signals for smart home material application. Mater. Today Proc. 2022, 49, 2955–2961. [Google Scholar] [CrossRef]
- Tian, T.; Wang, L.; Luo, M.; Sun, Y.; Liu, X. ResNet-50 based technique for EEG image characterization due to varying environmental stimuli. Comput. Methods Programs Biomed. 2022, 225, 107092. [Google Scholar] [CrossRef] [PubMed]
- Kora, P.; Meenakshi, K.; Swaraja, K.; Rajani, A.; Raju, M.S. EEG based interpretation of human brain activity during yoga and meditation using machine learning: A systematic review. Complement. Ther. Clin. Pract. 2021, 43, 101329. [Google Scholar] [CrossRef] [PubMed]
- Sharif, M.S.; Theeng Tamang, M.R.; Fu, C. Predicting the Health Impacts of Commuting Using EEG Signal Based on Intelligent Approach. In Proceedings of the 2021 International Conference on Innovation and Intelligence for Informatics, Computing, and Technologies (3ICT), Zallaq, Bahrain, 29–30 September 2021; IEEE: Zallaq, Bahrain, 2021; pp. 386–391. [Google Scholar] [CrossRef]
- Uchida, S.; Maloney, T.; Feinberg, I. Sigma (12–16 Hz) and beta (20–28 Hz) EEG discriminate NREM and REM sleep. Brain Res. 1994, 659, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Zea, A.; Lopez, J.D.; Smith, K.; Trujillo, N.; Parra, M.A.; Escudero, J. Phenotyping Ex-Combatants From EEG Scalp Connectivity. IEEE Access 2018, 6, 55090–55098. [Google Scholar] [CrossRef]
- Hsu, Y.L.; Yang, Y.T.; Wang, J.S.; Hsu, C.Y. Automatic sleep stage recurrent neural classifier using energy features of EEG signals. Neurocomputing 2013, 104, 105–114. [Google Scholar] [CrossRef]
- Patnaik, S.; Moharkar, L.; Chaudhari, A. Deep RNN learning for EEG based functional brain state inference. In Proceedings of the 2017 International Conference on Advances in Computing, Communication and Control (ICAC3), Mumbai, India, 1–2 December 2017; IEEE: Mumbai, India, 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Kim, C.; Sun, J.; Liu, D.; Wang, Q.; Paek, S. An effective feature extraction method by power spectral density of EEG signal for 2-class motor imagery-based BCI. Med. Biol. Eng. Comput. 2018, 56, 1645–1658. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, G.; Liu, J.; Wu, D.; Xu, W.; Wang, Z.; Ye, J.; Xia, M.; Hu, Y.; Tian, Y. Automatic Sleep Stage Classification With Single Channel EEG Signal Based on Two-Layer Stacked Ensemble Model. IEEE Access 2020, 8, 57283–57297. [Google Scholar] [CrossRef]
- Faisal, M.A.A.; Chowdhury, M.E.; Khandakar, A.; Hossain, M.S.; Alhatou, M.; Mahmud, S.; Ara, I.; Sheikh, S.I.; Ahmed, M.U. An investigation to study the effects of Tai Chi on human gait dynamics using classical machine learning. Comput. Biol. Med. 2022, 142, 105184. [Google Scholar] [CrossRef] [PubMed]
- Majhi, M.K.; Pradhan, B.K.; Sarkar, P.; Sivaraman, J.; Pal, K. Can statistical and entropy-based features extracted from ECG signals efficiently differentiate the cannabis-consuming women population from the non-consumer? Med. Hypotheses 2022, 167, 110952. [Google Scholar] [CrossRef]
- Rasheed, K.; Qayyum, A.; Qadir, J.; Sivathamboo, S.; Kwan, P.; Kuhlmann, L.; O’Brien, T.; Razi, A. Machine Learning for Predicting Epileptic Seizures Using EEG Signals: A Review. IEEE Rev. Biomed. Eng. 2021, 14, 139–155. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Garza, J.G.; Darfler, M.; Rounds, J.D.; Gao, E.; Kalantari, S. EEG-based investigation of the impact of room size and window placement on cognitive performance. J. Build. Eng. 2022, 53, 104540. [Google Scholar] [CrossRef]
- Zhang, R.; Jia, J.; Zhang, R. EEG analysis of Parkinson’s disease using time–frequency analysis and deep learning. Biomed. Signal Process. Control 2022, 78, 103883. [Google Scholar] [CrossRef]
- Zhuang, Z.; Wang, F.; Yang, X.; Zhang, L.; Fu, C.H.; Xu, J.; Li, C.; Hong, H. Accurate contactless sleep apnea detection framework with signal processing and machine learning methods. Methods 2022, 205, 167–178. [Google Scholar] [CrossRef]
- Parhi, K.K.; Ayinala, M. Low-Complexity Welch Power Spectral Density Computation. IEEE Trans. Circuits Syst. I Regul. Pap. 2014, 61, 172–182. [Google Scholar] [CrossRef]
- Gore, E.; Rathi, S. Surveying Machine Learning Algorithms On Eeg Signals Data For Mental Health Assessment. In Proceedings of the 2019 IEEE Pune Section International Conference (PuneCon), Pune, India, 18–20 December 2019; IEEE: Pune, India, 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Che Wan Fadzal, C.W.N.F.; Mansor, W.; Khuan, L.Y.; Mohamad, N.B.; Mahmoodin, Z.; Mohamad, S.; Amirin, S. Welch power spectral density of EEG signal generated from dyslexic children. In Proceedings of the 2014 IEEE REGION 10 SYMPOSIUM, Kuala Lumpur, Malaysia, 14–16 April 2014; IEEE: Kuala Lumpur, Malaysia, 2014; pp. 560–562. [Google Scholar] [CrossRef]
- Lai, D.; Heyat, M.B.B.; Khan, F.I.; Zhang, Y. Prognosis of Sleep Bruxism Using Power Spectral Density Approach Applied on EEG Signal of Both EMG1-EMG2 and ECG1-ECG2 Channels. IEEE Access 2019, 7, 82553–82562. [Google Scholar] [CrossRef]
- Kang, J.M.; Cho, S.E.; Moon, J.Y.; Kim, S.I.; Kim, J.W.; Kang, S.G. Difference in spectral power density of sleep electroencephalography between individuals without insomnia and frequent hypnotic users with insomnia complaints. Sci. Rep. 2022, 12, 2117. [Google Scholar] [CrossRef]
- Wang, L.; Deng, X.; Lv, X.; Liu, K.; Yang, Q.; Long, C. A WeChat Mini-program System with LSTM for The Emotional EEG Signal Recognition. In Proceedings of the 2020 2nd International Conference on Industrial Artificial Intelligence (IAI), Shenyang, China, 23–25 October 2020; IEEE: Shenyang, China, 2020; pp. 1–5. [Google Scholar] [CrossRef]
- Vuppalapati, C.; Raghu, N.; Veluru, P.; Khursheed, S. A System To Detect Mental Stress Using Machine Learning And Mobile Development. In Proceedings of the 2018 International Conference on Machine Learning and Cybernetics (ICMLC), Chengdu, China, 15–18 July 2018; IEEE: Chengdu, China, 2018; pp. 161–166. [Google Scholar] [CrossRef]
- You, Y.; Zhong, X.; Liu, G.; Yang, Z. Automatic sleep-stage classification: A light and efficient deep neural network model based on time, frequency and fractional Fourier transform domain features. Artif. Intell. Med. 2022, 127, 102279. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, K.; Wei, Y.; Guo, X.; Wen, J.; Luo, Y. Minimal EEG channel selection for depression detection with connectivity features during sleep. Comput. Biol. Med. 2022, 147, 105690. [Google Scholar] [CrossRef] [PubMed]
- Molin, N.L.; Molin, C.; Dalpatadu, R.J.; Singh, A.K. Prediction of obstructive sleep apnea using Fast Fourier Transform of overnight breath recordings. Mach. Learn. Appl. 2021, 4, 100022. [Google Scholar] [CrossRef]
- Chatterjee, R.; Bandyopadhyay, T.; Sanyal, D.K.; Guha, D. Dimensionality reduction of EEG signal using Fuzzy Discernibility Matrix. In Proceedings of the 2017 10th International Conference on Human System Interactions (HSI), Ulsan, Republic of Korea, 17–19 July 2017; IEEE: Ulsan, Republic of Korea, 2017; pp. 131–136. [Google Scholar] [CrossRef]
- Kadam, S.T.; Dhaimodker, V.M.; Patil, M.M.; Edla, D.R. EIQ: EEG based IQ test using wavelet packet transform and hierarchical extreme learning machine. J. Neurosci. Methods 2019, 322, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Bermúdez, G.; García-Laencina, P.J.; Roca-González, J.; Roca-Dorda, J. Efficient feature selection and linear discrimination of EEG signals. Neurocomputing 2013, 115, 161–165. [Google Scholar] [CrossRef]
- Zhang, J.; Yin, Z.; Chen, P.; Nichele, S. Emotion recognition using multi-modal data and machine learning techniques: A tutorial and review. Inf. Fusion 2020, 59, 103–126. [Google Scholar] [CrossRef]
- Adam, A.; Shapiai, M.I.; Mohd Tumari, M.Z.; Mohamad, M.S.; Mubin, M. Feature Selection and Classifier Parameters Estimation for EEG Signals Peak Detection Using Particle Swarm Optimization. Sci. World J. 2014, 2014, 973063. [Google Scholar] [CrossRef] [PubMed]
- Debarnot, U.; Perrault, A.A.; Sterpenich, V.; Legendre, G.; Huber, C.; Guillot, A.; Schwartz, S. Motor imagery practice benefits during arm immobilization. Sci. Rep. 2021, 11, 8928. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Ge, H.; Ma, W.; Miao, J. EEG feature selection method based on decision tree. Bio-Med. Mater. Eng. 2015, 26, S1019–S1025. [Google Scholar] [CrossRef]
- Duan, X.; Ying, S.; Yuan, W.; Cheng, H.; Yin, X. A Generative Adversarial Networks for Log Anomaly Detection. Comput. Syst. Sci. Eng. 2021, 37, 135–148. [Google Scholar] [CrossRef]
- Li, T.; Zhou, M. ECG Classification Using Wavelet Packet Entropy and Random Forests. Entropy 2016, 18, 285. [Google Scholar] [CrossRef]
- Utomo, O.K.; Surantha, N.; Isa, S.M.; Soewito, B. Automatic Sleep Stage Classification using Weighted ELM and PSO on Imbalanced Data from Single Lead ECG. Procedia Comput. Sci. 2019, 157, 321–328. [Google Scholar] [CrossRef]
- Annaby, M.; Said, M.; Eldeib, A.; Rushdi, M. EEG-based motor imagery classification using digraph Fourier transforms and extreme learning machines. Biomed. Signal Process. Control 2021, 69, 102831. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, W.; Liu, Y.; Wang, D.; Liu, B.; Shi, Y.; Gao, P. Feature Extracting of Weak Signal in Real-Time Sleeping EEG with Approximate Entropy and Bispectrum Analysis. In Proceedings of the 2009 3rd International Conference on Bioinformatics and Biomedical Engineering, Beijing, China, 11–16 June 2009; IEEE: Beijing, China, 2009; pp. 1–4. [Google Scholar] [CrossRef]
- Koh, J.; Ooi, C.P.; Lim-Ashworth, N.S.; Vicnesh, J.; Tor, H.T.; Lih, O.S.; Tan, R.S.; Acharya, U.; Fung, D.S.S. Automated classification of attention deficit hyperactivity disorder and conduct disorder using entropy features with ECG signals. Comput. Biol. Med. 2022, 140, 105120. [Google Scholar] [CrossRef] [PubMed]
- Tautan, A.M.; Rossi, A.C.; de Francisco, R.; Ionescu, B. Automatic Sleep Stage Detection: A Study on the Influence of Various PSG Input Signals. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; IEEE: Montreal, QC, Canada, 2020; pp. 5330–5334. [Google Scholar] [CrossRef]
- Zhou, D.; Xu, Q.; Wang, J.; Xu, H.; Kettunen, L.; Chang, Z.; Cong, F. Alleviating Class Imbalance Problem in Automatic Sleep Stage Classification. IEEE Trans. Instrum. Meas. 2022, 71, 1–12. [Google Scholar] [CrossRef]
- Efe, E.; Ozsen, S. CoSleepNet: Automated sleep staging using a hybrid CNN-LSTM network on imbalanced EEG-EOG datasets. Biomed. Signal Process. Control 2023, 80, 104299. [Google Scholar] [CrossRef]
- Xu, Q.; Zhou, D.; Wang, J.; Shen, J.; Kettunen, L.; Cong, F. Convolutional Neural Network Based Sleep Stage Classification with Class Imbalance. In Proceedings of the 2022 International Joint Conference on Neural Networks (IJCNN), Padua, Italy, 18–26 July 2022; IEEE: Padua, Italy, 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Patterson, J.; Gibson, A. Deep Learning A Practitioner’s Approach; y O’Reilly Media, Inc.: Sebastopol, CA, USA, 2017. [Google Scholar]
- Qiu, S.; Zhao, H.; Jiang, N.; Wang, Z.; Liu, L.; An, Y.; Zhao, H.; Miao, X.; Liu, R.; Fortino, G. Multi-sensor information fusion based on machine learning for real applications in human activity recognition: State-of-the-art and research challenges. Inf. Fusion 2022, 80, 241–265. [Google Scholar] [CrossRef]
- Singh, P.; Singh, N.; Singh, K.K.; Singh, A. Chapter 5—Diagnosing of disease using machine learning. In Machine Learning and the Internet of Medical Things in Healthcare; Singh, K.K., Elhoseny, M., Singh, A., Elngar, A.A., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 89–111. [Google Scholar] [CrossRef]
- Shehab, M.; Abualigah, L.; Shambour, Q.; Abu-Hashem, M.A.; Shambour, M.K.Y.; Alsalibi, A.I.; Gandomi, A.H. Machine learning in medical applications: A review of state-of-the-art methods. Comput. Biol. Med. 2022, 145, 105458. [Google Scholar] [CrossRef]
- Guo, K.; Mei, H.; Xie, X.; Xu, X. A Convolutional Neural Network Feature Fusion Framework with Ensemble Learning for EEG-based Emotion Classification. In Proceedings of the 2019 IEEE MTT-S International Microwave Biomedical Conference (IMBioC), Nanjing, China, 6–8 May 2019; IEEE: Nanjing, China, 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Shatte, A.; Hutchinson, D.; Teague, S. Machine learning in mental health: A systematic scoping review of methods and applications. Psychol. Med. 2019, 49, 1426–1448. [Google Scholar] [CrossRef]
- Wang, J.; Chen, Y.; Hao, S.; Peng, X.; Hu, L. Deep learning for sensor-based activity recognition: A survey. Pattern Recognit. Lett. 2019, 119, 3–11. [Google Scholar] [CrossRef]
- Sathya, R.; Abraham, A. Comparison of Supervised and Unsupervised Learning Algorithms for Pattern Classification. Int. J. Adv. Res. Artif. Intell. 2013, 2, 1–80. [Google Scholar] [CrossRef]
- Schmidt, F.; Suri-Payer, F.; Gulenko, A.; Wallschlager, M.; Acker, A.; Kao, O. Unsupervised Anomaly Event Detection for Cloud Monitoring Using Online Arima. In Proceedings of the 2018 IEEE/ACM International Conference on Utility and Cloud Computing Companion (UCC Companion), Zurich, Switzerland, 17–20 December 2018; IEEE: Zurich, Switzerland, 2018; pp. 71–76. [Google Scholar] [CrossRef]
- Garcia-Ceja, E.; Riegler, M.; Nordgreen, T.; Jakobsen, P.; Oedegaard, K.J.; Tørresen, J. Mental health monitoring with multi-modal sensing and machine learning: A survey. Pervasive Mob. Comput. 2018, 51, 1–26. [Google Scholar] [CrossRef]
- Nash, C.; Nair, R.; Naqvi, S.M. Machine Learning and ADHD Mental Health Detection—A Short Survey. In Proceedings of the 2022 25th International Conference on Information Fusion (FUSION), Linköping, Sweden, 4–7 July 2022; IEEE: Linköping, Sweden, 2022; pp. 1–8. [Google Scholar] [CrossRef]
- Sarkar, A.; Singh, A.; Chakraborty, R. A deep learning-based comparative study to track mental depression from EEG data. Neurosci. Inform. 2022, 2, 100039. [Google Scholar] [CrossRef]
- Liu, F.T.; Ting, K.M.; Zhou, Z.H. Isolation-Based Anomaly Detection. ACM Trans. Knowl. Discov. Data 2012, 6, 1–39. [Google Scholar] [CrossRef]
- Moura, M.d.C.; Zio, E.; Lins, I.D.; Droguett, E. Failure and reliability prediction by support vector machines regression of time-series data. Reliab. Eng. Syst. Saf. 2011, 96, 1527–1534. [Google Scholar] [CrossRef]
- Stranges, S.; Tigbe, W.; Gómez-Olivé, F.X.; Thorogood, M.; Kandala, N.B. Sleep Problems: An Emerging Global Epidemic? Findings From the INDEPTH WHO-SAGE Study Among More Than 40,000 Older Adults From 8 Countries Across Africa and Asia. Sleep 2012, 35, 1173–1181. [Google Scholar] [CrossRef]
- Rajagopalan, S.S.; Bhardwaj, S.; Panda, R.; Reddam, V.R.; Ganne, C.; Kenchaiah, R.; Mundlamuri, R.C.; Kandavel, T.; Majumdar, K.K.; Parthasarathy, S.; et al. Machine learning detects EEG microstate alterations in patients living with temporal lobe epilepsy. Seizure 2018, 61, 8–13. [Google Scholar] [CrossRef]
- Thamaraimanalan, T.; Mohankumar, M.; Anandakumar, H.; Deepha, M.; Priya, U.H.; Priya, G.B.; Devi, M.A. Machine Learning based Patient Mental Health Prediction using Spectral Clustering and RBFN Algorithms. In Proceedings of the 2022 8th International Conference on Advanced Computing and Communication Systems (ICACCS), Coimbatore, India, 25–26 March 2022; IEEE: Coimbatore, India, 2022; pp. 1840–1843. [Google Scholar] [CrossRef]
- van der Velden, B.H.; Kuijf, H.J.; Gilhuijs, K.G.; Viergever, M.A. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med. Image Anal. 2022, 79, 102470. [Google Scholar] [CrossRef]
- Ahuja, R.; Banga, A. Mental Stress Detection in University Students using Machine Learning Algorithms. Procedia Comput. Sci. 2019, 152, 349–353. [Google Scholar] [CrossRef]
- He, Z.; Xu, X.; Deng, S. Discovering cluster-based local outliers. Pattern Recognit. Lett. 2003, 24, 1641–1650. [Google Scholar] [CrossRef]
- Hosseini, M.P.; Hosseini, A.; Ahi, K. A Review on Machine Learning for EEG Signal Processing in Bioengineering. IEEE Rev. Biomed. Eng. 2021, 14, 204–218. [Google Scholar] [CrossRef]
- Suthaharan, S. Support Vector Machine. In Machine Learning Models and Algorithms for Big Data Classification. Integrated Series in Information Systems; Springer: Boston, MA, USA, 2016; Volume 36. [Google Scholar]
- Khatun, S.; Morshed, B.I.; Bidelman, G.M. A Single-Channel EEG-Based Approach to Detect Mild Cognitive Impairment via Speech-Evoked Brain Responses. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Raut, K.; Patil, J.; Wade, S.; Tinsu, J. Mental Health and Personality Determination using Machine Learning. In Proceedings of the 2022 7th International Conference on Communication and Electronics Systems (ICCES), Coimbatore, India, 22–24 June 2022; IEEE: Coimbatore, India, 2022; pp. 1231–1236. [Google Scholar] [CrossRef]
- Subhani, A.R.; Mumtaz, W.; Saad, M.N.B.M.; Kamel, N.; Malik, A.S. Machine Learning Framework for the Detection of Mental Stress at Multiple Levels. IEEE Access 2017, 5, 13545–13556. [Google Scholar] [CrossRef]
- Raj, A. The Perfect Recipe for Classification UsingLogistic Regression. Available online: https://towardsdatascience.com/the-perfect-recipe-for-classification-using-logistic-regression-f8648e267592 (accessed on 21 February 2023).
- Shen, C.; Lin, H.; Fan, X.; Chu, Y.; Yang, Z.; Wang, J.; Zhang, S. Biomedical event trigger detection with convolutional highway neural network and extreme learning machine. Appl. Soft Comput. 2019, 84, 105661. [Google Scholar] [CrossRef]
- Pawar, D.; Dhage, S. EEG-based covert speech decoding using random rotation extreme learning machine ensemble for intuitive BCI communication. Biomed. Signal Process. Control 2023, 80, 104379. [Google Scholar] [CrossRef]
- Cecaj, A.; Lippi, M.; Mamei, M.; Zambonelli, F. Comparing Deep Learning and Statistical Methods in Forecasting Crowd Distribution from Aggregated Mobile Phone Data. Appl. Sci. 2020, 10, 6580. [Google Scholar] [CrossRef]
- Guillot, A.; Sauvet, F.; During, E.H.; Thorey, V. Dreem Open Datasets: Multi-Scored Sleep Datasets to compare Human and Automated sleep staging. IEEE Trans. Neural Syst. Rehabilit. Eng. 2020, 28, 1955–1965. [Google Scholar] [CrossRef]
- Greff, K.; Srivastava, R.K.; Koutník, J.; Steunebrink, B.R.; Schmidhuber, J. LSTM: A Search Space Odyssey. IEEE Trans. Neural Netw. Learn. Syst. 2017, 28, 2222–2232. [Google Scholar] [CrossRef]
- Aydin, O.; Guldamlasioglu, S. Using LSTM networks to predict engine condition on large scale data processing framework. In Proceedings of the 2017 4th International Conference on Electrical and Electronic Engineering (ICEEE), Ankara, Turkey, 8–10 April 2017; IEEE: Ankara, Turkey, 2017; pp. 281–285. [Google Scholar] [CrossRef]
- Provotar, O.I.; Linder, Y.M.; Veres, M.M. Unsupervised Anomaly Detection in Time Series Using LSTM-Based Autoencoders. In Proceedings of the 2019 IEEE International Conference on Advanced Trends in Information Theory (ATIT), Kyiv, Ukraine, 18–20 December 2019; IEEE: Kyiv, Ukraine, 2019; pp. 513–517. [Google Scholar] [CrossRef]
- Shi, X.; Chen, Z.; Wang, H.; Yeung, D.Y.; Wong, W.k.; Woo, W.c. Convolutional LSTM Network: A Machine Learning Approach for Precipitation Nowcasting. In Proceedings of the 28th International Conference on Neural Information Processing Systems, Montreal, QC, Canada, 7–12 December 2015; p. 9. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Bhojanapalli, S.; Yun, C.; Rawat, A.S.; Reddi, S.; Kumar, S. Low-Rank Bottleneck in Multi-head Attention Models. In Proceedings of the 37th International Conference on Machine Learning, Virtual, 13–18 July 2020; pp. 864–873. [Google Scholar]
- Zhou, X.; Liang, W.; Wang, K.I.K.; Wang, H.; Yang, L.T.; Jin, Q. Deep-Learning-Enhanced Human Activity Recognition for Internet of Healthcare Things. IEEE Internet Things J. 2020, 7, 6429–6438. [Google Scholar] [CrossRef]
- Murad, A.; Pyun, J.Y. Deep Recurrent Neural Networks for Human Activity Recognition. Sensors 2017, 17, 2556. [Google Scholar] [CrossRef]
- Inoue, M.; Inoue, S.; Nishida, T. Deep recurrent neural network for mobile human activity recognition with high throughput. Artif. Life Robot. 2018, 23, 173–185. [Google Scholar] [CrossRef]
- Liu, L. Objects detection toward complicated high remote basketball sports by leveraging deep CNN architecture. Future Gener. Comput. Syst. 2021, 119, 31–36. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Erdaş, Ç.B.; Güney, S. Human Activity Recognition by Using Different Deep Learning Approaches for Wearable Sensors. Neural Process. Lett. 2021, 53, 1795–1809. [Google Scholar] [CrossRef]
- Nath, R.K.; Thapliyal, H.; Caban-Holt, A. Machine Learning Based Stress Monitoring in Older Adults Using Wearable Sensors and Cortisol as Stress Biomarker. J. Signal Process. Syst. 2022, 94, 513–525. [Google Scholar] [CrossRef]
- Wu, N.; Green, B.; Ben, X.; O’Banion, S. Deep Transformer Models for Time Series Forecasting: The Influenza Prevalence Case. arXiv 2020, arXiv:2001.08317. [Google Scholar]
- Guillot, A.; Thorey, V. RobustSleepNet: Transfer Learning for Automated Sleep Staging at Scale. IEEE Trans. Neural Syst. Rehabil. Eng. 2021, 29, 1441–1451. [Google Scholar] [CrossRef]
- Munir, M.; Siddiqui, S.A.; Chattha, M.A.; Dengel, A.; Ahmed, S. FuseAD: Unsupervised Anomaly Detection in Streaming Sensors Data by Fusing Statistical and Deep Learning Models. Sensors 2019, 19, 2451. [Google Scholar] [CrossRef]
- Thongsuwan, S.; Jaiyen, S.; Padcharoen, A.; Agarwal, P. ConvXGB: A new deep learning model for classification problems based on CNN and XGBoost. Nucl. Eng. Technol. 2021, 53, 522–531. [Google Scholar] [CrossRef]
- Choi, S.H.; Yoon, H.; Kim, H.S.; Kim, H.B.; Kwon, H.B.; Oh, S.M.; Lee, Y.J.; Park, K.S. Real-time apnea-hypopnea event detection during sleep by convolutional neural networks. Comput. Biol. Med. 2018, 100, 123–131. [Google Scholar] [CrossRef]
- Kavi, R.; Kulathumani, V.; Rohit, F.; Kecojevic, V. Multiview fusion for activity recognition using deep neural networks. J. Electron. Imaging 2016, 25, 043010. [Google Scholar] [CrossRef]
- Hssayeni, M.D.; Jimenez-Shahed, J.; Burack, M.A.; Ghoraani, B. Ensemble deep model for continuous estimation of Unified Parkinson’s Disease Rating Scale III. Biomed. Eng. OnLine 2021, 20, 32. [Google Scholar] [CrossRef] [PubMed]
- Mutegeki, R.; Han, D.S. A CNN-LSTM Approach to Human Activity Recognition. In Proceedings of the 2020 International Conference on Artificial Intelligence in Information and Communication (ICAIIC), Fukuoka, Japan, 19–21 February 2020; IEEE: Fukuoka, Japan, 2020; pp. 362–366. [Google Scholar] [CrossRef]
- Mekruksavanich, S.; Jitpattanakul, A. LSTM Networks Using Smartphone Data for Sensor-Based Human Activity Recognition in Smart Homes. Sensors 2021, 21, 1636. [Google Scholar] [CrossRef] [PubMed]
- Na, Y.; Kim, D.; Kim, D.K.; Lee, J.G. Evaluation of OSA Patient Sleep Stage Classification Performance Using a Multi-Channel PSG Dataset. In Proceedings of the 2022 IEEE International Conference on Consumer Electronics-Asia (ICCE-Asia), Yeosu, Republic of Korea, 17–20 December 2022; IEEE: Yeosu, Republic of Korea, 2022; pp. 1–4. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, H.; Yang, B. Automatic Sleep Arousals Detection From Polysomnography Using Multi-Convolution Neural Network and Random Forest. IEEE Access 2020, 8, 176343–176350. [Google Scholar] [CrossRef]
- Li, F.; Yan, R.; Mahini, R.; Wei, L.; Wang, Z.; Mathiak, K.; Liu, R.; Cong, F. End-to-end sleep staging using convolutional neural network in raw single-channel EEG. Biomed. Signal Process. Control 2021, 63, 102203. [Google Scholar] [CrossRef]
- Howe-Patterson, M.; Pourbabaee, B.; Benard, F. Automated Detection of Sleep Arousals From Polysomnography Data Using a Dense Convolutional Neural Network. In Proceedings of the 2018 Computing in Cardiology Conference (CinC), Maastricht, The Netherlands, 23–26 September 2018. [Google Scholar] [CrossRef]
- Yamazaki, K.; Vo-Ho, V.K.; Bulsara, D.; Le, N. Spiking Neural Networks and Their Applications: A Review. Brain Sci. 2022, 12, 863. [Google Scholar] [CrossRef] [PubMed]
- Doborjeh, Z.; Doborjeh, M.; Taylor, T.; Kasabov, N.; Wang, G.Y.; Siegert, R.; Sumich, A. Spiking Neural Network Modelling Approach Reveals How Mindfulness Training Rewires the Brain. Sci. Rep. 2019, 9, 6367. [Google Scholar] [CrossRef]
- Pfeiffer, M.; Pfeil, T. Deep Learning With Spiking Neurons: Opportunities and Challenges. Front. Neurosci. 2018, 12, 774. [Google Scholar] [CrossRef]
- Saputra, N.H.; Nafi’Iyah, N. Identification of Human Stress Based on EEG Signals Using Machine Learning. In Proceedings of the 2022 1st International Conference on Information System & Information Technology (ICISIT), Virtual, 27–28 July 2022; IEEE: Yogyakarta, Indonesia, 2022; pp. 176–180. [Google Scholar] [CrossRef]
- Bashar, K.; Chiaki, I.; Yoshida, H. Human identification from brain EEG signals using advanced machine learning method EEG-based biometrics. In Proceedings of the IEEE EMBS Conference on Biomedical Engineering and Sciences, Lyon, France, 4–8 December 2016; p. 5. [Google Scholar]
- Abdul Hamid, D.S.B.; Goyal, S.; Bedi, P. Integration of Deep Learning for Improved Diagnosis of Depression using EEG and Facial Features. Mater. Today Proc. 2021; in press. [Google Scholar] [CrossRef]
- Troncoso-García, A.; Martínez-Ballesteros, M.; Martínez-Álvarez, F.; Troncoso, A. Explainable machine learning for sleep apnea prediction. Procedia Comput. Sci. 2022, 207, 2930–2939. [Google Scholar] [CrossRef]
- Sharma, G.; Parashar, A.; Joshi, A.M. DepHNN: A novel hybrid neural network for electroencephalogram (EEG)-based screening of depression. Biomed. Signal Process. Control 2021, 66, 102393. [Google Scholar] [CrossRef]
- Acar, E. Unraveling Diagnostic Biomarkers of Schizophrenia Through Structure-Revealing Fusion of Multi-Modal Neuroimaging Data. Front. Neurosci. 2019, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, S.J.; Parrish, L.; Jiang, H.; Zhang, D.W.; Williams, V.; Li, S. Aiding diagnosis of childhood attention-deficit/hyperactivity disorder of the inattentive presentation: Discriminant function analysis of multi-domain measures including EEG. Biol. Psychol. 2021, 161, 108080. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, D.; Nariai, H.; Hussain, S.A.; Sankar, R.; Salamon, N.; Krueger, D.A.; Sahin, M.; Northrup, H.; Bebin, E.M.; Wu, J.Y. Visual and semi-automatic non-invasive detection of interictal fast ripples: A potential biomarker of epilepsy in children with tuberous sclerosis complex. Clin. Neurophysiol. 2018, 129, 1458–1466. [Google Scholar] [CrossRef] [PubMed]
- Mencar, C.; Gallo, C.; Mantero, M.; Tarsia, P.; Carpagnano, G.E.; Foschino Barbaro, M.P.; Lacedonia, D. Application of machine learning to predict obstructive sleep apnea syndrome severity. Health Inform. J. 2020, 26, 298–317. [Google Scholar] [CrossRef]
- Xu, D.; Wang, Y.; Meng, Y.; Zhang, Z. An Improved Data Anomaly Detection Method Based on Isolation Forest. In Proceedings of the 2017 10th International Symposium on Computational Intelligence and Design (ISCID), Hangzhou, China, 9–10 December 2017; IEEE: Hangzhou, China, 2017; pp. 287–291. [Google Scholar] [CrossRef]
- Satapathy, S.K.; Kondaveeti, H.K. Prognosis of Sleep Stage Classification Using Machine Learning Techniques Applied on Single-channel of EEG signal of both Healthy Subjects and Mild Sleep effected Subjects. In Proceedings of the 2021 International Conference on Artificial Intelligence and Machine Vision (AIMV), Gandhinagar, India, 28–30 June 2021; IEEE: Gandhinagar, India, 2021; pp. 1–7. [Google Scholar] [CrossRef]
- Zhu, G.; Li, Y.; Wen, P. Analysis and Classification of Sleep Stages Based on Difference Visibility Graphs From a Single-Channel EEG Signal. IEEE J. Biomed. Health Inform. 2014, 18, 1813–1821. [Google Scholar] [CrossRef]
- Salman, A.A.; Kumar, D.M.S. Introducing Confusion Matrix and Accuracy in Disease Prediction on Liver Using Machine Learning. Int. J. Comput. Sci. Trends Technol. (IJCST) 2020, 8, 5–9. [Google Scholar]
- Rahman, A.; Chowdhury, M.E.H.; Khandakar, A.; Kiranyaz, S.; Zaman, K.S.; Reaz, M.B.I.; Islam, M.T.; Kadir, M.A. Multimodal EEG and Keystroke Dynamics Based Biometric System Using Machine Learning Algorithms. IEEE Access 2021, 9, 19. [Google Scholar] [CrossRef]
- Zhou, D.; Wang, J.; Hu, G.; Zhang, J.; Li, F.; Yan, R.; Kettunen, L.; Chang, Z.; Xu, Q.; Cong, F. SingleChannelNet: A model for automatic sleep stage classification with raw single-channel EEG. Biomed. Signal Process. Control 2022, 75, 103592. [Google Scholar] [CrossRef]
- Kemp, B.; Olivan, J. European data format ‘plus’ (EDF+), an EDF alike standard format for the exchange of physiological data. Clin. Neurophysiol. 2003, 114, 1755–1761. [Google Scholar] [CrossRef]
- Korompili, G.; Amfilochiou, A.; Kokkalas, L.; Mitilineos, S.A.; Tatlas, N.A.; Kouvaras, M.; Kastanakis, E.; Maniou, C.; Potirakis, S.M. PSG-Audio, a scored polysomnography dataset with simultaneous audio recordings for sleep apnea studies. Sci. Data 2021, 8, 197. [Google Scholar] [CrossRef]
- Le Quy, T.; Roy, A.; Iosifidis, V.; Zhang, W.; Ntoutsi, E. A survey on datasets for fairness-aware machine learning. WIREs Data Min. Knowl. Discov. 2022, 12, e1452. [Google Scholar] [CrossRef]
- Ketola, E.C.; Barankovich, M.; Schuckers, S.; Ray-Dowling, A.; Hou, D.; Imtiaz, M.H. Channel Reduction for an EEG-Based Authentication System While Performing Motor Movements. Sensors 2022, 22, 39156. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.R.; Fiedler, P.; Kuhlmann, L.; Liley, D.; Vasconcelos, B.; Fonseca, C.; Tamburro, G.; Comani, S.; Lui, T.K.Y.; Tse, C.Y.; et al. Multi-Center Evaluation of Gel-Based and Dry Multipin EEG Caps. Sensors 2022, 22, 8079. [Google Scholar] [CrossRef] [PubMed]


| Digital Database | Search String Used | Total Articles Collected |
|---|---|---|
| IEEE Xplore Access | (“EEG” OR “ECG” OR “EOG” OR “EMG” OR “PSG”) AND “Machine Learning” AND “Mental Health” | 41 |
| Science Direct | (“EEG” OR “ECG” OR “EOG” OR “EMG” OR “PSG”) AND “Machine Learning” AND “Mental Health” | 944 |
| MDPI | (“EEG” OR “ECG” OR “EOG” OR “EMG” OR “PSG”) AND “Machine Learning” AND “Mental Health” | 26 |
| PubMed | (“EEG” OR “ECG” OR “EOG” OR “EMG” OR “PSG”) AND “Machine Learning” AND “Mental Health” | 76 |
| Dataset | Description | Source |
|---|---|---|
| Epileptic seizure developed by University of Bonn, Germany | Sampling frequency = 173.61 Hz; No. of persons = 5 (healthy) + 5 (Unhealthy); Total duration of a segment = 23.6 s; No. of trails/channels in a class = 100; Data size in this study = 200 × 4097 | [75] |
| Schizophrenia EEG dataset collected by the Institute of Psychiatry and Neurology in Warsaw, Poland | Sampling frequency = 250 Hz; No. of persons = 14 (healthy) + 14 (Unhealthy); Epoch size = 60 s × 250 Hz = 15,000; Data size in this study = [19 × (14 + 14)] × 15,000 = 532 × 15,000 | [75] |
| Sleep-EDF (S-EDF) (Scored by 1 sleep expert) | Sampling frequency = 100 Hz; No. of persons = 8; Epoch length = 30 s; Data size = 15,139 | [29,38,76] |
| Sleep-EDF (Expanded) (SE-EDF) (Scored by 1 sleep expert) | Sampling frequency = 100 Hz; No. of persons = 20 Epoch length = 30 s; Data size =40,100 | [29,38,76] |
| Laboratory for Neurophysiology and NeuroComputer Interfaces of M. V. Lomonosov Moscow State University | Sampling frequency = 128 Hz; No. of persons= 45 (schizophrenic) + 39 (Normal); Data size = 16 × 7680; Matrix with 1344 instances | [77] |
| The Epilepsy Ecosystem dataset | Sampling frequency = 400 Hz; No. of persons = 3 | [78] |
| The CHB-MIT dataset | Sampling frequency = 256 Hz; No. of persons = 23 | [78] |
| The BCI competition-II Dataset-III | Sampling frequency = 128 Hz | [79,80] |
| Test Set of SHHS1 Test Set of SHHS2 | Sampling frequency for ECG = 125 Hz in SHHS1 while ECG for SHHS2 = 250 Hz; No. of persons= 5793 for SHHS1 and 2651 for SHHS2 | [24,81,82] |
| MESA by National Sleep Research Resource | Sampling frequency = 256 Hz for ECG; No. of persons = 2056 | [81,83] |
| The SLPDB database | Sampling frequency = 250 Hz; No. of persons = 16 | [81] |
| Apnea-ECG dataset | Sampling frequency = 128 Hz; No. of persons = 57 men + 13 women); Epoch length = 60 s; Segments = 17,045 | [82,84] |
| The MIT-BIH polysomnography dataset | Sampling frequency = 250 Hz; No. of persons = 16; Epoch length = 30 s | [85] |
| The Massachusetts General Hospital (MGH Dataset) Sleep Laboratory | Sampling frequency = 200 Hz; Epoch length = 30 s | [24] |
| DREAMER dataset | Sampling frequency = 128 Hz for EEG and 256 Hz for ECG No. of persons = 23 | [76,82,86,87] |
| Haaglanden Medisch Centrum Sleep Center Database (HMC) | Sampling frequency = 256 Hz; No. of persons = 85 male + 66 female | [82] |
| Sleep Telemetry Study (Telemetry) | Sampling frequency = 200 Hz; No. of persons = 22 subjects (male and female) | [82] |
| ISRUC-SLEEP dataset (ISRUC) | Sampling frequency = 100 Hz; No. of persons = 100 subjects (55 male and 45 female) | [82] |
| National Institute of Mental Health of the Czech Republic (NIMH-CZ). | Sampling frequency = 250 Hz; No. of persons = 18 | [23] |
| DAIC-WOZ depression dataset | Sampling frequency = 16,000 Hz; No. of persons = 189 Subjects (54 % male and 46 % female ) | [5] |
| Montreal Archive of Sleep Studies (MASS) | Sampling frequency = 256 Hz; No. of persons = 97 male + 103 female | [69,88] |
| Department of Epileptology at Bonn University | Sampling frequency = 256 Hz; No. of persons = 23 subjects | [89] |
| Bands | Frequency (Hz) | Amplitude () | Activities |
|---|---|---|---|
| Delta | 0–4.5 | 20–100 | Deep sleep |
| Theta | 4–8 | 10 | Light sleep |
| Alpha | 8–13 | 2–100 | Calm or relaxed |
| Beta | 15–22 | 5–10 | Alert |
| Gamma | >30 | - | Hyperactive |
| Feature Extraction Techniques | Signal Type | Reference |
|---|---|---|
| Adaptive auto-regressive (AAR) | EEG-Motor-Imagery | [79] |
| Adaptive auto-regressive Fuzzy discernibility matrix (first adaptation) | EEG-Motor-Imagery | [124] |
| Random asynchronous particle swarm optimization | Eye Movement EEG | [128] |
| Least angle regression + the direct leave-one-out error estimation by the PRESS statistic | Motor-Imagery | [129] |
| Principal component analysis + decision-tree-based feature ranking (C4.5) | Motor-Imagery | [129,130,131] |
| Wavelet packet decomposition + approximation entropy + one-dimensional real-valued particle-swarm optimization | Motor-Imagery, Emotional Recognition | [132,133] |
| Common spatial model (CSP) | Motor-Imagery | [134] |
| Discrete wavelet decomposition (DWT) in five frequency bands, combined with wavelet entropy | Motor-Imagery, Emotional Recognition | [21,76,135] |
| Differential entropy (DE) | Motor-Imagery | [136] |
| Model | Application | Data Used | Accuracy | Year Ref. |
|---|---|---|---|---|
| LR | EEG abnormalities of micro-states in temporal lobe epilepsy (TLE) | Privately sourced dataset from a tertiary institute | 66.70% | 2018 [156] |
| Mental depression from EEG dataset | emotions.csv available on the Kaggle website | 96.60% | 2022 [152] | |
| Emotion Recognition | DREAMER (discrete emotion recognition) | 94.49% | 2021 [86] | |
| KNN | EGG, (stress and emotion classification | 97.00% | 2022 [200] | |
| Obstructive sleep apnea (OSA), ECG and SPO2 signals | PhysioNet Sleep Apnea Database | 95.08% | 2017 [71] | |
| SVM | EEG image data and emotion classification | SEED dataset | 56.00% | 2022 [99] |
| Obstructive sleep apnea (OSA), ECG and SPO2 signals | PhysioNet Sleep Apnea Database | 96.64% | 2017 [71] | |
| EEG sleep quality | Sleep-EDF Database | 91.40% | 2019 [28] | |
| Imaging and EEG data for ADHD | ADHD-200 dataset | 97.60% | 2022 [151] | |
| Human recognition EEG | EMOTIV INSIGHT dataset | 94.44% | 2016 [201] | |
| mental stress detection using EEG signal | mental arithmetic tasks database | 97.26% | 2022 [56] | |
| EEG-dimensionality reduction | Dataset III of BCI competition II | 81.40% | 2017 [124] | |
| motor imagery EEG signal | The BCI competition-II Dataset-III | 78.57% | 2019 [79] | |
| Identification of chronic alcohol users from ECG signals | NIMHANS- ECG dataset | 87.50% | 2017 [43] | |
| Sleep quality measurement | Sleep-EDF Database | 93.50% | 2019 [28] | |
| Mental depression from EEG dataset | emotions.csv available on the Kaggle website | 95.89% | 2022 [152] | |
| Detection of schizophrenia from EEG data | EEG dataset from NNCI M. V. Lomonosov Moscow State University | 53.50% | 2022 [77] | |
| ResNet-50 | EEG image data and emotion classification | SEED Dataset | 85.11% | 2022 [99] |
| CNN | EEG-sleep stage using multi-scale dual-attention | Sleep-EDF Database | 96.70% | 2022 [29] |
| Mental depression from EEG dataset | emotions.csv available on the Kaggle website | 49.82% | 2022 [152] | |
| Automatic sleep scoring | Multiple EGG dataset was used for this work | 74.17% | 2021 [82] | |
| Emotion recognition | DREAMER (discrete emotion recognition) | 99.90% | 2021 [86] | |
| ELM | Identification of chronic alcohol users from ECG signals | NIMHANS- ECG Dataset | 94.64% | 2017 [43] |
| MLP | Mental depression from EEG dataset | emotions.csv available on the Kaggle website | 76.43% | 2022 [152] |
| RNN | Mental depression from EEG dataset | emotions.csv available on the Kaggle website | 93.90% | 2022 [152] |
| RNN with LSTM | Mental depression from EEG dataset | emotions.csv available on the Kaggle website | 97.65% | 2022 [152] |
| Detection of schizophrenia from EEG Data | EEG dataset from NNCI M. V. Lomonosov Moscow State University | 98.00% | 2022 [77] | |
| Insomnia detection | MASS Dataset-EEG, EOG, EMG, ECG, and respiratory signals | 79.20% | 2021 [88] | |
| Depression using EEG | BCI project for EEG signal and frontal facial data | 99.66% | 2021 [202] | |
| CNN–LSTM | Automatic sleep scoring | Multiple EGG dataset was used for this work | 80.17% | 2021 [82] |
| Sleep apnea | Apnea-ECG dataset | 97.21% | 2022 [84] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehiabhi, J.; Wang, H. A Systematic Review of Machine Learning Models in Mental Health Analysis Based on Multi-Channel Multi-Modal Biometric Signals. BioMedInformatics 2023, 3, 193-219. https://doi.org/10.3390/biomedinformatics3010014
Ehiabhi J, Wang H. A Systematic Review of Machine Learning Models in Mental Health Analysis Based on Multi-Channel Multi-Modal Biometric Signals. BioMedInformatics. 2023; 3(1):193-219. https://doi.org/10.3390/biomedinformatics3010014
Chicago/Turabian StyleEhiabhi, Jolly, and Haifeng Wang. 2023. "A Systematic Review of Machine Learning Models in Mental Health Analysis Based on Multi-Channel Multi-Modal Biometric Signals" BioMedInformatics 3, no. 1: 193-219. https://doi.org/10.3390/biomedinformatics3010014
APA StyleEhiabhi, J., & Wang, H. (2023). A Systematic Review of Machine Learning Models in Mental Health Analysis Based on Multi-Channel Multi-Modal Biometric Signals. BioMedInformatics, 3(1), 193-219. https://doi.org/10.3390/biomedinformatics3010014







